Abstract
(α3)2(β4)3 is the most abundant type of neuronal nicotinic ACh receptor (nAChR) mediating cholinergic actions on the autonomic nervous system. Studies to refine or devise drugs selectively acting on (α3)2(β4)3 receptors would benefit from a detailed description of the hitherto unclear agonist-binding domain.
The present study reports a three-dimensional model for the ligand-binding domain (LBD) of this receptor either in its unoccupied or agonist-bound conformation. The receptor model was based on the structure of the acetylcholine-binding protein (AChBP), and was obtained using molecular modelling techniques.
ACh, nicotine and cytisine (full agonists), muscarine (a selective agonist of muscarinic ACh receptors) and the allosteric modulator eserine were docked into the binding pockets of the receptor model. Simulated agonist–receptor complexes were compared with the agonist-binding complex of the AChBP, as well as of the (α4)2(β2)3 type of nAChR, which is the commonest in the brain.
Agonist docking identified discrete amino-acid residues of the β subunits important for pharmacological selectivity of nAChRs. The predicted affinity of muscarine for the nAChR was low, suggesting the present model to be suitable for effective discrimination of nicotinic agonist binding versus nonselective cholinergic binding. Furthermore, the current model outlined a potential binding site for the allosteric modulator eserine, the site of action of which has remained elusive.
The present LBD model of the receptor in its free state provides a novel structural framework to interpret experimental observations and a useful template for future investigations to develop (α3)2(β4)3-selective ligands.
Keywords: Nicotinic acetylcholine receptor, molecular modelling, docking, allosteric binding site, muscarine, nicotine, cytisine, eserine
Introduction
Neuronal nicotinic acetylcholine receptors (nAChRs) belong to a superfamily of ligand-gated ion channels (Barnard, 1996). Their structure consists of five membrane-spanning subunits arranged around an aqueous central channel (Itier & Bertrand, 2001). A variety of receptor subclasses are known to exist, depending on different subunit assemblies (α1–α10, β1–β4, δ, γ and ɛ; Changeux, 1990). The current interest in nAChRs stems from the fact that their functional deficit is thought to be involved in neuropsychiatric disorders such as Alzheimer's disease, Parkinson's disease, epilepsy and schizophrenia (Paterson & Nordberg, 2000).
Among nAChRs, (α3)2(β4)3 is expressed by autonomic neurons (Lindstrom, 1997), and has also been found in the mammalian brain, where its function remains poorly understood (Sudweeks & Yakel, 2000). In autonomic ganglia, one β subunit is often replaced by α5, a structural characteristic which confers stronger receptor desensitization (Groot-Kormelink et al., 2001). There are detailed functional differences between (α3)2(β4)3 receptors and (α4)2(β2)3 receptors, the latter being the predominant type expressed in the mammalian brain (Lindstrom, 1997; Paterson & Nordberg, 2000). In particular, (α3)2(β4)3 receptors possess lower Ca2+ permeability (Haghighi & Cooper, 2000) and desensitize faster (Chavez-Noriega et al., 1997) when activated by an agonist. The pharmacological properties of (α3)2(β4)3 are also different, because, on such receptors, cytisine is less potent than on (α4)2(β2)3 receptors (Chavez-Noriega et al., 1997). Dihydro-β-erythroidine is a much more potent blocker of (α4)2(β2)3 than of (α3)2(β4)3 receptors, whereas the snail conotoxin AuIB is a selective antagonist of (α3)2(β4)3 receptors (Luo et al., 1998). While previous studies have indicated that the β subunits confer agonist selectivity to the nAChRs (Luetje & Patrick, 1991; Cohen et al., 1995; Parker et al., 1998; 2001), the precise identification of all the structural determinants of the β subunits responsible for the pharmacological properties of α3β4 receptors remains incompletely understood.
The α3 and β4 subunits share a topology common to all the other nAChR subunits (Unwin, 1993; 1995; Beroukhim & Unwin, 1995; Hucho et al., 1996). They are composed of: (i) a large N-terminal extracellular domain (also called the ligand-binding domain, LBD), which is believed to be located at the interface between α and neighboring non-α subunits (Corringer et al., 2000); (ii) four hydrophobic transmembrane regions (M1–M4), of which M2, and to a much lesser extent M1, is believed to contribute to the formation of the cationic channel (Le Novere et al., 1999); (iii) one intracellular domain joining M3 and M4; (iv) a small extracellular C-terminal domain.
The recent resolution of the X-ray structure (Brejc et al., 2001) of the acetylcholine-binding protein (AChBP) from the snail Lymnaea stagnalis (Smit et al., 2001) has paved the way to the construction of structural models of the nicotinic LBDs. Since almost all AChBP residues relevant for ligand binding are conserved within the nAChR family (Brejc et al., 2001), models of the (α4)2(β2)3 receptor and (α7)5 receptor, commonly found in the mammalian central nervous system, have recently been built by exploiting the structural similarity to AChBP (Le Novere et al., 2002).
Using the X-ray structure of AChBP as a template, we built a model of the (α3)2(β4)3 LBD in both the free state and in complex with cholinergic ligands such as acetylcholine (ACh), nicotine and cytisine, which are full agonists on (α3)2(β4)3 receptors (Di Angelantonio et al., 2002). In addition, to further test the validity of our simulations concerning nicotinic receptors, we investigated the binding of muscarine, which is the prototypical muscarinic agent. Finally, we were interested in modelling the binding of the typical allosteric modulator eserine to explore the allosteric binding region of nicotinic receptors.
For the sake of comparison, the binding mode of these ligands was also investigated on the (α4)2(β2)3 structural model (Le Novere et al., 2002) and AChBP X-ray structure (Brejc et al., 2001). Structural data were also validated by the experimental results obtained from the native receptor of the Torpedo electric organ, which is a standard model for molecular analysis of nicotinic receptors (Karlin, 2002).
Methods
Structural models
α3β4 model
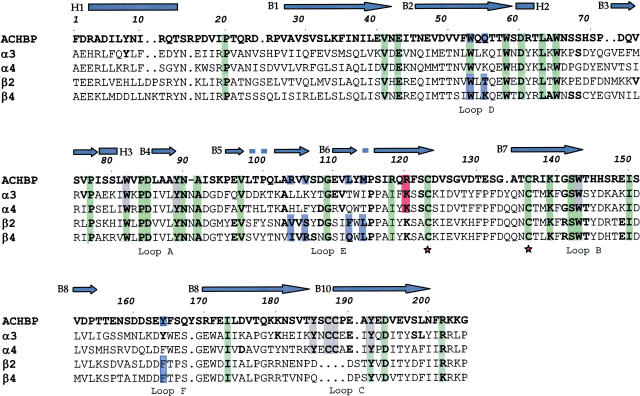
Using the CLUSTALX program (Thompson et al., 1997), the sequence of AChBP was aligned with those of the LBDs of α3 and β4 subunits (Figure 1). The α4 and β2 subunit LBDs were also aligned (see Figure 1) to confirm correspondence with the data recently reported (Le Novere et al., 2002; Schapira et al., 2002).
Figure 1.
Sequence alignment between AChBP and amino-terminal domains of rat α3, α4, β2 and β4 nAChR subunits. In this figure, the numbering of amino-acid residues refers to AChBP only. The top line (blue) presents the secondary structure of AChBP: bars indicate α-helices and arrows β-sheets. Green, gray, blue and red colors indicate residues present in all subunits, residues forming the agonist-binding site, those forming the antagonist-binding site, and residue interacting with the allosteric ligand, respectively. Asterisks indicate the beginning and the end of the Cys loop. The loops indicated in the bottom line are referred to residues belonging to the agonist-binding site.
The three-dimensional model of (α3)2(β4)3 LBD (α3β4 hereafter) was built using the program MODELLER (Sali & Blundell, 1993). The model with the highest MODELLER scores (Sanchez & Sali, 1997) was refined by imposing first symmetry constraints for the α and the β subunits and by loop structural improvements, and then energy minimization along with a short molecular dynamics (MD) run. The (α4)2(β2)3 LBD model (α4β2 hereafter; Le Novere et al., 2002) was instead downloaded from the ligand-gated ion channel database (http://www.pasteur.fr/recherche/banques/LGIC/), and its structure was minimized following the same protocol. While preparing the present manuscript, a model of the α3β4 receptor has been made available at the ligand-gated ion channel database. The main difference between that model and ours is based on the symmetry constraints, which were imposed to our model only. It should also be noted that the web-based model does not address the issue of agonist binding which is examined in the present study.
