Abstract
We describe a series of transcriptional activators generated by adding amino acids (eight in one case, six in another) to fragments of the yeast Saccharomyces cerevisiae activator Gal4 that dimerize and bind DNA. One of the novel activating regions identified by this procedure is unusual, compared with previously characterized yeast activating regions, in the following ways: it works more strongly than does Gal4's natural activating region as assayed in yeast; it is devoid of acidic residues; and several lines of evidence suggest that it sees targets in the yeast transcriptional machinery at least partially distinct from those seen by Gal4's activating region.
Transcriptional activating regions are defined as protein sequences that, when tethered to DNA near a promoter, activate transcription by contacting targets in the transcriptional machinery. Activating regions found in a wide array of eukaryotic activators, including the yeast activators Gal4, GCN4, and the herpesvirus activator VP16, are characterized by an excess of acidic residues. Each can work in a wide array of eukaryotes and, where tested, hydrophobic residues interspersed between the acidic residues have been found to be essential for activity (1–4). Deletion analyses of some of these activating regions have shown that the efficiency with which they work is more or less proportional to the length of the activating region, suggesting that each bears reiterations of functional regions (5, 6). In contrast to these acidic activators, mammalian activators (e.g., SP1, CTF) that lack excess negative charge work only weakly, if at all, in yeast (7–11).
A variety of lines of evidence indicate that activating regions such as those found in Gal4 work by recruiting the transcriptional machinery to DNA (5, 12–14). According to this idea, these activating regions merely have to perform a “glue-like” function, and in principle any of a variety of activator-machinery interactions could suffice for, or be involved in, activation. Consistent with this idea, an array of potential targets in the transcriptional machinery have been found to interact with activating regions in vitro, including TATA box-binding protein (TBP), TFIIB, SRB4, certain TBP-associated factors, TFIIH, and the Ada-Gcn5 and Swi/Snf complexes, and for some of these interactions there are corresponding genetic experiments that support the idea that they are physiologically relevant (15–25).
It has proved surprisingly easy to generate new activating regions. Thus, a short peptide (called AH) designed to form an amphipathic helix, with acidic residues along one surface and hydrophobic residues along another, is functional (26), as are an array of peptides encoded by random fragments of the Escherichia coli genome. The latter peptides, which varied from 12 to 99 residues in length, were invariably acidic and worked up to about 30% as effectively as did intact Gal4 when bound to four Gal4 binding sites upstream of the gene (27). Selection for improved activating variants of one of Gal4's natural activating region invariably increased negative charge by eliminating positively charged residues (28), and a similar result was obtained with a glutamine-rich mammalian activator as studied in yeast (29).
“Activator bypass” experiments illustrate a mechanism of gene activation that dispenses with any activating region such as those described above. Thus, for example, a protein comprising a DNA binding domain fused to a component of the transcriptional machinery often can activate transcription of a yeast gene bearing the appropriate DNA binding sites nearby (12). Examples of transcriptional machinery components that work in such constructs include Gal11, any of several suppressor of RNA polymerase B (SRB) proteins, and TBP (30–34). In these experiments, evidently, the transcriptional machinery is brought directly to the nearby promoter by the additional protein-DNA interaction provided by the fusion protein. Activation by one of these so-called “nonclassical” activators can be distinguished from that mediated by the typical activator in several ways. For example, overexpression of an activating region can inhibit (“squelch”) activation by another classical activator, but such overexpression does not affect activation by the nonclassical activator (31). The nonclassical activators that function in yeast usually do so with an efficiency much more dependent on promoter architecture than do classical activators (35). And unlike classical activators, nonclassical activators, when tested on their own, work poorly or not at all in mammalian cells, but can work synergistically with other, classical, activators bound to DNA sites nearby (ref. 36 and references therein).
We show here that random addition of 8 aa, in one experiment, and 6 aa in another, to DNA binding fragments of Gal4, yields yeast gene activators at high frequency. The activating regions are not particularly rich in acidic residues and one, totally devoid of such residues, works more powerfully than intact Gal4, and significantly more strongly than does any previously described synthetic activating sequence. We describe experiments that argue that this activating region sees a target(s) in the yeast transcriptional machinery distinct from those seen by classical activating regions.
Materials and Methods
Yeast Strains and Plasmids.
