Abstract
Bombesin (BN), originally isolated from amphibians, is structurally related to a family of BN-like peptides found in mammals, which include gastrin-releasing peptide (GRP) and neuromedin B (NMB). These peptides have important effects on secretion, smooth muscle contraction, metabolism and behavior. Here we report cloning and characterization of two subtypes of BN-like peptide receptors in Aves.
The amino-acid sequence of chick GRP-R (chGRP-R) is highly identical with mammalian and amphibian GRP-R, and this receptor showed high affinity for GRP, BN and synthetic bombesin agonist, [D-Phe6, β-Ala11, Phe13, Nle14]bombesin(6–14) ([FAFNl]BN(6–14)). The chGRP-R gene was localized to chicken chromosome 1q23distal-q24proximal, where chick homologs of other human X-linked genes have also been mapped.
ChBRS-3.5, having sequence similarities to both mammalian bombesin-like peptide receptor subtype-3 and amphibian bombesin-like peptide receptor subtype-4, showed high affinity for [FAFNl]BN(6–14), moderate affinity for BN, but low affinity for both GRP and NMB.
Expression of both receptors was detected in brain, but only chGRP-R was expressed in gastrointestinal (GI) tissues.
When expressed in Chinese hamster ovary K1 cells, these receptors mediate intracellular calcium mobilization upon agonist stimulation. These results suggest that a novel BN peptide may occur in Aves as an endogenous ligand for chBRS-3.5.
The receptor sequences responsible for ligand selectivities were discussed and this knowledge about avian BN-like peptide receptors will help us to understand the molecular basis for agonist sensitivities of BN-like peptide receptors.
Keywords: Bombesin, Aves, chick, BRS-3, BB4, gastrin-releasing peptide, GRP, NMB
Introduction
Bombesin (BN) is an amidated tetradecapeptide originally isolated from skin of the European frog, Bombina bombina, as an agent that has an ability to induce smooth muscle contraction (Anastasi et al., 1971). Two peptides, gastrin-releasing peptide (GRP) and neuromedin B (NMB), which have similarities with BN, have been purified subsequently from the porcine stomach and spinal cord, and these two peptides represent the only mammalian BN-related peptides cloned so far (McDonald et al., 1979; Minamino et al., 1983). GRP and NMB are widely distributed in the brain, lung, exocrine and endocrine organs and gastrointestinal (GI) tract, and mimic various physiological effects induced by BN (for a review, see Lebacq-Verheyden et al., 1990). These include smooth muscle contraction, exocrine and endocrine secretion, induction of grooming and scratching, anorexia, hyperglycemia and hypothermic effects.
The high-affinity receptors for these peptides (GRP-R and NMB-R) as well as a structurally related receptor, bombesin receptor subtype-3 (BRS-3), have been cloned in mammals (Spindel et al., 1990; Wada et al., 1991; Fathi et al., 1993). These receptors are G-protein coupled and expressed in the brain and other organs, notably in GI tract, with the exception of BRS-3 which is expressed in the central nervous system (Wada et al., 1992; Ohki-Hamazaki et al., 1997a) and the uterus of guinea pig (Gorbulev et al., 1992). Production and analysis of mice lacking GRP-R, NMB-R or BRS-3 revealed that these receptors have overlapping as well as distinct physiological roles. BRS-3, and to a less significant extent GRP-R, play a role in the regulation of metabolism and adiposity (Ohki-Hamazaki et al., 1997b; Hampton et al., 1998). Although the hypothermic effect is regulated by both GRP-R and NMB-R, contraction of smooth muscle in the stomach is controlled exclusively by GRP-R (Wada et al., 1997; Ohki-Hamazaki et al., 1999). In addition, NMB-R is shown to be involved in stress response through modulation of the 5-HT system (Yamano et al., 2002).
GRP and BN are structurally similar, but are distinct peptides (Nagalla et al., 1992). Frogs have GRP and several forms of BN in the brain or in GI tract. A high-affinity BN-like peptide receptor to one type of BN, [Phe13] BN, was cloned from amphibian brain and designated as bombesin-like peptide receptor subtype-4 (BB4) (Nagalla et al., 1995). These results suggested that mammals might have additional BN-like peptide receptors that have high affinity for a certain form of BN, yet to be identified. In fact, the endogenous ligand for BRS-3 has not yet been discovered. To address this question, we attempted to isolate BN-like peptide receptors from chick brain. Phylogenetically, amphibians and amniotes are included in the same taxon of Tetrapoda and birds and mammals are evolutionary descendants of the ancestral amniote lineage. Therefore, knowledge of the BN-like peptide receptors in chick will provide useful information about the evolution of BN-like peptide receptors as well as about the molecular basis for ligand selectivity of each receptor subtype by comparing the amino-acid sequences.
Methods
Cloning of chick BN-like peptide receptors
Oligo (dT) primed cDNA was synthesized from 200 ng chick brain poly-A+RNA (BD Biosciences Clontech, Palo Alto, CA, U.S.A.) using the Superscript II Preamplification system (Gibco-BRL Life Technologies, Gaithersburg, MD, U.S.A.) and used as a template for polymerase chain reactions (PCR). To isolate chick BN-like peptide receptors, two degenerate primers, BNF11 (5′-CCT CGG T(GCT)G GGG T(TGA)T C(GATC) G T(GC)T TCA C-3′) and BNR10 (5′-GAG CAA AGG GGT TTA C(AG)C A(AG)G A(GA)T T-3′), which correspond to nucleotides at the third and seventh transmembrane domains (TM) of BN-like peptide receptors of various species were designed. PCR was carried out in a 50 μl reaction mixture containing 3 μl of template, 0.4 μM of each primer, 0.2 mM dNTPs and 2.5 U KOD Dash DNA polymerase (TOYOBO, Osaka, Japan), with the GeneAmp PCR System 9700 (Perkin-Elmer Applied Biosystems, Foster City, CA, U.S.A.). Thermocycling conditions consist of 94°C for 10 s, followed by 30 cycles of 94°C for 30 s, 53°C for 2 s, and 74°C for 2 min, then a final extension at 72°C for 7 min.
A resulting PCR product (550 bp) was excised and purified from agarose gel, and fragment termini were made blunt and phosphorylated using a BKL Kit (TAKARA, Tokyo, Japan) and then ligated to the EcoRV site of pBluescriptIISK- (pBSK: Stratagene, La Jolla, CA, U.S.A.). Competent cells were transformed with the ligated vectors. Resultant colonies were isolated and cultured overnight in 3 ml LB medium. Plasmid DNAs were purified with a Quantum Prep Plasmid Miniprep Kit (BIO-RAD, Hercules, CA, U.S.A.). Sequencing reactions were performed using the dideoxy chain termination method according to the protocol followed by the ABI Prism Big Dye Terminator Reaction Kit (Perkin-Elmer Applied Biosystems) with M13 forward (5′-GTT TTC CCA GTC ACG AC-3′) and reverse primers (5′-CAG GAA ACA GCT ATG AC-3′). The ABI377 automated sequencer (Perkin-Elmer Applied Biosystems) was used. Partial sequences of two candidates, named chGRP-R and chBRS3.5, were obtained.
