Abstract
Hepatocyte growth factor (HGF) and vascular endothelial cell growth factor (VEGF) are two potent endothelial mitogens with demonstrated angiogenic activities in animal models of therapeutic angiogenesis. Several recent studies suggest that these growth factors may act synergistically, although the mechanism of this interaction is not understood. Changes in the gene expression profile of human umbilical vein endothelial cells treated with HGF, VEGF or the combination of the two were analyzed with high-density oligonucleotide arrays, representing approximately 22,000 genes. Notably, the genes significantly up- and downregulated by VEGF versus HGF exhibited very little overlap, indicating distinct signal transduction pathways. The combination of HGF and VEGF markedly increased the number of significantly up- and downregulated genes. At 4 h, the combination of the two growth factors induced a number of chemokine and cytokines and their receptors (IL-8, IL-6, IL-11, CCR6, CXCR1,CXC1 and IL17RC), numerous genes involved in growth factor signal transduction (egr-1, fosB, grb10, grb14,MAP2K3,MAP3K8, MAPKAP2,MPK3, DUSP4 and DUSP6), as well as a number of other growth factors (PDGFA, BMP2, Hb-EGF, FGF16, heuregulin beta 1, c-kit ligand, angiopoietin 2 and angiopoietin 4 and VEGFC). In addition, the VEGF receptors neuropilin-1 and flt-1 were also upregulated. At 24 h, a clear ‘cell cycle' signature is noted, with the upregulated expression of various cell cycle control proteins and gene involved in the regulation of mitosis and mitotic spindle assembly. The receptor for HGF, c-met, is also upregulated. These data are consistent with the hypothesis that the combination of HGF and VEGF results in the cooperative upregulation of a number of different molecular pathways leading to a more robust proliferative response, that is, growth factor(s), receptors, molecules involved in growth factor signal transduction, as well as, at later time points, upregulation of the necessary cellular proteins required for cells to escape cell cycle arrest and enter the cell cycle.
Keywords: Endothelium, vascular endothelial growth factor, angiogenesis, hepatocyte growth factor, gene expression
Introduction
There is considerable interest in the use of various growth factors, administered either as proteins or a genes, to induce angiogenesis to treat ischemic coronary and peripheral vascular disease. Growth factors, including vascular endothelial growth factor (VEGF), fibroblast growth factor (FGF), and hepatocyte growth factor (HGF) have been evaluated by many investigators, and at least in animal models of ischemic vascular disease showed promising effects (Baffour et al., 1992; 2000; Chleboun & Martins, 1994; Asahara et al., 1995; Baumgartner & Isner, 1998; Bush et al., 1998; Stark et al., 1998; Ferrara & Alitalo, 1999; Hayashi et al., 1999; Lazarous et al., 2000; Yang & Feng, 2000). Thus far, however, clinical trials using either intravascular or extravascular means of protein delivery have been disappointing, with only, at best, modest improvements in perfusion or clinical outcome (Henry et al., 2003; Khan et al., 2003).
While numerous studies evaluating the potential of gene-based therapy to deliver growth factors are now underway, the majority of these are evaluating the effects of a single factor. However, there are several studies that indicate that a combination of two or more growth factors may be far more effective than either factor alone (Pepper et al., 1992; Van Belle et al., 1998; Xin et al., 2001).
HGF and VEGF are potent endothelial mitogens, mitogens, and morphogens (Morimoto et al., 1991; Bussolino et al., 1992; Nakamura et al., 1996; Rosen et al., 1997; Ferrara, 1999). Data from several laboratories have shown that in vitro, the combination of HGF and VEGF results in a much more robust proliferative and chemotactic response than either growth factor alone (Van Belle et al., 1998; Xin et al., 2001). In three-dimensional collagen gels, neither HGF nor VEGF alone are sufficient to induce human endothelial cell survival and tubulogenesis, yet the combination of the two growth factors will (Xin et al., 2001). In vivo studies also suggest that combining HGF and VEGF can induce a more robust angiogenic response (Van Belle et al., 1998; Xin et al., 2001) than either growth factor alone.
The mechanism for the synergistic interactions of HGF and VEGF remains unclear. Van Belle et al. (1998) suggested that one of the effects of HGF in vivo was to induce VEGF production by surrounding smooth muscle cells. However, Sengupta et al. (2003) reported that HGF induced angiogenesis in vivo independently of VEGF. Wojta et al. (1999) found that HGF also increased the expression of VEGF and PAI-1 in human keratinocytes, and the VEGF receptor KDR (flk-1) in human endothelial cells.
Gene expression profiling offers the opportunity to assess rapidly the molecular pathways activated by growth factors, cytokines, and other stimuli. The effects of VEGF on endothelial gene expression have been described by several groups. However, the effects of HGF, or the combination of HGF and VEGF on endothelial gene expression are not well defined.
To address the molecular interactions of the HGF and VEGF signaling pathways, we evaluated endothelial mRNA expression using Affymetrix oligonucleotide arrays, examing the expression of over 20,000 genes at 4 and 24 h.
Methods
Cell culture
Human umbilical endothelial cells (HUVEC) were obtained from Clonetics (Cambrex Bioscience Walkersville, MD, U.S.A.). For the present study, three independent cell lots, each derived from three different pools (3–4 donors per pool) of umbilical cords, were used for replicates of each experimental condition. All cells were cultured under rigidly standardized conditions, with great care taken to ensure that the identical lot numbers of media, serum, growth factors, other supplements, and tissue culture plastic were used. The profiling experiments were performed on nearly confluent (95%) HUVEC that had been incubated in starvation medium: M199 containing 1 × ITS (insulin–transferrin–selenium-A), 2 mM glutamine, 100 U ml−1 pennicillin and 100 μg ml−1 streptomycin (all from Invitrogen Corp.; Carlsbad, CA, U.S.A.), and 1% fetal bovine serum (Tissue Culture Biologicals; Tulare, CA, U.S.A.) for 18 h. (This enables identification of growth factor-induced genes without the background caused by high concentrations of fetal bovine serum.) At the initiation of the experiments, the media was changed to fresh starvation medium with or without addition of VEGF (100 ng ml−1), HGF (100 ng ml−1), or a combination of both, and the cells incubated for either an additional 4 or 24 h. The concentrations of VEGF and HGF used were maximal effective doses (based on preliminary proliferation assays); that is, addition of higher concentrations of VEGF or HGF did not elicit any additional biological responses. At the termination of the experiments, 15 ml of Trizol (Invitrogen) was added and samples stored frozen at −80°C. RNA was subsequently extracted following the manufacturer's instructions. Total RNA isolated in this way further purified using RNeasy Mini kits as described in the product manual (Qiagen Inc.; Valencia, CA, U.S.A.).
mRNA extraction, affymetrix microarrays, and data analysis
DNAse-treated total RNA, 5 μg, was converted to cRNA and fragmented cRNA was hybridized to arrays (U133A) as per the manufacturer's suggested protocol (Affymetrix, Santa Clara, CA, U.S.A.). Data were analyzed with the MASv5 (Affymetrix) and Rosetta Resolver (Rosetta Biosoftware). Array results that met manufacturer's (Affymetrix) recommended quality criteria were imported into Rosetta Resolver (Roberts et al., 2000). Replicate hybridizations or profiles were combined to create ratio experiments using the Rosetta Resolver system as described (Stoughton & Dai, 2002). System processing consists of interchip normalization and nonlinear error correction (Schadt, 2002, #978). Ratios are calculated from the combined replicate profiles.
Results
There is very little overlap in the genes up- or downregulated by HGF and VEGF
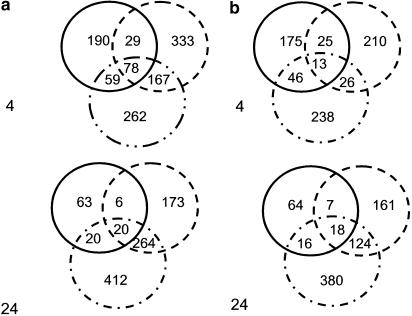
To identify the genes up- and downregulated by HGF and VEGF, the data from the three independent experiments were combined and ratios of HGF versus basal and VEGF versus basal at 4 and 24 h were generated (Table 1). Significantly regulated (both up- and downregulated) probesets with a log ratio of 0.17 (corresponding to 1.5-fold change) and a P-value ⩽0.1 were identified in the experimental ratios (time-matched basal conditions versus treated). These somewhat conservative and ‘arbitrary' cuts based on both the fold and significance of the change in expression are useful methods to analyze genes of interest rapidly. Unexpectedly, there was minimal overlap in the profiles of gene expression elicited by the two growth factors. For example, at 4 h, VEGF treatment resulted in the upregulated total of 607 different probe sets, which represent 432 different genes (since several genes were represented by multiple probesets). HGF upregulated 356 probe sets. However, only 107 different probe sets were common to the two treatment paradigms. Similar conclusions are derived from the 24 h plot of upregulated genes and the 4- and 24-h plot of downregulated genes (Figure 1).
Table 1.
Number of significantly regulated probesets (log ratio ⩾0.15, P-value ⩽0.1) in HUVEC treated with HGF, VEGF or the combination of the two growth factors
| Treatment | 4 h upregulated | 4 h downregulated | 24 h upregulated | 24 h downregulated |
|---|---|---|---|---|
| HGF | 356 | 259 | 109 | 105 |
| VEGF | 607 | 274 | 463 | 310 |
| HGF+VEGF | 566 | 323 | 716 | 142 |
Figure 1.
Venn diagram representation of those genes significantly upregulated (a) or downregulated (b) by HGF (solid circle), VEGF (dashed circle) or the combination of the two growth factors (circle with dashes and dots) at 4 h (top) and 24 h (bottom).
There are a number of genes that demonstrate additivity or synergy in expression levels when HGF and VEGF are combined
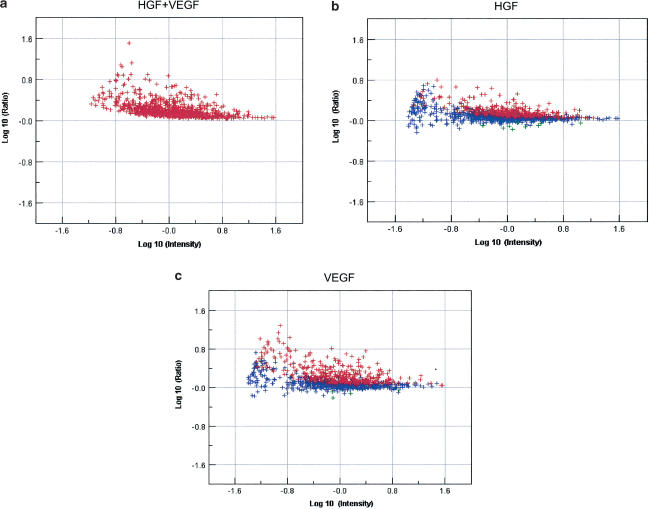
Figure 1 also illustrates the additive to synergistic interactions of HGF and VEGF. For example, at 4 h, the combination of HGF and VEGF resulted in the identification of 566 significantly upregulated probesets, 262 of which were unique to the combination of the two growth factors. In Figure 2, the expression data associated with the 566 probesets upregulated by the combination of HGF and VEGF (at 4 h) are compared to their regulation in the presence of HGF and VEGF individually versus basal ratio experiments. There are almost twice as many genes regulated by the combination treatment of HGF and VEGF together than either one alone.
Figure 2.
Dot plot representation of the expression of 566 probes upregulated by more than 1.5-fold, P<0.1 at 4 h, in the presence of the combination of HGF and VEGF at 4 h. The same probesets were compared to the expression data for HGF alone (b) and VEGF alone (a) at 4 h. Blue denotes not significantly changed from basal and red, significantly upregulated over basal expression.
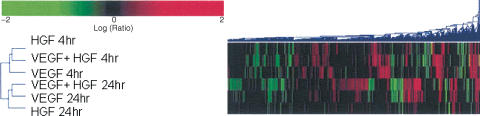
Figure 3 shows an agglomerative cluster of the different probe sets. The ‘black' bars denote ‘no data' and represent probesets that did not meet the statistical cutoff of P<0.1. This figure illustrates the differences in the global gene expression profiles elicited by HGF versus VEGF; moreover, it also suggests that the effects of HGF predominate at 4 h and those of VEGF predominate at 24 h, when cells are treated with the combination of the two growth factors.
Figure 3.
Agglomerative cluster of genes significantly upregulated or downregulated by HGF, VEGF, or the combination of the two growth factors at 4 h and 24 h. Black indicates ‘not detected' or did not reach statistical significance (P>0.1); green represents downregulated, and red, upregulated.
What are the major pathways induced by the combination of HGF and VEGF?
Individual genes are represented by ‘probe sets' on the Affymetrix oligonucleotide array. Details on the probe design and sequence information, reproducibility, and oligonucleotide array analysis are available on the manufacturer's web site (www.affymetrix.com). A ‘probe set' consists of 11 perfect match and 11 mismatch 25 mers representing each transcript. For each probe designed to be perfectly complementary to a target sequence, a partner probe is generated that is identical except for a single base mismatch in its center. These probe pairs, called the perfect match probe (PM) and the mismatch probe (MM), allow the quantitation and subtraction of signals caused by nonspecific cross-hybridization. The difference in hybridization signals between the partners, as well as their intensity ratios, serves as indicators of specific target abundance. The probe sets are selected based on their predicted hybridization properties, and filtered for specificity to reduce the potential for cross-hybridizing with similar, but unrelated sequences. To obtain a complete picture of a gene's activity, some probes are selected from regions shared by multiple splice or polyadenylation variants. In other cases, unique probes that distinguish between variants are favored. Interprobe distance is also factored into the selection process. Probes are 3′-biased to match the target generation characteristics of the amplification method, but are also widely spaced to sample various regions of each transcript and provide robustness of detection.