The following yeast strains and plasmids used in this study have been described: JPY9∷RJR227 (MATα, ura3–52, trp1Δ63, Leu2Δ1, his3Δ200, lys2Δ385, gal4Δ11, Δura3∷RJR227), a yeast strain harboring GAL1-lacZ gene bearing five consensus 17 mers of Gal4 binding sites 191 bp upstream of TATA box (5); YAG23 (MATα, his3–11, his3–15, leu2–3, leu2–112, canR, ura3Δ5, Δtrp1, Δpho4∷ura3Δ5, Δpho80∷HIS3) (37); BJ2168 (MATα, ura3–52, leu2–3, trp1Δ901, prb1–1122, pep4–3, prc1–407, gal2) (38); pG5E4, a DNA template for in vitro transcription assay bearing five Gal4 sites 23 bp upstream of E4 TATA box (39), and pRJR217, an autonomously replicating sequence-centromeric plasmid containing Gal4(1–100) driven by β-actin promoter (5).
Library Synthesis and Screening.
The random peptides fused to Gal4 DNA binding domain was generated by DNA synthesis. The oligonucleotides used are the following: oligo 1, 5′-CAT GCC ATG GCA AAG CTA CTG TCT TCT-3′; oligo 2, 5′-GTG TCG ACT ANN NNN NNN NNN NNN NNN NNN NNN NAT CTT GTA CAA ATA ATC C-3′; oligo 3, 5′-GTG TCG ACT ANN NNN NNN NNN NNN NNN NTC CAA ACG CGT TAT ACG-3′; oligo 4, 5′-GCT TAC TGC TTT TTT CTT CCC AAG ATC GAA AAT TTA CTG AAT TAA CCA TGG CAA AGC-3′; and oligo 5, 5′-CGA AGT AAC TTC AAA AGT ATC AAA AGT ATG GAA ACT TCA AAT GTT GTG TCG ACT A-3′.
The double-stranded DNA encoding Gal4(1–100)+8aa and Gal4(1–100)+(840–850)+6aa were generated by PCR with appropriate DNA template using oligos 1 and 2 and oligos 1 and 3, respectively. The PCR products then were extended by PCR using oligos 4 and 5 to add an additional 45 bp to each ends that share homology with the vector plasmid pRJR217. The resulting PCR fragment then were cotransformed with a prelinearized vector pRJR217 (NcoI, SalI cut) into yeast JPY9∷RIR227 bearing GAL1-lacZ reporter. The random peptide library was generated in yeast by homologue recombination as described by Lehming et al. (40). The resulting Gal4(1–100) + peptide fusion genes were expressed in yeast driven by β-actin promoter. The colonies were transferred onto a nitrocellulose filter, which then were laid on a 5-bromo-4-chloro-3-indolyl β-d-galactoside plate as described (27). Blue colonies were selected and the plasmids were rescued from yeast and sequenced. Plasmids encoding the activator were confirmed by retransformation back into JPY9∷RJR227. The activating potentials were determined by β-galactosidase (β-gal) assay (41). All assays were done in triplicate at least three times. The SDs of the data are less than 20%.
Mutagenesis.
The mutants of Gal4(1–100)+P201 were constructed by site-directed PCR mutagenesis. All mutants were essentially confirmed by DNA sequencing. The expression of all mutants in yeast were measured by gel mobility shift assay from the whole yeast extracts using a 32P-labeled double-stranded DNA oligonucleotide (5′-TCC GGA GGA CTG TCC TCC GGT-3′) containing a single Gal4 site as a probe (5).
Protein Purification and in Vitro Transcription.
Gal4(1–100) and its derivatives were overexpressed in E. coli BL21(DE3) and were purified as described (42, 43). The yeast holoenzyme was purified from a protease-deficient yeast strain BJ2168, and in vitro transcription assays were carried out as described (44, 45).
Coimmunoprecipitation.
Plasmids expressing Gal4(1–100) and its derivatives were incubated with the 35S methionine-supplemented transcription-translation mix from Promega. The resulting radioactive translation products were incubated with 1 μg of each of the baculovirus-purified Flag-tagged -TBP, -SRB4, and -SRB10 in a 500-μl reaction volume of MTB buffer (50 mM Hepes, pH 7.5/100 mM K-glutamate/25 mM Mg-acetate/5 mM EGTA/10% glycerol/0.01% NP-40) (15). After a 3-hr incubation, 30 μl of anti-Flag-Sepharose (2 μg of the anti-Flag mAb) was added, and the tubes were nutated for an additional 90 min at 4°C. The reaction was stopped, and the beads were washed five times with 500 μl of MTB buffer. The precipitated material was resuspended in SDS/PAGE sample buffer, heated at 95°C for 5 min, and resolved on a 10% SDS-Tricine gel (46).