Based on these partial sequences, 5′- and 3′-rapid amplifications of cDNA ends (RACE) were performed with the aid of the SMART™ RACE cDNA Amplification Kit (BD Biosciences Clontech). First-strand cDNA for 5′- and 3′-RACE were synthesized from 100 ng chick brain polyA+ RNA (BD Biosciences Clontech) according to the manufacturer's protocol. The primers used for 5′- and 3′-RACE reactions were GRP201 (5′-GCA TGG GAT GCT TGG ATC TCC ATT GGT-3′) and GRP203 (5′-CAT TTC ATT GCC AGC ATC TGT GCT CGG-3′), respectively for chick GRP-R (chGRP-R), and BRS46 (5′-ATA CCA TGG AGA CAA TCC AGA CA-3′) and BRS49 (5′-ACG GTC ATG CCC GTA AGC AGA TTG AAT C-3′), respectively, for chick BRS-3.5 (chBRS-3.5). The reaction mixture was prepared as indicated by the manufacturer. The cycling parameters were as follows: 20 cycles of 94°C for 5 s, 68°C for 10 s and 72°C for 3 min, followed by the second reaction consisting of 20 cycles of 94°C for 5 s, 60°C for 10 s and 72°C for 3 min. These products were purified by agarose gel electrophoresis, extracted from gel slices and directly sequenced. All of the sequence data obtained were aligned and sequences in the coding regions of both chGRP-R and chBRS-3.5 were determined. GENETYX software was used for genetic information processing.
Phylogenetic analysis
Predicted receptor amino-acid sequences were aligned using Clustal W and were used to construct phylogenetic trees according to the UPGMA method. Phylogenetic and evolutionary analyses were conducted using MEGA version 2.1.
Chromosome preparation and fluorescence in situ hybridization (FISH)
The fluorescence in situ hybridization (FISH) method was used to determine chromosomal location of the chicken GRP-R gene. Splenocyte culture and chromosome preparation were performed as described by Matsuda & Chapman (1995) and Suzuki et al. (1999). Mitogen-stimulated splenocyte culture was synchronized by thymidine block, and the incorporation of 5-bromodeoxyuridine during the late replication stage was made for differential replication staining after the release from excessive thymidine. The chromosomal slides were exposed to UV light after staining with Hoechst 33258.
The chromosome slides were hardened at 65°C for 2 h and then denatured at 70°C in 70% formamide/2 × SSC and dehydrated in a 70–85–100% ethanol series at 4°C. The 1.4 kb cDNA fragment inserted in pBSK was labeled with biotin 16-dUTP (Roche Diagnostics) following the manufacturer's protocol. The labeled DNA fragment was ethanol-precipitated with salmon sperm DNA and E. coli tRNA, and then denatured at 75°C for 10 min in 100% formamide. The denatured probe was mixed with an equal volume of hybridization solution resulting in a final concentration of 50% formamide, 2 × SSC, 10% dextran sulfate, and 2 μg μl−1 bovine serum albumin (BSA) (Sigma-Aldrich Co., St Louis, MO, U.S.A.). In all, 20 μl of this hybridization buffer containing 250 ng labeled DNA was added to the denatured slide, covered with parafilm and incubated overnight at 37°C. The slides were washed for 20 min in 50% formamide/2 × SSC at 37°C, and in 2 × SSC and 1 × SSC for 20 min each at room temperature. After rinsing in 4 × SSC, the slides were incubated under coverslips with goat anti-biotin antibodies (Vector Laboratories) at a 1 : 500 dilution in 1% BSA/4 × SSC for 1 h at 37°C. The slides were washed with 4 × SSC, 0.1% Nonidet P-40 in 4 × SSC and 4 × SSC for 5 min each and then stained with fluorescein-coupled donkey anti-goat IgG (Nordic Immunology) at a 1 : 500 dilution for 1 h at 37°C. After washing with 4 × SSC, 0.1% Nonidet P-40 in 4 × SSC, 4 × SSC for 10 min on the shaker, the slides were rinsed with 2 × SSC and stained with 0.50 μg ml−1 propidium iodide. The chromosome slides were analyzed with a Nikon fluorescence microscope using Nikon filter sets B-2A and UV-2A. Kodak Ektachrome ASA100 films were used for microphotography.
Reverse Transcription (RT) PCR
Total RNAs from embryonic and hatched chick tissues were prepared using TRIzol (Invitrogen, Carlsbad, CA, U.S.A.). For day 4 embryos (i.e., 4 days after the beginning of incubation at 37.8°C; E4), the brain and the body regions were dissected and processed. For E8 and E12 embryos, the whole brain and GI tract were removed separately. Tissues from E16 and E18 embryos were dissected and processed for RNA preparation. Single-stranded cDNA was synthesized from 3 μg of total RNA with the Superscript Preamplification System (Invitrogen). The cDNA was subjected to 35 cycles of PCR (94°C 30 s, 61°C 2 s, 74°C 30 s) using 2.5 U of KOD Dash (TOYOBO), 1 × KOD buffer, 0.2 mM dNTPs and specific primers for either chGRP-R (GRP205: CAT GAG AAA CGT CCC CAA TTT GTT and GRP207: GAA TGC CAG GAT CCG AGC AC) or chBRS-3.5 (BRS412: CCT TTC CAG GAT TAG AAA TAC TGT GCA C and BRS413: CAA TCT GCT TAC GGG CAT GAC C). A control reaction was performed using two primers for chick β-actin gene (forward: GTG ATG AAG CCC AGA GCA AAA G and reverse: AGG GTC CGG ATT CAT CGT ACT C) using the same PCR condition except for the annealing temperature, which was 63°C instead of 61°C. A volume of 10 μl of each reaction mixture was resolved by agarose gel electrophoresis and photographed.
Stable cell lines
The chGRP-R cDNAs were obtained from chick brain cDNA by PCR using the high-fidelity polymerase KOD-plus (TOYOBO) and two primers located just up- and down-stream of the open reading frame, chGRPR-1 (5′-TAT GAC TTT TAA ACT CTG GCG ACT G-3′) and chGRPR-2 (5′-TCT GAG TGC ATC AGA CTT CCT TGC-3′). Similarly, chBRS-3.5 was amplified with chBRS3.5-2 (5′-GCA AGC TAA ATA TTA GAC AAT TTT TTC ATC-3′) and chBRS3.5-3 (5′-GGC AGT AGT GAC TTG ACT GCA AAC GC-3′). The resulting PCR products were cloned into the SmaI site of pBSK. Both strands of each clone were sequenced at least two times.
To generate eukaryotic expression vectors, the chGRP-R fragment was cloned into pBSK. The cloned cDNA plasmids were digested with HindIII and NotI, and the insert fragment was ligated into pcDNA3 (Invitrogen). The chBRS-3.5 fragment was similarly digested with BamHI and EcoRI, and inserted into pcDNA3. The sequences of the inserts were verified and the plasmids were purified with the Endofree Qiagen Maxi Kit (Qiagen, Hilden, Germany) prior to transfection.
Chinese hamster ovary K1 (CHO-K1) cells were seeded in a six-well plate at a density of 3 × 105 per well and grown overnight in Dulbecco's modified Eagle's medium (DMEM; Sigma-Aldrich Co.) supplemented with 10% (v v−1) fetal calf serum (Mitsubishi Chemical corporation, Tokyo, Japan), penicillin (100 U ml−1, Sigma) and streptomycin (100 μg ml−1, Sigma). All cells were maintained at 37°C in a 5% CO2 atmosphere. The following morning cells were transfected with the plasmids (10 μg) containing either pcDNA3-chGRP-R or pcDNA3-chBRS-3.5 using 4 μl LipofectAMINETM (Invitrogen) in Opti-MEM (Gibco-BRL) and DMEM for 5 h. At the end of the incubation period, the medium was replaced with DMEM containing 20% fetal calf serum, penicillin and streptomycin. A 24 h after transfection, cells were cultivated in DMEM with fetal calf serum, penicillin, streptomycin and 1000 μg ml−1 Geneticin (Gibco-BRL) to select for stable transfectants. Selection continued for about 3 weeks. Selected clones were expanded and split (1 : 30–50) every 5–7 days at confluence. Individual clones were screened for high receptor expression using a binding assay described below.