There are various approaches to identify pathways or cellular processes activated by cytokines and growth factors when using genome scale profiling. One method is group or cluster genes based on their patterns of expression. Another method is to group or cluster genes based on their proposed functions or interactions with other genes. As is evident in Tables 2 and 3, a number of genes are represented several times by different probe sets, and the probe sets were designed from different Genbank or REFSEQ accession numbers. Using the Netaffyx web site (www.affymetrix.com), probe subsets can be readily generated based on the annotation associated with each probeset (e.g. Kegg pathways, Gene Ontology, PRO domains, etc.). To obtain further molecular insights into the possible mechanism(s) of VEGF/HGF interactions, a union subset of probesets was generated from those identified as significantly upregulated when HGF and VEGF were combined (note this is the total set, not the set exclusive to the combination of the two growth factors). The keywords used were ‘cell proliferation'; ‘apoptosis', ‘receptors', and ‘growth factors', and the intersection of these probe sets with the subset of genes identified as upregulated from the combination of HGF and VEGF determined at 4 and 24 h determined.
Table 2.
Genes significantly upregulated (>1.5-fold, P<0.1) by the combination of HGF and VEGF at 4 h
| Sequence derived from | Gene symbol | Gene description | Ratio of HGF+VEGF basal | P-value | Ratio of VEGF basal | P-value | Ratio of HGF basal | P-value |
|---|---|---|---|---|---|---|---|---|
| AK023795.1 | ADAMTS1 | A disintegrin-like and metalloprotease (reprolysin type) with thrombospondin type 1 motif, 1 | 4.6 | <0.01 | 4.8 | <0.01 | 0.9 | 0.69 |
| AB003476.1 | AKAP12 | A kinase (PRKA) anchor protein (gravin) 12 | 1.6 | <0.01 | 1.2 | 0.31 | 1.6 | <0.01 |
| M90360.1 | AKAP13 | A kinase (PRKA) anchor protein 13 | 1.6 | 0.04 | 2.0 | <0.01 | 1.3 | 0.35 |
| NM_001147.1 | ANGPT2 | Angiopoietin 2 | 3.2 | <0.01 | 5.1 | <0.01 | 2.8 | <0.01 |
| AF187858.1 | ANGPT2 | Angiopoietin 2 | 3.2 | <0.01 | 4.6 | <0.01 | 2.7 | <0.01 |
| NM_015985.1 | ANGPT4 | Angiopoietin 4 | 3.0 | 0.08 | 0.9 | 0.85 | 0.7 | 0.52 |
| NM_016109.1 | ANGPTL4 | Angiopoietin-like 4 | 7.6 | <0.01 | 4.6 | <0.01 | 1.7 | 0.41 |
| NM_012099.1 | ASE-1 | CD3-epsilon-associated protein; antisense to ERCC-1 | 1.8 | 0.02 | 1.3 | 0.35 | 1.7 | 0.06 |
| NM_001673.1 | ASNS | Asparagine synthetase | 1.5 | 0.02 | 1.3 | 0.25 | 1.5 | 0.07 |
| AF095192.1 | BAG2 | BCL2-associated athanogene 2 | 1.7 | <0.01 | 1.1 | 0.8 | 1.4 | 0.23 |
| NM_004049.1 | BCL2A1 | BCL2-related protein A1 | 6.3 | <0.01 | 2.0 | 0.23 | 2.8 | 0.09 |
| AA583044 | BMP2 | Bone morphogenetic protein 2 | 2.0 | <0.01 | 2.1 | <0.01 | 1.7 | <0.01 |
| NM_001200.1 | BMP2 | Bone morphogenetic protein 2 | 1.9 | <0.01 | 1.7 | <0.01 | 1.4 | <0.01 |
| NM_001717.1 | BNC | Basonuclin | 1.9 | 0.09 | 1.6 | 0.32 | 1.6 | 0.37 |
| NM_025195.1 | C8FW | Phosphoprotein regulated by mitogenic pathways | 2.1 | <0.01 | 1.5 | 0.08 | 1.5 | <0.01 |
| NM_005795.1 | CALCRL | Calcitonin receptor-like | 1.5 | <0.01 | 1.2 | 0.17 | 1.0 | 0.89 |
| U17473.1 | CALCRL | Calcitonin receptor-like | 1.5 | 0.01 | 1.2 | 0.32 | 1.1 | 0.71 |
| NM_016557.1 | CCRL1 | Chemokine (C–C motif) receptor-like 1 | 7.2 | <0.01 | 5.8 | <0.01 | 1.8 | 0.01 |
| AV700298 | CD44 | CD44 antigen (homing function and Indian blood group system) | 2.0 | 0.06 | 2.3 | 0.06 | 1.9 | 0.09 |
| NM_003672.1 | CDC14A | CDC14 cell division cycle 14 homolog A (S. cerevisiae) | 1.9 | 0.01 | 2.2 | 0.01 | 1.8 | 0.1 |
| AF115544.1 | CDKN2A | Cyclin-dependent kinase inhibitor 2A(melanoma, p16, inhibits CDK4) | 4.3 | 0.07 | 3.5 | 0.16 | 1.4 | 0.81 |
| BC000059.1 | CELSR1 | Cadherin, EGF LAG seven-pass G-typereceptor 1 (flamingo homolog, Drosophila) | 2.5 | 0.05 | 2.5 | 0.14 | 1.7 | 0.52 |
| NM_021797.1 | CHIA | Eosinophil chemotactic cytokine | 3.0 | 0.01 | 1.0 | 0.93 | 2.2 | 0.38 |
| NM_000748.1 | CHRNB2 | Cholinergic receptor, nicotinic, beta polypeptide 2 (neuronal) | 6.8 | 0.03 | 2.1 | 0.7 | 3.0 | 0.6 |
| NM_013246.1 | CLC | Cardiotrophin-like cytokine; neurotrophin-1/B-cell-stimulating factor-3 | 2.6 | 0.1 | 1.5 | 0.58 | 1.9 | 0.29 |
| NM_001842.1 | CNTFR | Ciliary neurotrophic factor receptor | 2.8 | 0.02 | 2.3 | 0.08 | 1.9 | 0.63 |
| NM_004750.1 | CRLF1 | Cytokine receptor-like factor 1 | 5.6 | 0.03 | 0.7 | 0.71 | 3.1 | 0.44 |
| D83702.1 | CRY1 | Cryptochrome 1 (photolyase-like) | 1.9 | <0.01 | 1.9 | <0.01 | 1.1 | 0.42 |
| BC005921.1 | CSH1 | Chorionic somatomammotropin hormone 1 (placental lactogen) | 4.0 | 0.01 | 2.0 | 0.5 | 3.2 | 0.13 |
| U83410.1 | CUL2 | Cullin 2 | 2.0 | 0.02 | 1.7 | 0.26 | 2.0 | 0.16 |
| L01639.1 | CXCR4 | Chemokine (C–X–C motif) receptor 4 | 3.0 | <0.01 | 2.8 | <0.01 | 4.2 | <0.01 |
| AJ224869 | CXCR4 | Chemokine (C–X–C motif) receptor 4 | 2.4 | <0.01 | 2.5 | <0.01 | 3.0 | <0.01 |
| AF348491.1 | CXCR4 | Chemokine (C–X–C motif) receptor 4 | 1.9 | <0.01 | 1.7 | 0.04 | 2.2 | 0.02 |
| BC003637.1 | DDIT3 | DNA-damage-inducible transcript 3 | 1.9 | <0.01 | 3.3 | <0.01 | 1.6 | 0.1 |
| AL050069.1 | DOK5 | Docking protein 5 | 3.1 | <0.0 | 1.5 | 0.16 | 1.0 | 0.81 |
| NM_000796.1 | DRD3 | Dopamine receptor D3 | 2.6 | 0.02 | 1.5 | 0.38 | 2.5 | 0.13 |
| M60278 | DTR | Diphtheria toxin receptor (heparin-binding epidermal growth factor-like growth factor) | 2.6 | <0.01 | 3.8 | <0.01 | 0.7 | <0.01 |
| NM_001945.1 | DTR | Diphtheria toxin receptor (heparin-binding epidermal growth factor-like growth factor) | 2.2 | <0.01 | 3.1 | <0.01 | 0.7 | 0.04 |
| BC002671.1 | DUSP4 | Dual specificity phosphatase 4 | 9.3 | <0.01 | 8.5 | <0.01 | 3.8 | 0.11 |
| NM_001394.2 | DUSP4 | Dual specificity phosphatase 4 | 5.8 | <0.01 | 6.5 | <0.01 | 2.8 | 0.03 |
| BC005047.1 | DUSP6 | Dual specificity phosphatase 6 | 7.2 | <0.01 | 6.3 | <0.01 | 3.2 | 0.03 |
| BC003143.1 | DUSP6 | Dual specificity phosphatase 6 | 3.1 | <0.01 | 2.5 | <0.01 | 1.7 | <0.01 |
| BC003143.1 | DUSP6 | Dual specificity phosphatase 6 | 2.9 | <0.01 | 2.7 | <0.01 | 1.7 | <0.01 |
| NM_001964.1 | EGR1 | Early growth response 1 | 4.7 | <0.01 | 5.8 | <0.01 | 4.4 | <0.01 |
| NM_012153.1 | EHF | ets homologous factor | 2.3 | 0.05 | 1.1 | 0.88 | 1.4 | 0.64 |
| BF445047 | EMP1 | Epithelial membrane protein 1 | 3.2 | <0.01 | 1.9 | <0.01 | 1.9 | <0.01 |
| NM_001423.1 | EMP1 | Epithelial membrane protein 1 | 2.1 | <0.01 | 1.5 | <0.01 | 1.6 | <0.01 |
| NM_001423.1 | EMP1 | Epithelial membrane protein 1 | 1.7 | <0.01 | 1.5 | <0.01 | 1.4 | <0.01 |
| NM_004438.1 | EPHA4 | EphA4 | 3.1 | 0.06 | 1.8 | 0.2 | 2.2 | 0.18 |
| NM_001993.2 | F3 | Coagulation factor III (thromboplastin, tissue factor) | 7.8 | <0.01 | 19.1 | <0.01 | 0.8 | 0.84 |
| NM_012306.1 | FAIM2 | Fas apoptotic inhibitory molecule 2 | 3.2 | 0.02 | 2.5 | 0.05 | 3.0 | <0.01 |
| NM_003868.1 | FGF16 | Fibroblast growth factor 16 | 2.1 | 0.03 | 1.6 | 0.12 | 1.1 | 0.57 |
| U01134.1 | FLT1 | fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor) | 13.5 | <0.01 | 4.3 | <0.01 | 2.7 | 0.01 |
| NM_006732.1 | FOSB | FBJ murine osteosarcoma viral oncogene homolog B | 7.4 | <0.01 | 1.2 | 0.9 | 1.0 | 1 |
| BG251266 | FOSL1 | FOS-like antigen 1 | 1.9 | <0.01 | 1.4 | <0.01 | 1.3 | 0.02 |
| NM_005261.1 | GEM | GTP binding protein overexpressed in skeletal muscle | 1.5 | 0.01 | 1.3 | 0.34 | 1.0 | 0.99 |
| U35399.1 | GPR4 | G protein-coupled receptor 4 | 1.7 | 0.08 | 1.4 | 0.37 | 1.8 | 0.04 |
| AL554008 | GPR56 | G protein-coupled receptor 56 | 1.9 | <0.01 | 1.1 | 0.68 | 1.5 | 0.03 |
| U66065.1 | GRB10 | Growth factor receptor-bound protein 10 | 1.7 | <0.01 | 1.2 | 0.01 | 1.6 | <0.01 |
| D86962.1 | GRB10 | Growth factor receptor-bound protein 10 | 1.7 | <0.01 | 1.3 | 0.03 | 1.5 | <0.01 |
| NM_004490.1 | GRB14 | Growth factor receptor-bound protein 14 | 1.6 | <0.01 | 1.1 | 0.47 | 1.7 | <0.01 |
| NM_021643.1 | GS3955 | GS3955 protein | 1.9 | 0.09 | 1.8 | 0.08 | 0.8 | 0.58 |
| AI761561 | HK2 | Hexokinase 2 | 3.5 | <0.01 | 1.5 | 0.06 | 2.6 | <0.01 |
| NM_000872.2 | HTR7 | 5-Hydroxytryptamine (serotonin) receptor 7 (adenylate cyclase-coupled) | 2.6 | 0.1 | 2.1 | 0.14 | 1.7 | 0.49 |
| AA284705 | ICAM1 | Intercellular adhesion molecule 1 (CD54), Human rhinovirus receptor | 1.8 | 0.07 | 2.0 | 0.03 | 1.5 | 0.31 |
| NM_000201.1 | ICAM1 | Intercellular adhesion molecule 1 (CD54), Human rhinovirus receptor | 1.8 | <0.01 | 2.6 | <0.01 | 1.1 | 0.22 |
| AI608725 | ICAM1 | Intercellular adhesion molecule 1 (CD54), Human rhinovirus receptor | 1.7 | <0.01 | 1.8 | <0.01 | 1.0 | 0.86 |
| M31159.1 | IGFBP3 | Insulin-like growth factor binding protein 3 | 5.5 | <0.01 | 2.5 | <0.01 | 0.8 | 0.65 |
| NM_000641.1 | IL11 | Interleukin 11 | 3.4 | <0.01 | 2.2 | 0.04 | 1.7 | 0.22 |
| BF112057 | IL-17RC | Interleukin 17 receptor C | 4.5 | 0.06 | 2.3 | 0.44 | 2.1 | 0.5 |
| NM_000600.1 | IL6 | Interleukin 6 (interferon, beta 2) | 1.7 | <0.01 | 3.1 | <0.01 | 1.1 | 0.59 |
| AF043337.1 | IL8 | Interleukin 8 | 2.3 | <0.01 | 3.2 | <0.01 | 0.9 | 0.68 |
| NM_000584.1 | IL8 | Interleukin 8 | 1.9 | <0.01 | 2.1 | 0.02 | 1.0 | 0.88 |
| NM_002192.1 | INHBA | Inhibin, beta A (activin A, activin AB alpha polypeptide) | 1.6 | 0.04 | 2.2 | 0.04 | 1.0 | 0.94 |
| NM_002201.2 | ISG20 | Interferon-stimulated gene 20 k Da | 6.8 | 0.03 | 3.3 | 0.35 | 1.9 | 0.65 |
| U88964 | ISG20 | Interferon-stimulated gene 20 k Da | 1.9 | <0.01 | 1.2 | 0.47 | 1.4 | 0.18 |
| NM_002203.2 | ITGA2 | integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) | 1.9 | <0.01 | 1.5 | 0.14 | 1.7 | <0.01 |
| AV733308 | ITGA6 | integrin, alpha 6 | 1.6 | <0.01 | 1.3 | 0.11 | 1.1 | 0.41 |
| AA215854 | ITGB1 | Integrin, beta 1 (Fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) | 2.6 | 0.04 | 3.3 | 0.04 | 2.2 | 0.11 |
| NM_002214.1 | ITGB8 | Integrin, beta 8 | 4.1 | <0.01 | 2.6 | 0.04 | 2.8 | <0.01 |
| BC002630.1 | ITGB8 | Integrin, beta 8 | 1.7 | 0.08 | 0.9 | 0.82 | 1.1 | 0.86 |
| L38019.1 | ITPR1 | Inositol 1,4,5-triphosphate receptor, type 1 | 4.5 | 0.04 | 4.9 | <0.01 | 2.4 | 0.35 |
