Abstract
In a recent paper, patients with a progressive juvenile-onset hereditary cataract have been reported to have a point mutation in the human γD crystallin gene (Stephan, D. A., Gillanders, E., Vanderveen, D., Freas-Lutz, D., Wistow, G., Baxevanis, A. D., Robbins, C. M., VanAuken, A., Quesenberry, M. I., Bailey-Wilson, J., et al. (1999) Proc. Natl. Acad. Sci. USA 96, 1008–1012). This mutation results in the substitution of Arg-14 in the native protein by a Cys residue. It is not understood how this mutation leads to cataract. We have expressed recombinant wild-type human γD crystallin (HGD) and its Arg-14 to Cys mutant (R14C) in Escherichia coli and show that R14C forms disulfide-linked oligomers, which markedly raise the phase separation temperature of the protein solution. Eventually, R14C precipitates. In contrast, HGD slowly forms only disulfide-linked dimers and no oligomers. These data strongly suggest that the observed cataract is triggered by the thiol-mediated aggregation of R14C. The aggregation profiles of HGD and R14C are consistent with our homology modeling studies that reveal that R14C contains two exposed cysteine residues, whereas HGD has only one. Our CD, fluorescence, and differential scanning calorimetric studies show that HGD and R14C have nearly identical secondary and tertiary structures and stabilities. Thus, contrary to current views, unfolding or destabilization of the protein is not necessary for cataractogenesis.
In the hereditary, juvenile-onset cataract described by Stefan et al. (1), the lens, which is clear at birth, develops punctate opacities progressively, such that by two years of age the cataract is readily detectable, and matures by early childhood or adolescence. The punctate opacities seen in this cataract are in the nucleus and inner cortex, regions of the lens that are enriched in the γ-crystallins. In the human lens, only two members of the γ-crystallin family, γC and γD, are expressed in appreciable amounts, and only γD crystallin continues to be expressed until late childhood (2, 3). In affected individuals, a single point mutation has been identified in the γD crystallin gene that corresponds to the substitution of Arg-14 by a Cys. The identification of this mutation and the parallel between the time course of the pathology and the physiological expression of human γD crystallin strongly implicate the Arg-14 → Cys mutant of γD in the development of this cataract. However, the molecular mechanism invoked to explain the observed opacity has been speculative (1).
In the past, it has not been possible to conduct detailed studies on human γD crystallin because of the difficulty of obtaining sufficient quantities of pure protein from young, normal human lenses (4). Therefore, to characterize the normal protein thoroughly and investigate the mechanism by which the Arg-14 → Cys mutation in γD could lead to cataract, we cloned and expressed human γD crystallin and its Arg-14 → Cys mutant in Escherichia coli. Both the wild-type recombinant γD crystallin (HGD) and its Arg-14 → Cys mutant (R14C) folded efficiently in E. coli and accumulated as soluble proteins. We isolated and purified the HGD and R14C proteins and determined their solution properties. Our results suggest that the disulfide-crosslinked oligomerization of R14C is responsible for the observed cataract. Furthermore, such oligomerization occurs without significant change in protein structure, conformation, and stability.
Materials and Methods
Cloning, Expression, and Isolation of Proteins.