Binding assay
The procedure has already been reported (Kris et al., 1987). Briefly, cells were seeded into 24-well tissue culture plates coated with fibronectin (5 μg cm−2, Sigma) and grown to confluence. The cells were washed three times with binding buffer that consists of DMEM containing 20 mM HEPES (Sigma), 100 μg ml−1 bacitracin (Sigma) and 0.2% BSA (Sigma). For saturation analysis, various concentrations (from 50 pM to 1 nM) of [125I][D-Tyr6, β-Ala11, Phe13, Nle14]bombesin(6–14) ([125I][YAFNl]BN(6–14); Perkin-Elmer Life Sciences, Boston, MA, U.S.A.) were added to binding buffer with or without 1 μM unlabeled [D-Phe6, β-Ala11, Phe13, Nle14]bombesin(6–14) ([FAFNl]BN(6–14); PHOENIX Pharmaceuticals, Belmont, CA, U.S.A.) and then incubated for 30 min at 37°C. The cells were washed twice with ice-cold binding buffer and three times with ice-cold PBS (Sigma) and then solubilized with 1 N NaOH. Total cell-associated radioactivity was determined in a γ-counter. Nonspecific binding was defined as the amount of [125I][YAFNl]BN(6–14) bound to CHO-K1-transfected cells in the presence of 1 μM [FAFNl]BN(6–14), and was subtracted from the total binding for determination of the specific binding. All binding values represented saturable binding. Binding data were analyzed using a radioligand binding analysis program (GraphPad PRISM) and Kd values were determined. For displacement analysis, cells were incubated with 40 pM [125I][YAFNl]BN(6–14) for 30 min at 37°C without or with various (from 1 × 10−10 to 5 × 10−6 M) concentrations of unlabeled peptides; Bombesin (BACHEM California, Torrance, CA, U.S.A.), GRP(14–27) (BACHEM California), porcine NMB10 (PENINSULA Laboratories, San Carlos, CA, U.S.A.) or [FAFNl]BN(6–14) (Figure 1). The dose–inhibition curves assessing the potency of peptides to inhibit binding of [125I][YAFNl]BN(6–14) to CHO-K1 expressing chick BN-like peptide receptors were analyzed by a curve-fitting program (SOFTmax Pro) and found to be best fit to four logistic parameters. IC50 were calculated using this program, and Ki values were obtained from the IC50 value using the Cheng–Prusoff equation (Ki=IC50/(1+[ligand]/Kd)).
Figure 1.
Amino-acid sequences of amphibian BN, chGRP, C-terminal portion (14–27) of pig and human GRP, synthetic BN analog ([FAPNl]BN(6–14)) and porcine NMB10.
Measurement of intracellular calcium ([Ca2+])
To measure changes in cytosolic calcium, transfected cells were plated on 35-mm glass-bottomed plastic dishes (MatTek, Ashland, MA, U.S.A.) at a density of 1 × 105 cells per well. After 24 h of culture, the cells were washed with buffer (145 mM NaCl, 2 mM CaCl2, 3 mM KCl, 1 mM MgCl2, 5 mM glucose, 10 mM HEPES, pH 7.4) and incubated with 10 μM fluo-3/AM (Molecular Probes, Eugene, OR, U.S.A.) in the buffer for 15 min at 37°C under reduced light condition. After washing with the buffer, the dish was mounted on the stage of confocal laser scanning microscope (FLUOVIEW 500; Olympus, Tokyo, Japan). Fluo-3 was excited at 488 nm with an argon laser and fluorescence was detected at 526 nm. Agonist diluted in the buffer was directly added to the glass-bottom area of the dish.
Results
Identifying and cloning of chick BN-like peptide receptors
Chick brain poly A+ RNA was reverse transcribed and amplified with degenerate primers. The primers were designed based on the published sequences of mammalian and amphibian BN-like peptide receptors. We chose the most conserved regions, TM3 and 7, to determine the sequence of the degenerate primers. The 550 bp PCR product was cloned into the pBSK and sequenced. Two different sequences showing identities to known BN-like peptide receptors were obtained. One candidate clone showed high identity to GRP-R and we named it chGRP-R. Another one having significant identities to both BRS-3 and BB4 was designated as chBRS-3.5. Based on these partial sequences, primers for both 5′ and 3′RACE were designed. For chGRP-R, a 1 kb single band was obtained in each 5′ and 3′RACE and the putative chGRP-R sequence in its coding region was determined (Figure 2). 5′ and 3′RACE for chBRS-3.5 generated 0.8 and 1 kb band, respectively, and the sequence of the whole coding region was determined (Figure 3). The chGRP-R and chBRS-3.5 cDNAs were 1164 and 1191 bp long, encoded a protein of 387 and 396 amino acids with a calculated molecular mass of 43.7 and 44.2 kDa, respectively, in the absence of any post-translational modifications.
Figure 2.
Nucleotide and predicted amino-acid sequences of chGRP-R (Database accession: AY225966). Putative TM domains are underlined and numbered. Location of potential N-linked glycosylation sites (arrowed) and PKC phosphorylation sites (starred) are shown. Boxed cysteine residues indicate a putative palmitoylation site. Two cysteine residues possibly forming disulfide bonds are double-lined.
Figure 3.
Nucleotide and predicted amino-acid sequences of chick BRS-3.5 (Database accession: AY225967). For an explanation of symbols, see Figure 1 legend.
Hydropathy analysis of the predicted chGRP-R and chBRS-3.5 proteins exhibited seven hydrophobic regions with an extracellular amino (N-) terminal end followed by three interdigitated intracellular and extracellular loops and a carboxy (C-) terminal intracellular domain, consistent with a seven transmembrane structure typical of members of the G-protein-coupled receptor superfamily. As shown in Figure 2 and Figure 3, there are four sites of potential N-linked glycosylation (Asn-X-Ser/Thr). Three of them are present within the extracellular N-terminal domain of both chGRP-R (Asn20, Asn24 and Asn28; arrowed) and chBRS-3.5 (Asn10, Asn18 and Asn29; arrowed), and one site within the second extracellular loop (Asn192 and Asn196, respectively; arrowed). These two receptors contain three Cys residues that are highly conserved among G-protein-coupled receptors: one in the first extracellular loop (Cys115 and Cys119, respectively; double-lined) and another in the second extracellular loop (Cys198 and Cys202, respectively; double-lined), and finally within the intracellular C-terminal domain (Cys341,342 and Cys346,347, respectively; boxed). Consensus sequences of a potential protein kinase C (PKC) phosphorylation site (Ser/Thr-X-Arg/Lys) are well conserved in both proteins at the site between TM5 and 6 (Ser260 and Ser264, respectively; starred) and within the C-terminal cytoplasmic region (Ser330,362, Thr357 of chGRP-R and Ser335 of chBRS-3.5, respectively; starred).
When the predicted amino-acid sequences are compared within two avian receptors, the homology between chGRP-R and chBRS-3.5 is 60%' higher than that among mouse receptors (48% between moGRP-R and moBRS-3, 55% between moGRP-R and moNMB-R and 46% between moNMB-R and moBRS-3). As depicted in Figure 4, amino acids are well conserved in the TM domains, especially in TM2, 3, 6 and 7. In contrast, the N- and C-terminal domains are less preserved. The putative chGRP-R amino-acid sequence is also compared with the mouse, rat and human GRP-R. As shown in Figure 5, sequence similarity is high except for the N-terminal region and the overall amino-acid identity is more than 80% between chick and mouse, rat or human (Table 1). The similarity between chBRS-3.5 and mammalian BRS-3 is 66–69% (Table 1), and the sequence disparities are evident in the N-, C-terminal regions and in the third extracellular loop (Figure 6). The putative chBRS-3.5 protein has 74% identity to frog BB4, higher than the identity between chBRS-3.5 and mammalian BRS-3.