| NM_002224.1 | ITPR3 | Inositol 1,4,5-triphosphate receptor, type 3 | 1.7 | 0.03 | 0.9 | 0.64 | 1.1 | 0.57 |
| AL137000 | KIAA0970 | KIAA0970 protein | 1.7 | 0.05 | 1.9 | <0.01 | 1.1 | 0.81 |
| AF119835.1 | KITLG | KIT ligand | 4.3 | <0.01 | 2.2 | 0.1 | 1.4 | 0.53 |
| NM_000899.1 | KITLG | KIT ligand | 2.8 | <0.01 | 1.8 | 0.01 | 1.3 | 0.15 |
| NM_012302.1 | LPHH1 | Latrophilin 1 | 2.5 | <0.01 | 2.1 | <0.01 | 1.7 | <0.01 |
| NM_017522.1 | LRP8 | Low-density lipoprotein receptor-related protein 8, apolipoprotein e receptor | 1.9 | 0.02 | 1.0 | 0.97 | 1.4 | 0.42 |
| AA780381 | MAP2K3 | Mitogen-activated protein kinase kinase 3 | 5.9 | <0.01 | 3.5 | <0.01 | 3.2 | <0.01 |
| AA780381 | MAP2K3 | Mitogen-activated protein kinase kinase 3 | 2.6 | <0.01 | 1.6 | <0.01 | 1.5 | <0.01 |
| NM_002756.1 | MAP2K3 | Mitogen-activated protein kinase kinase 3 | 2.5 | <0.01 | 1.5 | 0.02 | 1.5 | <0.01 |
| NM_005204.1 | MAP3K8 | Mitogen-activated protein kinase kinase kinase 8 | 2.4 | 0.03 | 3.8 | <0.01 | 0.9 | 0.73 |
| NM_004759.1 | MAPKAPK2 | Mitogen-activated protein kinase activated protein kinase 2 | 1.5 | <0.01 | 1.4 | <0.01 | 1.0 | 0.7 |
| NM_000381.1 | MID1 | Midline 1 (Opitz/BBB syndrome) | 1.5 | <0.01 | 1.7 | <0.01 | 1.2 | 0.3 |
| AL545921 | MPHOSPH10 | M-phase phosphoprotein 10 (U3 small nucleolar ribonucleoprotein) | 1.5 | 0.02 | 1.5 | 0.02 | 1.1 | 0.44 |
| NM_002467.1 | MYC | V-myc myelocytomatosis viral oncogene homolog (avian) | 1.5 | <0.01 | 1.0 | 0.67 | 1.2 | 0.07 |
| BF337329 | NAB2 | NGFI-A binding protein 2 (EGR1 binding protein 2) | 10.0 | <0.01 | 7.4 | <0.01 | 5.1 | 0.01 |
| U08015.1 | NFATC1 | Nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 | 1.7 | <0.01 | 1.1 | 0.19 | 0.9 | 0.58 |
| AW027545 | NFATC1 | Nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 | 1.7 | 0.07 | 1.2 | 0.57 | 0.9 | 0.73 |
| M55643.1 | NFKB1 | Nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 (p105) | 1.5 | 0.01 | 1.3 | 0.23 | 1.1 | 0.26 |
| NM_006170.1 | NOL1 | Nucleolar protein1, 120 k Da | 1.6 | <0.01 | 1.1 | 0.59 | 1.3 | <0.01 |
| D21262.1 | NOLC1 | Nucleolar and coiled-body phosphoprotein 1 | 2.3 | <0.01 | 1.4 | 0.01 | 1.9 | <0.01 |
| NM_000271.1 | NPC1 | Niemann–Pick disease, type C1 | 1.6 | <0.01 | 1.0 | 0.76 | 1.3 | 0.05 |
| D49728.1 | NR4A1 | Nuclear receptor subfamily 4, group A, member 1 | 7.9 | <0.01 | 8.1 | <0.01 | 1.6 | 0.79 |
| NM_002135.1 | NR4A1 | Nuclear receptor subfamily 4, group A, member 1 | 6.6 | <0.01 | 8.5 | 0.03 | 1.0 | 0.98 |
| S77154.1 | NR4A2 | Nuclear receptor subfamily 4, group A, member 2 | 3.6 | <0.01 | 2.6 | 0.02 | 0.8 | 0.63 |
| NM_006186.1 | NR4A2 | Nuclear receptor subfamily 4, group A, member 2 | 2.4 | 0.01 | 2.3 | 0.06 | 1.0 | 0.95 |
| U12767.1 | NR4A3 | Nuclear receptor subfamily 4, group A,member 3 | 3.1 | <0.01 | 3.5 | 0.26 | 1.3 | 0.84 |
| AF146343.1 | NR5A2 | Nuclear receptor subfamily 5, group A, member 2 | 4.3 | <0.01 | 4.2 | <0.01 | 2.6 | <0.01 |
| AF228413.1 | NR5A2 | Nuclear receptor subfamily 5, group A, member 2 | 3.2 | <0.01 | 2.6 | 0.09 | 2.4 | 0.07 |
| NM_003822.1 | NR5A2 | Nuclear receptor subfamily 5, group A, member 2 | 2.4 | 0.09 | 2.2 | 0.11 | 1.9 | 0.18 |
| NM_005010.1 | NRCAM | Neuronal cell adhesion molecule | 1.9 | <0.01 | 1.7 | <0.01 | 1.7 | <0.01 |
| NM_013960.1 | NRG1 | Neuregulin 1 | 1.5 | 0.08 | 1.0 | 0.93 | 1.2 | 0.59 |
| NM_012377.1 | OR7C2 | Olfactory receptor, family 7, subfamily C, member 2 | 5.4 | 0.04 | 1.5 | 0.58 | 1.3 | 0.42 |
| X03795.1 | PDGFA | Platelet-derived growth factor alpha polypeptide | 4.1 | 0.03 | 1.4 | 0.61 | 1.7 | 0.44 |
| NM_002607.1 | PDGFA | Platelet-derived growth factor alpha polypeptide | 3.0 | <0.01 | 1.1 | 0.44 | 1.8 | <0.01 |
| NM_007169.1 | PEMT | Phosphatidylethanolamine N-methyltransferase | 1.7 | 0.1 | 1.3 | 0.67 | 1.5 | 0.4 |
| AA576961 | PHLDA1 | Pleckstrin homology-like domain, family A, member 1 | 1.7 | <0.01 | 1.3 | 0.17 | 1.1 | 0.56 |
| NM_006875.1 | PIM2 | pim-2 oncogene | 1.7 | 0.02 | 1.6 | <0.01 | 1.0 | 0.85 |
| NM_002658.1 | PLAU | Plasminogen activator, urokinase | 1.8 | <0.01 | 1.3 | 0.05 | 0.7 | <0.01 |
| K03226.1 | PLAU | Plasminogen activator, urokinase | 1.7 | <0.01 | 1.1 | 0.53 | 0.6 | <0.01 |
| AY029180.1 | PLAUR | Plasminogen activator, urokinase receptor | 7.8 | <0.01 | 3.3 | 0.03 | 2.8 | 0.03 |
| U08839.1 | PLAUR | Plasminogen activator, urokinase receptor | 3.3 | <0.01 | 1.7 | <0.01 | 1.6 | <0.01 |
| NM_025179.1 | PLXNA2 | Plexin A2 | 1.8 | 0.01 | 1.7 | <0.01 | 1.3 | 0.17 |
| AI688418 | PLXNA2 | Plexin A2 | 1.6 | <0.01 | 1.4 | <0.01 | 1.2 | <0.01 |
| NM_002674.1 | PMCH | Pro-melanin-concentrating hormone | 17.0 | <0.01 | 5.6 | <0.01 | 2.1 | 0.47 |
| L03203.1 | PMP22 | Peripheral myelin protein 22 | 2.8 | <0.01 | 2.0 | <0.01 | 1.6 | <0.01 |
| NM_003967.1 | PNR | Putative neurotransmitter receptor | 3.5 | 0.1 | 1.1 | 0.97 | 0.4 | 0.58 |
| BC000535.1 | PPAN | Peter pan homolog (Drosophila) | 2.3 | 0.08 | 1.3 | 0.49 | 1.3 | 0.49 |
| AF014403.1 | PPAP2A | Phosphatidic acid phosphatase type 2A | 1.8 | <0.01 | 2.8 | <0.01 | 1.1 | 0.34 |
| AB000888.1 | PPAP2A | Phosphatidic acid phosphatase type 2A | 1.8 | <0.01 | 2.5 | <0.01 | 1.1 | 0.62 |
| AB000889.1 | PPAP2B | Phosphatidic acid phosphatase type 2B | 6.3 | <0.01 | 4.8 | <0.01 | 1.7 | <0.01 |
| AL576654 | PPAP2B | Phosphatidic acid phosphatase type 2B | 4.4 | <0.01 | 3.5 | <0.01 | 1.5 | <0.01 |
| BC005961.1 | PTHLH | Parathyroid hormone-like hormone | 13.2 | <0.01 | 2.8 | 0.07 | 6.0 | <0.01 |
| NM_002820.1 | PTHLH | Parathyroid hormone-like hormone | 7.8 | <0.01 | 1.7 | 0.28 | 2.9 | <0.01 |
| J03580.1 | PTHLH | Parathyroid hormone-like hormone | 3.6 | 0.01 | 1.0 | 0.96 | 1.7 | 0.32 |
| NM_002849.1 | PTPRR | Protein tyrosine phosphatase, receptor type, R | 2.3 | 0.05 | 1.4 | 0.63 | 1.7 | 0.41 |
| BE615277 | PVR | Poliovirus receptor | 1.5 | <0.01 | 1.3 | 0.29 | 1.1 | 0.59 |
| NM_000319.1 | PXR1 | Peroxisome receptor 1 | 1.5 | 0.05 | 1.2 | 0.34 | 1.2 | 0.2 |
| AI817041 | RDC1 | G protein-coupled receptor | 1.7 | <0.01 | 1.5 | 0.12 | 0.9 | 0.45 |
| NM_002923.1 | RGS2 | Regulator of G-protein signalling 2, 24 k Da | 4.5 | <0.01 | 2.1 | <0.01 | 1.5 | <0.01 |
| BF062629 | RIS1 | Ras-induced senescence 1 | 7.6 | <0.01 | 1.8 | 0.26 | 2.8 | 0.02 |
| NM_002575.1 | SERPINB2 | Serine (or cysteine) proteinase inhibitor, clade B (ovalbumin), member 2 | 3.5 | <0.01 | 1.5 | 0.13 | 2.5 | <0.01 |
| NM_003012.2 | SFRP1 | Secreted frizzled-related protein 1 | 2.0 | 0.02 | 1.2 | 0.51 | 1.3 | 0.37 |
| AF017987.1 | SFRP1 | Secreted frizzled-related protein 1 | 1.7 | 0.05 | 1.1 | 0.8 | 1.0 | 0.84 |
| NM_005627.1 | SGK | Serum/glucocorticoid regulated kinase | 2.0 | <0.01 | 2.1 | <0.01 | 1.2 | 0.24 |
| AF153330.1 | SLC19A2 | Solute carrier family 19 (thiamine transporter), member 2 | 1.7 | 0.01 | 1.3 | 0.11 | 1.4 | 0.09 |
| NM_005415.2 | SLC20A1 | Solute carrier family 20 (phosphate transporter), member 1 | 2.8 | <0.01 | 1.4 | <0.01 | 1.9 | <0.01 |
| AW452623 | SLC7A1 | Solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 | 2.1 | <0.01 | 2.1 | 0.01 | 1.2 | 0.51 |
| NM_003045.1 | SLC7A1 | Solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 | 2.0 | 0.02 | 1.9 | 0.08 | 0.8 | 0.37 |
| NM_014720.1 | SLK | Ste20-related serine/threonine kinase | 1.5 | 0.04 | 1.3 | 0.08 | 1.1 | 0.36 |
| AB004903.1 | SOCS2 | Suppressor of cytokine signaling 2 | 2.3 | 0.05 | 1.4 | 0.55 | 1.7 | 0.29 |
| NM_003877.1 | SOCS2 | Suppressor of cytokine signaling 2 | 1.8 | 0.06 | 1.1 | 0.74 | 0.9 | 0.77 |
| NM_003155.1 | STC1 | Stanniocalcin 1 | 30.9 | <0.01 | 7.8 | <0.01 | 1.4 | 0.44 |
| AI300520 | STC1 | Stanniocalcin1 | 12.9 | <0.01 | 3.6 | 0.03 | 0.8 | 0.47 |
| U46768.1 | STC1 | Stanniocalcin 1 | 11.0 | <0.01 | 4.3 | <0.01 | 1.1 | 0.87 |
| NM_005990.1 | STK10 | Serine/threonine kinase 10 | 1.9 | 0.01 | 1.9 | 0.08 | 1.1 | 0.68 |
| BE550452 | SYN47 | Homer, neuronal immediate early gene, 1B | 2.6 | 0.09 | 2.5 | 0.12 | 2.5 | 0.07 |
| NM_015727.1 | TACR1 | Tachykinin receptor 1 | 3.4 | 0.1 | 2.7 | 0.21 | 3.4 | 0.05 |
| J04152 | TACSTD2 | Tumor-associated calcium signal transducer 2 | 1.9 | <0.01 | 1.1 | 0.5 | 1.1 | 0.55 |
| D38081 | TBXA2R | Thromboxane A2 receptor | 1.5 | 0.04 | 0.9 | 0.55 | 1.3 | 0.1 |
| NM_030751.1 | TCF8 | Transcription factor 8 (represses interleukin 2 expression) | 1.9 | 0.02 | 1.5 | 0.15 | 1.6 | 0.14 |
| NM_000361.1 | THBD | Thrombomodulin | 4.9 | <0.01 | 3.4 | <0.01 | 1.4 | 0.04 |
| NM_000361.1 | THBD | Thrombomodulin | 4.9 | <0.01 | 3.1 | 0.06 | 1.3 | 0.7 |
| NM_003254.1 | TIMP1 | Tissue inhibitor of metalloproteinase 1 (erythroid potentiating activity, collagenase inhibitor) | 1.7 | <0.01 | 1.2 | 0.1 | 1.4 | <0.01 |
| NM_003823.1 | TNFRSF6B | Tumor necrosis factor receptor superfamily, member 6b, decoy | 1.6 | <0.01 | 0.9 | 0.77 | 1.3 | 0.21 |
| NM_005118.1 | TNFSF15 | Tumor necrosis factor (ligand) superfamily, member 15 | 2.2 | <0.01 | 1.6 | 0.16 | 1.3 | 0.5 |
| NM_016179.1 | TRPC4 | Transient receptor potential cation channel, subfamily C, member 4 | 2.6 | 0.09 | 2.5 | 0.09 | 3.2 | <0.01 |
| AF001294.1 | TSSC3 | Tumor suppressing subtransferable candidate 3 | 1.6 | <0.01 | 1.0 | 0.84 | 1.5 | 0.04 |
| U58111.1 | VEGFC | Vascular endothelial growth factor C | 1.7 | <0.01 | 1.4 | 0.01 | 1.3 | 0.11 |
| AI983115 | WSX1 | Class I cytokine receptor | 3.8 | 0.02 | 1.9 | 0.43 | 3.1 | 0.06 |
| NM_004843.1 | WSX1 | Class I cytokine receptor | 2.2 | 0.09 | 1.0 | 0.94 | 1.4 | 0.44 |
| NM_005283.1 | XCR1 | Chemokine (C motif) receptor 1 | 3.2 | 0.03 | 2.8 | 0.27 | 3.2 | 0.15 |
| NM_016164.2 | XLKD1 | Extracellular link domain containing 1 | 2.2 | 0.06 | 2.8 | 0.07 | 0.6 | 0.58 |
| U87460.1 | Putative endothelin receptor type B-like protein [Homo sapiens], mRNA sequence | 5.1 | <0.01 | 4.2 | <0.01 | 2.4 | 0.19 | |
| NM_004367.1 | Chemokine (C–C motif) receptor 6; chemokine (C–C) receptor 6; G proteincoupled receptor 29; seven-transmembrane receptor, lymphocyte, 22; chemokine receptor-like 3 [Homo sapiens], mRNA sequence | 2.3 | 0.09 | 1.7 | 0.3 | 1.4 | 0.6 | |
| AA058828 | Soluble vascular endothelial cell growth factor receptor (A49636) | 1.7 | 0.03 | 2.6 | 0.01 | 1.5 | 0.02 |
Table 3.