Ligand structure
For nicotine, cytisine and eserine, which are relatively rigid molecules, the X-ray structures at highest resolution were considered (Cambridge Structural Database (CSD) entries DOXSIS (Barlow et al., 1986), FITPIHO2 (Barlow & Johnson, 1989) and ESEIN10 (Pauling & Petcher, 1973), respectively). In contrast, for ACh and muscarine, which are more flexible molecules, all X-ray structures were considered (CSD entries ACHOLC (Herdklotz & Sass, 1970), ACCHOBII (Svinning & Sourum, 1975), ACHTPB (Datta et al., 1980) and GEBMEF (Frydenvang et al., 1988) for ACh; HABNON (Frydenvang, 1990) and HABNUT (Frydenvang, 1990) for muscarine). The basic nitrogens were assumed to have a positively charged protonated form.
Ligand–receptor complexes
ACh, nicotine, cytisine, muscarine and eserine were docked into α3β4, α4β2 and AChBP. In order to reduce the computational time, only single pairs of subunits were considered. This approach was taken because the ligand-binding sites are located at the interface of subunit pairs (+)(−) or αβ (Karlin, 2002). The agonist-binding site was defined as the region which includes the residues listed in Results and (within a sphere of 6 Å from the center of mass of these residues) all other groups possessing at least one non-hydrogen atom. The allosteric agonist-binding site was defined by residues within a 14 Å sphere centered on the center mass of residues αLys122/+Lys119 (Results).
First, the binding pockets were relaxed by performing a short MD simulation with water molecules and, afterwards, the ligands were added by molecular docking techniques (see below).
Computational details
The AMBER (Cornell et al., 1995) and TIP3P (Jorgensen et al., 1983) force fields were used to describe the interatomic potential energy functions of proteins and water, respectively. The topology and force-field parameters of the ligands were assigned from equivalent atom types present in the AMBER databases (Case et al., 1997). RESP atomic partial charges of the ligands were calculated using the standard AMBER procedure (Bayly et al., 1993; Case et al., 1997). For ACh and muscarine, the partial charges were the averages for all conformers considered here.
MD simulations were performed using the AMBER program (Case et al., 1997). The temperature and pressure were kept constant by coupling the systems to a Berendsen bath algorithm (Berendsen et al., 1984) with 1.0 ps relaxation time. van der Waals interactions were truncated up to a spherical, residue-based cutoff of 12 Å. Electrostatic interactions were calculated using the Ewald particle mesh method (Essman et al., 1995). The dielectric constant was set to 1. The time-step integration of the Newton equation of motion was set to 1.5 fs.
Basic receptor model
In all, 1000 steps of steepest descent followed by 2000 steps of conjugate-gradient algorithm were first carried out. Then, the position of the side chains was relaxed by 30 ps of MD at a low temperature (50 K), while the backbone was harmonically constrained (force constant 10 kcal mol−1 Å−2). Note that we used MD simulations only to optimize the structure locally. Thus, the temperature of 50 K is not a physical parameter.
Ligand–receptor complexes
MD simulation of the receptor capped with a water sphere of 15 Å radius around the binding site was performed for 50 ps. Next, cytisine (that is the largest agonist considered here) or eserine (an allosteric modulator) was docked inside its respective binding pocket (see below for docking details), followed by another 50 ps MD. All calculations were run while simulating a low constant temperature (50 K) and constraining the position of the backbone atoms. The final MD structure of the receptors was used for docking studies.
The DOCK program (Ewing & Kuntz, 1997) was used for docking the ligands to the receptors. Binding sites were parameterized by a two-step procedure: (i) constructing the molecular surfaces of the protein sites using Connolly's algorithm (Connolly, 1983). A probe atom of 1.0 Å radius and a dot density of 4 points Å−1 were used. (ii) Filling the cavity with water molecules using spheres of variable radii (1.0–4.0 Å).
For each ligand and each receptor, the following procedure was followed. (i) Four docking runs were performed starting from different initial conditions and treating the ligand as a flexible molecule. Each run supplied 50–200 possible configurations for each ligand/receptor pair differing in their DOCK scores (DSs; Ewing & Kuntz, 1997). (ii) The conformations obtained for each complex were clustered in several classes, depending on their similarity in terms of r.m.s.d.: each class comprised structures differing by an r.m.s.d. value for the non-hydrogen atoms lower than 2 Å from its neighboring conformations. Classes were ranked on the basis of their DSs (when two classes possessed equivalent DSs, the most populated class was considered first). (iii) For each ligand/receptor complex, the structure with the highest DS was chosen (Table 1 ). (iv) A sphere of 100 water molecules around the ligand was added to each complex. (v) Finally, the complexes were relaxed through a short (30 ps) MD simulation at room temperature. The receptor residues were constrained with a force constant K=0.01 kcal mol−1 Å−2 when the distance from the ligand was less than 10 Å, with K=1.0 kcal mol−1 Å−2 between 10 and 20 Å, and with K=10.0 kcal mol−1 Å−2 at distances larger than 20 Å.
Table 1.
Subunit/subunit interactions: distances (Å) between residues forming salt bridgesa
| α3β4 | AChPB | ||||
|---|---|---|---|---|---|
| Pair of residues | Pair of residues | ||||
| αArg17(Nδ) | βGlu1(Oγ) | 2.7 | +Glu148(Oγ) | −Arg2(Nδ) | 2.8 |
| αArg78(Nδ) | βGlu2(Oγ) | 2.8 | +Glu148(Oγ) | −Arg103(Nδ) | 2.9 |
| βGlu10(Oγ) | αArg13(Nδ) | 3.1 | +Glu148(Oγ) | −Arg2(Nδ) | 2.8 |
| βAsp88(Oδ) | αLys104(Nɛ) | 2.8 | +Glu148(Oγ) | −Arg103(Nδ) | 2.9 |
Salt bridges crossing α/β (+/−) and β/α (−/+) interfaces in α3β4 and AChBP. Interactions mainly involved not-aligned residues.
Electrostatic calculations
Electrostatic potentials on the molecular surface of the three receptors (α3β4, α4β2 and of AChBP) were calculated by solving the Poisson–Boltzmann equation. The calculations were carried out assuming room temperature and a 0.150 mM salt concentration using the Delphi program (Gilson & Honig, 1988; Honig & Nicholls, 1995). Dielectric constants were set to 80 and 2, respectively, for the solvent and solute.
Results
α3β4 Receptor structure
The sequence identities of α3 and β4 subunits with AChBP were found to be 22 and 20%, respectively, and the alignment shows that most conserved residues were hydrophobic (Figure 1). While the present data are compatible with those of Brejc et al. (2001) and Le Novere et al. (2002) for α4 and β2 subunits, they are, however, significantly different from those of Schapira et al. (2002).
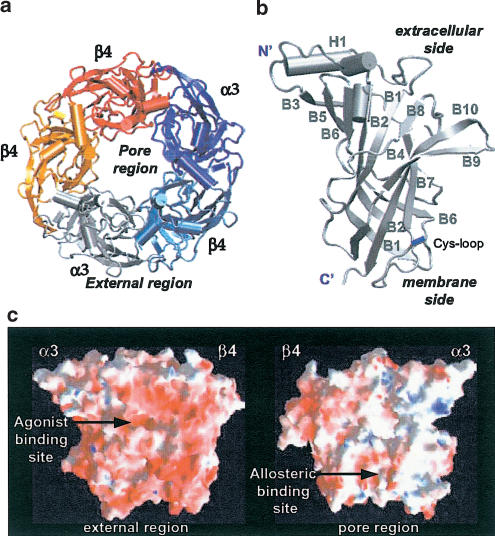
The resulting 3D structural model (structure available at http://www.sissa.it/sbp/bc/publications/publications_5.html) was a barrel of 80 Å diameter and 63 Å height with a central irregular pore (10–15 Å; Figure 2a). The single subunit structure (exemplified with the α3 subunit; Figure 2b) did not differ largely from that of the template, as expected from the relatively low number of mismatches in the alignment. Only small differences were observed in the β4 subunits; in particular, the α-helix H1 was shorter, B6 comprised two components and the loop between B8 and B9 was longer than the one of the template.
Figure 2.