Figure 4.
Comparison of predicted amino-acid sequences of two BN-R subtypes in chick. Asterisks designate identical amino-acid residues in the two sequences.
Figure 5.
Alignment of predicted GRP-R amino-acid sequences of chick, human, mouse and rat. Identical residues in the four sequences are boxed.
Table 1.
Predicted amino-acid identity (%) of BN-like peptide receptor subtypes
| BB4 | BRS-3.5 | BRS-3 | GRP-R | NMB-R | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| fr | Chick | mo | ra | gp | sh | hu | Chick | mo | ra | hu | mo | ra | hu | |
| ChGRP-R | 60 | 60 | 59 | 60 | 61 | 60 | 63 | — | 80 | 80 | 82 | 60 | 60 | 60 |
| ChBRS-3.5 | 74 | — | 67 | 67 | 66 | 68 | 69 | 60 | 58 | 59 | 58 | 52 | 52 | 52 |
Receptor sequences have been reported as follows: Frog (fr) BB4 (Nagalla et al., 1995), mouse (mo) BRS-3 (Ohki-Hamazaki et al., 1997a), rat (ra) BRS-3 (Liu et al., 2002), guinea pig (gp) BRS-3 (Gorbulev et al., 1992), sheep (sh) BRS-3 (Whitley et al., 1999), human (hu) BRS-3 (Fathi et al., 1993), mouse GRP-R (Spindel et al., 1990), rat GRP-R (Wada et al., 1992), human GRP-R (Corjay et al., 1991), mouse NMB-R (Ohki-Hamazaki et al., 1997a), rat NMB-R (Wada et al., 1991), human NMB-R (Corjay et al., 1991).
Figure 6.
Comparison of deduced amino-acid sequences of chick BRS-3.5 with mammalian BRS-3 (a), or with frog (fr) BB4 (b). gp, guinea pig; hu, human; mo, mouse; ra, rat; sh, sheep. Identical residues are boxed. References for each sequence are indicated in Table 1.
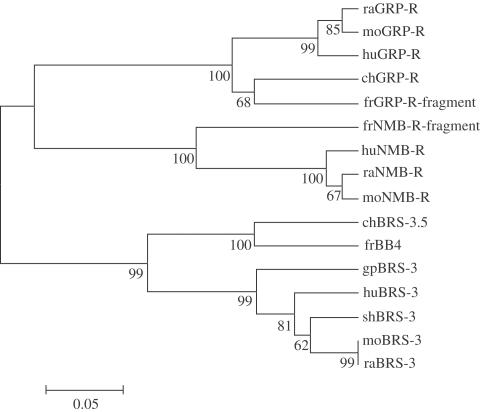
Phylogenetic analysis of all BN-like peptide receptors found in the database demonstrated three main branches: GRP-R, NMB-R and BB4/BRS-3 families (Figure 7). ChGRP-R is a component of the vertebrate BN-R family, together with the GRP-R subtype. ChBRS-3.5, although a member of BN-R family, is not a member of GRP-R or NMB-R subtype, but a member of BRS-3/BB4 subtype. This analysis also revealed that chBN-R genes are more closely related to frog BN-R rather than mammalian BN-R.
Figure 7.
Phylogenetic analysis of BN-like peptide receptors. Putative amino-acid sequences were aligned and the alignments were used to construct phylogenetic trees according to the UPGMA criteria. Numbers next to branch points are bootstrap values based on 1000 resamplings. The evolutionary distance was described in terms of substitutions per site. Amino-acid sequences of fr (frog) GRP-R and NMB-R proteins are from Nagalla et al. (1995). Other references and abbreviations are explained in Table 1.
Chromosomal localization of the chGRP-R gene
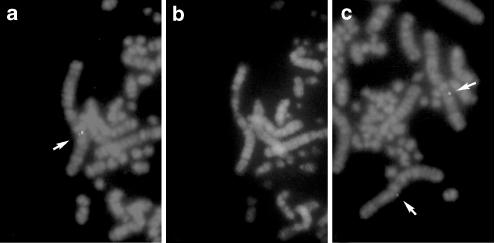
Chromosomal location of the chGRP-R gene was determined by FISH (Figure 8). The chGRP-R was localized to chicken chromosome 1q23distal–q24proximal (ChickGBASE; http://www.thearkdb.org/browser?species=chicken, 2002). This region, chicken chromosome 1q had been shown to contain five chicken homologs of human X-linked genes, MAOA, OTC, NROB1, ZFX and SCML2 (Schmid et al., 2000; Burt, 2002).
Figure 8.
Chromosomal localization of the chGRP-R using a cDNA clone as a biotinylated probe. Arrows indicates the hybridization signals. The metaphase spreads were photographed with Nikon B-2A (a, c) and UV-2A (b) filters. R- and G-banded patterns are demonstrated in (a, c) and (b), respectively.
Expression pattern of chBN-R
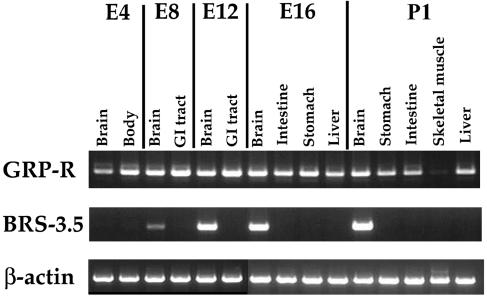
Tissue distribution and developmental changes in expression of chBN-Rs are analyzed by the RT–PCR method (Figure 9). Striking differences are found between the tissue distribution patterns of these two receptors. Significant expression of chGRP-R mRNA was detected in the brain, GI tissue and liver, but not in skeletal muscle. The expression could be detected after E4 until at least at P1. In contrast, chBRS-3.5 expression is limited to the brain, and expression could not be detected in the early E4 embryo.
Figure 9.
Tissue-specific expression of two chick BN-like peptide receptors are analyzed by RT–PCR. E, embryonic day; P, posthatching day.
Functional analysis of chBN-R
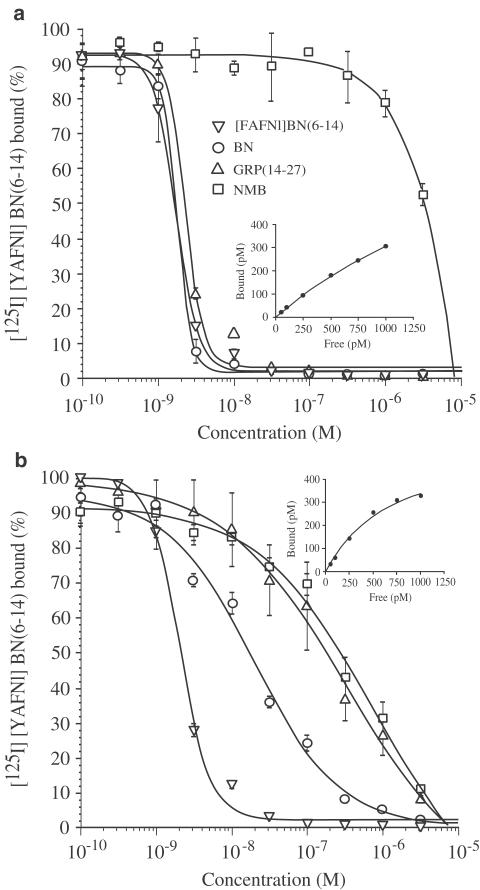
In order to study the pharmacological properties of the chick BN-like peptide receptors, two independent cell lines stably expressing chGRP-R or chBRS-3.5 were established. CHO-K1 cells lacking expression of endogenous BN-like peptide receptors (Katsuno et al., 1999) were transfected with chGRP-R or chBRS-3.5 expression vector using lipofection, and G418-resistant colonies were expanded. To select transfected cell lines expressing high levels of receptors, clones were screened by comparing their ability to bind 40 pM [125I][YAFNl]BN(6–14), which has high affinity for rat GRP-R, rat NMB-R, human BRS-3 as well as for frog BB4 (Pradhan et al., 1998). [125I][YAFNl]BN(6–14) bound to both chGRP-R and chBRS-3.5 in a saturable manner. In the presence of 1 μM of non-radiolabeled ligand, only nonspecific binding was observed, which was proportional to the ligand concentration and lower than 10% of the total binding. Analysis of these data indicated that the ligand possessed a higher affinity for chBRS-3.5 (Kd=0.73 nM) than chGRP-R (Kd=2.57 nM) (Figure 10, insets).