Genes significantly upregulated (>1.5-fold, P<0.1) by the combination of HGF and VEGF at 24 h
| Sequence derived from | Gene symbol | Description | Ratio of HGF–VEGF/basal | P-value | Ratio of VEGF/basal | P-value | Ratio of HGF/basal | P-value |
|---|---|---|---|---|---|---|---|---|
| NM_000014.3 | A2M | Alpha-2-macroglobulin | 28.2 | <0.01 | 19.5 | <0.01 | 2.5 | 0.29 |
| NM_000675.2 | ADORA2A | Adenosine A2a receptor | 2.2 | <0.01 | 1.5 | 0.16 | 1.3 | 0.29 |
| NM_001621.2 | AHR | Aryl hydrocarbon receptor | 1.7 | <0.01 | 1.3 | 0.07 | 1.3 | <0.01 |
| AF187858.1 | ANGPT2 | Angiopoietin 2 | 5.9 | <0.01 | 3.8 | <0.01 | 1.9 | <0.01 |
| NM_001147.1 | ANGPT2 | Angiopoietin 2 | 6.0 | <0.01 | 4.1 | <0.01 | 1.7 | 0.05 |
| AF007150.1 | ANGPTL2 | Angiopoietin-like 2 | 1.9 | <0.01 | 1.9 | <0.01 | 1.1 | 0.33 |
| NM_012098.1 | ANGPTL2 | Angiopoietin-like 2 | 2.2 | 0.04 | 1.6 | 0.23 | 1.4 | 0.45 |
| AF007150.1 | ANGPTL2 | Angiopoietin-like 2 | 1.6 | <0.01 | 1.4 | 0.11 | 1.0 | 0.79 |
| NM_006305.1 | ANP32A | Acidic (leucine-rich) nuclear phosphoprotein 32 family, member A | 1.5 | 0.01 | 1.0 | 0.87 | 1.3 | 0.15 |
| AF149794.1 | APAF1 | Apoptotic protease activating factor | 1.5 | <0.01 | 1.0 | 0.86 | 1.0 | 0.93 |
| AB000815.1 | ARNTL | Aryl hydrocarbon receptor nuclear translocator-like | 1.6 | 0.06 | 1.3 | 0.72 | 1.6 | 0.35 |
| NM_006716.1 | ASK | Activator of S phase kinase | 1.8 | 0.03 | 1.4 | 0.18 | 1.1 | 0.79 |
| AI735391 | BIKE | BMP-2-inducible kinase | 2.4 | 0.08 | 2.1 | 0.09 | 1.8 | 0.29 |
| AB028869.1 | BIRC5 | Baculoviral IAP repeat-containing 5 (survivin) | 1.8 | <0.01 | 1.3 | 0.14 | 1.2 | 0.11 |
| AA648913 | BIRC5 | Baculoviral IAP repeat-containing 5 (survivin) | 2.6 | <0.01 | 1.9 | 0.08 | 1.5 | 0.16 |
| NM_001168.1 | BIRC5 | Baculoviral IAP repeat-containing 5 (survivin) | 2.2 | <0.01 | 1.7 | <0.01 | 1.3 | 0.17 |
| NM_016098.1 | BRP44L | Brain protein 44-like | 1.8 | <0.01 | 1.5 | 0.01 | 1.1 | 0.22 |
| AF043294.2 | BUB1 | BUB1 budding uninhibited by benzimidazoles 1 homolog (yeast) | 2.3 | <0.01 | 1.9 | <0.01 | 1.3 | 0.17 |
| NM_001211.2 | BUB1B | BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) | 2.0 | <0.01 | 1.7 | <0.01 | 1.2 | 0.21 |
| W72082 | C1QR1 | Complement component 1 q subcomponent receptor 1 | 1.5 | 0.08 | 1.3 | 0.32 | 1.1 | 0.72 |
| AF098158.1 | C20orf1 | Chromosome 20 open reading frame 1 | 2.1 | <0.01 | 1.6 | 0.04 | 1.3 | 0.2 |
| U17473.1 | CALCRL | Calcitonin receptor-like | 1.8 | <0.01 | 1.1 | 0.56 | 1.4 | <0.01 |
| NM_005795.1 | CALCRL | Calcitonin receptor-like | 1.5 | <0.01 | 1.0 | 0.87 | 1.3 | 0.05 |
| N25325 | CALM1 | Calmodulin 1 (phosphorylase kinase delta) | 1.6 | <0.01 | 1.3 | 0.15 | 1.0 | 0.96 |
| BF439983 | CASP8 | Caspase 8 apoptosis-related cysteine protease | 1.9 | 0.06 | 1.8 | 0.04 | 1.6 | 0.24 |
| NM_004166.1 | CCL14 | Chemokine (C–C motif) ligand 14 | 2.6 | <0.01 | 1.5 | 0.03 | 1.9 | <0.01 |
| NM_006273.2 | CCL7 | Chemokine (C–C motif) ligand 7 | 3.2 | 0.08 | 1.0 | 0.99 | 1.0 | 1 |
| AF112857.1 | CCNE2 | Cyclin E2 | 2.2 | <0.01 | 1.6 | 0.08 | 1.0 | 0.88 |
| U17105.1 | CCNF | Cyclin F | 2.8 | 0.01 | 1.4 | 0.48 | 1.0 | 0.97 |
| AF064103.1 | CDC14A | CDC14 cell division cycle 14 homolog a (S. cerevisiae) | 2.5 | <0.01 | 1.9 | 0.13 | 1.8 | 0.19 |
| AF064103.1 | CDC14A | CDC14 cell division cycle 14 homolog a (S. cerevisiae) | 3.5 | <0.01 | 2.5 | 0.16 | 1.6 | 0.55 |
| NM_003672.1 | CDC14A | CDC14 cell division cycle 14 homolog A (S. cerevisiae) | 2.2 | <0.01 | 1.7 | 0.02 | 1.1 | 0.75 |
| AL524035 | CDC2 | Cell division cycle 2 G1 to S and G2 to M | 2.5 | <0.01 | 2.0 | <0.01 | 1.2 | 0.07 |
| NM_001786.1 | CDC2 | Cell division cycle 2 G1 to S and G2 to M | 2.5 | <0.01 | 1.9 | <0.01 | 1.2 | 0.26 |
| D88357.1 | CDC2 | Cell division cycle 2 G1 to S and G2 to M | 3.0 | <0.01 | 2.1 | <0.01 | 1.3 | 0.32 |
| NM_001255.1 | CDC20 | CDC20 cell division cycle 20 homolog (S. cerevisiae) | 1.9 | <0.01 | 1.5 | 0.04 | 1.5 | 0.06 |
| AI343459 | CDC25A | Cell division cycle 25A | 3.4 | <0.01 | 3.1 | 0.1 | 1.3 | 0.68 |
| NM_021873.1 | CDC25B | Cell division cycle 25B | 1.6 | <0.01 | 1.5 | 0.01 | 1.1 | 0.39 |
| NM_001790.2 | CDC25C | Cell division cycle 25C | 1.6 | <0.01 | 1.4 | 0.07 | 1.1 | 0.59 |
| NM_003504.1 | CDC45L | CDC45 cell division cycle 45-like (S. cerevisiae) | 2.1 | <0.01 | 2.1 | 0.02 | 1.0 | 0.94 |
| U77949.1 | CDC6 | CDC6 cell division cycle 6 homolog (S. cerevisiae) | 2.0 | <0.01 | 1.6 | 0.14 | 0.9 | 0.55 |
| NM_001254.1 | CDC6 | CDC6 cell division cycle 6 homolog (S. cerevisiae) | 1.9 | <0.01 | 1.8 | <0.01 | 1.0 | 0.9 |
| AB012305.1 | CDK2 | Cyclin-dependent kinase 2 | 1.9 | <0.01 | 1.3 | 0.45 | 1.3 | 0.51 |
| U17074.1 | CDKN2C | Cyclin-dependent kinase inhibitor 2C (p18 inhibits CDK4) | 2.6 | 0.08 | 1.8 | 0.58 | 1.9 | 0.32 |
| NM_001262.1 | CDKN2C | Cyclin-dependent kinase inhibitor 2C (p18 inhibits CDK4) | 1.8 | 0.01 | 1.4 | 0.18 | 1.3 | 0.42 |
| AF213033.1 | CDKN3 | Cyclin-dependent kinase inhibitor 3 (CDK2-associated dual specificity phosphatase) | 1.6 | 0.03 | 1.3 | 0.3 | 1.3 | 0.25 |
| U30872.1 | CENPF | Centromere protein F, 350/400 ka (mitosin) | 1.6 | <0.01 | 1.3 | 0.06 | 1.3 | 0.08 |
| NM_005196.1 | CENPF | Centromere protein F, 350/400 ka (mitosin) | 1.9 | <0.01 | 1.5 | 0.03 | 1.2 | 0.28 |
| AF041461.1 | CFLAR | CASP8 and FADD-like apoptosis regulator | 1.7 | 0.01 | 0.9 | 0.51 | 1.1 | 0.49 |
| NM_003879.1 | CFLAR | CASP8 and FADD-like apoptosis regulator | 1.5 | <0.01 | 1.0 | 0.98 | 1.0 | 0.83 |
| AF009619.1 | CFLAR | CASP8 and FADD-like apoptosis regulator | 1.5 | 0.02 | 0.9 | 0.66 | 1.0 | 0.88 |
| X06130.1 | CHC1 | Chromosome condensation 1 | 1.7 | 0.01 | 1.0 | 0.9 | 1.2 | 0.45 |
| NM_001826.1 | CKS1B | CDC28 protein kinase regulatory subunit 1B | 1.9 | <0.01 | 1.5 | <0.01 | 1.3 | 0.14 |
| AF053640.1 | CSE1L | CSE1 chromosome segregation 1-like (yeast) | 1.7 | <0.01 | 1.5 | 0.01 | 1.2 | 0.14 |
| AF053641.1 | CSE1L | CSE1 chromosome segregation 1-like (yeast) | 1.5 | <0.01 | 1.3 | 0.05 | 1.1 | 0.26 |
| NM_022646.1 | CSH2 | Chorionic somatomammotropin hormone 2 | 3.3 | 0.09 | 1.7 | 0.63 | 1.6 | 0.64 |
| AJ224869 | CXCR4 | Chemokine (C–X–C motif) receptor 4 | 3.1 | <0.01 | 2.7 | <0.01 | 1.4 | <0.01 |
| L01639.1 | CXCR4 | Chemokine (C–X–C motif) receptor 4 | 4.5 | <0.01 | 3.2 | <0.01 | 1.6 | <0.01 |
| AF348491.1 | CXCR4 | Chemokine (C–X–C motif) receptor 4 | 4.1 | <0.01 | 2.8 | <0.01 | 1.4 | <0.01 |
| U33833.1 | DDX11 | DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 11 (CHL1-like helicase homolog, S. cerevisiae) | 2.0 | <0.01 | 1.4 | 0.42 | 0.7 | 0.65 |
| NM_004399.1 | DDX11 | DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 11 (CHL1-like helicase homolog, S. cerevisiae) | 1.5 | 0.04 | 1.9 | 0.03 | 1.0 | 0.73 |
| R60068 | DDX3 | DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 3 | 1.5 | <0.01 | 0.9 | 0.73 | 1.3 | 0.09 |
| NM_001930.2 | DHPS | Deoxyhypusine synthase | 1.6 | 0.05 | 1.8 | 0.13 | 1.4 | 0.3 |
| NM_007309.1 | DIAPH2 | Diaphanous homolog 2 (Drosophila) | 2.0 | <0.01 | 2.3 | <0.01 | 1.1 | 0.47 |
| NM_006729.1 | DIAPH2 | Diaphanous homolog 2 (Drosophila) | 1.9 | <0.01 | 2.3 | <0.01 | 1.0 | 0.83 |
| AJ010395 | DKC1 | Dyskeratosis congenita 1, dyskerin | 1.6 | 0.1 | 1.3 | 0.48 | 1.2 | 0.64 |
| BC003541.1 | DOK4 | Docking protein 4 | 1.5 | 0.08 | 1.8 | 0.11 | 1.4 | 0.32 |
| NM_012145.1 | DTYMK | Deoxythymidylate kinase (thymidylate kinase) | 2.5 | 0.03 | 2.2 | 0.13 | 1.3 | 0.5 |
| NM_001394.2 | DUSP4 | Dual specificity phosphatase 4 | 3.5 | <0.01 | 2.9 | 0.03 | 1.4 | 0.3 |
| BC002671.1 | DUSP4 | Dual specificity phosphatase 4 | 3.4 | <0.01 | 2.3 | 0.11 | 1.5 | 0.31 |