α3β4 structure obtained by comparative modelling and MD calculations. (a) Top view of the pentameric structure. The model has the shape of a barrel of 80 Å diameter and 63 Å height, with a central irregular pore of variable diameter (10–15 Å). (b) Secondary structure of the α3 subunit. It is composed of one α helix (H1) and 10 β sheets (B1–B10). Two very small α helices are also present. (c) Color-coded calculated electrostatic potential at the surface of one α3β4 pair of subunits. The external and pore sides are shown, outlining the amphipatic profile of the nicotinic receptor N-terminal domain. The prevalence of negatively charged residues is apparent. The binding site location of full and allosteric agonists is indicated within the external and pore region, respectively. Red, blue and white colors indicate the negative, positive and neutral regions.
As usual for cytoplasmatic domains, hydrophobic residues were mainly located in the core region of the subunits. In contrast, the charged residues, mainly distributed over the external side of the pentamer, were not well conserved even within the α3β4 and α4β2 receptors despite high sequence identity (62 and 69%, between α and β subunits, respectively). The hydrophobic core region and the negatively charged protein surface of the external side are highlighted by plotting their electrostatic potential (Figure 2c).
The subunit interfaces were formed entirely by loops on one side and mostly by β sheets on the opposite side. The interface residues were poorly conserved between nAChR subunits and AChBP, or within the nAChR family. In particular, the salt bridges crossing the α3/β4 interfaces mostly involved nonhomologous residues, and are listed (with their distance values) in Table 1.
The 13-residue Cys loop, which is known to be important for the complete nAChR assembly (Green & Wanamaker, 1998), was characterized by the presence of a disulfide bridge between the two β-sheets B6 and B7 (Figure 2b). Whereas the Cys loop of α3β4 was hydrophobic, in contrast, the 12-Cys loop of AChBP was mostly hydrophilic. Segments 1–20 and 152–170 (Figure 1) were also not homologous with those of AChBP. The accuracy of the model in those regions was therefore expected to be lower than for the general structure.
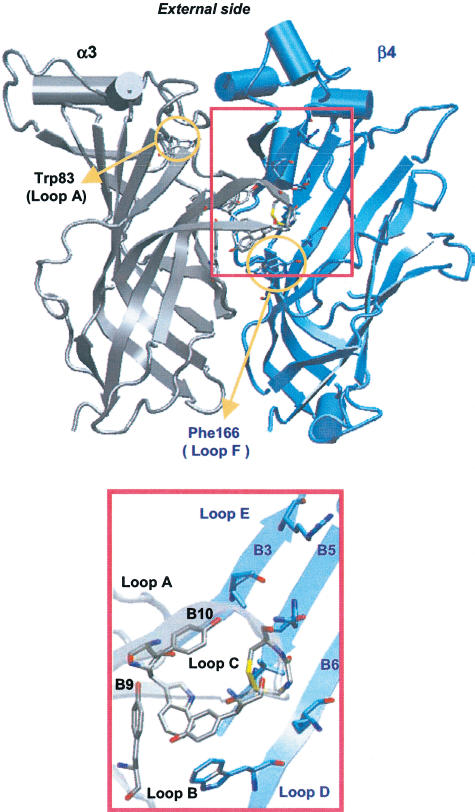
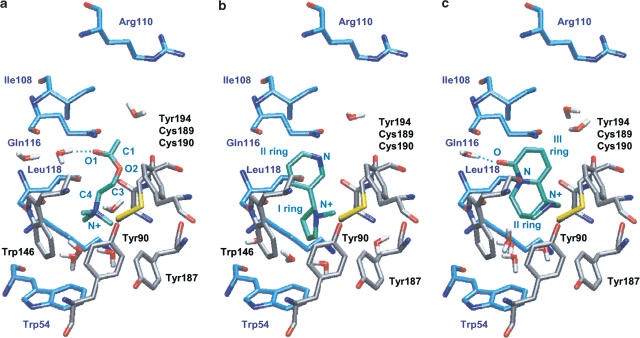
As expected, the agonist-binding site was the same as in the structure of the template used for the homology model calculations. It was located at the interface between α3 and β4 subunits, and mostly composed of aromatic residues (Figure 3). At the binding site, six amino acids of the α3 subunit (αTyr90 of loop A, αTrp146 of loop B and αTyr187/αCys189/αCys190/αTyr194 of loop C) formed the principal binding component (made up of the α subunit of nAChRs; Karlin, 2002). The complementary binding component (made up of the β subunit; Karlin, 2002) included five residues on the β4 subunit (βTrp54 of loop D and βIle108/βArg110/βGln116/βLeu118 of loop E). The residues of the α subunit were highly conserved within the nAChR family and AChBP. Conversely, only βTrp54 was conserved within the β subunits (see loop D in Figure 1).
Figure 3.
α3β4 agonist-binding site. Seven residues of the α subunit (gray) on loops A, B and C formed the principal component of the binding site, while five residues of the β subunit (blue) on loops D and E formed the complementary component. This arrangement yields a small hydrophobic cavity on the external side at the interface between α and β subunits.
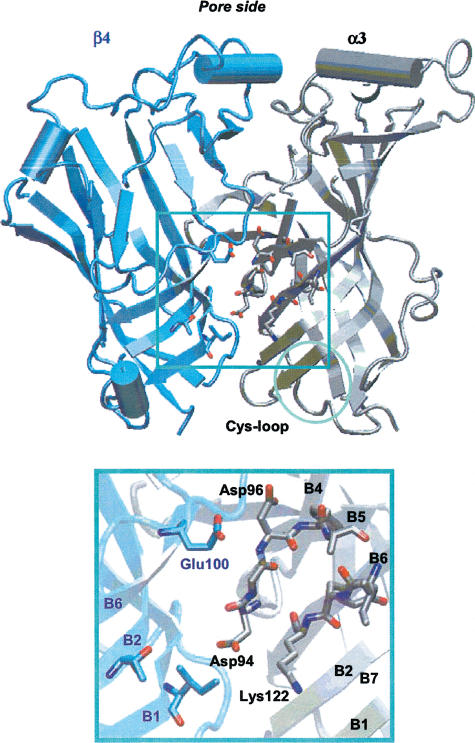
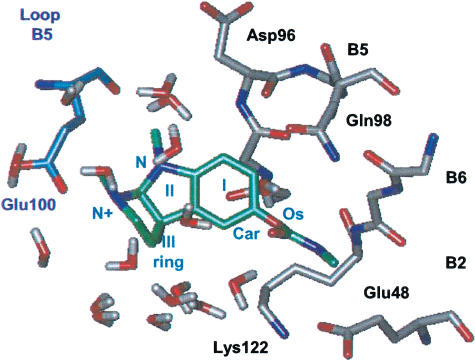
The location of the allosteric binding site(s) is not clearly determined (Arias, 2000). Photoaffinity labelling experiments have shown eserine to label α1Lys125 of the Torpedo receptor (Schrattenholz et al., 1993). On the basis of the current subunit alignment (see Figure 1), the residue α1Lys125 of the Torpedo receptor corresponded to αLys122 for the nAChRs and +Arg119 for the AChBP, which could be found on β-sheet B6 of the nAChR α subunits or the corresponding AChBP (+) subunit (Figure 4). Unlike the external location of the agonist-binding site (see Figure 3), the allosteric binding site was internally located between β-sheets B2 and B5, thus not far from the subunit interface (Figure 4).
Figure 4.
α3β4 allosteric modulator-binding site. The suggested binding region is located around αLys122, between segments of β-sheets B1, B2, B6 and B5 on the α subunit (gray) and β-sheets B1, B2 and B6 of the β subunit (blue). Residues involved in eserine binding are shown as sticks, and are observed within the pore region.
We defined the allosteric binding site on nAChRs as the region within 14 Å from αLys122. This was mostly composed of four segments of the αα subunit, namely αVal38–αAsp41 (β-sheet B1), αAsn44–αAsn50 (β-sheet B2), αLeu89–αPhe97 (β-sheet B5) and αAla119–αCys125 (β-sheet B6), and some residues of the β component facing the subunit interface (on β-sheets B1, B2 and B6). In the α3β4 structural model, αLys122 made a salt bridge with α3Glu48 (β-sheet B2), substituted by α4Asp39 (β-sheets B1) and +Asp46 (β-sheets B2) in α4β2 and AChBP, respectively.