Figure 10.
Abilities of BN-like peptides to inhibit binding of [125I][YAFNl]BN(6–14) to chick GRP-R (a) or chick BRS-3.5 (b) expressed in CHO-K1 cells. Cells expressing either receptor were incubated with 40 pM [125I][YAFNl]BN(6–14) and the indicated concentrations of peptides for 30 min at 37°C. The mean of at least two experiments is shown. Each experiment includes two replicates. [125I][YAFNl]BN(6–14) binding affinity was determined for each cell line in saturation analysis as described under Methods. Saturation binding curves are shown in the insets.
Using these two cell lines, the affinity of chick BN-like peptide receptors to the BN and BN-like peptides was determined in the competition experiments measuring the displacement of [125I][YAFNl]BN(6–14) in the presence of unlabeled peptides. ChGRP-R showed high affinity for [FAFNl]BN(6–14), BN and GRP, with Ki values of 1.7, 1.8 and 2.3 nM, respectively. But this receptor showed more than 1000-fold lower affinity for NMB (Figure 10a). In contrast, rank affinities for chBRS-3.5 were [FAFNl]BN(6–14) (Ki=2.0 nM)>BN (Ki=19.1 nM)>GRP (Ki=318 nM)>NMB (Ki=678 nM), demonstrating that this receptor had high affinity for [FAFNl]BN(6–14), moderate affinity for BN, but lower affinity for both GRP and NMB (Figure 10b).
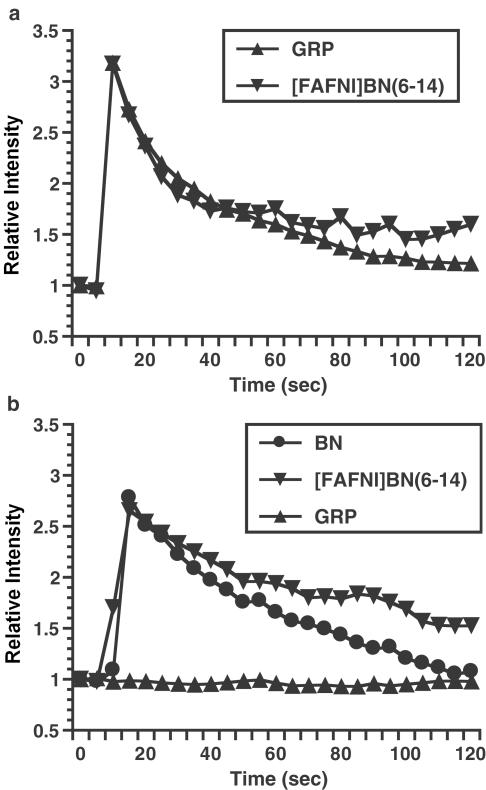
We further evaluated the functional activity of chGRP-R and chBRS-3 in the calcium mobilization assay. When CHO cells stably expressing chGRP-R were incubated with calcium indicator Fluo-3 and stimulated with either 10 nM GRP or 10 nM [FAFNl]BN(6–14), a transient three-fold increase in intracellular calcium concentration was observed (Figure 11a). Similarly, both 10 nM BN and 10 nM [FAFNl]BN(6–14) elicited strong response in CHO cells expressing chBRS-3.5 (Figure 11b). Since the cells are rapidly desensitized after agonist challenge, we could not perform dose–response analysis in detail by using a confocal microscope. However, we confirmed that GRP failed to elicit calcium response at 10 nM (Figure 11b), although 1 μM GRP was able to increase intracellular calcium (data not shown). These results showed that these receptors when expressed on the cell surface can initiate an appropriate signal transduction cascade.
Figure 11.
Intracellular calcium mobilization in CHO-K1 cells expressing chick GRP-R (a) or chick BRS-3.5 (b). Cells were labeled with calcium indicator Fluo-3/AM, stimulated with 10 nM of the indicated agonists and fluorescence was measured over 120 s.
Discussion
In this paper, we reported for the first time, the cloning and characterization of two BN-like peptide receptors in birds. By exploiting the PCR technology using degenerative primers, chick GRP-R and BRS-3.5 were cloned. The amino-acid sequence of chGRP-R was highly homologous to mammalian and amphibian GRP-R and in addition, its pharmacological properties, which were characterized by high affinity for [FAFNl]BN(6–14) (Ki=1.7 nM), BN (Ki=1.8 nM) and GRP (Ki=2.3 nM), resembled GRP-Rs cloned and characterized to date. For example, rat pancreatic acinar cells expressing GRP-R show a Ki value of 0.99 nM for [FAFNl]BN(6–14) (Pradhan et al., 1998), mouse GRP-R-transfected BALB3T3 has Ki values of 0.9 and 3.1 nM for BN and GRP, respectively (Benya et al., 1994), and these values for human GRP-R transfected BALB3T3 are 1.4 and 6.2 (Benya et al., 1995). ChGRP-R was distributed in the brain and GI tissue, which is a common feature of GRP-R of mammals, amphibians and Aves (Battey et al., 1994; Nagalla et al., 1995). Chick GRP-R gene was mapped to chicken chromosome 1q23distal–q24proximal, the region where the similarity to the human X chromosome had been determined in previous studies (Schmid et al., 2000; Burt, 2002). In contrast, chBRS-3.5 had high similarity to amphibian BB4 and to a lesser extent, to mammalian BRS-3. ChBRS-3.5 had high affinity for the synthetic peptide [FAFNl]BN(6–14) (Ki=2.0 nM), moderate affinity for BN (Ki=19.1 nM) and low affinity for both GRP (Ki=318 nM) and NMB (Ki=678 nM). These pharmacological properties of chBRS-3.5 were reminiscent of BB4 (Ki values are 0.41 nM for [FAFNl]BN(6–14), 14 nM for BN, 79 nM for GRP and 11 nM for NMB) rather than BRS-3 (Ki values are 8.9 nM for [FAFNl]BN(6–14), >10,000 nM for BN and GRP and 4800 nM for NMB in human BRS-3-transfected BALB3T3), suggesting that the chBRS-3.5 belonged to the BB4 family (Mantey et al., 1997; Katsuno et al., 1999). This hypothesis was further supported by phylogenetic analysis based on amino-acid sequence similarity. Expression of chBRS-3.5 mRNA was limited to the brain, a characteristic shared by both BB4 and BRS-3 (Nagalla et al., 1995; Ohki-Hamazaki et al., 1997a). Upon binding of agonist to both receptors, we observed mobilization of intracellular calcium, which showed that these receptors were able to initiate a signal transduction cascade.