The human γD crystallin coding sequence was amplified from a human fetal lens cDNA library by using forward (5′-GCC ATG GGG AAG ATC ACC CTC TAC- 3′) and reverse (5′-AGG ATC CAA ATT AAG AAA CAA CAA AGG AG- 3′) primers based on the published genomic sequence (5). The PCR product was cloned in the EcoRV site of pBluescript vector. Both strands were sequenced on an Applied Biosystems Prism automatic sequencer with pBluescript T3 and T7 primers. Only one difference with the published sequence was noted: codon 17 reads TAC instead of TAT. This sequence change is silent and may represent a naturally occurring polymorphism. The insert was recloned in a pET3a vector NcoI/BamH in which the NdeI site had been replaced with a NcoI site. The recombinant DNA was transformed into BL21(DE3)pLysS cells (Stratagene). For the overexpression of γD crystallin, the bacterial cultures were grown at 37°C to an absorbance at 600 nm (A600) of 1. Expression of γ-crystallins was induced by the addition of isopropyl 1-thio-d-galactopyranoside to a final concentration of 0.5 mM, and the cultures were grown for an additional 5–6 h. Cells were pelleted by centrifugation, and the pellet was resuspended in lysis buffer (50 mM Tris⋅HCl containing 25 mM NaCl and 2 mM EDTA, pH 8), to which Complete protease inhibitor (Roche Molecular Biochemicals) was added at 1 tablet per 30 ml. The cell suspension was lysed with lysozyme (250 μg/ml) followed by five cycles of a rapid freeze-thaw procedure that involved freezing in liquid nitrogen followed by thawing in a water bath set at 30°C. To this suspension, DNase (1 mg/ml) was added, followed by centrifugation at 48,400 × g. Both the supernatant and pellet were tested for the presence of crystallins by using SDS/PAGE. The crystallins fractionated almost exclusively (>95%) into the supernatant. The supernatant was subjected first to size exclusion (SE), followed by cation-exchange chromatography (6). The final product was analyzed by using electrospray ionization mass spectroscopy. The concentration of human γD was determined by using an extinction coefficient of 41.4 mM−1⋅cm−1 at 280 nm (7). The same extinction coefficient was used for R14C.
Preparation of the R14C Mutant.
To introduce a Cys residue in place of Arg-14, the following oligonucleotide primers were made (Life Technologies, Grand Island, NY): 5′-CCA GGG CTG CCA CTA CGA ATG CAG CAG C-3′ as the forward primer and 5′-GCT GCT GCA TTC GTA GTG GCA GCC CTG G-3′ as the reverse primer. Site-directed mutagenesis was performed with the QuickChange site-directed mutagenesis kit (Stratagene). The plasmid DNA obtained after mutagenesis was sequenced with the T7 promoter primer and was found to contain the desired mutation but no other sequence changes. Mutant protein was expressed and isolated as described above for HGD.
SE-HPLC.
The HGD and R14C proteins were subjected to SE-HPLC on a Superdex 200HR FPLC column (Amersham Pharmacia) at a flow rate of 1 ml/min. Proteins were eluted isocratically with 0.1 M sodium phosphate buffer (pH 7.1) containing 0.02% sodium azide (8).
Electrospray Ionization Mass Spectroscopy.
Mass spectrometry was performed at the Biopolymers Lab at the Center for Cancer Research at Massachusetts Institute of Technology. Four separate preparations of HGD and R14C gave an average mass of 20,610 ± 2 and 20,556 ± 2 units, respectively. These results agree with those of previously published work (9) to within 3 mass units and are consistent with the Arg-14 → Cys replacement in HGD. The N-terminal protein sequence of the first 18 residues of R14C (determined at the Biopolymers Lab at Massachusetts Institute of Technology) confirmed this replacement.
Phase Separation Measurements.
The phase diagram (shown in Fig. 3) was obtained by the cloud point and temperature quench methods by using published procedures (6).
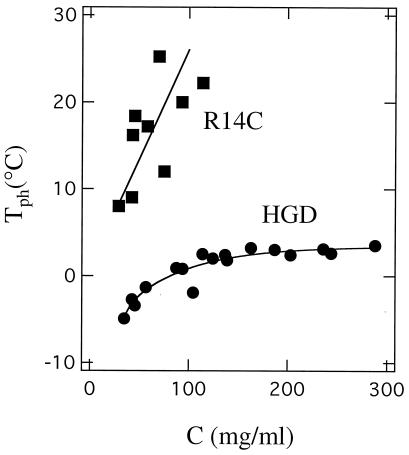
Figure 3.