| BC003143.1 | DUSP6 | Dual specificity phosphatase 6 | 1.7 | <0.01 | 1.2 | 0.15 | 1.0 | 0.66 |
| BC005047.1 | DUSP6 | Dual specificity phosphatase 6 | 2.3 | <0.01 | 1.3 | 0.23 | 1.1 | 0.69 |
| BC003143.1 | DUSP6 | Dual specificity phosphatase 6 | 1.5 | <0.01 | 1.2 | 0.03 | 1.0 | 0.96 |
| AV702405 | EBP | Emopamil binding protein (sterol isomerase) | 1.7 | <0.01 | 1.3 | 0.15 | 1.2 | 0.14 |
| NM_006579.1 | EBP | Emopamil binding protein (sterol isomerase) | 1.7 | <0.01 | 1.5 | 0.01 | 1.1 | 0.59 |
| AF061192.1 | ED1 | Ectodermal dysplasia 1, anhidrotic | 3.6 | 0.08 | 1.9 | 0.59 | 1.5 | 0.76 |
| NM_001964.1 | EGR1 | Early growth response 1 | 1.5 | 0.01 | 1.7 | 0.08 | 1.4 | 0.18 |
| BF445047 | EMP1 | Epithelial membrane protein 1 | 1.7 | <0.01 | 1.6 | 0.04 | 1.0 | 0.97 |
| NM_001424.1 | EMP2 | Epithelial membrane protein 2 | 1.7 | <0.01 | 1.6 | 0.02 | 1.1 | 0.72 |
| NM_001776.1 | ENTPD1 | Ectonucleoside triphosphate diphosphohydrolase 1 | 7.1 | <0.01 | 5.9 | <0.01 | 2.4 | 0.08 |
| U87967.1 | ENTPD1 | Ectonucleoside triphosphate diphosphohydrolase 1 | 8.1 | <0.01 | 4.4 | 0.03 | 2.0 | 0.43 |
| AV717590 | ENTPD1 | Ectonucleoside triphosphate diphosphohydrolase 1 | 2.6 | <0.01 | 1.8 | 0.08 | 1.1 | 0.77 |
| NM_007036.2 | ESM1 | Endothelial cell-specific molecule 1 | 2.1 | <0.01 | 2.4 | <0.01 | 1.0 | 0.43 |
| NM_005238.1 | ETS1 | v-ets erythroblastosis virus E26 oncogene homolog 1 (avian) | 1.6 | 0.01 | 1.1 | 0.82 | 1.1 | 0.76 |
| NM_004629.1 | FANCG | Fanconi anemia, complementation group G | 1.7 | <0.01 | 1.7 | <0.01 | 1.2 | 0.24 |
| NM_004117.1 | FKBP5 | FK506 binding protein 5 | 6.9 | <0.01 | 3.3 | 0.07 | 3.7 | 0.03 |
| U01134.1 | FLT1 | fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor) | 2.6 | 0.01 | 0.9 | 0.85 | 1.1 | 0.76 |
| NM_021953.1 | FOXM1 | Forkhead box M1 | 3.0 | <0.01 | 2.2 | <0.01 | 1.3 | 0.34 |
| NM_002039.1 | GAB1 | GRB2-associated binding protein 1 | 1.6 | 0.02 | 1.3 | 0.45 | 1.0 | 1 |
| NM_022560.1 | GH1 | Growth hormone 1 | 3.0 | 0.1 | 3.5 | 0.03 | 3.5 | 0.1 |
| NM_022559.1 | GH1 | Growth hormone 1 | 4.1 | 0.1 | 2.7 | 0.22 | 2.8 | 0.31 |
| NM_022561.1 | GH1 | Growth hormone 1 | 4.6 | 0.04 | 2.8 | 0.36 | 1.1 | 0.83 |
| NM_015895.1 | GMNN | Geminin DNA replication inhibitor | 1.8 | <0.01 | 1.8 | <0.01 | 1.0 | 0.81 |
| NM_006572.1 | GNA13 | Guanine nucleotide binding protein (G protein) alpha 13 | 2.6 | <0.01 | 1.3 | 0.55 | 2.1 | <0.01 |
| AB018301.1 | GPR116 | G protein-coupled receptor 116 | 3.3 | <0.01 | 5.4 | <0.01 | 1.6 | 0.35 |
| AB018301.1 | GPR116 | G protein-coupled receptor 116 | 1.9 | 0.1 | 2.2 | 0.07 | 1.2 | 0.59 |
| NM_005282.1 | GPR4 | G protein-coupled receptor 4 | 2.4 | <0.01 | 3.1 | 0.03 | 0.9 | 0.82 |
| U35399.1 | GPR4 | G protein-coupled receptor 4 | 2.0 | 0.01 | 1.8 | 0.17 | 1.0 | 0.98 |
| NM_005308.1 | GPRK5 | G protein-coupled receptor kinase 5 | 1.9 | <0.01 | 1.3 | 0.35 | 1.3 | 0.31 |
| W93728 | GUCY1B3 | Guanylate cyclase 1 soluble, beta 3 | 2.8 | <0.01 | 1.4 | 0.26 | 1.4 | 0.13 |
| AF020340.1 | GUCY1B3 | Guanylate cyclase 1 soluble, beta 3 | 2.4 | <0.01 | 1.3 | 0.34 | 1.1 | 0.77 |
| NM_006101.1 | HEC | Highly expressed in cancer, rich in leucine heptad repeats | 2.2 | <0.01 | 1.9 | 0.05 | 1.4 | 0.18 |
| NM_018063.1 | HELLS | Helicase lymphoid-specific | 4.1 | <0.01 | 3.0 | 0.09 | 0.8 | 0.83 |
| NM_012485.1 | HMMR | Hyaluronan-mediated motility receptor (RHAMM) | 2.2 | <0.01 | 1.8 | 0.01 | 1.5 | 0.1 |
| U29343.1 | HMMR | Hyaluronan-mediated motility receptor (RHAMM) | 1.9 | <0.01 | 1.4 | 0.02 | 1.3 | 0.14 |
| NM_002158.1 | HTLF human T-cell leukemia virus enhancer factor | 1.9 | 0.06 | 1.7 | 0.22 | 1.6 | 0.36 | |
| BE964655 | HUMGT198 A | GT198 complete ORF | 2.1 | 0.02 | 2.7 | 0.04 | 1.1 | 0.84 |
| NM_001552.1 | IGFBP4 | Insulin-like growth factor binding protein 4 | 2.1 | <0.01 | 1.9 | <0.01 | 1.2 | 0.45 |
| NM_002189.1 | IL15RA | Interleukin 15 receptor alpha | 1.7 | 0.03 | 1.9 | 0.06 | 1.1 | 0.76 |
| NM_002183.1 | IL3RA | Interleukin 3 receptor alpha (low affinity) | 6.8 | 0.06 | 8.7 | <0.01 | 1.1 | 0.95 |
| M96651.1 | IL5RA | Interleukin 5 receptor alpha | 2.8 | 0.01 | 2.5 | 0.04 | 1.3 | 0.6 |
| NM_002184.1 | IL6ST | Interleukin 6 signal transducer (gp130 oncostatin M receptor) | 2.1 | <0.01 | 1.3 | 0.42 | 1.5 | <0.01 |
| BE300521 | INSIG1 | Insulin-induced gene 1 | 1.6 | <0.01 | 1.0 | 0.87 | 1.2 | 0.16 |
| BE300521 | INSIG1 | Insulin-induced gene 1 | 1.6 | 0.02 | 0.8 | 0.63 | 1.3 | 0.28 |
| NM_005542.1 | INSIG1 | Insulin-induced gene 1 | 1.5 | 0.02 | 0.9 | 0.8 | 1.1 | 0.68 |
| AI247494 | IRS3L | Insulin receptor substrate 3-like | 2.0 | 0.01 | 1.8 | <0.01 | 1.3 | 0.19 |
| AV733308 | ITGA6 | Integrin alpha 6 | 4.0 | <0.01 | 2.2 | <0.01 | 1.3 | <0.01 |
| NM_000210.1 | ITGA6 | Integrin alpha 6 | 2.6 | <0.01 | 2.1 | <0.01 | 1.2 | 0.21 |
| NM_014288.1 | ITGB3BP | Integrin beta 3 binding protein (beta3-endonexin) | 1.5 | <0.01 | 1.4 | 0.07 | 1.1 | 0.37 |
| AF002256.1 | KIR2DL4 | Killer cell immunoglobulinlike receptor two domains, long cytoplasmic tail 4 | 1.7 | <0.01 | 1.4 | 0.41 | 1.5 | 0.23 |
| AF119835.1 | KITLG | KIT ligand | 3.9 | 0.02 | 2.8 | 0.06 | 2.9 | 0.02 |
| NM_004523.2 | KNSL1 | Kinesin-like 1 | 2.4 | <0.01 | 1.7 | <0.01 | 1.3 | 0.1 |
| AC002301 | KNSL4 | Kinesin-like 4 | 2.2 | <0.01 | 1.4 | 0.43 | 1.3 | 0.36 |
| AY026505.1 | KNSL6 | Kinesin-like 6 (mitotic centromere-associated kinesin) | 1.8 | <0.01 | 1.3 | 0.11 | 1.2 | 0.24 |
| U63743.1 | KNSL6 | Kinesin-like 6 (mitotic centromere-associated kinesin) | 1.8 | <0.01 | 1.5 | <0.01 | 1.1 | 0.39 |
| NM_020242.1 | KNSL7 | Kinesin-like 7 | 2.0 | 0.02 | 1.7 | 0.17 | 1.1 | 0.73 |
| NM_004690.2 | LATS1 | LATS large tumor suppressor homolog 1 (Drosophila) | 1.5 | 0.03 | 1.4 | 0.11 | 1.3 | 0.51 |
| S70123.1 | LDLR | Low-density lipoprotein receptor (familial hypercholesterolemia) | 2.3 | <0.01 | 1.1 | 0.82 | 1.2 | 0.38 |
| AI861942 | LDLR | Low-density lipoprotein receptor (familial hypercholesterolemia) | 2.1 | 0.02 | 1.1 | 0.9 | 1.1 | 0.73 |
| NM_013296.1 | LGN | LGN protein | 5.8 | <0.01 | 3.3 | 0.18 | 2.7 | 0.3 |
| W05463 | LOC51275 | Apoptosis-related protein PNAS-1 | 1.5 | 0.02 | 1.3 | 0.27 | 1.1 | 0.62 |
| NM_017522.1 | LRP8 | Low-density lipoprotein receptor-related protein 8, apolipoprotein e receptor | 1.7 | 0.02 | 1.3 | 0.29 | 1.0 | 0.83 |
| NM_002349.1 | LY75 | Lymphocyte antigen 75 | 3.7 | <0.01 | 3.8 | <0.01 | 1.5 | 0.56 |
| NM_002358.2 | MAD2L1 | MAD2 mitotic arrest deficient-like 1 (yeast) | 2.0 | <0.01 | 1.6 | <0.01 | 1.2 | 0.21 |
| NM_005903.1 | MADH5 | MAD mothers against decapentaplegic homolog 5 (Drosophila) | 1.7 | 0.09 | 1.6 | 0.32 | 1.4 | 0.54 |
| AA780381 | MAP2K3 | Mitogen-activated protein kinase kinase 3 | 1.7 | 0.04 | 3.4 | 0.02 | 0.9 | 0.84 |
| Z25432.1 | MAPK14 | Mitogen-activated protein kinase 14 | 1.6 | 0.06 | 0.8 | 0.65 | 1.3 | 0.43 |
| NM_021960.1 | MCL1 | Myeloid cell leukemia sequence 1 (BCL2-related) | 1.6 | <0.01 | 1.0 | 0.9 | 1.3 | 0.1 |
| NM_004526.1 | MCM2 | MCM2 minichromosome maintenance deficient 2, mitotin (S. cerevisiae) | 1.7 | <0.01 | 1.6 | <0.01 | 1.0 | 0.9 |
| AA807529 | MCM5 | MCM5 minichromosome maintenance deficient 5 cell division cycle 46 (S. cerevisiae) | 2.0 | <0.01 | 2.3 | <0.01 | 1.1 | 0.65 |
| NM_006739.1 | MCM5 | MCM5 minichromosome maintenance deficient 5 cell division cycle 46 (S. cerevisiae) | 2.1 | <0.01 | 1.7 | 0.1 | 1.0 | 0.83 |
| NM_002389.1 | MCP | Membrane cofactor protein (CD46 trophoblastlymphocyte cross-reactive antigen) | 1.7 | <0.01 | 1.1 | 0.74 | 1.2 | 0.01 |