Results
Isolation of Activators.
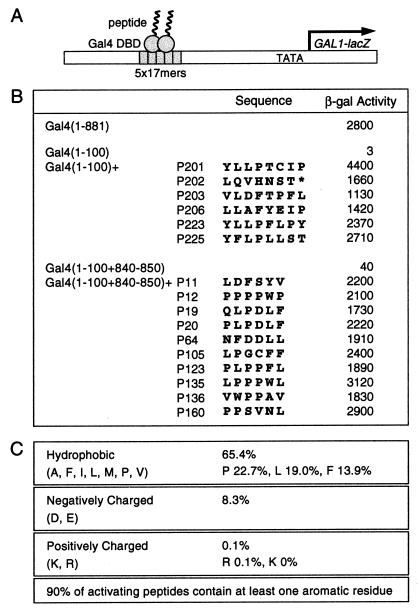
The scheme used to isolate activators is diagrammed in Fig. 1A. Approximately 0.1% of the molecules bearing 8 aa added in random order to Gal4(1–100) encoded strong activators as judged by colony color. Gal4(1–100), which lacks the Gal4 activating regions, comprises a DNA binding and dimerization region, and, as shown, does not function as an activator. Fig. 1B lists the sequences of several of the 8-mers that confer the activating function to Gal4(1–100). None of these sequences is found encoded in the current database, which includes the entire genomes of E. coli and S. cerevisiae. One of these activators, Gal4(1–100)+P201, activated transcription to a higher level than did intact Gal4. We repeated this experiment by adding six residues in random order to Gal4(1–100)+(840–850). This fragment bears, in addition to the DNA binding and dimerization regions, a small part of Gal4's activating region II and, as shown, activated transcription very weakly. Strong activators were recovered at a frequency similar to that observed in the first experiment, and several of the activators (P11 and P12) activated about as efficiently as did intact Gal4. No obvious consensus sequence describes these peptides, but their compositions are not random. Thus, considering both the 8-mers and the 6-mers, over 65% of the residues are hydrophobic and about 8% are acidic; only one positively charged residue was recovered, and that in an activator significantly weaker than any of those whose sequence is shown (Fig. 1C).
Figure 1.
Isolation of Gal4-fused peptide activators. (A) A lacZ reporter bearing five consensus 17-bp Gal4 binding sites 191 bp upstream of the GAL1 TATA box. (B) Sequences of activating regions and their transcriptional activities in yeast as assayed by using the reporter in A. (C) Amino acid compositions of the peptides.
Mutagenesis.
We performed a series of directed mutagenesis experiments on the strongest activator, Gal4(1–100)+P201. Fig. 2A shows that two constructs bearing the eight residues of Gal4(1–100)+P201, but in scrambled order, were virtually inactive. Fig. 2B shows that substitution with arginine at any position in the 8-mer, except at the carboxyl terminus, abolished activity. This result indicates that our failure to find positively charged residues in any of our strong activating regions is not accidental. The carboxyl terminal proline evidently plays no role as it can be deleted without affecting function, whereas further deletion, as shown in Fig. 2C, abolishes activity. Fig. 2D shows that deletion of any single residue among the carboxyl five residues of Gal4(1–100) either abolishes activation or reduces activation by 10-fold. The experiment of Fig. 2E shows that each of the residues in the 8-mer can be replaced by alanine without strong loss of activity with the exception of tyrosine at position 1. Extension of this analysis to the five carboxyl residues of Gal4(1–100) suggests that the identities of two hydrophobic residues (F97 and V98) are crucial for function, but each of the others, including the aspartate residue at position 100, can be replaced by alanine without loss of function. All mutants were detectable by gel shift analyses performed with whole-cell extracts. To the extent that the levels varied, the most active derivatives were correspondingly less abundant, perhaps because of increased proteolysis elicited by the stronger activating regions (ref. 47 and W. Tansey, personal communication).
Figure 2.
Activities of mutants of Gal4(1–100)+P201. (A) Scrambled P201. (B) Arginine substitutions. (C) Carboxyl-terminal truncations. (D) Single internal deletions in the region of Gal4(96–100). (E) Alanine substitutions.
