Abstract
A mouse homeobox gene, Nkx-1.2, (previously termed Sax-1) that is closely related to the Drosophila NK-1/S59 gene was cloned, and genomic DNA and cDNA were sequenced. Nine Nkx-1.2 cDNA clones were found that correspond to three species of Nkx-1.2 mRNA that are formed by alternative splicing at conventional 5′ donor and 3′ acceptor splice sites; however, seven cDNA clones were found that correspond to three species of Nkx-1.2 mRNA from testes that have novel TG/AC 5′ and 3′ splice sites. The consensus splice sequences are: 5′ donor, CC↓TGGAAG; 3′ acceptor, ACTTAC↓. Predicted amino acid sequences suggest that some transcripts may be translated into proteins that lack part or all of the homeodomain. At least three bands of Nkx-1.2 mRNA were found in RNA from the testes. Nkx-1.2 mRNA was shown to be present in postmeiotic germ cells of the testis and in mature spermatozoa. Nkx-1.2 mRNA also was found in regions of the adult cerebral cortex, hippocampus, diencephalon, pons/medulla, and cerebellum. Nkx-1.2 mRNA was found in embryos in highest abundance in 10-day embryos; the mRNA levels decrease during further development. Nkx-1.2 mRNA also was found in discrete zones of the embryonic mesencephalon and myelencephalon.
Keywords: Sax-1, transcription factors, embryo, brain, spermatozoa
The Drosophila NK-1/S59 homeobox gene (1, 2) encodes a homeodomain protein that is expressed about 5 hr after fertilization in mesodermal cells that develop as a subset of muscle founder cells (2). In addition, NK-1/S59 is expressed in a subset of cells in the developing central nervous system and a small region of the midgut (2). The NK-1/S59 homeodomain has been highly conserved during evolution, and close homologs have been identified in Caenorhabditis elegans (3), honeybee (4), flatworm (5), chicken (6), mouse (7, 8), and humans (9). In the mouse, two homologs of NK-1 have been described: Nkx-1.1 (7), also named Sax-2 (10), and Nkx-1.2, previously termed Nkx-1.1 (11) or Sax-1 (8). The patterns of expression of Nkx-1.2/Sax-1 in the mouse (8) and Sax-1 in the chick embryo (12) have been described. In both species Sax-1 mRNA is expressed in the ectoderm aligned with the primitive streak. Later in development, the mRNA extends into the spinal part of the neural plate, with the last somite formed as the anterior border of Sax-1 expression throughout somitogenesis. Around midgestation, Sax-1 is expressed in the hindbrain, spinal cord, and more anterior parts of the brain, including the mesencephalon, tegmentum, diencephalon, and pretectum. Based on the expression pattern, the mouse Nkx-1.2/Sax-1 protein was hypothesized to be involved in specification of the posterior neuroectoderm and formation of the posterior part of the neural plate and, later in development, in specification of subsets of neurons within more anterior areas of the central nervous system (8), similar to a role proposed for the Drosophila homolog NK-1/S59 (2).
In the present paper we describe mouse Nkx-1.2 genomic DNA and Nkx-1.2 cDNA corresponding to six species of Nkx-1.2 mRNA formed by alternative splicing. Nucleotide sequence alignments between Nkx-1.2 genomic and cDNA clones revealed expected mRNA splice sites as well as noncanonical sequences for 5′ donor and 3′ acceptor mRNA splice sites. In addition, we show that Nkx-1.2 mRNA is expressed in adult brain, postmeiotic male germ cells, and spermatozoa. The analysis of Nkx-1.2 expression also was extended to later stages of mouse embryogenesis.
Materials and Methods
Library Screening and Gene Cloning.
A DNA fragment, 120 bp in length, amplified by PCR from mouse genomic DNA and encompassing part of the Nkx-1.2 homeobox was a gift from Yongsok Kim (National Institutes of Health). A genomic DNA library from BALB/cAn mouse liver DNA partially digested with EcoRI and cloned in the EcoRI site of EMBL-4 was used. Approximately 106 plaque-forming units were screened in duplicate by hybridization under moderate stringency with a single-stranded Nkx-1.2 DNA probe labeled with [32P] by primer extension catalyzed by the Klenow fragment of DNA polymerase I (New England Biolabs).
Reverse-Transcription–PCR (RT-PCR).
First-strand cDNA was synthesized from total or poly(A+) RNA samples previously treated with RNase-free DNase I (Life Technologies, Rockville, MD), using oligo(dT)12–18 primers and SuperScript II RT (Life Technologies). RNA was removed by incubation with Escherichia coli RNase H (Life Technologies), and aliquots of (−) strand cDNA were subjected to PCR with gene-specific primers and either Taq or Ultma DNA polymerase (Boehringer Mannheim), or TaKaRa LA Taq (Oncor) or eLONGase Enzyme Mix (Life Technologies).