The phase diagram of HGD and R14C. Ascending limbs of the coexistence curves (plots of Tph versus protein concentration) for HGD and R14C proteins. The marked increase in the Tph of R14C is a result of protein aggregation caused by intermolecular disulfide crosslinking as in bovine γB (19).
Quasielastic Light Scattering.
The onset of aggregation, hydrodynamic radii, and evolution of the size distribution of particles were studied by quasielastic light scattering with a 144-channel Langley–Ford (Amherst, MA) model 1097 correlator and a Spectra-Physics model 164 argon laser. Further discussion on this subject can be found elsewhere (10, 11).
Modeling.
The structures of HGD and its R14C mutant were modeled based on the structures of bovine γD (chain A and B; PDB structure ELP; resolution 1.95 Å; ref. 12) and bovine γB (PDB structure 4GCR; resolution 1.47 Å; ref. 13) by using the method of Peitsch (14–16). The human γD protein sequence shares 87% identity with bovine γD and 78% identity with bovine γB in the primary structure (17, 18). The comparative modeling was carried out in the automated internet server Swiss Model (www.expasy.ch), and the structures were displayed by using the personal computer version of the swiss-pdb viewer.
CD Spectra.
CD spectra were obtained with an Aviv Associates (Lakewood, NJ) model 202 spectrometer. Protein concentrations of 0.5 mg/ml and 0.1 mg/ml were used for near- and far-UV CD spectra respectively. Far-UV spectra are normalized with respect to the concentration of peptide bonds, whereas near-UV spectra are normalized with respect to protein concentration.
Fluorescence Spectra.
These were measured in a Hitachi F-4500 spectrometer by using an excitation wavelength of 290 nm. The excitation and emission slits were set to 5 nm. Spectra were measured by using the same cuvette and identical protein concentrations (0.1 mg/ml) for HGD and R14C. The R14C spectra were taken with and without the addition of 20 mM DTT.
Differential Scanning Calorimetry (DSC).
DSC measurements were made at Microcal (Amherst, MA) with a VP-DSC instrument. Protein concentrations were 0.1 mg/ml. Data were normalized for protein concentration. DSC data have not been used for quantitative determination of Tm or ΔH, because the thermal transition was not completely reversible.
Results and Discussion
Homology Modeling.