| D84105.1 | MCP | Membrane cofactor protein (CD46 trophoblastlymphocyte cross-reactive antigen) | 1.8 | <0.01 | 1.0 | 0.98 | 1.2 | 0.11 |
| NM_014791.1 | MELK | Maternal embryonic leucine zipper kinase | 1.8 | <0.01 | 1.8 | <0.01 | 1.0 | 0.79 |
| NM_005924.1 | MEOX2 | Mesenchyme homeo box 2 (growth arrest-specific homeo box) | 3.3 | <0.01 | 2.1 | 0.23 | 1.6 | 0.47 |
| BG170541 | MET | Met proto-oncogene (hepatocyte growth factor receptor) | 1.7 | <0.01 | 1.4 | 0.27 | 1.3 | 0.25 |
| X54559.1 | MET | Met proto-oncogene (hepatocyte growth factor receptor) | 1.6 | <0.01 | 1.0 | 0.84 | 1.2 | 0.39 |
| BC005043.1 | MGC31957 | Hypothetical protein MGC31957 | 2.2 | 0.02 | 1.3 | 0.57 | 1.4 | 0.22 |
| BF001806 | MKI67 | Antigen identified by monoclonal antibody Ki-67 | 2.1 | <0.01 | 1.7 | 0.01 | 1.0 | 0.76 |
| Z69744 | MLL | Myeloid/lymphoid or mixedlineage leukemia (trithorax homolog Drosophila) | 1.7 | <0.01 | 1.2 | 0.44 | 1.4 | 0.03 |
| NM_016195.1 | MPHOSPH1 | M-phase phosphoprotein 1 | 1.9 | 0.01 | 1.8 | 0.2 | 1.2 | 0.58 |
| NM_006540.1 | NCOA2 | Nuclear receptor coactivator 2 | 2.0 | 0.05 | 2.0 | 0.3 | 2.1 | 0.01 |
| NM_002497.1 | NEK2 | NIMA (never in mitosis gene a)-related kinase 2 | 1.6 | 0.08 | 1.5 | 0.16 | 1.3 | 0.44 |
| Z25425.1 | NEK2 | NIMA (never in mitosis gene a)-related kinase 2 | 1.7 | 0.03 | 1.3 | 0.53 | 1.3 | 0.54 |
| Z25434.1 | NEK3 | NIMA (never in mitosis gene a)-related kinase 3 | 1.7 | 0.01 | 1.3 | 0.34 | 1.3 | 0.3 |
| NM_007361.1 | NID2 | Nidogen 2 (osteonidogen) | 2.1 | <0.01 | 2.4 | <0.01 | 1.0 | 0.83 |
| AF146343.1 | NR5A2 | Nuclear receptor subfamily 5, group a member 2 | 3.2 | 0.08 | 3.1 | 0.06 | 1.7 | 0.27 |
| AF228413.1 | NR5A2 | Nuclear receptor subfamily 5, group a member 2 | 4.3 | <0.01 | 3.0 | 0.07 | 1.4 | 0.62 |
| NM_003822.1 | NR5A2 | Nuclear receptor subfamily 5, group a member 2 | 1.9 | 0.08 | 1.5 | 0.38 | 1.1 | 0.84 |
| AF145712.1 | NRP1 | Neuropilin 1 | 2.0 | <0.01 | 1.1 | 0.77 | 1.5 | 0.09 |
| NM_003999.1 | OSMR | Oncostatin M receptor | 1.7 | 0.07 | 1.3 | 0.49 | 1.2 | 0.59 |
| NM_005746.1 | PBEF | Pre-B-cell colony-enhancing factor | 1.6 | <0.01 | 1.2 | 0.17 | 1.2 | 0.05 |
| BF575514 | PBEF | Pre-B-cell colony-enhancing factor | 1.5 | <0.01 | 1.2 | 0.12 | 1.1 | 0.1 |
| BC001422.1 | PGF | Placental growth factor, vascular endothelial growth factor-related protein | 1.5 | <0.01 | 1.7 | <0.01 | 1.2 | 0.06 |
| AA805318 | PIK3CB | Phosphoinositide-3-kinase, catalytic beta polypeptide | 1.7 | 0.09 | 1.1 | 0.73 | 1.4 | 0.3 |
| NM_004203.1 | PKMYT1 | Membrane-associated tyrosine- and threoninespecific cdc2-inhibitory kinase | 1.7 | 0.03 | 1.7 | 0.1 | 1.0 | 0.97 |
| NM_005030.1 | PLK | Polo-like kinase (Drosophila) | 1.6 | 0.02 | 1.3 | 0.21 | 1.1 | 0.82 |
| NM_025179.1 | PLXNA2 | Plexin A2 | 1.6 | 0.04 | 0.9 | 0.8 | 1.0 | 0.94 |
| NM_002674.1 | PMCH | Pro-melanin-concentrating hormone | 10.7 | <0.01 | 6.6 | <0.01 | 0.5 | 0.46 |
| BC002715.1 | PPARD | Peroxisome proliferative activated receptor delta | 2.7 | 0.06 | 2.1 | 0.21 | 0.8 | 0.79 |
| NM_003981.1 | PRC1 | Protein regulator of cytokinesis 1 | 2.4 | <0.01 | 2.0 | <0.01 | 1.3 | 0.24 |
| AF100763.1 | PRKAA1 | Protein kinase AMPactivated, alpha 1 catalytic subunit | 1.5 | <0.01 | 1.0 | 0.98 | 1.0 | 0.76 |
| NM_000315.1 | PTH | Parathyroid hormone | 1.8 | 0.08 | 1.9 | 0.07 | 2.2 | 0.07 |
| AF074979.1 | RGS20 | Regulator of G-protein signalling 20 | 2.6 | <0.01 | 2.7 | <0.01 | 1.2 | 0.59 |
| BF062629 | RIS1 | RAS-induced senescence 1 | 5.9 | <0.01 | 10.5 | <0.01 | 3.2 | 0.27 |
| NM_003035.1 | SIL | TAL1 (SCL) interrupting locus | 1.5 | 0.03 | 1.5 | 0.05 | 1.0 | 0.99 |
| BC001441.1 | SKP2 | S-phase kinase-associated protein 2 (p45) | 1.7 | <0.01 | 1.1 | 0.74 | 1.3 | 0.2 |
| NM_005983.1 | SKP2 | S-phase kinase-associated protein 2 (p45) | 3.7 | 0.05 | 1.1 | 0.76 | 0.9 | 0.89 |
| NM_006444.1 | SMC2L1 | SMC2 structural maintenance of chromosomes 2-like 1 (yeast) | 1.9 | <0.01 | 1.5 | <0.01 | 1.2 | 0.11 |
| AU154486 | SMC2L1 | SMC2 structural maintenance of chromosomes 2-like 1 (yeast) | 2.2 | <0.01 | 1.7 | 0.16 | 1.3 | 0.25 |
| BG035761 | SOCS3 | Suppressor of cytokine signaling 3 | 3.0 | 0.07 | 2.5 | 0.26 | 0.7 | 0.71 |
| NM_001049.1 | SSTR1 | Somatostatin receptor 1 | 3.1 | 0.05 | 2.0 | 0.39 | 1.1 | 0.88 |
| NM_003155.1 | STC1 | Stanniocalcin 1 | 22.8 | <0.01 | 2.6 | 0.02 | 0.8 | 0.53 |
| NM_005990.1 | STK10 | Serine/threonine kinase 10 | 2.3 | <0.01 | 2.4 | <0.01 | 1.0 | 0.84 |
| AB015718 | STK10 | Serine/threonine kinase 10 | 1.5 | <0.01 | 1.7 | <0.01 | 1.0 | 0.9 |
| AB011446.1 | STK12 | Serine/threonine kinase 12 | 1.9 | <0.01 | 1.6 | 0.06 | 1.1 | 0.51 |
| AL043646 | STK18 | Serine/threonine kinase 18 | 5.0 | <0.01 | 4.9 | <0.01 | 2.9 | 0.11 |
| NM_014264.1 | STK18 | Serine/threonine kinase 18 | 3.0 | <0.01 | 2.1 | 0.05 | 1.2 | 0.66 |
| NM_003158.1 | STK6 | Serine/threonine kinase 6 | 1.8 | <0.01 | 1.4 | 0.14 | 1.2 | 0.32 |
| NM_003600.1 | STK6 | Serine/threonine kinase 6 | 1.7 | <0.01 | 1.4 | 0.1 | 1.2 | 0.36 |
| NM_003242.1 | TGFBR2 | Transforming growth factor, beta receptor II (70/80 k Da) | 1.5 | 0.02 | 0.9 | 0.61 | 1.1 | 0.28 |
| NM_000361.1 | THBD | Thrombomodulin | 2.8 | <0.01 | 2.6 | <0.01 | 1.1 | 0.78 |
| NM_000361.1 | THBD | Thrombomodulin | 5.6 | <0.01 | 5.4 | <0.01 | 1.2 | 0.79 |
| AF153687.1 | TNFRSF10B | Tumor necrosis factor receptor superfamily member 10b | 1.8 | <0.01 | 0.9 | 0.58 | 1.2 | 0.17 |
| AF012536.1 | TNFRSF10C | Tumor necrosis factor receptor superfamily member 10c, decoy without an intracellular domain | 2.2 | <0.01 | 1.5 | 0.16 | 1.3 | 0.36 |
| NM_003841.1 | TNFRSF10C | Tumor necrosis factor receptor superfamily member 10c, decoy without an intracellular domain | 2.1 | <0.01 | 1.4 | 0.09 | 1.1 | 0.44 |
| BF664114 | TNFRSF5 | Tumor necrosis factor receptor superfamily member 5 | 2.3 | 0.01 | 1.9 | 0.03 | 0.8 | 0.49 |
| NM_001250.1 | TNFRSF5 | Tumor necrosis factor receptor superfamily member 5 | 4.7 | <0.01 | 2.8 | 0.02 | 1.1 | 0.65 |
| M15565.1 | TRA | T cell receptor alpha locus | 1.9 | 0.07 | 1.7 | 0.02 | 1.0 | 0.98 |
| NM_005879.1 | TRIP | TRAF interacting protein | 1.7 | 0.09 | 2.0 | 0.02 | 1.2 | 0.66 |
| NM_004237.1 | TRIP13 | Thyroid hormone receptor interactor 13 | 1.7 | <0.01 | 1.6 | 0.06 | 1.1 | 0.7 |
| NM_003318.1 | TTK | TTK protein kinase | 2.1 | <0.01 | 1.5 | 0.08 | 1.3 | 0.19 |
| AJ003062.1 | TUBGCP3 | Tubulin gamma complex-associated protein 3 | 1.6 | 0.03 | 1.3 | 0.39 | 1.2 | 0.65 |
| NM_007019.1 | UBE2C | Ubiquitin-conjugating enzyme E2C | 1.7 | <0.01 | 1.4 | 0.04 | 1.1 | 0.57 |
| NM_007063.1 | VRP | Vascular Rab-GAP/TBC containing | 2.0 | <0.01 | 1.7 | 0.01 | 1.2 | 0.12 |
| AI983115 | WSX1 | Class I cytokine receptor | 3.9 | <0.01 | 2.7 | 0.12 | 2.3 | 0.16 |
| NM_016164.2 | XLKD1 | Extracellular link domain containing 1 | 3.4 | 0.01 | 1.6 | 0.47 | 1.2 | 0.81 |
| Z25433.1 | Protein-serine/threonine kinase (Homo sapiens), mRNA sequence | 4.5 | <0.01 | 2.3 | 0.32 | 2.5 | 0.06 | |
| X07868 | Homo sapiens cDNA:FLJ22066 fis clone HEP10611 mRNA sequence | 2.9 | <0.01 | 3.8 | <0.01 | 1.4 | 0.23 | |
| AA058828 | Soluble vascular endothelial cell growth factor receptor (A49636) | 1.9 | <0.01 | 1.6 | 0.03 | 1.1 | 0.45 | |
| AA351360 | KIAA0585 protein (Homo sapiens) (AB011157) | 2.0 | <0.01 | 1.4 | 0.74 | 1.2 | 0.86 |
The resulting subsets of genes for the 4 and 24 h time points, respectively, are provided in Tables 2, and 3 with annotation of the Genbank accession number and gene abbreviation, and corresponding ratio and P-values, for the HGF versus basal, VEGF versus basal, and HGF plus VEGF versus basal experiments.