DNA Sequencing.
Cloned DNA fragments obtained by library screening or PCR were subcloned in pBluescript II KS+ (Stratagene), and both strands were sequenced with a Perkin–Elmer/Applied Biosystem 373A DNA sequencer using fluorescent dideoxynucleotides. Sequence analysis and assembly was performed by using the Wisconsin Sequence Analysis Package (GCG).
Nkx-1.2 Chromosomal Mapping.
The Nkx-1.2 gene was mapped by analysis of progeny from the crosses (NFS/N × Mus spretus) × M. spretus or C58/J (13).
RNase Protection Assay, Northern Analysis, and in Situ Hybridization.
Mouse embryos at the indicated developmental stages or organs from adult mice were dissected in ice-cold PBS. Tissues for in situ hybridization were embedded immediately in Tissue-Tek OCT compound and frozen in a mixture of 2-methylbutane/dry ice; otherwise, tissues were weighed, frozen, and stored at −80°C until needed. Total RNA was prepared by a modified version of the Chomczynski and Sacchi method (14) (Totally RNA system, Ambion, Austin, TX), and poly(A)+ RNA was purified by chromatography on oligo(dT) cellulose spin columns following the manufacturer's instructions (5 Prime → 3 Prime). For RNase protection assays, a 32P-labeled antisense Nkx-1.2 RNA probe was synthesized from a linearized recombinant plasmid DNA template using T7 RNA polymerase (MaxiScript system, Ambion), and poly(A)+ RNA samples were processed according to the RPA II system (Ambion). For Northern analyses, MTN blots were purchased from CLONTECH and hybridized according to the manufacturer's instructions with an Nkx-1.2 DNA probe labeled with [32P] by random priming catalyzed by the FPLCpure Klenow fragment (Oligolabeling system, Amersham Pharmacia Biotech). For in situ hybridization, serial sections (10–12 μm thick) were cut from frozen mouse embryos or brains in a cryostat at −20°C and collected on slides coated with poly(l-lysine). Sections were fixed and hybridized with RNA probes labeled with [35S] or [33P], as described by Hogan et al. (15). Paraffin-embedded sections of testes from adult mice were purchased from Novagen (Hybrid-Ready tissues) and processed for hybridization with digoxigenin-labeled RNA probes as described by Dijkman et al. (16). Antisense and sense Nkx-1.2 RNA were synthesized from linearized DNA templates either by incorporating uridine 5′ [α-35S]thiotriphosphate catalyzed by T3 or T7 RNA polymerase (Ambion), or by incorporating DIG-11-UTP in the presence of T7 RNA polymerase (DIG/Genius 4 system, Boehringer Mannheim).
Mouse Spermatozoa.
Approximately 1.5 × 108 spermatozoa were recovered from the lumen of the vas deferens and from the epididymis of six 12- to 15-week-old mice as described (17), collected by centrifugation, and frozen at −80°C.
Results and Discussion
Cloning Nkx-1.2 Genomic DNA.
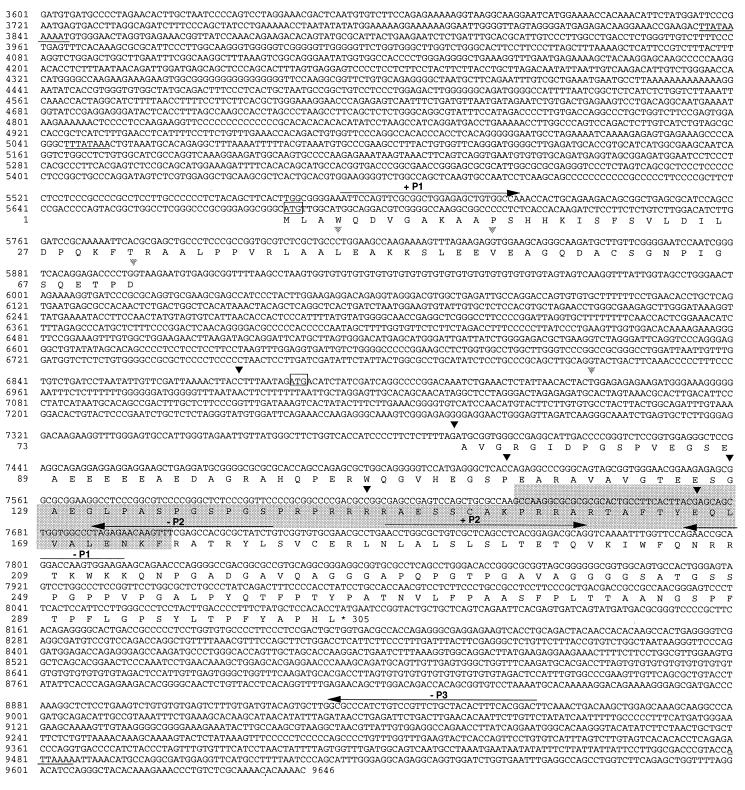
Two recombinant Nkx-1.2 clones were isolated by screening a mouse genomic DNA library in EMBL-4. Analysis of restriction maps obtained with EcoRI, BamHI, HindIII, SalI, EcoRI plus BamHI, EcoRI plus HindIII, or EcoRI plus SalI showed that the two clones partially overlap. By sequencing restriction fragments of DNA subcloned in pBlueScript, a Nkx-1.2 contig, 9,646 bp in length, was identified and deposited in GenBank. The nucleotide sequence shown in Fig. 1 is a composite of nucleotide residues 3601–9646 of Nkx-1.2 genomic DNA and a Nkx-1.2 cDNA contig that was constructed by assembling two partially overlapping cDNA clones obtained by RT-PCR with mouse embryo poly(A)+ RNA and primer pairs +P1 × −P1 and −P2 × −P3. The deduced amino acid sequence also is shown. Comparison of the nucleotide sequences of Nkx-1.2 genomic DNA and cDNA showed that the Nkx-1.2 gene is composed of two exons that encode a deduced protein of 305 amino acid residues that is identical to Sax-1 (8). However, the nucleotide sequence previously