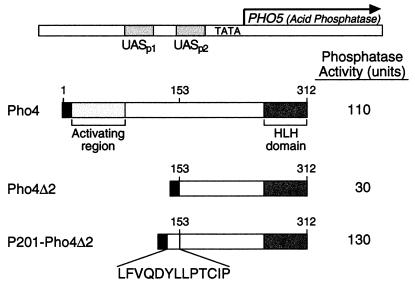
These results, taken together, are consistent with the notion that some aspect of structure is required for the activating function of Gal4(1–100)+P201. One possible such structure is shown, at least in part, in Fig. 3A. The figure shows that the carboxyl five residues of Gal4(1–100) plus the first three residues of the 8-mer can be drawn as an alpha-helix with the hydrophobic residues F97, V98, and Y1 of the 8-mer exposed along one face. As predicted by the idea that such a helix is relevant, substitution of Q99 or D100 with alanine has no effect on activity, whereas substitution with proline, a helix breaker, abolishes activity (Fig. 3B). Fig. 4 shows that the activation domain consisting of P201 plus five residues from Gal4 also functions in the context of a different protein and a different promoter. In particular, the figure shows that the artificial activation domain can substitute for the natural activation domain of the yeast Pho4 activator.
Figure 3.
A hypothetical helical structure of GAL4(1–100)+P201. (A) A putative helical wheel formed by GAL4(1–100)+P201. (B) Effects of proline substitution at Q99 and D100.
Figure 4.
Activity of a fragment of Gal4(1–100)+P201 inserted in place of the natural activating region of the yeast activator Pho4. Cells were grown and assayed for acid phosphatase assay as described by Svaren et al. (37).
Activation in Vitro.
The experiment of Fig. 5 shows that Gal4(1–100)+P201 activated transcription directed by the yeast RNA polymerase II holoenzyme purified according to Koleske et al. (44) and supplemented with TBP and TFIIE. The activation observed was equivalent to that elicited by Gal4-VP16 (see Fig. 5). The results indicate that both of these activators interact with targets in the holoenzyme. Both of these activators also stimulated transcription efficiently in yeast nuclear extracts (not shown). Also shown is the somewhat weaker activation mediated by Gal4(1–100)+(840–850)+P64.
Figure 5.
Activation by Gal4-peptides in vitro using purified yeast holoenzyme. The holoenzyme (1 μg) was supplemented with TBP (1 pmol), TFIIE (0.5 pmol), and DNA template shown above (100 ng) in the absence or presence of Gal4-VP16 (50 ng), Gal4(1–100)+P201 (20 ng), or Gal4(1–100)+(840–850)+P64 (20 ng). Arrow indicates the E4 extension product.
Target(s) of P201.
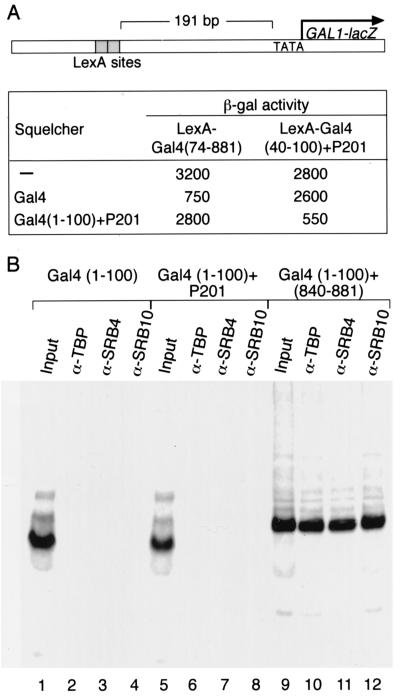
The experiment of Fig. 6A suggests that the artificial activator Gal4(1–100)+P201 and native Gal4 interact with different targets in the transcriptional machinery. Thus, an excess of Gal4(1–100)+P201 inhibits (squelches) activation by LexA-Gal4(40–100)+P201, and an excess of Gal4 inhibits activation by LexA-Gal4, presumably because excess activator competes with promoter-bound activator for interaction sites in the transcriptional machinery. The artificial activator, however, does not squelch activation by LexA-Gal4, and vice versa, indicating that they do not compete for the same sites.
Figure 6.