While there are a number of the same genes represented on both the 4 and 24 h lists, careful perusal of the lists indicates significant differences in the ongoing molecular events at these time points. For example, the 4 h list contains a number of chemokines and chemokine receptors (IL-8, CCR6, CXCR4, and CXC1), and cytokines and cytokine receptors (IL-6, -11, and IL17RC). Another notable feature of the 4 h list is the upregulated expression of a number of genes playing an important role in growth factor signal transduction, including egr-1, fos B, grb10, grb14, MAP2K3, MAP3K8, MAPKAP2, MPK3, DUSP4 and 6, which may play a role in amplifying the growth factor response. A number of genes with some relationship to calcium homeostasis are also upregulated, including stanniocalcin 1, calcitonin gene-related peptide, and parathyroid hormone-related protein precursor. Another notable signature are the number of G-protein-coupled receptors (in addition to the chemokine receptors) and related signaling proteins, which are also upregulated, including “G-protein-coupled receptor-induced protein GIG2” (C8FW), RGS2, D(3) dopamine receptor, serotonin receptor 7, GPR4, GPR37, thromboxane A2 receptor, and lectomedin 1. Several growth factors including PDGF A chain, BMP2, Hb-EGF, FGF-16, heuregulin-beta 1, and c-kit ligand are induced, as well as the potent endothelial mitogen, VEGF-C, and an inhibitor of endothelial cell proliferation, VEGI (TNFSF15). ADAMTS1 (which is upregulated to a similar extent by both HGF and VEGF and is not further upregulated by the combination) is known to inhibit endothelial proliferation by binding and sequestering VEGF. Interestingly, the ligand ‘cardiotrophin like cytokine' and its receptor ‘ciliary neurotrophic factor receptor alpha precursor' show very similar regulation and suggest a potential role for this pathway in the angiogenesis induced by HGF and VEGF. Angiopoietin 2, which is believed to play a modulatory role in angiogenesis and lymphangiogenesis (Kim et al., 2000; Veikkola & Alitalo, 2002; Satchell & Mathieson, 2003), is upregulated by both VEGF and HGF, although there appears to be no additive interaction between the two growth factors on the expression of this gene. The related angiopoietin 4 is also upregulated at 4 h, and a recent report suggests this factor may also have angiogenic activities (Zhu et al., 2002). The VEGF receptors, neuropilin-1 and flt-1 are also upregulated. Also notable is the upregulated expression of a number of orphan nuclear receptors (NURR1/NR4A2, NR4A3, NR5A2).
Several of the genes in the HGF+VEGF subset are involved in the regulation of hematopoietic precursor cells, which may be related to the recruitment and survival of angiopoietic precursor cells to sites of ongoing angiogenesis. For example, kit ligand is able to augment the proliferation of both myeloid and lymphoid hematopoietic progenitors in bone marrow cells (Smith et al., 2001; Duarte & Franf, 2002; Heike & Nakahata, 2002), and can also mediate cell adhesion (Pesce et al., 1997; Bendall et al., 1998; Ashman, 1999; Mitsunari et al., 1999; Shimizu et al., 2001). Interleukin 11 stimulates the proliferation of hematopoietic stem cells and megakaryocyte progenitor cells. (Turner et al., 1996; Nandurkar et al., 1998; Lazzari et al., 2001; Momose et al., 2002). The cellular adhesion molecules ICAM-1 and CD44 may also play a role in the recruitment of progenitor cells.
A review of the list of genes identified as upregulated at 24 h reveals a strong ‘cell cycle' signature, with the upregulation of a number of genes involved in the regulation of the cell cycle, including the cyclins E2 and F, the dual specificity phosphatase CDC14A, the protein kinase CDC2, as well as other related cell cycle control proteins including CDC20, CDC25a,b, and c, CDC6, CDK2, CKS1b, and CDKN2C, and genes involved in the regulation of mitosis including BUB1 (mitotic checkpoint serine–threonine-protein kinase), mitotic spindle assembly checkpoint protein (MAD2L1), DNA replication licensing factor (MCM5), proliferating cell nuclear antigen KI67, and the kinases PLK1, STK 6,12, and 18. Related to this control of the cell cycle ‘theme' there is also upregulation of genes involved in the regulation of apoptosis, including CSE1l, BIRC5 (survivin), apoptotic protease-activating factor (APAF1), and procaspase 8 (CASP8). At 24 h, there is also upregulation of the HGF receptor, c-met, and the flt-1 ligand, placental growth factor.
These data are consistent with the hypothesis that the combination of HGF and VEGF provides a strong push to move cells from quiescence into the cell cycle. At earlier time points, a number of important steps in receptor tyrosine kinase signaling and downstream activation of mitogen-activated protein kinase pathways are upregulated, as well as the mRNA for a number of important growth factors and receptors with a potential role in the proliferative response to the two growth factors. At a later time point (24 h), the sequelae of these early events are apparent with the clear signal that the cells have progressed from cell cycle arrest and are actively undergoing mitosis. These data are also consistent with previous published observations that the combination of HGF and VEGF are additive to synergistic on endothelial cell proliferation (Van Belle et al., 1998).
To validate the expression of some of the mRNAs identified in this study, we performed independent analysis of HGF- and VEGF-treated HUVEC mRNA (from three different endothelial isolates (i.e. different from those used for the array studies)) using real-time PCR (Taqman) analysis as previously described (Kahn et al., 2000; Gerritsen et al., 2002; Yang et al., 2002). Data were normalized to the housekeeping gene cyclophilin. As shown in Table 4, all four of the genes evaluated exhibited alterations in gene expression, consistent with the data obtained from the oligonucleotide array analysis. In addition, we measured the protein levels by ELISA of one of the more highly regulated, and ‘synergistic' genes, that is, the secreted protein STC1. At 24 h, the levels of STC-1 in basal, HGF- or VEGF-treated cell supernatants were below the level of detection (<0.02 ng ml−1). However, when cells were treated with the combination of HGF and VEGF, STC-1 levels were markedly increased (2.8±0.2 ng ml−1).
Table 4.
Taqman validation
| Gene | Ratio of fold change/cyclophilin | Ratio of fold change/cyclophilin |
|---|---|---|
| 4 h | 24 h | |
| ANGPT2 | 15.91±1.83 | 18.73±0.88 |
| CXCR4 | 1.02±1.00 | 10.50±1.95 |
| NID2 | 1.89±0.76 | 9.62±1.64 |
| STC1 | 11.00±11.50 | 60.71±4.93 |
Discussion
Using rigorous and tightly controlled experimental conditions, tightly controlled biological replicates, and multiple comparisons, gene expression profiling using the Affymetrix oligonucleotide technology combined with software analysis packages can yield reliable, highly validated results. For example, using a similar approach, Gerritsen and co-workers identified over 1000 differentially expressed genes as regulated in a three-dimensional collagen gel model of endothelial differentiation (Gerritsen et al., 2002; 2003). Several hundred of these genes were selected for further evaluation by an independent method (RT–PCR, Taqman) and greater than 95% of the genes identified were shown to be regulated in a manner suggested by the results from the oligonucleotide array technology.
In the present study, we have identified discrete subsets of genes that were upregulated by HGF, VEGF, and the combination of the two growth factors. Examination of the list of genes upregulated by VEGF provided further confirmation of the method, since many of the genes identified (e.g. DUSP6, stanniocalcin, CXCR4, FGF16, angiopoeitin 2, Flt-1, Nurr44, nidogen2, melanin concentrating hormone, stanniocalcin-1) have been reported in earlier studies by Yang et al., Bell and co-workers, Abe and others (Abe & Sato, 2001; Bell et al., 2001; Yang et al., 2002). Similarly, although the literature on HGF-induced endothelial cell gene expression is limited; upregulated expression of ets-1, CD44, thymosin B4, and downregulated expression of occludin 1 has been previously noted (Jiang et al., 1999; Oh et al., 2002; Recio & Merling, 2003; Tomita et al., 2003). We found that probe sets corresponding to these same genes were also identified as ‘significantly' regulated. The present study shows for the first time, the additive to synergistic interactions of HGF with VEGF on endothelial gene expression, the differential effects of HGF versus VEGF on endothelial cell gene expression, and moreover, provides the first large-scale gene expression analysis of HGF-induced gene expression in endothelial cells.
There are known differences in the actions of HGF versus VEGF. For example, in addition to eliciting endothelial proliferation, VEGF also induces increased vascular permeability in vivo, and increased expression of fenestrae in vitro. HGF does not demonstrate these activities. HGF was originally called ‘scatter factor' based on its ability to include scattering of polarized epithelial cells, an activity not shared by VEGF. The effects of VEGF are primarily restricted to endothelial cells, due to the limited expression of the receptors KDR and flt-1 (although the expression of flt-1 has been described on cells of monocytic lineage and on smooth muscle cells). In contrast, the c-met receptor is expressed in epithelial cells, various tumor cells, keratinocytes, smooth muscle cells, hepatocytes, and endothelial cells.
This study clearly demonstrates that HGF and VEGF signal through independent pathways in endothelial cells. Since both HGF and VEGF are upregulated at sites of pathological angiogenesis (e.g. tumors, rheumatoid arthritis, diabetic retinopathy), this raises the possibility that successful antiangiogenic therapies may require antagonism of multiple pathways of angiogenic growth factor signaling. These observations also suggest that further evaluation of growth factor combinations in ‘therapeutic angiogenesis' indications should be encouraged.
Acknowledgments
We express our sincere appreciate to Irina Agoulnik, Alan Carpino, Jean Courtemanche, Eric Koenig, Stephen Tirrell, and Trudy Wilson of the Department of Applied Research Technologies and Services at Millennium (Cambridge, MA, U.S.A.) for their assistance in performing the oligonucleotide array studies.
Abbreviations
- FGF
fibroblast growth factor
- HGF
hepatocyte growth factor
- HUVEC
human umbilical vein endothelial cells
- PAI-1
plasminogen activator inhibitor-1
- VEGF
vascular endothelial growth factor
References
- ABE M., SATO Y. cDNA microarray analysis of the gene expression profile of VEGF-activated human umbilical vein endothelial cells. Angiogenesis. 2001;4:289–298. doi: 10.1023/a:1016018617152. [DOI] [PubMed] [Google Scholar]
- ASAHARA T., BAUTERS C., ZHENG L.P., TAKESHIT A.S., BUNTING S., FERRARA N., SYMES J.F., ISNER J.M. Synergistic effect of vascular endothelial growth factor and basic fibroblast growth factor on angiogenesis in vivo. Circulation. 1995;92:II365–II371. doi: 10.1161/01.cir.92.9.365. [DOI] [PubMed] [Google Scholar]
- ASHMAN L.K. The biology of stem cell factor and its receptor C-kit. Int. J. Biochem. Cell. Biol. 1999;31:1037–1051. doi: 10.1016/s1357-2725(99)00076-x. [DOI] [PubMed] [Google Scholar]
- BAFFOUR R., BERMAN J., GARB J.L., RHEE S.W., KAUFMAN J., FRIEDMANN P. Enhanced angiogenesis and growth of collaterals by in vivo administration of recombinant basic fibroblast growth factor in a rabbit model of acute lower limb ischemia:dose–response effect of basic fibroblast growth factor. J. Vasc. Surg. 1992;16:181–191. [PubMed] [Google Scholar]
- BAFFOUR R., GARB J.L., KAUFMAN J., BERMAN J., RHEE S.W., NORRIS M.A., FRIEDMANN P. Angiogenic therapy for the chronically ischemic lower limb in a rabbit model. J. Surg. Res. 2000;93:219–229. doi: 10.1006/jsre.2000.5980. [DOI] [PubMed] [Google Scholar]
- BAUMGARTNER I., ISNER J.M. Stimulation of peripheral angiogenesis by vascular endothelial growth factor (VEGF) Vasa. 1998;27:201–206. [PubMed] [Google Scholar]
- BELL S.E., MAVILA A., SALAZAR R., BAYLESS K.J., KANAGALA S., MAXWELL S.A., DAVIS G.E. Differential gene expression during capillary morphogenesis in 3D collagen matrices:regulated expression of genes involved in basement membrane matrix assembly,cell cycle progression,cellular differentiation and G-protein signaling. J. Cell Sci. 2001;114:2755–2773. doi: 10.1242/jcs.114.15.2755. [DOI] [PubMed] [Google Scholar]