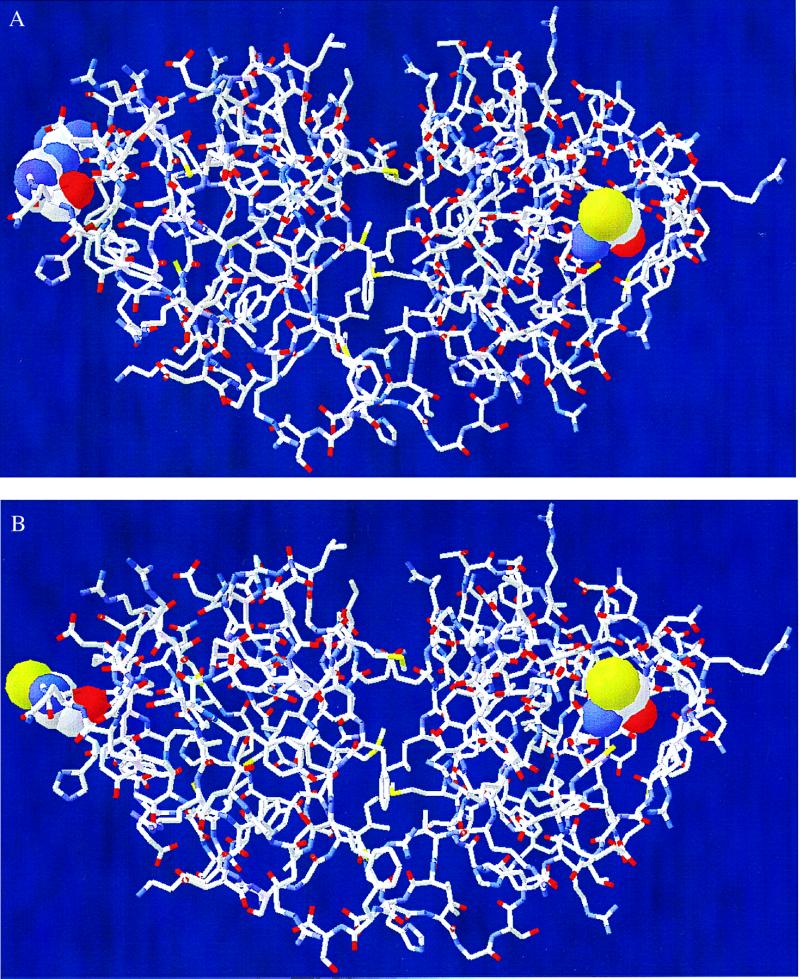
Our three-dimensional homology modeling studies performed according to the method of Peitsch (14–16) show that, among the six cysteine residues of HGD, only Cys-110 has a significant portion (10%) of its area exposed to solvent (Fig. 1A). When Arg-14 is replaced with Cys in the mutant R14C (Fig. 1B), our model shows that Cys-14 is about 37% exposed. Thus, as a result of the introduction of the surface Cys-14, two reactive sites become available in the mutant protein as compared with one (Cys-110) in the normal protein. Therefore, oxidation of Cys-110 in HGD is expected to lead only to dimerization of the protein, whereas in the mutant, the exposed Cys-14 together with Cys-110 can initiate further aggregation to higher oligomers.
Figure 1.
Models of the three-dimensional structure of HGD (A) and R14C (B) based on bovine γD and γB crystallins (12, 13). Model of R14C shows two reactive sites (Cys-14 and Cys-110) compared with one (Cys-110) in HGD.
Aggregation and Its Effect on Phase Separation.
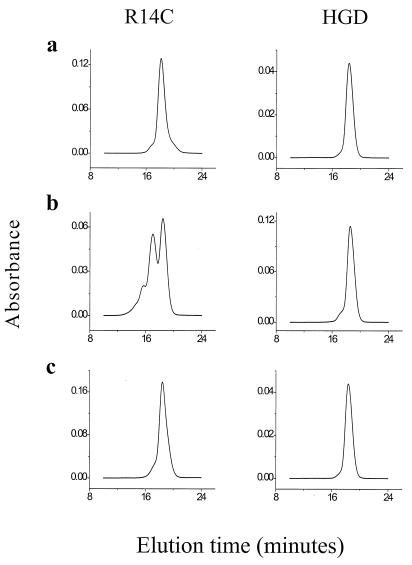
The results from our modeling studies are consistent with the SE-HPLC profiles of the two proteins (Fig. 2). Even at low protein concentration, when freshly prepared, the R14C mutant contains a small amount (4%) of dimers, whereas HGD is monomeric (Fig. 2a). After 24 h at neutral pH, the mutant has a significant accumulation of dimers (40%), trimers (10%), and higher oligomers (5%). In contrast, during the same period, HGD formed only a small proportion (5%) of dimers (Fig. 2b). In fact, even after prolonged incubation at high concentrations (≈400 mg/ml), no oligomers beyond the dimer were seen in several HGD samples. Addition of DTT reduces either protein back to the monomer form (Fig. 2c). Thus, we deduce that the dimers in HGD as well as the oligomers in R14C are formed by the oxidation of cysteine thiols to intermolecular disulfide crosslinks.
Figure 2.
(a) SE-HPLC profiles of HGD and R14C at pH 7 when freshly prepared in 0.1 M sodium phosphate buffer. R14C shows 4% dimers, whereas HGD is monomeric. (b) After 24 h at pH 7, R14C shows a marked increase in the proportion of dimers (40%) and oligomers (10% trimers and 5% higher oligomers), whereas HGD shows only a few dimers (5%). (c) After the addition of 20 mM DTT and a further 24 h, both proteins become monomeric.