reported (8) for the 5′ end of Sax-1 cDNA (nucleotides 1–73) differs from the nucleotide sequence of genomic Nkx-1.2 DNA. This discrepancy might be explained by the presence of an untranslated 5′ exon in Sax-1 cDNA. However, further sequence comparison failed to identify the 73 nucleotide residues in about 5 kb of 5′ flanking Nkx-1.2 genomic DNA; instead, the reverse, complementary nucleotide sequence was found near of the 3′ end of Nkx-1.2 genomic DNA (8843–8907 residues in Fig. 1), and in PCR products obtained with several primer pairs and mouse genomic DNA, mouse embryo cDNA, or brain cDNA (data not shown). These data suggest that the initial 73-nt residue sequence previously reported (8) at the 5′ terminus of Sax-1 cDNA is a cloning artifact.
Figure 1.
Nucleotide sequence and predicted amino acid sequence of Nkx-1.2. A portion of the Nkx-1.2 genomic DNA sequence is aligned to the nucleotide sequence and predicted amino acid sequence of an Nkx-1.2 cDNA constructed by assembling two overlapping cDNA clones obtained by RT-PCR with primer pairs +P1 × −P1 and +P2 × −P3. The positions of primers used for RT-PCR experiments are indicated by arrows. Putative TATA boxes and a polyadenylation signal are underlined; putative translation initiation sites are enclosed in boxes. Inverted triangles indicate splice sites shown in Fig. 3.
The mouse genome contains two homeobox genes with homeodomains that are highly related to the Drosophila NK-1 homeodomain, i.e., NKx-1.1 (7) and Nkx-1.2. The amino acid sequences of the two homeodomains differ by only two amino acid residues and both are conservative amino acid substitutions. However, only 34% of the 38 amino acid residues that can be compared that flank the homeodomain are identical.
Mouse Nkx-1.2 originally was named Nkx-1.1 by Rovescalli et al. (18) and Yamada et al. (11) based on the homology of the amino acid sequence of the homeodomain to the Drosophila NK-1 homeodomain. cDNA corresponding to a different but related homeobox gene was reported subsequently and also was named Nkx-1.1 (7). To avoid confusion, Gruss and his colleagues (8) renamed their Nkx-1.1 cDNA Sax-1 because the amino acid sequence of the mouse homeodomain is closely related to the Sax-1 homeodomain of the chicken (12), previously named Chox-3 (6). We have renamed the mouse Nkx-1.1 gene that we have been characterizing and the mouse Sax-1 gene, Nkx-1.2, to comply with the recommended nomenclature for mouse homeobox genes, to indicate the homology between the mouse Nkx-1.1 and Nkx-1.2 homeodomains and the Drosophila NK-1 homeodomain, and to avoid further confusion in nomenclature.
Chromosomal Location of the Nkx-1.2 Gene.
Southern analysis of genomic DNA from the progeny of (NFS/N × Mus spretus) × M. spretus or C58/J crosses with a 500-bp DNA probe encoding the Nkx-1.2 homeodomain and the C-terminal region of the protein identified fragments of >28 kb in M. spretus DNA and 9.3 kb in NFS/N. Inheritance of the polymorphic DNA fragments in the progeny of the genetic cross was compared with the inheritance of marker loci, and the Nkx-1.2 gene was mapped to chromosome 7, with no recombination with Oat in 100 mice. A bacterial artificial chromosome (BAC) clone containing Nkx-1.2 genomic DNA isolated from a mouse genomic DNA library also contains part of Oat (data not shown); therefore, Nkx-1.2 is located within 100–300 kb (i.e., the average size of BAC library DNA inserts) of Oat. Nkx-5.1/Hmx3 and Nkx-5.2/Hmx2, two mouse homeobox genes moderately related to Drosophila NK-1 (58% identical amino acid sequence in the homeodomain), and NK-3 (60% identical amino acid residues in the homeodomain) also have been mapped to this same region of chromosome 7 (19), suggesting that there may be a cluster of homeobox genes here. Nkx-1.1 (also referred to as Sax-2) has been mapped to mouse chromosome 5 (7, 10).