After the discovery of BN in frog, antiserum against this peptide was applied to the tissues from other species including birds, revealing immunopositive cells in the endocrine tissue of chick. These studies indicated that the avian proventriculus was an abundant source of BN (Erspamer et al., 1978; Timson et al., 1979; Vaillant et al., 1979). Subsequently, McDonald et al. (1980) succeeded in purifying GRP from chicken proventricular tissue. Its sequence showed the expected C-terminal homology with BN, and identical N and C termini (18 identities out of 27 residues) with porcine GRP (Figure 1). Following intravenous or intracerebroventricular injections, avian GRP had a satiety effect on chick and turkey (Savory & Hodgkiss, 1984; Denbow, 1989). The effectiveness of GRP upon intracerebroventricular injection suggested that this peptide localized not only in the peripheral endocrine organ, as demonstrated earlier by immunohistochemistry, but also in brain. In fact, Vaillant et al. (1979) found substantial quantities of BN-like immunoreactivity in turkey brain; Cassone & Moore (1987) reported the existence of BN-like reactivity in the suprachiasmatic region of house sparrow and Berk & Smith (1994) described projection of BN-immuno-reactive cells to the vagal complex of the pigeon. More recently, Dubbeldam & den Boer-Visser (2000) studied in detail the distribution of GRP immunoreactivity in the brain of the dove. These investigations suggested that receptors for BN-like peptides, in particular GRP, should exist in the avian brain, and here we provide evidence for GRP-R and BRS-3.5 expressions in the brain.
ChGRP-R and chBRS-3.5 retain many structural features that are signatures for members of the G-protein-coupled receptor, and which are common in BN-like peptide receptors. First, three possible N-linked glycosylation sites are present within the extracellular N-terminal domain and also within the second extracellular loop. Second, Cys residues that may serve to stabilize the structure of the receptor are conserved: two Cys residues in the first and second extracellular loops, which can form an intramolecular disulfide bridge constraining the conformation of extracellular domain, and a Cys residue located in the intracellular C-terminal region, which may be important in anchoring the receptor to the plasma membrane. And finally, potential PKC phosphorylation sites are located between TM5 and 6, and within the C-terminal cytoplasmic region. These latter sites along with other Ser and Thr residues that could be phosphorylated with other kinases may be important in desensitization and internalization of the receptor upon agonist binding (Benya et al., 1993; Kroog et al., 1995).
The human Grpr is linked to chromosome Xp22.2–p22.13 (reference or NCBI database: http://www.ncbi.nlm.nih.gov/). Five chicken homologs of human X-linked genes, MAOA, OTC, NROB1, ZFX and SCML2, have been mapped to chicken chromosome 1q (Schmid et al., 2000; Burt, 2002), the region where the chGRP-R was localized by FISH. Therefore, the chGRP-R is mapped to the chromosomal region where conserved linkage homology with human chromosome X has been demonstrated (Schmid et al., 2000). This result confirms that the GRP-R is phylogenically conserved and provides additional genetic evidence to confirm that the Z chromosome, one of the avian sex chromosome, and the mammalian X chromosome had not evolved from a common ancestral linkage group. Concerning chBRS-3.5, we tried to elucidate its chromosomal location using the same method, but unfortunately, we could not define it because of the high background.
When comparing the pharmacological properties of chGRP-R and chBRS-3.5, an important difference should be noted. Although the chGRP-R has high affinities for BN, [FAFNl]BN(6–14) and also for GRP, chBRS-3.5 has high affinity only for [FAFNl]BN(6–14), and affinities for BN and GRP are 10- and 100-folds lower, respectively. These results demonstrate that chBRS-3.5 can discriminate BN and GRP, despite the remarkable structural similarity between these two peptides. This implies that the endogenous ligand for chBRS-3.5 may be some form of BN, probably the chick type, like frog BB4 having [Phe13]BN for the endogenous ligand. In support of this notion, the similarity between frog GRP-R and BB4 is 59% (Nagalla et al., 1995), the value almost equivalent to the similarity between chGRP-R and chBRS-3.5 (60%).
Akeson et al. (1997) reported four amino acids required for high-affinity BN binding. All of these residues, Gln121, Pro199, Arg288 and Ala308 of mouse GRP-R are conserved in equivalent position in chGRP-R, and in chBRS-3.5 except for Ala308. These results coincide with the high affinity of chGRP-R and moderate affinity of chBRS-3.5 for BN. Moreover, two residues, Phe185 and Ala198, have been shown to be indispensable for the selectivity of mouse GRP-R for GRP (Tokita et al., 2002). These residues are conserved in both chGRP-R and chBRS-3.5 that showed selectivity for GRP rather than NMB. Affinity for NMB was not significant in both chGRP-R and chBRS-3.5, consistent with the lack of Ile216 of rat NMB-R in equivalent positions of these receptors (Fathi et al., 1993). However, it should be noted that the low affinity for NMB cannot be explained solely by this fact because BB4 has a high affinity for NMB in spite of the lack of this amino-acid residue (Katsuno et al., 1999). We have not yet succeeded in isolating chick NMB-R, and it is not known whether the birds have NMB as well as NMB-R.
Recently, cloning and characterization of rat BRS-3 revealed that this receptor does not have high affinity for [FAFNl]BN(6–14) (Liu et al., 2002). The amino acids located in the third extracellular loop were divergent between human and rat BRS-3 and by constructing chimeric molecules, they reported that the amino-acid difference of this domain was indeed the main cause of the poor affinity. Alignment of chBRS-3.5 with mammalian BRS-3 revealed major amino-acid differences in the N-terminal domain and in the third extracellular loop. Since the difference in the N-terminal region is conspicuous between chBRS-3.5 and human BRS-3, this domain is probably not the main factor controlling the affinity for [FAPNl]BN(6–14). Therefore, it may be the third extracellular loop that is important for high-affinity binding to [FAPNl]BN(6–14).
The phylogenetic tree indicates that frog BB4, chick BRS-3.5 and mammalian BRS-3 had a close evolutionary history, but the group for frog BB4 and chick BRS-3.5, and mammalian BRS-3 group, evolved differently after a certain point in time. Our results suggest that the endogenous ligand for BRS-3 may have an amino-acid sequence significantly different from BN.
Acknowledgments
This work was supported by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan and the Japan Society for the Promotion of Science.