Interaction with targets in the transcriptional machinery. (A) The effect of overexpression of one activator on the activity of another. The overexpressed activator is called the squelcher, and the reporter is as indicated. (B) Interaction of Gal4-peptides with TBP, SRB4, and SRB10. Labeled Gal4(1–100), Gal4(1–100)+P201, or Gal4(1–100)+(840–881) were incubated with flag-tagged targets of Gal4 and then tested for their ability to coimmunoprecipitate with the targets (15). Input lane contains 10% of the TnT material used in each coimmunoprecipitation.
A series of biochemical and genetic experiments have indicated that TBP, SRB4, and SRB10 are physiologically relevant targets of the ordinary activating region of Gal4 (refs. 5, 15, and 16; A.Z.A., S. S. Koh, Z. Zaman, R. A. Young, and M.P.) unpublished results). The experiment of Fig. 6B shows that Gal4(1–100)+P201 fails to interact with any of these proteins in vitro as assayed by coimmunoprecipitation experiments under conditions in which the interactions with Gal4(1–100+840–881) are readily apparent. Also consistent with the idea that Gal4(1–100)+P201 interacts with targets different from those seen by Gal4 is the finding that mutants of SRB4 that suppress weak activating derivatives of Gal4 fail to suppress the activating defect of mutants of Gal4(1–100)+P201 (S.S. Koh and A.Z.A., unpublished results).
Discussion
Our results reinforce an earlier surmise that it is remarkably easy to generate new activating regions by attaching peptide sequences to a DNA binding domain. In one case described here, addition of eight residues (the carboxyl terminal of which can be deleted without loss of function) suffices to convert an inactive DNA binding dimer of Gal4 to an activator [Gal4(1–100)+P201] that is more powerful than natural Gal4. Our mutagenesis analyses indicate that the five carboxyl terminal residues of Gal4(1–100) participate in forming the activating region. A peptide comprising these five residues plus the 8-mer from P201 also functions as a strong activating region when inserted in place of Pho4's activating region.
Our strongest activator, Gal4(1–100)+P201, like most of the other activators described here, differs from those isolated by Ma and Ptashne (27) and from previously described natural yeast activators in its lack of acidic residues. We suspect this difference may be accounted for by the different Gal4 DNA binding fragments used for activator isolation in the two experiments. The ones used here, for example, might more efficiently display short peptides for interaction with the transcriptional machinery. It is also possible that the E. coli genome encodes few short hydrophobic peptides that would confer the activating function, and hence such sequences would not have been recovered in the experiment of Ma and Ptashne (27). P201 is novel in that it is a nonacidic activating region that works with high efficiency in yeast. Like previously described yeast activating regions, however, this and other activating regions described here bear few or no positively charged residues; they, like their natural counterparts, are relatively rich in hydrophobic residues; and a tyrosine residue in one of them (P201) was found to be critical for function.
Our experiments suggest that our strongest activator, Gal4(1–100)+P201, contacts a target or targets in the yeast transcriptional machinery, but these targets differ, at least in part, from those seen by the classical activator Gal4. Thus whereas the natural Gal4 activating region interacts with the three seemingly well-defined targets TBP, SRB4, and SRB10, Gal4(1–100)+P201 interacts with none of these, and the results of squelching experiments are consistent with the notion that different targets are seen in vivo as well. Moreover, unlike Gal4 and other natural yeast activators, but like certain nonclassical activators, Gal4(1–100)+P201 fails to activate transcription in mammalian cells, but can work synergistically with other, classical activators bound to DNA sites nearby (data not shown).
Our findings are consistent with the idea that there may be many ways to interact with the transcriptional machinery to effect recruitment and thereby activate transcription in yeast. This notion does not imply that all such interactions will be physiologically equivalent, however. For example, as we will show elsewhere, Gal4(1–100)+P201, like other artificial (nonclassical) activators (e.g., the fusion protein LexA-Gal11) is unusually sensitive to TUP-mediated repression compared with classical activators (Z. Zaman, A.Z.A., and M.P. unpublished data).
Acknowledgments
We thank Maryse Adam and Keylen Chee for technical assistance; Sang Seok Koh, Zaf Zaman, and William Tansey for sharing unpublished data; Luc Gaudreau, Julián Nevado, and the members of Ptashne lab for valuable discussion; W. Tansey, D. Dorsett, K. Scotto, and A. Gann for comments on the manuscript, and Renate Hellmiss for illustrations. This study was funded by the Ludwig Foundation and a grant from the National Institutes of Health (to M.P.). X.L was supported by the Markey Foundation, and A.Z.A was supported by the Helen Hay Whitney Foundation.