Ligand-binding modes
Selected ligands were docked on the α3β4 and the α4β2 structural models (Le Novere et al., 2002) and the AChBP X-ray structure (Brejc et al., 2001). The docking results obtained for each ligand–receptor pair were analyzed according to two properties: (1) the number and the rank order of the classes for all binding conformations, (2) the structural properties of the selected ligand/receptor complexes. Table 2 lists all the main interactions of cholinergic ligand molecules with discrete motifs of the α3β4 and α4β2 receptors as well as the AChBP. On the basis of such data, it was possible to provide a detailed reconstruction of the binding interaction of various ligands with the α3β4 receptor, indicating the spatial arrangement of these agonists within the binding region (Figure 5). Binding of different ligands was qualitatively evaluated on the basis of their DS (Table 3 ) and their electrostatic properties. Comparison was also made with experimental data carried out on Torpedo nicotinic receptors.
Table 2.
Ligand/receptor distances (Å) in the final MD structures
| Ligand | α3β4 | α4β2 | AChPB | |||
|---|---|---|---|---|---|---|
| Acetylcholine | ||||||
| N+ | αTrp146(ring) | 4.8 | αTrp146(ring) | 4.5 | +Tyr184(ring) | 4.8 |
| αTyr90(OH) | 4.3 | αTyr90(OH) | 4.7 | +Tyr191(ring) | 4.6 | |
| αTrp146(O) | 4.3 | αTrp146(O) | 4.2 | −Trp52(ring) | 4.6 | |
| 3H2O(O) | <5.0 | H2O(O) | 3.9 | +Tyr88(OH) | 4.3 | |
| C4 | αCys189(S) | 3.5 | — | — | — | — |
| C3 | αTrp146(ring) | 3.7 | αCys189(S) | 3.7 | +Trp142(ring) | 3.5 |
| C1 | βLeu118(Cδ) | 4.4 | βPhe116(ring) | 4.0 | — | — |
| O | H2O(O) | 2.8 | H2O(O) | 3.1 | — | — |
| Nicotine | ||||||
| N+ | αTyr194(ring) | 4.8 | αTrp146(ring) | 4.8 | −Trp52(ring) | 4.8 |
| αTyr90(OH) | 4.4 | αTyr90(OH) | 4.3 | — | — | |
| H2O(O) | 4.8 | αTrp146(O) | 4.2 | — | — | |
| — | — | 3H2O(O) | <5.0 | — | — | |
| Ring (I) | αTrp146(ring) | 3.6 | αTyr187(ring ⊥) | 3.4 | +Tyr88(ring ⊥) | 4.7 |
| αCys189(S) | 3.5 | — | — | +Trp142(ring) | 4.7 | |
| — | — | — | — | +Tyr184(ring ⊥) | 4.3 | |
| — | — | — | — | +Tyr191(ring) | 4.2 | |
| N | βLeu118(O) | 2.8 | — | — | +Tyr191(OH) | 3.2 |
| αTyr194(OH) | 3.2 | — | — | — | — | |
| Ring (II) | αTrp146(O) | 2.8 | βPhe116(ring) | 3.4 | +Cyx187(S) | 3.4 |
| βLeu118(Cδ) | 4.0 | αCys189(S) | 3.9 | −Met113(S) | 3.9 | |
| Cytisine | ||||||
| N+ | αTyr194(ring) | 3.8 | αTyr194(ring) | 3.4 | +Tyr88(OH) | 3.1 |
| αTyr90(OH) | 4.3 | αTyr90(OH) | 3.1 | +Ser141(O) | 3.6 | |
| αSer145(O) | 4.2 | αTrp146(O) | 3.1 | H2O(O) | 4.4 | |
| αTrp146(O) | 3.1 | H2O(O) | 4.7 | — | — | |
| Ring (I) | — | — | αTyr187(ring ⊥) | 4.0 | +Tyr88(ring ⊥) | 3.7 |
| — | — | — | — | +Tyr184(ring) | 4.2 | |
| — | — | — | — | +Tyr191(ring ⊥) | 4.2 | |
| Ring(I) | αTrp146(ring ⊥) | 3.7 | αTrp146(ring ⊥) | 3.8 | −Trp52(ring ⊥) | 3.8 |
| αCys190(S) | 4.0 | βLeu118(Cδ) | 3.4 | +Trp142(O) | 2.9 | |
| Ring(III) | βLeu118(Cδ) | 3.6 | βPhe116(ring) | 4.0 | +Cyx188(S) | 4.0 |
| βIle108(Cδ) | 3.3 | αCys190(S) | 3.5 | −Met113(S) | 3.9 | |
| O | H2O(O) | 3.2 | H2O(O) | 2.7 | +Trp142(ring) | 2.9 |
Note that the term (ring) after an amino-acid residue denotes either the interaction of the ligand N+ atom with the corresponding amino-acid ring (π-cation type of interaction) or the π–π planar interaction between the ring structure of nicotine (or cytisine) and the corresponding ring structure of the amino-acid residue on the receptor subunit. When one ring interacts orthogonally to the other, this reaction is indicated as ring ⊥.
Figure 5.
Simplified scheme of agonist binding to the α3β4 receptor. In (a—c), the ligand molecule is depicted in green. The conformation of the receptor structure binding ACh (a), nicotine (b) or cytisine (c) is represented along the barrel axis of the pentameric receptor. Note that all side chains are in the same position.
Table 3.
DOCK scores of ligand/receptor complexes
| Ligand | α3β4 DS | α4β2 DS | AChPB DS |
|---|---|---|---|
| Acetylcholine | −26 | −26 | −28 |
| Nicotine | −26 | −20 | −23 |
| Cytisine | −33 | −34 | −31 |
| Muscarine | −8 | −14 | −17 |
| Eserine | −25 | −23 | −26 |
DS are expressed as kcal mol−1 (Ewing & Kuntz, 1997).
In particular, ACh bound in a similar fashion to the three receptor types, in accordance with experimental data, which predicted the same selective orientation (Corringer et al., 2000; Karlin, 2002). Two main binding modes were identified, sharing a common position for the quaternary ammonium group, but differing for the orientation of the tail within the binding pocket. On α3β4 and α4β2 receptors, the quaternary ammonium group formed π–cation interaction with the aromatic ring of αTrp146 (Figure 5a; Table 2), in accordance with previous evidence (Zhong et al., 1998). The group was also stabilized by long-range electrostatic interactions with the αTyr90 side chain, as previously predicted by photoaffinity labelling experiments (Cohen et al., 1991; Sine et al., 1994), and by water molecules present in the binding pocket. Three other aromatic residues, αTyr187, αTyr194 and βTrp54, capped the α3β4- and α4β2-binding pocket over the ACh positively charged moiety at 5–6 Å distance. The pharmacological effect of mutation of these residues is directly associated with reduced ACh-binding affinity (Sine et al., 1994; Corringer et al., 1999). In the AChBP, the ammonium group formed π–cation interactions with +Tyr184, +Tyr191 and −Trp52 aromatic side chains, and electrostatic interactions with +Tyr88. In these cases, no interaction with water was detected.
The methyl group of ACh ester moiety established hydrophobic interactions with the αCys189–αCys190 loop, in accordance with chemical modification experiments (Karlin, 1969), and with aromatic rings (α3Tyr194, β2Phe116 and +Trp142). αTyr194 has been predicted to play a role in binding the ACh ester group (Grutter et al., 2000). The carbonyl oxygen of the ester moiety formed a hydrogen bond with a water molecule in the case of α3β4 and α4β2 receptors (Figure 5a and Table 2).
Nicotine was predicted to bind to both nAChR models similarly with respect to the ammonium group, but with a different orientation of the aromatic rings. Usually, the aromatic nitrogen of nicotine replaces the ester carbonyl of ACh in ligand/receptor adducts (Beers & Reich, 1970). This was actually observed for its binding to the α4β2 receptor. In the case of α3β4, we obtained mainly two binding modes of nicotine with equivalent DSs: the expected one and the one with the aromatic ring rotated by 180° (40 and 50% of configurations, respectively). Since the DSs of the two conformations were equivalent, we suggest that both conformations can occur at the binding site. On α3β4 and α4β2 receptors, the ammonium group formed a π–cation interaction with α3Tyr194 and α4Trp146, and long-range electrostatic interaction with the αTyr90 side chain (Figure 5b and Table 2), in accordance with the experimentally characterized residues essential for nicotine binding (Galzi et al., 1991; Kearney et al., 1996). The ligand also interacted with one to three water molecules. van der Waals interactions stabilized the first and second rings in both nicotinic receptors. On α3β4 receptors, the nitrogen atom of the second ring formed a hydrogen bond with αTyr194(OH). The major difference between nicotine binding to α3β4 or α4β2 receptor subtypes was due to the presence of phenylalanine or glutamine in β2 or β4 subunits, respectively. In fact, while β2Phe116 stabilized its second ring by hydrophobic interactions, β4Gln116 did not play a role in nicotine binding.