Abbreviations
- BB4
bombesin-like peptide receptor subtype-4
- BN
bombesin
- BRS-3
bombesin-like peptide receptor subtype-3
- CHO-K1
Chinese hamster ovary K1 cells
- DMEM
Dulbecco's modified Eagle's medium
- [FAFNl]BN(6–14)
[D-Phe6, β−Ala11, Phe13, Nle14]bombesin(6–14)
- FISH
fluorescence in situ hybridization
- GI
gastrointestinal
- GRP
gastrin-releasing peptide
- NMB
neuromedin B
- PBSK
pBluescriptIISK(−)
- PCR
polymerase chain reactions
- RACE
rapid amplifications of cDNA ends
- RT
reverse transcription
- TM
transmembrane domain
- [YAFNl]BN(6–14)
[D-Tyr6, β-Ala11, Phe13, Nle14]bombesin(6–14)
References
- AKESON M., SAINZ E., MANTEY S.A., JENSEN R.T., BATTEY J.F. Identification of four amino acids in the gastrin-releasing peptide receptor that are required for high affinity agonist binding. J. Biol. Chem. 1997;272:17405–17409. doi: 10.1074/jbc.272.28.17405. [DOI] [PubMed] [Google Scholar]
- ANASTASI A., ERSPAMER V., BUCCI M. Isolation and structure of bombesin and alytesin, 2 analogous active peptides from skin of the European amphibians Bombina and Alytes. Experimentia. 1971;27:166–167. doi: 10.1007/BF02145873. [DOI] [PubMed] [Google Scholar]
- BATTEY J., WADA E., WRAY S. Bombesin receptor gene expression during mammalian development. Ann. N. Y. Acad. Sci. 1994;739:244–252. doi: 10.1111/j.1749-6632.1994.tb19826.x. [DOI] [PubMed] [Google Scholar]
- BENYA R.V., FATHI Z., BATTEY J.F., JENSEN R.T. Serines and threonines in the gastrin-releasing peptide receptor carboxyl terminus mediate internalization. J. Biol. Chem. 1993;268:20285–20290. [PubMed] [Google Scholar]
- BENYA R.V., FATHI Z., KUSUI T., PRADHAN T., BATTEY J.F., JENSEN R.T. Gastrin-releasing peptide receptor-induced internalization, down-regulation, desensitization, and growth: possible role for cyclic AMP. Mol. Pharmacol. 1994;46:235–245. [PubMed] [Google Scholar]
- BENYA R.V., KUSUI T., PRADHAN T.K., BATTEY J.F., JENSEN R.T. Expression and characterization of cloned human bombesin receptors. Mol. Pharmacol. 1995;47:10–20. [PubMed] [Google Scholar]
- BERK M.L., SMITH S.E. Local and commissural neuropeptide-containing projections of the nucleus of the solitary tract to the dorsal vagal complex in the pigeon. J. Comp. Neurol. 1994;347:369–396. doi: 10.1002/cne.903470305. [DOI] [PubMed] [Google Scholar]
- BURT D.W. Origin and evolution of avian microchromosomes. Cytogenet. Genome Res. 2002;96:97–112. doi: 10.1159/000063018. [DOI] [PubMed] [Google Scholar]
- CASSONE V.M., MOORE R.Y. Retinohypothalamic projection and suprachiasmatic nucleus of the house sparrow, Passer domesticus. J. Comp. Neurol. 1987;266:171–182. doi: 10.1002/cne.902660204. [DOI] [PubMed] [Google Scholar]
- CORJAY M.H., DOBRZANSKI D.J., VIALLET J., SHAPIRA H., WORLAND P.J., SAUSVILLE E.A., BATTEY J. Two distinct Bombesin receptor subtypes are expressed and functional in human lung carcinoma cells. J. Biol. Chem. 1991;266:18771–18779. [PubMed] [Google Scholar]
- DENBOW D.M. Centrally and peripherally administered bombesin decreases food intake in turkeys. Peptides. 1989;10:275–279. doi: 10.1016/0196-9781(89)90030-2. [DOI] [PubMed] [Google Scholar]
- DUBBELDAM J.L., DEN BOER-VISSER A.M. Distribution of gastrin-releasing peptide immunoreactivity in the brain of the collared dove (Streptopelia decaocto) Cell Tissue Res. 2000;300:139–151. doi: 10.1007/s004410000191. [DOI] [PubMed] [Google Scholar]
- ERSPAMER V., MELCHIORRI P., ERSPAMER C.F., NEGRI L. Polypeptides of the amphibian skin active on the gut and their mammalian counterparts. Adv. Exp. Med. Biol. 1978;106:51–64. doi: 10.1007/978-1-4684-7248-6_6. [DOI] [PubMed] [Google Scholar]
- FATHI Z., CORJAY M.H., SHAPIRA H., WADA E., BENYA R.V., JENSEN R.T., VIALLET J., SAUSVILLE E.A., BATTEY J. BRS-3: a novel bombesin receptor subtype selectively expressed in testis and lung carcinoma cells. J. Biol. Chem. 1993;268:5979–5984. [PubMed] [Google Scholar]
- GORBULEV V., AKHUNDOVA A., BUCHNER H., FAHRENHOLZ F. Molecular cloning of a new bombesin receptor subtype expressed in uterus during pregnancy. Eur. J. Biochem. 1992;208:405–410. doi: 10.1111/j.1432-1033.1992.tb17201.x. [DOI] [PubMed] [Google Scholar]
- HAMPTON L.L., LADENHEIM E.E., AKESON M., WAY J.M., WEBER H.C., SUTLIFF V.E., JENSEN R.T., WINE L.J., ARNHEITER H., BATTEY J. Loss of bombesin-induced feeding suppression in gastrin-releasing peptide receptor-deficient mice. Proc. Natl. Acad. Sci. U.S.A. 1998;95:3188–3192. doi: 10.1073/pnas.95.6.3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KATSUNO T., PRADHAN T.K., RYAN R.R., MANTEY S.A., HOU W., DONOHUE P.J., AKESON M.A., SPINDEL E.R., BATTEY J.F., COY D.H., JENSEN R.T. Pharmacology and cell biology of the bombesin receptor subtype 4 (BB4-R) Biochemistry. 1999;38:7307–7320. doi: 10.1021/bi990204w. [DOI] [PubMed] [Google Scholar]
- KRIS R.M., HAZAN R., VILLINES J., MOODY T.W., SCHLESSINGER J. Identification of the bombesin receptor on murine and human cells by cross-linking experiments. J. Biol. Chem. 1987;262:11215–11220. [PubMed] [Google Scholar]
- KROOG G.S., SAINZ E., WORLAND P.J., AKESON M.A., BENYA R.V., JENSEN R.T., BATTEY J.F. The gastrin releasing peptide receptor is rapidly phosphorylated by a kinase other than protein kinase C after exposure to agonist. J. Biol. Chem. 1995;270:8217–8224. doi: 10.1074/jbc.270.14.8217. [DOI] [PubMed] [Google Scholar]
- LEBACQ-VERHEYDEN A.M., TREPEL J., SAUSVILLE E.A., BATTEY J.Bombesin and gastrin-releasing peptide: neuropeptides, secretogogues, and growth factors Handbook of Experimental Pharmacology. 1990Berlin: Springer-Verlag; 71–124.ed. Roberts, A.B. pp [Google Scholar]
- LIU J., LAO Z.J., ZHANG J., SCHAEFFER M.T., JIANG M.M., GUAN X.M., VAN DER PLOEG L.H., FONG T.M. Molecular basis of the pharmacological difference between rat and human bombesin receptor subtype-3 (BRS-3) Biochemistry. 2002;41:8954–8960. doi: 10.1021/bi0202777. [DOI] [PubMed] [Google Scholar]
- MANTEY S.A., WEBER H.C., SAINZ E., AKESON M., RYAN R.R., PRADHAN T.K., SEARLES R.P., SPINDEL E.R., BATTEY J.F., COY D.H., JENSEN R.T. Discovery of a high affinity radioligand for the human orphan receptor, bombesin receptor subtype 3, which demonstrates that it has a unique pharmacology compared with other mammalian bombesin receptors. J. Biol. Chem. 1997;272:26062–26071. doi: 10.1074/jbc.272.41.26062. [DOI] [PubMed] [Google Scholar]
- MATSUDA Y., CHAPMAN V.M. Application of fluorescence in situ hybridization in genome analysis of the mouse. Electrophoresis. 1995;16:261–272. doi: 10.1002/elps.1150160142. [DOI] [PubMed] [Google Scholar]