Abbreviations
- TBP
TATA box-binding protein
- SRB
suppressor of RNA polymerase B
- β-gal
β-galactosidase
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF223921–AF223936).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.040573197.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.040573197
References
- 1.Triezenberg S J. Curr Opin Genet Dev. 1995;5:190–196. doi: 10.1016/0959-437x(95)80007-7. [DOI] [PubMed] [Google Scholar]
- 2.Lin J, Chen J, Elenbaas B, Levine A J. Genes Dev. 1994;8:1235–1246. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- 3.Drysdale C M, Duenas E, Jackson B M, Reusser U, Braus G H, Hinnebusch A G. Mol Cell Biol. 1995;15:1220–1233. doi: 10.1128/mcb.15.3.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ptashne M, Gann A A. Nature (London) 1990;346:329–331. doi: 10.1038/346329a0. [DOI] [PubMed] [Google Scholar]
- 5.Wu Y, Reece R J, Ptashne M. EMBO J. 1996;15:3951–3963. [PMC free article] [PubMed] [Google Scholar]
- 6.Hope I A, Mahadevan S, Struhl K. Nature (London) 1988;333:635–640. doi: 10.1038/333635a0. [DOI] [PubMed] [Google Scholar]
- 7.Ponticelli A S, Pardee T S, Struhl K. Mol Cell Biol. 1995;15:983–988. doi: 10.1128/mcb.15.2.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao H, Jeang K T. J Biol Chem. 1998;273:22873–22876. doi: 10.1074/jbc.273.36.22873. [DOI] [PubMed] [Google Scholar]
- 9.Kim T K, Roeder R G. J Biol Chem. 1993;268:20866–20869. [PubMed] [Google Scholar]
- 10.Kunzler M, Braus G H, Georgiev O, Seipel K, Schaffner W. EMBO J. 1994;13:641–645. doi: 10.1002/j.1460-2075.1994.tb06302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang D, Childs G. J Biol Chem. 1998;273:6868–6877. doi: 10.1074/jbc.273.12.6868. [DOI] [PubMed] [Google Scholar]
- 12.Ptashne M, Gann A. Nature (London) 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 13.Ptashne M, Gann A. Curr Biol. 1998;8:R812–R822. doi: 10.1016/s0960-9822(07)00508-8. [DOI] [PubMed] [Google Scholar]
- 14.Keaveney M, Struhl K. Mol Cell. 1998;1:917–924. doi: 10.1016/s1097-2765(00)80091-x. [DOI] [PubMed] [Google Scholar]
- 15.Koh S S, Ansari A Z, Ptashne M, Young R A. Mol Cell. 1998;1:895–904. doi: 10.1016/s1097-2765(00)80088-x. [DOI] [PubMed] [Google Scholar]
- 16.Melcher K, Johnston S A. Mol Cell Biol. 1995;15:2839–2848. doi: 10.1128/mcb.15.5.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stringer K F, Ingles C J, Greenblatt J. Nature (London) 1990;345:783–786. doi: 10.1038/345783a0. [DOI] [PubMed] [Google Scholar]
- 18.Ingles C J, Shales M, Cress W D, Triezenberg S J, Greenblatt J. Nature (London) 1991;351:588–590. doi: 10.1038/351588a0. [DOI] [PubMed] [Google Scholar]
- 19.Lin Y S, Ha I, Maldonado E, Reinberg D, Green M R. Nature (London) 1991;353:569–571. doi: 10.1038/353569a0. [DOI] [PubMed] [Google Scholar]
- 20.Goodrich J A, Hoey T, Thut C J, Admon A, Tjian R. Cell. 1993;75:519–530. doi: 10.1016/0092-8674(93)90386-5. [DOI] [PubMed] [Google Scholar]
- 21.Roberts S G, Ha I, Maldonado E, Reinberg D, Green M R. Nature (London) 1993;363:741–744. doi: 10.1038/363741a0. [DOI] [PubMed] [Google Scholar]
- 22.Xiao H, Pearson A, Coulombe B, Truant R, Zhang S, Regier J L, Triezenberg S J, Reinberg D, Flores O, Ingles C J, et al. Mol Cell Biol. 1994;14:7013–7024. doi: 10.1128/mcb.14.10.7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thut C J, Chen J L, Klemm R, Tjian R. Science. 1995;267:100–104. doi: 10.1126/science.7809597. [DOI] [PubMed] [Google Scholar]