Likewise, nicotine binding to AChBP was stabilized by van der Waals interactions mostly involving aromatic side chains. A π–cation interaction with −Trp52 was also observed (Table 2). The importance of this residue for nicotinic receptors has been predicted by mutagenesis experiments reporting the reduction in agonist binding and agonist-induced channel activation (Chiara et al., 1998).
In the case of cytisine, only two conformers with similar orientation within the three binding sites were obtained. This observation could be partially due to the rigidity of the agonist skeleton. The cytisine ammonium group bound the same region as ACh or nicotine did (Figure 5c and Table 2), forming a π–cation interaction with the ring of αTyr194. A H-bond between the cytisine oxygen and one water molecule was observed on α3β4 and α4β2 receptors, but not on the AChBP. Within the nAChRs, several main chain and side chain oxygen atoms stabilized the ammonium group through long-range electrostatic interactions (Table 2). Furthermore, several hydrophobic residues made van der Waals contacts with the three cytisine rings. Among these residues, on the β4 subunit, the highly conserved Ile108 (Dayhoff, 1978) interacted with cytisine; conversely, on the β2 subunit, the conserved Val108 (Dayhoff, 1978) did not establish any relevant contact with this agonist.
Our findings provide the first clue to cytisine binding to nicotinic receptors, thus supplying a working model for designing future mutagenesis and photoaffinity labelling experiments.
Muscarine has minimal affinity for nicotinic receptors and would, therefore, not be expected to bind them in a specific manner. For these reasons, muscarine provides a useful negative control for modelling studies. Therefore, the binding of muscarine to the agonist-binding site was investigated. For each receptor, different modes of binding were found, each one with small DSs (Table 3). In detail, the ligand was found to be (i) unable to bind inside the agonist-binding pocket of the α4β2 model, although it could be positioned at the external surface; (ii) positioned inside the agonist-binding site of AChBP and α3β4, with many different orientations of the ammonium group and of the tetrahydrofuran. Taken together, the low DSs and distinct binding modes are consistent with a very low affinity of muscarine for nicotinic receptors, and are in agreement with its well-known lack of agonist activity on nAChRs.
Eserine was docked to a region 14 Å around αLys122 (+Arg119), which is indeed a residue belonging to the binding site (Schrattenholz et al., 1993). The DSs were similar for the three receptors (Table 3). In all the cases, eserine displayed a very similar conformation to the crystal structure (Pauling & Petcher, 1973), characterized by the complete planarity of the carbamate moiety and Car–Os twisted out of plane (Figure 6) to avoid steric interactions between the carbamate and the aryl ring.
Figure 6.
Simplified scheme of eserine binding to the α3β4 receptor. Eserine is shown in green. The conformation of the receptor structure binding eserine is shown along the barrel axis of the pentameric receptor. Water molecules at less than 3.5 Å from the molecule are shown.
As far as eserine binding was concerned, this process was receptor-selective. In fact, in all receptors, the eserine hydrophobic ring (ring I in Figure 6) lined the β-sheet B5 of the α subunit, but the carbamate tail and the ammonium group were differently oriented within the binding sites. In detail, on the α3β4 receptor, the carbamate moiety was located in proximity of αGlu48 (β-sheet B2) and αLys122 (β-sheet B6), and the charged group was stabilized by either water molecules or βGlu100 (β-sheet B5). In the case of the α4β2 receptor, the carbamate tail did not form specific interactions and the ammonium group was stabilized by either water molecules or αAsp96 (β-sheet B5). Finally, for AChBP, electrostatic interactions were slightly higher (Table 3). In fact, not only the carbamate tail and ammonium group were stabilized by −Glu95/−Arg119 (β-sheet B5/B6) and +Glu95 (β-sheet B5)/water molecules, respectively, but also the nitrogen of ring II (Figure 6) interacted with the protein through +Asp39 (β-sheet B1) and +Asp48 (β-sheet B2) side chains.
Discussion
The principal results of the present study are the provision of the first structural model of α3β4-binding domain and a computational description of the agonist interaction with its binding site.
Three-dimensional model of the α3β4 receptor
Although the overall fold of the α3β4 model was similar to the one of the AChBP template (Brejc et al., 2001) and α4β2 model (Le Novere et al., 2002), it is, however, important to point out that there were significant differences between the α4β2 model and the present α3β4 model.
In the α3β4 model, the agonist-binding site appeared to be located on the external side of the protein and to be made up of a conserved core of aromatic residues on the α and β subunits (loops A–D), and several amino acids from the nonconserved loops E and F, located on the β subunits. The latter may confer individual pharmacological properties to this particular receptor subtype (Corringer et al., 2000) as certain regions of β subunits are believed to be important for agonist selectivity (Cohen et al., 1995; Parker et al., 1998; 2001; Luetje & Patrick, 1991).
In the present model, the residue αTrp83 (loop A), which was identified by labelling studies on the Torpedo receptor as generally important for agonist binding (Galzi et al., 1990), was located outside the binding pocket (Figure 3), and was involved in hydrophobic core formation, perhaps to stabilize the binding site structure. Experimental studies on Torpedo receptors suggest an additional residue (on loop F; see Figure 1) to be important for agonist binding (Czajkowski & Karlin, 1995). In our alignment, this would correspond to βPhe169, which was far away from the putative binding site (Figure 3). Note, however, that the relatively low resolution of the crystal structure in this region makes the structural analysis difficult (Brejc et al., 2001). Moreover, the loop F has low sequence conservation in the nicotinic family (Brejc et al., 2001) and, in the δ subunit of the Torpedo receptor, it corresponds to an aspartate (δAsp182). This and other aspartic residues present in the Torpedo receptor along (and near) loop F are reported to decrease the affinity for ACh (Czajkowski & Karlin, 1995; Martin et al, 1996). Their role, which is probably to stabilize the agonist cationic charge by long-range interactions, could not be fulfilled in the α3β4 receptors by the corresponding residues, because they were absent from β subunits.
In the present study, long-range electrostatic stabilization of the ACh charge was provided by two other aspartates from the principal component, namely αAsp149 (loop B) and αAsp197 (loop C), consistent with mutagenesis experiments (Sugiyama et al., 1996; Osaka et al., 1998). These residues were the source of the negative electrostatic potential which characterized the agonist-binding site, as detected by electrostatic calculations (Figure 3c) and as expected from experimental findings and theoretical studies (Stauffer & Karlin, 1994).
A putative binding site for the allosteric modulators was identified around αLys122, as suggested by photoaffinity labeling experiments (Schrattenholz et al., 1993). In particular, this site was located between β-sheets B2, B5 and B6 of the α subunit and β-sheets B5 and B6 of the β component. The region adjacent to αLys122 was relatively hydrophobic, as shown by the electrostatic potential plot in Figure 2c. The αLeu89–αPhe97 segment (β-sheet B5), which is completely conserved within the nAChR α subunits (cf. Figure 1), seemed to be the most suited to bind a molecule with a hydrophobic moiety within the region investigated here. In fact, it was an amphipathic segment in which nonpolar residues faced the external surface of the pore, and polar (or charged) side chains pointed toward the internal core of the subunit.
It has been suggested (Arias, 2000) that the binding pocket of allosteric effectors is located between the hydrophobic segments 118–124 and 130–137 of the Torpedo α1 subunit. This proposal was based on the presence of aromatic groups in most allosteric ligands so far investigated (Arias, 2000). However, our model reveals that this region is far away (more than 14 Å) from αLys122, making this hypothesis unsuitable for these nAChRs.
Modelling agonist binding to nicotinic receptors
Our study investigated the structural determinants for the complexes between the α3β4 receptor and selected ligands. The ligands examined in the present study included not only ACh and nicotine, whose interactions with α4β2 and α7 receptors have been previously reported (Le Novere et al., 2002; Schapira et al., 2002), but also cytisine (Di Angelantonio et al., 2000) and the allosteric modulator eserine, which does not directly activate receptors but enhances the action of the other full agonists (Albuquerque et al., 1997). Note that the effect of eserine is not mediated by muscarinic receptors as its enhancing action on nicotinic receptors is insensitive to the muscarinic antagonist atropine (Pereira et al., 1993).