Expression of Nkx-1.2 in Mouse Embryos and Adult Tissues.
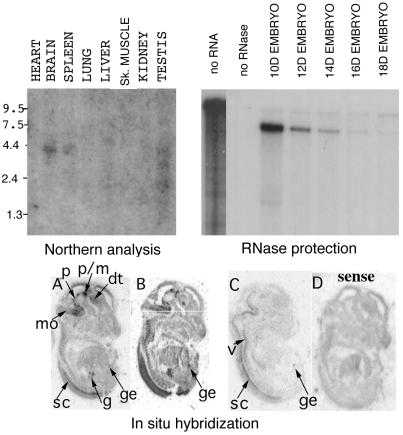
Northern analysis of poly(A)+ RNA from adult mouse organs (Fig. 2) revealed one major band of Nkx-1.2 mRNA, about 3.8 kb in length, in RNA from brain and spleen, and three bands of Nkx-1.2 mRNA, approximately 3.8, 2.4, and 2.0 kb in length, in RNA from testes; traces of smaller transcripts also were found in RNA from testes. Similarly, multiple Sax-1 transcripts were reported in P19 EC cells aggregated for 2 days in the presence of retinoic acid (8).
Figure 2.
Expression of Nkx-1.2 mRNA. (Upper Left) Northern analysis. mRNAs from adult mouse tissues blotted on Nylon membranes (CLONTECH) were hybridized following the manufacturer's instructions, with a 300-bp 32P-labeled DNA probe containing the Nkx-1.2 coding region after the homebox (residues 7823–8123 in Fig. 1). (Upper Right) RNase protection assay. Poly(A)+ RNA preparations were hybridized with a 32P-labeled Nkx-1.2 cRNA probe (nucleotides 7623–8123 in Fig. 1) and processed according to the RPA-II system instructions (Ambion). The autoradiogram was exposed overnight. (A–D) In situ hybridization. Serial sections from mouse embryos 14 days after fertilization were hybridized with a 500-nt residue (7623–8123 in Fig. 1), 35S-labeled Nkx-1.2 antisense (A–C) or sense (D) RNA probe, and slides were contact-exposed for 15 days. mo, medulla oblongata; p, pons; p/m, pons/mesencephalon boundary; dt, dorsal thalamus; sc, spinal cord; g, gut; ge, genital eminence; v, vertebrae.
Expression of Nkx-1.2 during mouse embryo development also was analyzed by RNase protection. The results (Fig. 2) show that Nkx-1.2 mRNA is most abundant in 10-day embryos and gradually decreases in abundance in 12- and 14-day embryos. Little Nkx-1.2 mRNA was detected in RNA from 18-day embryos. One prominent protected RNA fragment of the expected size (ca. 500 bp) corresponding to the full-length cRNA probe was observed. In addition, a less abundant, smaller (ca. 200–300 bp) protected fragment was detected, which also is more abundant at day 10 of intrauterine life, and may indicate alternative splicing.
The pattern of expression of Nkx-1.2 mRNA in mouse embryos was analyzed by in situ hybridization (Fig. 2 A-D). At 14 days after fertilization, Nkx-1.2 mRNA is found in the spinal cord, mesencephalon, and diencephalon. Interestingly, the hybridization signal in the mesencephalon appears in discrete areas, mostly in the medulla oblongata, pontine flexure, and pons, and at the level of the pons/midbrain boundary; in the diencephalon, Nkx-1.2 mRNA expression is detected mainly in the dorsal thalamus. This pattern is consistent with Nkx-1.2 mRNA expression in clusters of cells, possibly within neuronal nuclei, rather than in a continuous domain along the posterior-anterior axis. Our findings are essentially in agreement with in situ hybridization data obtained with whole-mount embryos at 11 days postcoitum (8). However, discrete clusters of Nkx-1.2 mRNA are not as apparent in 11-day whole-mount embryos as they are in tissue sections from 14-day mouse embryos. This observation may suggest that during development from days 10 to 14, Nkx-1.2 mRNA expression gradually becomes restricted to subsets of cells in the brain.
Cloning of Nkx-1.2 cDNAs.