- 24.Yudkovsky N, Logie C, Hahn S, Peterson C L. Genes Dev. 1999;13:2369–2374. doi: 10.1101/gad.13.18.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neely K E, Hassan A H, Wallberg A E, Steger D J, Cairns B R, Wright A P, Workman J L. Mol Cell. 1999;4:649–655. doi: 10.1016/s1097-2765(00)80216-6. [DOI] [PubMed] [Google Scholar]
- 26.Giniger E, Ptashne M. Nature (London) 1987;330:670–672. doi: 10.1038/330670a0. [DOI] [PubMed] [Google Scholar]
- 27.Ma J, Ptashne M. Cell. 1987;51:113–119. doi: 10.1016/0092-8674(87)90015-8. [DOI] [PubMed] [Google Scholar]
- 28.Gill G, Sadowski I, Ptashne M. Proc Natl Acad Sci USA. 1990;87:2127–2131. doi: 10.1073/pnas.87.6.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benuck M L, Li Z, Childs G. J Biol Chem. 1999;274:25419–25425. doi: 10.1074/jbc.274.36.25419. [DOI] [PubMed] [Google Scholar]
- 30.Barberis A, Pearlberg J, Simkovich N, Farrell S, Reinagel P, Bamdad C, Sigal G, Ptashne M. Cell. 1995;81:359–368. doi: 10.1016/0092-8674(95)90389-5. [DOI] [PubMed] [Google Scholar]
- 31.Farrell S, Simkovich N, Wu Y, Barberis A, Ptashne M. Genes Dev. 1996;10:2359–2367. doi: 10.1101/gad.10.18.2359. [DOI] [PubMed] [Google Scholar]
- 32.Chatterjee S, Struhl K. Nature (London) 1995;374:820–822. doi: 10.1038/374820a0. [DOI] [PubMed] [Google Scholar]
- 33.Klages N, Strubin M. Nature (London) 1995;374:822–823. doi: 10.1038/374822a0. [DOI] [PubMed] [Google Scholar]
- 34.Xiao H, Friesen J D, Lis J T. Mol Cell Biol. 1995;15:5757–5761. doi: 10.1128/mcb.15.10.5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gaudreau L, Keaveney M, Nevado J, Zaman Z, Bryant G O, Struhl K, Ptashne M. Proc Natl Acad Sci USA. 1999;96:2668–2673. doi: 10.1073/pnas.96.6.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nevado J, Gaudreau L, Adam M, Ptashne M. Proc Natl Acad Sci USA. 1999;96:2674–2677. doi: 10.1073/pnas.96.6.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Svaren J, Schmitz J, Horz W. EMBO J. 1994;13:4856–4862. doi: 10.1002/j.1460-2075.1994.tb06812.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohashi Y, Brickman J M, Furman E, Middleton B, Carey M. Mol Cell Biol. 1994;14:2731–2739. doi: 10.1128/mcb.14.4.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carey M, Lin Y S, Green M R, Ptashne M. Nature (London) 1990;345:361–364. doi: 10.1038/345361a0. [DOI] [PubMed] [Google Scholar]
- 40.Lehming N, McGuire S, Brickman J M, Ptashne M. Proc Natl Acad Sci USA. 1995;92:10242–10246. doi: 10.1073/pnas.92.22.10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rose M D, Winston F, Hieter P. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1990. [Google Scholar]
- 42.Ansari A Z, Reece R J, Ptashne M. Proc Natl Acad Sci USA. 1998;95:13543–13548. doi: 10.1073/pnas.95.23.13543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reece R J, Rickles R J, Ptashne M. Gene. 1993;126:105–107. doi: 10.1016/0378-1119(93)90596-u. [DOI] [PubMed] [Google Scholar]
- 44.Koleske A J, Chao D M, Young R A. Methods Enzymol. 1996;273:176–184. doi: 10.1016/s0076-6879(96)73018-5. [DOI] [PubMed] [Google Scholar]
- 45.Gaudreau L, Adam M, Ptashne M. Mol Cell. 1998;1:913–916. doi: 10.1016/s1097-2765(00)80090-8. [DOI] [PubMed] [Google Scholar]
- 46.Schagger H, von Jagow G. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 47.Molinari E, Gilman M, Natesan S. EMBO J. 1999;18:6439–6447. doi: 10.1093/emboj/18.22.6439. [DOI] [PMC free article] [PubMed] [Google Scholar]