Comparison of the ligand-α3β4 adducts with the ones in complex with the AChBP X-ray structure (Brejc et al., 2001), as well as with the α4β2 structural model (Le Novere et al., 2002), has helped to validate the binding mode of each molecule to these proteins.
The agonist (ACh, nicotine and cytisine) ammonium groups were predicted to bind within the same region of the binding pockets, as inferred from experimental studies (Karlin, 2002). This group may be stabilized by the negatively charged electrostatic potential generated mostly by αAsp86 (loop A), αAsp196 (loop C) and αAsp149 (loop B). Interestingly, the role of αAsp86, which is highly conserved within the ligand-gated ion channel family, had not been previously detected by experimental studies.
ACh- and nicotine-binding modes turned out to be in agreement with experimental data (Galzi et al., 1991; Kearney et al., 1996; Karlin, 2002) and similar to those obtained in previous molecular modelling studies (Le Novere et al., 2002; Schapira et al., 2002). However, our simulations pointed out the role of the solvent in agonist binding. This issue was not addressed by Le Novere et al. (2002), while Schapira et al. (2002) assumed only one water molecule around each ligand. A significant difference between ours and Shapira's model was the Cys loop–ACh interaction previously suggested by photoaffinity labeling (Grutter et al., 2000).
The present work also provided novel information on the cytisine-binding mode, since no mutagenesis or photoaffinity labeling data are currently available on this issue. In particular, we noted how cytisine interacted with Ile108 on the β4 subunit, while it did not bind the corresponding Val108 on the β2 subunit. Thus, it is possible that these differences might contribute to the differential affinity of cytisine towards α3β4 and α4β2 receptors (Chavez-Noriega et al., 1997).
Although the DSs of these three agonists were similar, the ranking order of DSs (cytisine>ACh=nicotine) for the α3β4 receptor was analogous to that for radioactive ligand-binding affinity (Di Angelantonio et al., 2000; Free et al., 2002). However, since the DS neglects entropic effects and estimates rather approximately the strength of the intermolecular interactions, correlating computational data with experimental ones requires caution. Further considerations restrain the application of data from three-dimensional models to the interpretation of the physiological properties of agonist-bound receptors. In fact, a model presents a static rather than a dynamic representation of drug–receptor interactions. In particular, after agonist binding, it is suggested that the physiological response due to channel opening is caused by movements triggered by the agonist at the binding site (Jones et al., 2001). Such a phenomenon cannot be described in these current three-dimensional models, whose value consists, instead, in providing a detailed view of binding regions which may be powerful tools for computational screening of potential new ligands to be tested experimentally.
To validate our model, muscarine, a selective agonist on muscarinic ACh receptors and ineffective on nicotinic receptors, was also docked and found to have a low DS.
The docking of the allosteric modulator eserine was located near α1Lys122, consistent with photoaffinity labelling experiments (Schrattenholz et al., 1993). In the nicotinic receptors investigated in the present study, the eserine hydrophobic ring lined the β-sheet B5 of the α subunit (see Figure 6). In particular, both in the α4β2 receptor and the AChBP, αAsp96 (β-sheet B5) appears to have an important role in eserine-binding stabilization. Since the β-sheet B5 is also involved in agonist binding through αTyr90 (β-sheet B5), we can hypothesize that this secondary structure element has a molecular role in the transfer of the eserine allosteric effects to the agonist-binding site. Future studies will be necessary to examine whether other allosteric modulators of nAChRs (Albuquerque et al., 1997) have analogous binding site properties like eserine.
In principle, the carbamate moiety of eserine could interact with the Lys side chain. However, this phenomenon was not observed and the distance between the two groups was 5 Å or larger. This result could be due to the simplified docking procedure, which cannot take into account a large re-equilibration of the protein upon ligand binding. In contrast, αLys122 (+Arg119) formed a salt bridge with a negatively charged side chain with the three receptors (α3Glu48, α4Asp39 or +Asp46 for α3β4, α4β2 or AChBP, respectively). The present results also indicate that an allosteric modulator can possess differential ability to facilitate nicotinic receptors depending on the subunit composition. Further experimental studies should address this issue.
The current model may, therefore, be viewed as a structural framework to interpret a variety of experimental observations, and it represents a useful template for future investigations to devise α3β4-selective drugs.
Acknowledgments
This work was supported by cofinanced grants from MIUR and INFM.
Abbreviations
- ACh
acetylcholine
- AChBP
acetylcholine-binding protein
- α3β4
(α3)2(β4)3 ligand binding domain
- α4β2
(α4)2(β2)3 ligand binding domain
- CSD
Cambridge structural database
- DS
DOCK scores
- ESP
electrostatic potential
- LBD
ligand-binding domain
- MD
molecular dynamics
- nAChRs
neuronal nicotinic receptors
References
- ALBUQUERQUE E.X., ALKONDON M., PEREIRA E.F., CASTRO N.G., SCHRATTENHOLZ A., BARBOSA C.T., BONFANTE-CABARCAS R., ARACAVA Y., EISENBERG H.M., MAELICKE A. Properties of neuronal nicotinic acetylcholine receptors: pharmacological characterization and modulation of synaptic function. J. Pharmacol. Exp. Ther. 1997;280:1117–1136. [PubMed] [Google Scholar]
- ARIAS H.R. Localization of agonist and competitive antagonist binding sites on nicotinic acetylcholine receptors. Neurochem. Int. 2000;36:595–645. doi: 10.1016/s0197-0186(99)00154-0. [DOI] [PubMed] [Google Scholar]
- BAYLY C., CIEPLACK P., CORNELL W., KOLLMAN P.A. A well-behaved electrostatic potential based method using charge restraints deriving atomic charges: the RESP model. J. Phys. Chem. 1993;97:10269–10280. [Google Scholar]
- BARLOW R.B., HOWARD J.A.K., JOHNSON O. Structure of nicotine monomethyl iodide and nicotine monohydrogen iodide. Acta Crystallogr. C. 1986;42:852. [Google Scholar]
- BARLOW R.B., JOHNSON O. Relations between structure and nicotine-like activity: X-ray crystal structure analysis of (−)-cytisine and (−)-lobeline hydrochloride and a comparison with (−)-nicotine and other nicotine-like compounds. Br. J. Pharmacol. 1989;98:799–808. doi: 10.1111/j.1476-5381.1989.tb14608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BARNARD E.A. The transmitter-gated channels: a range of receptor types and structures. Trends Pharmacol. Sci. 1996;17:305–309. [PubMed] [Google Scholar]
- BEERS W.H., REICH E. Structure and activity of acetylcholine. Nature. 1970;228:917–922. doi: 10.1038/228917a0. [DOI] [PubMed] [Google Scholar]
- BERENDSEN H.J.C., POSTMA J.P.M., VAN GUNSTEREN W.F., DINOLA A., HAAK J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- BEROUKHIM R., UNWIN N. Three-dimensional location of the main immunogenic region of the acetylcholine receptor. Neuron. 1995;15:323–331. doi: 10.1016/0896-6273(95)90037-3. [DOI] [PubMed] [Google Scholar]