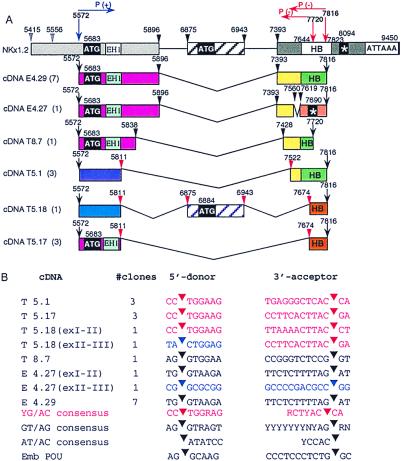
RT-PCR was used to clone Nkx-1.2 cDNA from 10- to 12-day mouse embryo RNA, adult mouse brain RNA, and testis RNA. One major PCR product, about 800 bp in length, was amplified from cDNA from mouse embryos and brain; whereas, RT-PCR with RNA from testis yielded three bands, approximately 800, 600, and 400 bp in length (data not shown). The amplified DNA fragments were subcloned and sequenced. Comparison of the nucleotide sequences of Nkx-1.2 genomic DNA and cDNAs revealed cDNAs that correspond to six species of Nkx-1.2 mRNA that are formed by alternative splicing (Fig. 3A). Eight Nkx-1.2 cDNA clones from embryo RNA and one clone from testis RNA correspond to three species of Nkx-1.2 mRNA that are formed by alternative splicing at conventional 5′ donor and 3′ acceptor sites; however, seven cDNA clones from adult testes mRNA correspond to three species of Nkx-1.2 mRNA that have novel 5′ and 3′ splice sites. The consensus sequence of the novel 5′ donor splice site is CC↓TGGAAG, and the consensus 3′ acceptor splice site is ACTTAC↓ (Fig. 3B). The consensus at the 3′ acceptor bears some resemblance to the consensus sequence at the 3′ acceptor site of the AT/AC family of introns, YCCAC↓; however the 5′ donor site differs markedly from that of the AT/AC family of introns, ↓ATATCC (20, 21). The presence of novel sequences at both the 5′ and 3′ splice sites suggests that the testes may contain unique splicing factors. A similar scenario has been depicted for the AT/AC family of introns, which are spliced via a unique mechanism involving splice factor U12 (22, 23). Further work is needed to determine the mechanism of splicing and the functional significance of the unique splicing patterns of Nkx-1.2 RNA and the tissue specificity of the splice products.
Figure 3.
(A) Nkx-1.2 gene structure, intron-exon junctions, and schematic representation of deduced Nkx-1.2 proteins. (B) Nucleotide sequences at the 5′ donor and 3′ acceptor splice sites of six species of Nkx-1.2 cDNA. Nkx-1.2 cDNA species cloned from embryo mRNA or testis mRNA are aligned to genomic Nkx-1.2 DNA. The first letter of each clone name (E or T) means that the cDNA clone was derived from embryo or adult testis poly(A)+ RNA, respectively. The number of clones isolated and sequenced for each species of cDNA is shown enclosed by parentheses. Noncanonical splice sites are indicated by red arrowheads; canonical splice sites are shown as black arrowheads. Horizontal arrows indicate the position of the primers used for the RT-PCR experiment. A star represents a termination codon.
An expressed sequence tag (EST) clone (ID number: mouse 906755 vi45F06.R1; GenBank accession number: AA522346) corresponding to a Mus musculus cDNA similar to Nkx-1.2/Sax-1, was obtained from the IMAGE consortium, and the complete nucleotide sequence was determined and deposited to GenBank. The EST clone, approximately 1,750 nt in length, contains most of the 3′ untranslated region of Nkx-1.2 cDNA and matches the nucleotide sequence 8613–9646 of genomic Nkx-1.2 DNA shown in Fig. 1. However, the nucleotide sequence of the EST clone extends further than the nucleotide sequence of Sax-1 cDNA and Nkx-1.2 genomic DNA shown in Fig. 1; hence, the EST clone has a poly(A) tail attached to a nucleotide residue that differs from the site predicted based on the sequence of Sax-1 cDNA (8).
The predicted amino acid sequences of six species of Nkx-1.2 proteins (Fig. 4) show that Nkx-1.2 transcripts found in both testes and embryo may be translated into proteins that lack the N-terminal portion of the homeodomain (cDNA clones T5.17 and T5.18) or all of the homeodomain (clone E4.27); two clones correspond to deduced proteins with alternative amino-terminal regions (clones T 5.18 and T5.1; the putative translational start site for clone T5.1 is unknown); whereas four clones encode proteins that share the same amino acid sequence at the N terminus and contain a conserved EH1/tinman motif, which reportedly functions as a transcriptional repressor (24). The C-terminal sequences of all of the predicted Nkx-1.2 proteins, with the exception of clone E4.27, are unknown, because the primers used for cloning correspond to sequences in the 5′ flanking region of the termination codon.
Figure 4.