- MCDONALD T.J., JORNVALL H., GHATEI M., BLOOM S.R., MUTT V. Characterization of an avian gastric (proventricular) peptide having sequence homology with the porcine gastrin-releasing peptide and the amphibian peptides bombesin and alytesin. FEBS Lettes. 1980;122:45–48. doi: 10.1016/0014-5793(80)80398-x. [DOI] [PubMed] [Google Scholar]
- MCDONALD T.J., JORNVALL H., NILSSON G., VAGNE M., GHATEI M., BLOOM S.R., MUTT V. Characterization of a gastrin releasing peptide from porcine non-antral gastric tissue. Biochem. Biophys. Res. Commun. 1979;90:227–233. doi: 10.1016/0006-291x(79)91614-0. [DOI] [PubMed] [Google Scholar]
- MINAMINO N., KANGAWA K., MATSUO H. Neuromedin B: a novel bombesin-like peptide identified in porcine spinal cord. Biochem. Biophys. Res. Commun. 1983;114:541–548. doi: 10.1016/0006-291x(83)90814-8. [DOI] [PubMed] [Google Scholar]
- NAGALLA S.R., BARRY B.J., CRESWICK K.C., EDEN P., TAYLOR J.T., SPINDEL E.R. Cloning of a receptor for amphibian [Phe13] bombesin distinct from the receptor for gastrin-releasing peptide: identification of a fourth bombesin receptor subtype (BB4) Proc. Natl. Acad. Sci. U.S.A. 1995;92:6205–6209. doi: 10.1073/pnas.92.13.6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NAGALLA S.R., GIBSON B.W., TANG D., REEVE J.R., SPINDEL E.R. Gastrin-releasing peptide (GRP) is not mammalian bombesin. Identification and molecular cloning of a true amphibian GRP distinct from amphibian in Bombina orientalis. J. Biol. Chem. 1992;267:6916–6922. [PubMed] [Google Scholar]
- OHKI-HAMAZAKI H., SAKAI Y., KAMATA K., OGURA H., OKUYAMA S., WATASE K., YAMADA K., WADA K. Functional properties of two bombesin-like peptide receptors revealed by the analysis of mice lacking neuromedin B receptor. J. Neurosci. 1999;19:948–954. doi: 10.1523/JNEUROSCI.19-03-00948.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OHKI-HAMAZAKI H., WADA E., WADA K. Cloning and expression of the neuromedin B receptor and the third subtype of bombesin receptor genes in the mouse. Brain Res. 1997a;762:165–172. doi: 10.1016/s0006-8993(97)00380-6. [DOI] [PubMed] [Google Scholar]
- OHKI-HAMAZAKI H., WATASE K., YAMAMOTO K., OGURA H., YAMAMOTO M., YAMADA K., MAENO H., IMAKI J., KIKUYAMA S., WADA E., WADA K. Mice lacking bombesin receptor subtype-3 develop metabolic defects and obesity. Nature. 1997b;390:165–169. doi: 10.1038/36568. [DOI] [PubMed] [Google Scholar]
- PRADHAN T.K., KATSUNO T., TAYLOR J.T., KIM S.H., RYAN R.R., MANTEY S.A., DONOHUE P.J., WEBER H.C., SAINZ E., BATTEY J.F., COY D.H., JENSEN R.T. Identification of a unique ligand which has high affinity for all four bombesin receptor subtypes. Eur. J. Pharm. 1998;343:275–287. doi: 10.1016/s0014-2999(97)01527-6. [DOI] [PubMed] [Google Scholar]
- SAVORY C.J., HODGKISS J.P. Influence of vagotomy in domestic fowls on feeding activity, food passage, digestibility and satiety effects of two peptides. Physiol. Behav. 1984;33:937–944. doi: 10.1016/0031-9384(84)90233-6. [DOI] [PubMed] [Google Scholar]
- SCHMID M., NANDA I., GUTTENBACH M., STEINLEIN C., HOEHN M., SCHARTL M., HAAF T., WEIGEND S., FRIES R., BUERSTEDDE J.M., WIMMERS K., BURT D.W., SMITH J., A'HARA S., LAW A., GRIFFIN D.K., BUMSTEAD N., KAUFMAN J., THOMSON P.A., BURKE T., GROENEN M.A., CROOIJMANS R.P., VIGNAL A., FILLON V., MORISSON M., PITEL F., TIXIER-BOICHARD M., LADJALI-MOHAMMEDI K., HILLEL J., MAKI-TANILA A., CHENG H.H., DELANY M.E., BURNSIDE J., MIZUNO S. First report on chicken genes and chromosomes 2000. Cytogenet. Cell Genet. 2000;90:169–218. doi: 10.1159/000056772. [DOI] [PubMed] [Google Scholar]
- SPINDEL E.R., GILADI E., BREHM P., GOODMAN R.H., SEGERSON T.P. Cloning and functional characterization of a complementary DNA encoding the murine fibroblast bombesin/gastrin-releasing peptide receptor. Mol. Endocrinol. 1990;4:1956–1963. doi: 10.1210/mend-4-12-1956. [DOI] [PubMed] [Google Scholar]
- SUZUKI T., KUROSAKI T., SHIMADA K., KANSAKU N., KUHNLEIN U., ZADWORNY D., AGATA K., HASHIMOTO A., KOIDE M., KOIKE M., TAKATA M., KUROIWA A., MINAI S., NAMIKAWA T., MATSUDA Y. Cytogenetic mapping of 31 functional genes on chicken chromosomes by direct R-banding FISH. Cytogenet. Cell Genet. 1999;87:32–40. doi: 10.1159/000015388. [DOI] [PubMed] [Google Scholar]
- TIMSON C.M., POLAK J.M., WHARTON J., GHATEI M.A., BLOOM S.R., USELLINI L., CAPELLA C., SOLCIA E., BROWN M.R., PEARSE A.G. Bombesin-like immunoreactivity in the avian gut and its localization to a distinct cell type. Histochemistry. 1979;61:213–221. doi: 10.1007/BF00496533. [DOI] [PubMed] [Google Scholar]
- TOKITA K., HOCART S.J., COY D.H., JENSEN R.T. Molecular basis of the selectivity of gastrin-releasing peptide receptor for gastrin-releasing peptide. Mol. Pharmacol. 2002;61:1435–1443. doi: 10.1124/mol.61.6.1435. [DOI] [PubMed] [Google Scholar]
- VAILLANT C., DOCKRAY G.J., WALSH J.H. The avian proventriculus is an abundant source of endocrine cells with bombesin-like immunoreactivity. Histochemistry. 1979;64:307–314. doi: 10.1007/BF00495031. [DOI] [PubMed] [Google Scholar]
- WADA E., WATASE K., YAMADA K., OGURA H., YAMANO M., INOMATA Y., EGUCHI J., YAMAMOTO K., SUNDAY M.E., MAENO H., MIKOSHIBA K., OHKI-HAMAZAKI H., WADA K. Generation and characterization of mice lacking gastrin-releasing peptide receptor. Biochem. Biophys. Res. Commun. 1997;239:28–33. doi: 10.1006/bbrc.1997.7418. [DOI] [PubMed] [Google Scholar]
- WADA E., WAY J.M., SHAPIRA H., KUSANO K., LEBACQ-VERHEYDEN A.M., COY D., JENSEN R.T., BATTEY J. cDNA cloning, characterization, and brain region-specific expression of a neuromedin B-preferring bombesin receptor. Neuron. 1991;6:421–430. doi: 10.1016/0896-6273(91)90250-4. [DOI] [PubMed] [Google Scholar]
- WADA E., WRAY S., KEY S., BATTEY J. Comparison of gene expression for two distinct bombesin receptor subtypes in postnatal rat central nervous system. Mol. Cell. Neurosci. 1992;3:446–460. doi: 10.1016/1044-7431(92)90056-8. [DOI] [PubMed] [Google Scholar]
- WHITLEY J.C., MOORE C., GIRAUD A.S., SHULKES A. Molecular cloning, genomic organization and selective expression of bombesin receptor subtype 3 in the sheep hypothalamus and pituitary. J. Molec. Endocrinol. 1999;21:107–116. doi: 10.1677/jme.0.0230107. [DOI] [PubMed] [Google Scholar]
- YAMANO M., OGURA H., OKUYAMA S., OHKI-HAMAZAKI H. Modulation of 5-HT system in mice with a targeted disruption of neuromedin B receptor. J. Neurosci. Res. 2002;68:59–64. doi: 10.1002/jnr.10194. [DOI] [PubMed] [Google Scholar]