- BENDALL L.J., MAKRYNIKOLA V., HUTCHINSON A., BIANCHI A.C., BRADSTOCK K.F., GOTTLIEB D.J. Stem cell factor enhances the adhesion of AML cells to fibronectin and augments fibronectin-mediated anti-apoptotic and proliferative signals. Leukemia. 1998;12:1375–1382. doi: 10.1038/sj.leu.2401136. [DOI] [PubMed] [Google Scholar]
- BUSH R.L., PEVEC W.C., NDOYE A., CHEUNG A.T., SASSE J., PEARSON D.N. Regulation of new blood vessel growth into ischemic skeletal muscle. J. Vasc. Surg. 1998;28:919–928. doi: 10.1016/s0741-5214(98)70070-9. [DOI] [PubMed] [Google Scholar]
- BUSSOLINO F., DI RENZO M.F., ZICHE M., BOCCHIETTO E., OLIVERO M., NALDINI L., GAUDINO G., TAMAGNONE L., COFFER A., COMOGLIO P.M. Hepatocyte growth factor is a potent angiogenic factor which stimulates endothelial cell motility and growth. J. Cell Biol. 1992;119:629–641. doi: 10.1083/jcb.119.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHLEBOUN J.O., MARTINS R.N. The development and enhancement of the collateral circulation in an animal model of lower limb ischaemia. Aust. NZJ. Surg. 1994;64:202–207. doi: 10.1111/j.1445-2197.1994.tb02179.x. [DOI] [PubMed] [Google Scholar]
- DUARTE R.F., FRANF D.A. The synergy between stem cell factor (SCF) and granulocyte colony-stimulating factor (G-CSF):molecular basis and clinical relevance. Leuk. Lymphoma. 2002;43:1179–1187. doi: 10.1080/10428190290026231. [DOI] [PubMed] [Google Scholar]
- FERRARA N. Role of vascular endothelial growth factor in the regulation of angiogenesis. Kidney Int. 1999;56:794–814. doi: 10.1046/j.1523-1755.1999.00610.x. [DOI] [PubMed] [Google Scholar]
- FERRARA N., ALITALO K. Clinical applications of angiogenic growth factors and their inhibitors. Nat. Med. 1999;5:1359–1364. doi: 10.1038/70928. [DOI] [PubMed] [Google Scholar]
- GERRITSEN M., SORIANO R., YANG S., ZLOT C., INGLE G., TOY K., WILLIAMS P. Branching out: a molecular fingerprint generated by Affymetrix Oligonucleotide arrays. Microcirculation. 2003;10:63–81. doi: 10.1038/sj.mn.7800170. [DOI] [PubMed] [Google Scholar]
- GERRITSEN M.E., SORIANO R., YANG S., INGLE G., ZLOT C., TOY K., WINER J., DRAKSHARAPU A., PEALE F., WU T.D., WILLIAMS P.M. In silico data filtering to identify new angiogenesis targets from a large in vitro gene profiling data set. Physiol. Genomics. 2002;10:13–20. doi: 10.1152/physiolgenomics.00035.2002. [DOI] [PubMed] [Google Scholar]
- HAYASHI S., MORISHITA R., NAKAMURA S., YAMAMOTO K., MORIGUCHI A., NAGANO T., TAIJI M., NOGUCHI H., MATSUMOTO K., NAKAMURA T., HIGAKI J., OGIHARA T. Potential role of hepatocyte growth factor, a novel angiogenic growth factor, in peripheral arterial disease:downregulation of HGF in response to hypoxia in vascular cells. Circulation. 1999;100:II301–II308. doi: 10.1161/circ.100.suppl_2.Ii-301. [DOI] [PubMed] [Google Scholar]
- HEIKE T., NAKAHATA T. Ex vivo expansion of hematopoietic stem cells by cytokines. Biochim. Biophys. Acta. 2002;1592:313–321. doi: 10.1016/s0167-4889(02)00324-5. [DOI] [PubMed] [Google Scholar]
- HENRY T.D., ANNEX B.H., MCKENDALL G.R., AZRIN M.A., LOPEZ J.J., GIORDANO F.J., SHAH P.K., WILLERSON J.T., BENZA R.L., BERMAN D.S., GIBSON C.M., BAJAMONDE A., RUNDLE A.C., FINE J., MCCLUSKEY E.R. The VIVA trial: vascular endothelial growth factor in ischemia for vascular angiogenesis. Circulation. 2003;107:1359–1365. doi: 10.1161/01.cir.0000061911.47710.8a. [DOI] [PubMed] [Google Scholar]
- JIANG W.G., MARTIN T.A., MATSUMOTO K., NAKAMURA T., MANSEL R.E. Hepatocyte growth factor/scatter factor decreases the expression of occludin and transendothelial resistance (TER) and increases paracellular permeability in human vascular endothelial cells. J. Cell Physiol. 1999;181:319–329. doi: 10.1002/(SICI)1097-4652(199911)181:2<319::AID-JCP14>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- KAHN J., MEHRABAN F., INGLE G., XIN X., BRYANT J., VEHAR G., SCHOENFELD J., GRIMALDI C., PEALE F., DRAKHARAPU A., LEWIN D., GERRITSEN M. Gene expression profiling in an in vitro model of angiogenesis. Am. J. Pathol. 2000;156:1887–1900. doi: 10.1016/S0002-9440(10)65062-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KHAN T.A., SELLKE F.W., LAHAM R.J. Therapeutic angiogenesis:protein-based therapy for coronary artery disease. Expert Opin. Pharmacother. 2003;4:219–226. doi: 10.1517/14656566.4.2.219. [DOI] [PubMed] [Google Scholar]
- KIM I., KIM J.H., MOON S.O., KWAK H.J., KIM N.G., KOH G.Y. Angiopoietin-2 at high concentration can enhance endothelial cell survival through the phosphatidylinositol 3′-kinase/Akt signal transduction pathway. Oncogene. 2000;19:4549–4552. doi: 10.1038/sj.onc.1203800. [DOI] [PubMed] [Google Scholar]
- LAZAROUS D.F., UNGER E.F., EPSTEIN S.E., STINE A., AREVALO J.L., CHEW E.Y., QUYYUMI A.A. Basic fibroblast growth factor in patients with intermittent claudication: results of a phase I trial. J. Am. Coll. Cardiol. 2000;36:1239–1244. doi: 10.1016/s0735-1097(00)00882-2. [DOI] [PubMed] [Google Scholar]
- LAZZARI L., LUCCHI S., REBULLA P., PORRETTI L., PUGLISI G., LECCHI L., SIRCHIA G. Long-term expansion and maintenance of cord blood haematopoietic stem cells using thrombopoietin, Flt3-ligand, interleukin (IL)-6 and IL-11 in a serum-free and stroma-free culture system. Br. J. Haematol. 2001;112:397–404. doi: 10.1046/j.1365-2141.2001.02528.x. [DOI] [PubMed] [Google Scholar]
- MITSUNARI M., HARADA T., TANIKAWA M., IWABE T., TANIGUCHI F., TERAKAWA N. The potential role of stem cell factor and its receptor c-kit in the mouse blastocyst implantation. Mol. Hum. Reprod. 1999;5:874–879. doi: 10.1093/molehr/5.9.874. [DOI] [PubMed] [Google Scholar]
- MOMOSE K., TAGUCHI K., SAITOH M., YASUDA S., MIYATA K. Effects of interleukin-11 on the hematopoietic action of granulocyte colony-stimulating factor. Arzneimittelforschung. 2002;52:857–861. doi: 10.1055/s-0031-1299981. [DOI] [PubMed] [Google Scholar]
- MORIMOTO A., OKAMURA K., HAMANAKA R., SATO Y., SHIMA N., HIGASHIO K., KUWANO M. Hepatocyte growth factor modulates migration and proliferation of human microvascular endothelial cells in culture. Biochem. Biophys. Res. Commun. 1991;179:1042–1049. doi: 10.1016/0006-291x(91)91924-2. [DOI] [PubMed] [Google Scholar]
- NAKAMURA Y., MORISHITA R., HIGAKI J., KIDA I., AOKI M., MORIGUCHI A., YAMADA K., HAYASHI S., YO Y., NAKANO H., MATSUMOTO K., NAKAMURA T., OGIHARA T. Hepatocyte growth factor is a novel member of the endothelium-specific growth factors:additive stimulatory effect of hepatocyte growth factor with basic fibroblast growth factor but not with vascular endothelial growth factor. J. Hypertens. 1996;14:1067–1072. doi: 10.1097/00004872-199609000-00004. [DOI] [PubMed] [Google Scholar]
- NANDURKAR H.H., ROBB L., BEGLEY C.G. The role of IL-II in hematopoiesis as revealed by a targeted mutation of its receptor. Stem Cells. 1998;16:53–65. doi: 10.1002/stem.5530160708. [DOI] [PubMed] [Google Scholar]
- OH I.S., SO S.S., JAHNG K.Y., KIM H.G. Hepatocyte growth factor upregulates thymosin beta4 in human umbilical vein endothelial cells. Biochem. Biophys. Res. Commun. 2002;296:401–405. doi: 10.1016/s0006-291x(02)00888-4. [DOI] [PubMed] [Google Scholar]
- PEPPER M.S., FERRARA N., ORCI L., MONTESANO R. Potent synergism between vascular endothelial growth factor and basic fibroblast growth factor in the induction of angiogenesis in vitro. Biochem. Biophys. Res. Commun. 1992;189:824–831. doi: 10.1016/0006-291x(92)92277-5. [DOI] [PubMed] [Google Scholar]
- PESCE M., DI CARLO A., DE FELICI M. The c-kit receptor is involved in the adhesion of mouse primordial germ cells to somatic cells in culture. Mech. Dev. 1997;68:37–44. doi: 10.1016/s0925-4773(97)00120-2. [DOI] [PubMed] [Google Scholar]
- RECIO J.A., MERLINO G. Hepatocyte growth factor/scatter factor induces feedback up-regulation of CD44v6 in melanoma cells through Egr-1. Cancer Res. 2003;63:1576–1582. [PubMed] [Google Scholar]
- ROBERTS C.J., NELSON B., MARTON M.J., STOUGHTON R., MEYER M.R., BENNETT H.A., HE Y.D., DAI H., WALKER W.L., HUGHES T.R., TYERS M., BOONE C., FRIEND S.H. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science. 2000;287:873–880. doi: 10.1126/science.287.5454.873. [DOI] [PubMed] [Google Scholar]
- ROSEN E.M., LAMSZUS K., LATERRA J., POLVERINI P.J., RUBIN J.S., GOLDBERG I.D.HGF/SF in angiogenesis Ciba Found. Symp. 1997212215–226.(discussion 227–229) [DOI] [PubMed] [Google Scholar]
- SATCHELL S.C., MATHIESON P.W. Angiopoietins: microvascular modulators with potential roles in glomerular pathophysiology. J. Nephrol. 2003;16:168–178. [PubMed] [Google Scholar]
- SCHADT E.E., LI C., ELLIS B., WONG W.H. Feature extraction and normalization algorithm for high-density oligonucleotide gene expression array data. J. Cell. Biochem. 2002;84:120–125. doi: 10.1002/jcb.10073. [DOI] [PubMed] [Google Scholar]
- SENGUPTA S., GHERARDI E., SELLERS L.A., WOOD J.M., SASISEKHARAN R., FAN T.P. Hepatocyte growth factor/scatter factor can induce angiogenesis independently of vascular endothelial growth factor. Arterioscler. Thromb. Vasc. Biol. 2003;23:69–75. doi: 10.1161/01.atv.0000048701.86621.d0. [DOI] [PubMed] [Google Scholar]
- SHIMIZU M., MINAKUCHI K., TSUDA A., HIROI T., TANAKA N., KOGA J., KIYONO H. Role of stem cell factor and c-kit signaling in regulation of fetal intestinal epithelial cell adhesion to fibronectin. Exp. Cell Res. 2001;266:311–322. doi: 10.1006/excr.2001.5221. [DOI] [PubMed] [Google Scholar]
- SMITH M.A., COURT E.L., SMITH J.G. Stem cell factor: laboratory and clinical aspects. Blood Rev. 2001;15:191–197. doi: 10.1054/blre.2001.0167. [DOI] [PubMed] [Google Scholar]
- STARK J., BAFFOUR R., GARB J.L., KAUFMAN J., BERMAN J., RHEE S., NORRIS M.A., FRIEDMANN P. Basic fibroblast growth factor stimulates angiogenesis in the hindlimb of hyperglycemic rats. J. Surg. Res. 1998;79:8–12. doi: 10.1006/jsre.1998.5392. [DOI] [PubMed] [Google Scholar]
- STOUGHTON R., DAI H.Statistical combining of cell expression profiles 2002. US Patent No.6, 351, 712
- TOMITA N., MORISHITA R., TANIYAMA Y., KOIKE H., AOKI M., SHIMIZU H., MATSUMOTO K., NAKAMURA T., KANEDA Y., OGIHARA T. Angiogenic property of hepatocyte growth factor is dependent on upregulation of essential transcription factor for angiogenesis, ets-1. Circulation. 2003;107:1411–1417. doi: 10.1161/01.cir.0000055331.41937.aa. [DOI] [PubMed] [Google Scholar]
- TURNER K.J., NEBEN S., WEICH N., SCHAUB R.G., GOLDMAN S.J. The role of recombinant interleukin 11 in megakaryocytopoiesis. Stem Cells. 1996;14:53–61. doi: 10.1002/stem.5530140707. [DOI] [PubMed] [Google Scholar]
- VAN BELLE E., WITZENBICHLER B., CHEN D., SILVER M., CHANG L., SCHWALL R., ISNER J.M. Potentiated angiogenic effect of scatter factor/hepatocyte growth factor via induction of vascular endothelial growth factor: the case for paracrine amplification of angiogenesis. Circulation. 1998;97:381–390. doi: 10.1161/01.cir.97.4.381. [DOI] [PubMed] [Google Scholar]
- VEIKKOLA T., ALITALO K. Dual role of Ang2 in postnatal angiogenesis and lymphangiogenesis. Dev. Cell. 2002;3:302–304. doi: 10.1016/s1534-5807(02)00231-9. [DOI] [PubMed] [Google Scholar]
- WOJTA J., KAUN C., BREUSS J.M., KOSHELNICK Y., BECKMANN R., HATTEY E., MILDNER M., WENINGER W., NAKAMURA T., TSCHACHLER E., BINDER B.R. Hepatocyte growth factor increases expression of vascular endothelial growth factor and plasminogen activator inhibitor-1 in human keratinocytes and the vascular endothelial growth factor receptor flk-1 in human endothelial cells. Lab. Invest. 1999;79:427–438. [PubMed] [Google Scholar]
- XIN X., YANG S., INGLE G., ZLOT C., RANGELL L., KOWALSKI J., SCHWALL R., FERRARA N., GERRITSEN M.E. Hepatocyte growth factor enhances vascular endothelial growth factor-induced angiogenesis in vitro and in vivo. Am. J. Pathol. 2001;158:1111–1120. doi: 10.1016/S0002-9440(10)64058-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YANG H.T., FENG Y. bFGF increases collateral blood flow in aged rats with femoral artery ligation. Am. J. Physiol. Heart Circ. Physiol. 2000;278:H85–H93. doi: 10.1152/ajpheart.2000.278.1.H85. [DOI] [PubMed] [Google Scholar]
- YANG S., TOY K., INGLE G., ZLOT C., WILLIAMS P.M., FUH G., LI B., DE VOS A., GERRITSEN M.E. Vascular endothelial growth factor-induced genes in human umbilical vein endothelial cells:relative roles of KDR and Flt-1 receptors. Arterioscler. Thromb. Vasc. Biol. 2002;22:1797–1803. doi: 10.1161/01.atv.0000038995.31179.24. [DOI] [PubMed] [Google Scholar]
- ZHU H., LI J., QIN W., YANG Y., HE X., WAN D., GU J. Cloning of a novel gene, ANGPTL4 and the functional study in angiogenesis. Zhonghua Yi Xue Za Zhi. 2002;82:94–99. [PubMed] [Google Scholar]