Deduced amino acid sequences of Nkx-1.2 proteins corresponding to six species of Nkx-1.2 mRNA from 10-day embryos or adult testis mRNA. Interesting motifs are enclosed in boxes; the EH1 motif is a conserved motif mediating transcriptional repression for engrailed, which is found in engrailed, goosecoid, NK-1, NK-2, and msh-class homeodomain proteins (24).
Localization of Nkx-1.2 mRNA in Male Germ Cells and Spermatozoa.
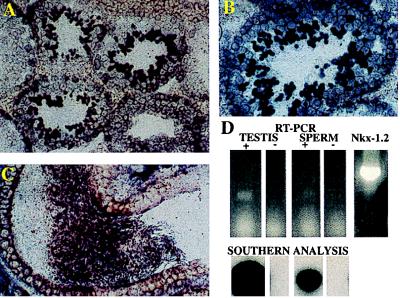
The localization of Nkx-1.2 mRNA in the testes of adult mice was studied by in situ hybridization. In Fig. 5 A and B, Nkx-1.2 mRNA is shown to be present in the germ cells of the seminiferous epithelium of the testis. The intensity of the hybridization signal indicates that the concentration of Nkx-1.2 mRNA is present in the postmitotic cells of the seminiferous epithelium; highest abundance was observed in the elongating spermatids and in a few cells within the lumen of the seminiferous tubule, which are testicular spermatozoa on their way to the efferent duct (25). Haploid-specific cDNAs have been isolated from testis (26). Although there is some evidence for mRNA synthesis in the haploid nucleus during spermatogenesis, as the spermatid differentiates to spermatozoon, RNA transcription decreases and terminates (26, 27). The presence of Nkx-1.2 mRNA in elongating spermatids and testicular spermatozoa therefore is consistent with Nkx-1.2 mRNA synthesis in diploid nuclei or in the early stages of spermatogenesis and accumulation of the transcripts in the haploid cells during spermiogenesis, which in mice lasts about 14 days. A similar mechanism has been demonstrated for the protamines, the major haploid specific mRNAs of the testis. Mammalian protamine mRNAs are synthesized during the early stages of spermatogenesis, and then are stored as RNA–protein complexes in round spermatids and translated in the elongating spermatids (26). In Fig. 5C is shown in situ hybridization of Nkx-1.2 in the duct of the epididymis, where maturation of spermatozoa is completed: numerous cells in the lumen, with the characteristic head and tail morphology of spermatozoa, display strong hybridization signals. The presence of Nkx-1.2 mRNA in spermatozoa also was demonstrated by RT-PCR (Fig. 5D) with RNA prepared from spermatozoa purified from the cauda epydidimi, followed by Southern blot analysis, subcloning, and sequencing the PCR products. These results show that at least one major species of Nkx-1.2 mRNA is present in spermatozoa. The nucleotide sequence of the transcript corresponds to the cDNA clone T5.17, which has novel splice sites and encodes a protein that lacks the first 10 amino acid residues of the homeodomain. The presence of specific mRNAs in mature spermatozoa has been reported (27, 28), such as RNA splice factors U1 and U2 small nuclear RNA, human protooncogene c-myc, mouse zinc-finger protein Zfp59, human β-actin, protamine-1 and -2, and transition protein 2 (27, 28). The functional significance of these species of mRNA in spermatozoa is unknown. They may represent the remnants of stored mRNAs from postmeiotically transcribed genes; however, the presence of RNA polymerase activity, pre-mRNA, and transcription-competent chromatin in the spermatozoon nucleus has been reported (29). In addition, specific mRNA from the male gamete may be associated with the highly transcriptionally active male pronucleus (30) and may play a role after fertilization. The time and site of Nkx-1.2 mRNA translation have not been determined; hence, it is not known whether Nkx-1.2 protein is present in spermatids or mature spermatozoa, nor is it known whether Nkx-1.2 protein regulates transcription in male germ cells. The signal transducer and activator of transcription, STAT4 protein, has been localized in the perinuclear theca of spermatozoa (31); because the theca depolymerizes in the cytoplasm of the fertilized egg, the possibility that sperm STAT4 may be a paternal regulator of the onset of zygotic transcription has been suggested (31).
Figure 5.
Nkx-1.2 gene expression in germ cells. (A–C) In situ hybridization of Nkx-1.2 mRNA in the seminiferous epithelium of the testis (A and B) and in the epididymal duct of the adult mouse (C). (D) RT-PCR amplification of Nkx-1.2 cDNA from testis and spermatozoa RNA. In situ hybridization was performed as described in the text. PCR amplification was performed with primers +P1 and −P1 shown in Fig. 1. PCR products were separated by size by agarose gel electrophoresis, and the DNA bands were transferred to nylon membranes. Southern analysis was performed by hybridization with a 32P-labeled oligodeoxynucleotide probe corresponding to primer −P2 shown in Fig. 1.