- BREJC K., VAN DIJK W.J., KLAASSEN R.V., SCHUURMANS M., VAN DER O.J., SMIT A.B., SIXMA T.K. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature. 2001;411:269–276. doi: 10.1038/35077011. [DOI] [PubMed] [Google Scholar]
- CASE D.A., PEARLMAN D.A., CALDWELL J.W., CHEATHAM T.E., III, ROSS W.S., SIMMERLING C.L., DARDEN T.A., MERZ K.M., STANTON R.V., CHENG A.L., VINCENT J.J., CROWLEY M.F., FERGUSON D.M., RADMER R.J., SINGH U.C., WEINER P.K., KOLLMAN P.A. University of California, San Francisco; 1997. AMBER 5. [Google Scholar]
- CHANGEUX J.P., The TIPS Lecture The nicotinic acetylcholine receptor: an allosteric protein prototype of ligand-gated ion channels. Trends Pharmacol. Sci. 1990;11:485–492. doi: 10.1016/0165-6147(90)90049-e. [DOI] [PubMed] [Google Scholar]
- CHAVEZ-NORIEGA L.E., CRONA J.H., WASHBURN M.S., URRUTIA A., ELLIOTT K.J., JOHNSON E.C. Pharmacological characterization of recombinant human neuronal nicotinic acetylcholine receptors hα2β2, hα2β4, hα3β2, hα3β4, hα4β2, hα4β4 and hα7 expressed in Xenopus oocytes. J. Pharmacol. Exp. Ther. 1997;280:346–356. [PubMed] [Google Scholar]
- CHIARA D.C., MIDDLETON R.E., COHEN J.B. Identification of tryptophan 55 as the primary site of [3H]nicotine photoincorporation in the gamma-subunit of the torpedo nicotinic acetylcholine receptor. FEBS Lett. 1998;423:223–226. doi: 10.1016/s0014-5793(98)00093-3. [DOI] [PubMed] [Google Scholar]
- COHEN B.N., FIGL A., QUICK M.W., LABARCA C., DAVIDSON N., LESTER H.A. Regions of beta 2 and beta 4 responsible for differences between the steady state dose–response relationships of the alpha 3 beta 2 and alpha 3 beta 4 neuronal nicotinic receptors. J. Gen. Physiol. 1995;105:745–764. doi: 10.1085/jgp.105.6.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COHEN J.B., SHARP S.D., LIU W.S. Structure of the agonist-binding site of the nicotinic acetylcholine receptor. [3H]acetylcholine identifies residues in the cation-binding subsite. J. Biol. Chem. 1991;266:23354–23364. [PubMed] [Google Scholar]
- CONNOLLY M.L. Analytical molecular surface calculation. J. Appl. Crystallogr. 1983;16:548–558. [Google Scholar]
- CORNELL W.D., CIEPLAK P., BAYLY C.I., GOULD I.R., MERZ K.M., JR, FERGUSON D.M., SPELLMEYER D.C., FOX T., CALDWELL J.W., KOLLMAN P.A. A second-generation force field for the simulation of proteins and nucleic acids. J. Chem. Phys. 1995;117:5179–5197. [Google Scholar]
- CORRINGER P.J., BERTRAND S., GALZI J.L., DEVILLERS-THIERY A., CHANGEUX J.P., BERTRAND D. Molecular basis of the charge selectivity of nicotinic acetylcholine receptor and related ligand-gated ion channels. Novartis Found. Symp. 1999;225:215–224. doi: 10.1002/9780470515716.ch14. [DOI] [PubMed] [Google Scholar]
- CORRINGER P.J., LE NOVERE N., CHANGEUX J.P. Nicotinic receptors at the amino acid level. Annu. Rev. Pharmacol. Toxicol. 2000;40:431–458. doi: 10.1146/annurev.pharmtox.40.1.431. [DOI] [PubMed] [Google Scholar]
- CZAJKOWSKI C., KARLIN A. Structure of the nicotinic receptor acetylcholine-binding site. Identification of acidic residues in the delta subunit within 0.9 Nm of the 5 alpha subunit-binding. J. Biol. Chem. 1995;270:3160–3164. doi: 10.1074/jbc.270.7.3160. [DOI] [PubMed] [Google Scholar]
- DATTA N., MONDAL P., PAULING P. Acta Crystallogr. B. 1980. p. 906.
- DAYHOFF M.O.A model of evolutionary change Proteins in Atlas of Protein Sequence and Structure 1978Washington: Georgetown University Medical Center, National Biomedical Research Foundation; 345–358.ed. Dayhoff, M.O. Vol. 5, Suppl. 3, pp [Google Scholar]
- DI ANGELANTONIO S., COSTA V., CARLONI P., NISTRI A. A novel class of peptides with facilitating action on neuronal nicotinic receptors of rat chromaffin cells in vitro: functional and molecular dynamics studies. Mol. Pharmacol. 2002;61:43–54. doi: 10.1124/mol.61.1.43. [DOI] [PubMed] [Google Scholar]
- DI ANGELANTONIO S., NISTRI A., MORETTI M., CLEMENTI F., GOTTI C. Antagonism of nicotinic receptors of rat chromaffin cells by N,N, N-trimethyl-1-(4-trans-stilbenoxy)-2-propylammonium iodide: a patch clamp and ligand binding study. Br. J. Pharmacol. 2000;129:1771–1779. doi: 10.1038/sj.bjp.0703264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ESSMAN U., PERERA L., BERKOWITZ M.L., DARDEN T., LEE H., PEDERSEN L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- EWING T.J.A., KUNTZ I.D. Critical evaluation of search algorithms for automated molecular docking and database screening. J. Comput. Chem. 1997;18:1175–1189. [Google Scholar]
- FREE R.B., BRYANT D.L., MCKAY S.B., KASER D.J., MCKAY D.B. [3H]epibatidine binding to bovine adrenal medulla: evidence for alpha3beta4* nicotinic receptors. Neurosci. Lett. 2002;318:98–102. doi: 10.1016/s0304-3940(01)02489-2. [DOI] [PubMed] [Google Scholar]
- FRYDENVANG K. J. Acta Crystallogr. C. 1990. p. 985.
- FRYDENVANG K., GRONBORG L., JENSEN B. Structures of acetylcholine picrate and methoxycarbonylcholine picrate hemihydrate. Acta Crystallogr. C. 1988;44:841–845. doi: 10.1107/s0108270188000186. [DOI] [PubMed] [Google Scholar]
- GALZI J.L., REVAH F., BLACK D., GOELDNER M., HIRTH C., CHANGEUX J.P. Identification of a novel amino acid alpha-tyrosine 93 within the cholinergic ligands-binding sites of the acetylcholine receptor by photoaffinity labeling. Additional evidence for a three-loop model of the cholinergic ligands-binding sites. J. Biol. Chem. 1990;265:10430–10437. [PubMed] [Google Scholar]
- GALZI J.L., BERTRAND D., DEVILLERS-THIERY A., REVAH F., BERTRAND S., CHANGEUX J.P. Functional significance of aromatic amino acids from three peptide loops of the alpha 7 neuronal nicotinic receptor site investigated by site-directed mutagenesis. FEBS Lett. 1991;294:198–202. doi: 10.1016/0014-5793(91)80668-s. [DOI] [PubMed] [Google Scholar]
- GILSON M.K., HONIG B. Calculation of the total electrostatic energy of a macromolecular system: solvation energies, binding energies, and conformational analysis. Proteins. 1988;4:7–18. doi: 10.1002/prot.340040104. [DOI] [PubMed] [Google Scholar]
- GREEN W.N., WANAMAKER C.P. Formation of the nicotinic acetylcholine receptor binding sites. J. Neurosci. 1998;18:5555–5564. doi: 10.1523/JNEUROSCI.18-15-05555.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GROOT-KORMELINK P.J., BOORMAN J.P., SIVILOTTI L.G. Formation of functional alpha3beta4alpha5 human neuronal nicotinic receptors in Xenopus oocytes: a reporter mutation approach. Br. J. Pharmacol. 2001;134:789–796. doi: 10.1038/sj.bjp.0704313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GRUTTER T., EHRET-SABATIER L., KOTZYBA-HIBERT F., GOELDNER M. Photoaffinity labeling of torpedo nicotinic receptor with the agonist [3H]DCTA: identification of amino acid residues which contribute to the binding of the ester moiety of acetylcholine. Biochemistry. 2000;39:3034–3043. doi: 10.1021/bi992393o. [DOI] [PubMed] [Google Scholar]
- HAGHIGHI A.P., COOPER E. A molecular link between inward rectification and calcium permeability of neuronal nicotinic acetylcholine α3β4 and α4β2 receptors. J. Neurosci. 2000;20:529–541. doi: 10.1523/JNEUROSCI.20-02-00529.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HERDKLOTZ J.K., SASS R.L. The crystal structure of acetylcholine chloride: a new conformation for acetylcholine. Biochem. Biophys. Res. Commun. 1970;40:583–588. doi: 10.1016/0006-291x(70)90942-3. [DOI] [PubMed] [Google Scholar]