Expression of Nkx-1.2 mRNA in the Adult Brain.
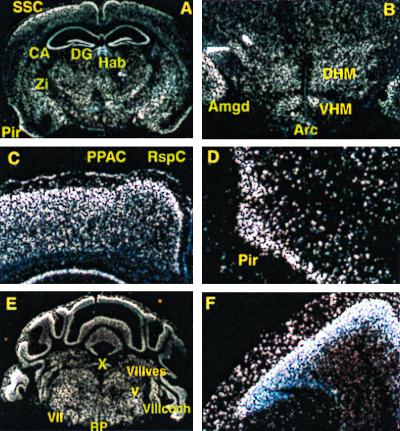
Cells expressing Nkx-1.2 were found by in situ hybridization scattered throughout the brain of adult mice (Fig. 6 A and B). However, areas with the most intense hybridization signals are as follows: retrosplenial cortex, posterior parietal associative cortex, somato-sensory cortex, piriform cortex, amygdaloid body, cornu ammoni, dentate gyrus, zona incerta, medial and lateral habenula, and dorsomedial, ventromedial and arcuate hypothalamic nuclei. In the cerebral cortex, Nkx-1.2 mRNA is most abundant in the external granular layer, which contains mostly neuronal cells, but little or no Nkx-1.2 mRNA is present in the molecular layer, which contains mostly neuroglia and few neurons (Fig. 6 C and D). Many nuclei were found in the pons that express Nkx-1.2 (Fig. 6B). Highest hybridization signals were detected in the dorsal motor nucleus vagus, ventral and dorsal cochlear nuclei, facial nucleus, trigeminal nucleus, vestibular nucleus, and the raphe pallidus. Nkx-1.2 mRNA is abundant in both the molecular and the granular layers of the cerebellum (Fig. 6 E and F); the hybridization signal is most intense at the boundary between the molecular and granular layers, where Purkinje neurons are most abundant.
Figure 6.
Nkx-1.2 gene expression in adult brain. (A) Whole brain, coronal. (B) Pons, diencephalon. (C) Cerebral cortex. (E) Piriform cortex. (F) Cerebellum; cerebellar cortex. In situ hybridization was performed as described in Materials and Methods using Nkx-1.2 RNA probes labeled with [33P] corresponding to an EcoRI–BamHI nucleotide sequence, approximately 850 bp in length, starting at the EcoRI site located at residues 8119–8123 in Fig. 1. Shown are dark-field images of sections hybridized with the antisense NKx-1.2 cRNA probe; little or no hybridization signal was obtained with the sense probe (data not shown). RspC, retrosplenial cortex; PPAC, posterior parietal associative cortex; SSC, somatosensory cortex; Pir, piriform cortex, Amgd, amygdaloid body; CA, cornu ammoni; DG, dentate gyrus; Zi, zona incerta; mHab and lHAb, medial and lateral habenula, respectively; DMH and VHM, dorsomedial and, ventromedial hypothalamic nucleus, respectively; Arc, arcuate nucleus; X, dorsal motor nucleus vagus; VIIIcoch, ventral and dorsal cochlear nuclei; VIII vest, vestibular nucleus; VII, facial nucleus; V, trigeminal nucleus; RP, raphe pallidus.
Acknowledgments
We thank Yongsok Kim for a DNA fragment encompassing part of the Nkx-1.2 homeobox and Vicky Guo for DNA sequencing and oligonucleotide synthesis.
Abbreviation
- RT-PCR
reverse-transcription–PCR
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. Nkx-1.2 genomic DNA, AF222443; cDNA, AF222444; cDNA E4.29, AF223361; cDNA E4.27, AF222445; cDNA T8.7, AF223362; cDNA T5.1, AF223363; cDNA T5.17, AF223675; cDNA T5.18, AF223674; cDNA EST, AF222442).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.030539397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.030539397
References
- 1.Kim Y, Nirenberg M. Proc Natl Acad Sci USA. 1989;86:7716–7720. doi: 10.1073/pnas.86.20.7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dohrmann C, Azpiazu N, Frasch M. Genes Dev. 1990;4:2098–2111. doi: 10.1101/gad.4.12a.2098. [DOI] [PubMed] [Google Scholar]
- 3.Hawkins N C, McGhee J D. Nucleic Acids Res. 1990;18:6101–6106. doi: 10.1093/nar/18.20.6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oliver G, Vispo M, Mailhos A, Martinez C, Sosa-Pineda B, Fielitz W, Ehrlich R. Gene. 1992;121:337–342. doi: 10.1016/0378-1119(92)90140-k. [DOI] [PubMed] [Google Scholar]
- 5.Walldorf U, Fleig R, Gehring W J. Proc Natl Acad Sci USA. 1989;86:9971–9975. doi: 10.1073/pnas.86.24.9971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rangini Z, Frumkin A, Shani G, Guttmann M, Eyal-Giladi H, Gruenbaum Y, Fainsod A. Gene. 1989;76:61–74. doi: 10.1016/0378-1119(89)90008-5. [DOI] [PubMed] [Google Scholar]
- 7.Bober E, Baum C, Braun T, Arnold H-H. Dev Biol. 1994;162:288–303. doi: 10.1006/dbio.1994.1086. [DOI] [PubMed] [Google Scholar]
- 8.Schubert F R, Fainsod A, Gruenbaum Y, Gruss P. Mech Dev. 1995;51:99–114. doi: 10.1016/0925-4773(95)00358-8. [DOI] [PubMed] [Google Scholar]
- 9.Moretti P, Simmons P, Thomas P, Haylock D, Rathjen P, Vadas M, D'Andrea R. Gene. 1994;144:213–219. doi: 10.1016/0378-1119(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 10.Chen X, Lufkin T. Mamm Genome. 1997;8:697–698. doi: 10.1007/s003359900541. [DOI] [PubMed] [Google Scholar]
- 11.Yamada G, Kioussi C, Schubert F R, Eto Y, Chowdhury K, Pituello F, Gruss P. Biochem Biophys Res Commun. 1994;199:552–563. doi: 10.1006/bbrc.1994.1264. [DOI] [PubMed] [Google Scholar]
- 12.Spann P, Ginsburg M, Rangini Z, Fainsod A, Eyal-Giladi H, Gruenbaum Y. Development (Cambridge, UK) 1994;120:1817–1828. doi: 10.1242/dev.120.7.1817. [DOI] [PubMed] [Google Scholar]
- 13.Adamson M C, Silver J, Kozak C A. Virology. 1991;183:778–781. doi: 10.1016/0042-6822(91)91010-e. [DOI] [PubMed] [Google Scholar]
- 14.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 15.Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the Mouse Embryo: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. pp. 344–351. [Google Scholar]
- 16.Dijkman H B P M, Mentzel S, de Jong A S, Assmann K J M. Biochemica. 1995;2:23–27. [Google Scholar]
- 17.Bellve' A R, Zheng W, Martinova Y S. Methods Enzymol. 1993;225:113–136. doi: 10.1016/0076-6879(93)25010-y. [DOI] [PubMed] [Google Scholar]
- 18.Rovescalli A C, Kim Y, Kim S, Ferrante J, Nirenberg M. Neuroscience. 1993;19:3326. [Google Scholar]
- 19.Wang W D, Lufkin T. Chromosome Res. 1997;5:501–502. doi: 10.1023/a:1018429316424. [DOI] [PubMed] [Google Scholar]
- 20.Jackson I J. Nucleic Acids Res. 1991;19:3795–3798. doi: 10.1093/nar/19.14.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall S L, Padgett R A. J Mol Biol. 1994;239:357–365. doi: 10.1006/jmbi.1994.1377. [DOI] [PubMed] [Google Scholar]
- 22.Hall S L, Padgett R A. Science. 1996;271:1716–1718. doi: 10.1126/science.271.5256.1716. [DOI] [PubMed] [Google Scholar]
- 23.Tarn W-Y, Steitz J A. Cell. 1996;84:801–811. doi: 10.1016/s0092-8674(00)81057-0. [DOI] [PubMed] [Google Scholar]
- 24.Smith S T, Jaynes J B. Development (Cambridge, UK) 1996;122:3141–3150. doi: 10.1242/dev.122.10.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leblond C P, Clermont Y. Ann NY Acad Sci. 1952;55:548–573. doi: 10.1111/j.1749-6632.1952.tb26576.x. [DOI] [PubMed] [Google Scholar]
- 26.Hecht N B. Dev Genet. 1995;16:95–103. doi: 10.1002/dvg.1020160202. [DOI] [PubMed] [Google Scholar]
- 27.Kramer J A, Krawetz S A. Mol Hum Reprod. 1997;3:473–478. doi: 10.1093/molehr/3.6.473. [DOI] [PubMed] [Google Scholar]
- 28.Wykes S M, Visscher D W, Krawetz S A. Mol Hum Reprod. 1997;3:15–19. doi: 10.1093/molehr/3.1.15. [DOI] [PubMed] [Google Scholar]
- 29.Miteva K, Valkov N, Goncharova-Peinova J, Kovachev K, Zlatarev S, Pironcheva G, Russev G. Microbios. 1995;84:91–96. [PubMed] [Google Scholar]
- 30.Aoki F, Worrad D M, Schultz R M. Dev Biol. 1997;181:296–307. doi: 10.1006/dbio.1996.8466. [DOI] [PubMed] [Google Scholar]
- 31.Herrada G, Wolgemuth D J. J Cell Sci. 1997;110:1543–1553. doi: 10.1242/jcs.110.14.1543. [DOI] [PubMed] [Google Scholar]