- HONIG B., NICHOLLS A. Classical electrostatics in biology and chemistry. Science. 1995;268:1144–1149. doi: 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
- HUCHO F., TSETLIN V.I., MACHOLD J. The emerging three-dimensional structure of a receptor. The nicotinic acetylcholine receptor. Eur. J. Biochem. 1996;239:539–557. doi: 10.1111/j.1432-1033.1996.0539u.x. [DOI] [PubMed] [Google Scholar]
- ITIER V., BERTRAND D. Neuronal nicotinic receptors: from protein structure to function. FEBS Lett. 2001;504:118–125. doi: 10.1016/s0014-5793(01)02702-8. [DOI] [PubMed] [Google Scholar]
- JONES M.V., JONAS P., SAHARA Y., WESTBROOK G.L. Microscopic kinetics and energetics distinguish GABAA receptor agonists from antagonists. Biophys. J. 2001;81:2660–2670. doi: 10.1016/S0006-3495(01)75909-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JORGENSEN W.L., CHANDRASEKHAR J., MADURA J.D. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- KARLIN A. Chemical modification of the active site of the acetylcholine receptor. J. Gen. Physiol. 1969;54:245S–264S. doi: 10.1085/jgp.54.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KARLIN A. Emerging structure of the nicotinic acetylcholine receptors. Nat. Rev. Neurosci. 2002;3:102–114. doi: 10.1038/nrn731. [DOI] [PubMed] [Google Scholar]
- KEARNEY P.C., NOWAK M.W., ZHONG W., SILVERMAN S.K., LESTER H.A., DOUGHERTY D.A. Dose–response relations for unnatural amino acids at the agonist binding site of the nicotinic acetylcholine receptor: tests with novel side chains and with several agonists. Mol. Pharmacol. 1996;50:1401–1412. [PubMed] [Google Scholar]
- LE NOVERE N., CORRINGER P.J., CHANGEUX J.P. Improved secondary structure predictions for a nicotinic receptor subunit: incorporation of solvent accessibility and experimental data into a two-dimensional representation. Biophys. J. 1999;76:2329–2345. doi: 10.1016/S0006-3495(99)77390-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LE NOVERE N., GRUTTER T., CHANGEUX J.P. Models of the extracellular domain of the nicotinic receptors and of Ago. Proc. Natl. Acad. Sci. U.S.A. 2002;99:3210–3215. doi: 10.1073/pnas.042699699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LINDSTROM J. Nicotinic acetylcholine receptors in health and disease. Mol. Neurobiol. 1997;15:193–222. doi: 10.1007/BF02740634. [DOI] [PubMed] [Google Scholar]
- LUETJE C.W., PATRICK J. Both alpha- and beta-subunits contribute to the agonist sensitivity of neuronal nicotinic acetylcholine receptors. J. Neurosci. 1991;11:837–845. doi: 10.1523/JNEUROSCI.11-03-00837.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LUO S., KULAK J.M., CARTIER G.E., JACOBSEN R.B., YOSHIKAMI D., OLIVERA B.M., MCINTOSH J.M. α-Conotoxin AuIB selectively blocks α3β4 nicotinic acetylcholine receptors and nicotine-evoked norepinephrine release. J. Neurosci. 1998;18:8571–8579. doi: 10.1523/JNEUROSCI.18-21-08571.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARTIN M., CZAJKOWSKI C., KARLIN A. The contributions of aspartyl residues in the acetylcholine receptor gamma and delta subunits to the binding of agonists and competitive antagonists. J. Biol. Chem. 1996;271:13497–13503. doi: 10.1074/jbc.271.23.13497. [DOI] [PubMed] [Google Scholar]
- OSAKA H., SUGIYAMA N., TAYLOR P. Distinctions in agonist and antagonist specificity conferred by anionic residues of the nicotinic acetylcholine receptor. J. Biol. Chem. 1998;273:12758–12765. doi: 10.1074/jbc.273.21.12758. [DOI] [PubMed] [Google Scholar]
- PARKER M.J., BECK A., LUETJE C.W. Neuronal nicotinic receptor beta2 and beta4 subunits confer large differences in agonist binding affinity. Mol. Pharmacol. 1998;54:1132–1139. [PubMed] [Google Scholar]
- PARKER M.J., HARVEY S.C., LUETJE C.W. Determinants of agonist binding affinity on neuronal nicotinic receptor beta subunits. J. Pharmacol. Exp. Ther. 2001;299:385–391. [PubMed] [Google Scholar]
- PATERSON D., NORDBERG A. Neuronal nicotinic receptors in the human brain. Prog. Neurobiol. 2000;61:75–111. doi: 10.1016/s0301-0082(99)00045-3. [DOI] [PubMed] [Google Scholar]
- PAULING P., PETCHER T.J. Neuromuscular blocking agents: structure and activity. Chem. Biol. Interact. 1973;6:351–365. doi: 10.1016/0009-2797(73)90056-2. [DOI] [PubMed] [Google Scholar]
- PEREIRA E.F., REINHARDT-MAELICKE S., SCHRATTENHOLZ A., MAELICKE A., ALBUQUERQUE E.X. Identification and functional characterization of a new agonist site on nicotinic acetylcholine receptors of cultured hippocampal neurons. J. Pharmacol. Exp. Ther. 1993;265:1474–1491. [PubMed] [Google Scholar]
- SALI A., BLUNDELL T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- SANCHEZ R., SALI A. Evaluation of comparative protein structure modeling by MODELLER-3. Proteins. 1997. pp. 50–58. [DOI] [PubMed]
- SCHAPIRA M., ABAGYAN R., TOTROV M. Structural model of nicotinic acetylcholine receptor isotypes bound to acetylcholine and nicotine. BMC Struct. Biol. 2002;2:1. doi: 10.1186/1472-6807-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHRATTENHOLZ A., GODOVAC-ZIMMERMANN J., SCHAFER H.J., ALBUQUERQUE E.X., MAELICKE A. Photoaffinity labeling of torpedo acetylcholine receptor by physostigmine. Eur. J. Biochem. 1993;216:671–677. doi: 10.1111/j.1432-1033.1993.tb18187.x. [DOI] [PubMed] [Google Scholar]
- SINE S.M., QUIRAM P., PAPANIKOLAOU F., KREIENKAMP H.J., TAYLOR P. Conserved tyrosines in the alpha subunit of the nicotinic acetylcholine receptor stabilize quaternary ammonium groups of agonists and curariform antagonists. J. Biol. Chem. 1994;269:8808–8816. [PubMed] [Google Scholar]
- SMIT A.B., SYED N.I., SCHAAP D., VAN MINNEN J., KLUMPERMAN J., KITS K.S., LODDER H., VAN DER SCHORS R.C., VAN ELK R., SORGEDRAGER B., BREJC K., SIXMA T.K., GERAERTS W.P. A glia-derived acetylcholine-binding protein that modulates synaptic transmission. Nature. 2001;411:261–268. doi: 10.1038/35077000. [DOI] [PubMed] [Google Scholar]
- STAUFFER D.A., KARLIN A. Electrostatic potential of the acetylcholine binding sites in the nicotinic receptor probed by reactions of binding-site cysteines with charged methanethiosulfonates. Biochemistry. 1994;33:6840–6849. doi: 10.1021/bi00188a013. [DOI] [PubMed] [Google Scholar]
- SUDWEEKS S.N., YAKEL J.L. Functional and molecular characterization of neuronal nicotinic ACh receptors in Rat CA1 hippocampal neurons. J. Physiol. 2000;527:515–528. doi: 10.1111/j.1469-7793.2000.00515.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SUGIYAMA N., BOYD A.E., TAYLOR P. Anionic residue in the alpha-subunit of the nicotinic acetylcholine receptor contributing to subunit assembly and ligand binding. J. Biol. Chem. 1996;271:26575–26581. doi: 10.1074/jbc.271.43.26575. [DOI] [PubMed] [Google Scholar]
- SVINNING T., SOURUM H. Acta Crystallogr. 1975. p. 1581.
- THOMPSON J.D., GIBSON T.J., PLEWNIAK F., JEANMOUGIN F., HIGGINS D.G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UNWIN N. Nicotinic acetylcholine receptor at 9 A resolution. J. Mol. Biol. 1993;229:1101–1124. doi: 10.1006/jmbi.1993.1107. [DOI] [PubMed] [Google Scholar]
- UNWIN N. Acetylcholine receptor channel imaged in the open state. Nature. 1995;373:37–43. doi: 10.1038/373037a0. [DOI] [PubMed] [Google Scholar]
- ZHONG W., GALLIVAN J.P., ZHANG Y., LI L., LESTER H.A., DOUGHERTY D.A. From ab initio quantum mechanics to molecular neurobiology: a cation–Pi binding site in the nicotinic receptor. Proc. Natl. Acad. Sci. U.S.A. 1998;95:12088–12093. doi: 10.1073/pnas.95.21.12088. [DOI] [PMC free article] [PubMed] [Google Scholar]