We showed earlier that intermolecular, disulfide-crosslinked aggregates (both directly S-S-linked and chemically crosslinked with a spacer) markedly affect another important property of the γ-crystallins, namely liquid–liquid phase separation (8, 19). In this phenomenon, the protein solution segregates spontaneously into protein-rich and protein-poor phases (20). Such phase separation has been implicated in several animal models of cataract (21–25). An increase in the phase separation temperature (Tph) is diagnostic of a cataractogenic change (19–26). We now show that aggregation of the R14C mutant of HGD has a pronounced effect on the Tph of the protein solution.
In Fig. 3, we present the ascending limb of the coexistence curves of HGD and R14C. The critical temperature (Tc; i.e., the maximum temperature on the coexistence curve) of HGD is about 3°C, making it a low-Tc protein (6) and not a high-Tc protein as has been claimed (1, 4). Fig. 3 also shows that the Tph of R14C is typically 20°C higher than that of HGD. This difference is almost entirely caused by the oligomerization of the mutant. In the presence of DTT, when both proteins are monomers, the Tph of the mutant is only about 4°C higher than that of HGD (data not shown). As is evident from Fig. 3, at protein concentrations greater than 100 mg/ml, the Tph of R14C is no longer measurable, because the mutant protein rapidly precipitates at high concentrations.
Structure and Stability.
To determine the effect of the Arg-14 → Cys mutation on the secondary and tertiary structure of HGD, we compared the fluorescence emission and the near- and far-UV CD spectra of HGD and R14C. We also measured the thermal stabilities of the two proteins by DSC.
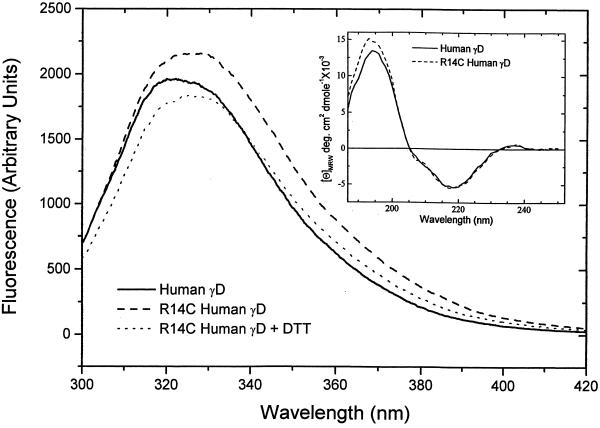
The fluorescence emission spectrum of γ-crystallins arises mainly from the four buried tryptophan residues that are invariant in all members of this family (27). The tryptophan residues, two in each domain, are excellent reporter groups that monitor the structural integrity of the protein. The fluorescence emission spectra of HGD and R14C are shown in Fig. 4. The λmax values for HGD and R14C are 325 nm and 329 nm, respectively, indicating that the tryptophan residues in both proteins are buried (27). These values are well within the range of those observed for all native bovine γ-crystallins (λmax from 324–335 nm; ref. 28). The small red shift in the λmax for R14C could be caused by the loss of hydrogen bonds (typically formed by the guanidinium group of Arg) after the substitution of Arg-14 by Cys. In Fig. 4, the difference in the fluorescence intensities between HGD and R14C is caused by the fact that, at pH 7, R14C is mainly (>75%) oligomeric. Addition of DTT monomerizes R14C and moves the spectrum closer to that of HGD. The DTT data are identical to those taken at pH 2, where R14C remains monomeric. Fig. 4 Inset shows the far-UV CD spectra of HGD and R14C. The CD spectrum in the far-UV region is an empirical measure of the secondary structure of a protein. The data show that there is no significant difference between HGD and R14C in secondary structure, within the limits of experimental uncertainty.
Figure 4.
Fluorescence emission spectra (excitation wavelength = 290 nm) of HGD and R14C in 0.1 M sodium phosphate buffer at pH 7 at a protein concentration of 0.1 mg/ml. Spectra of R14C were taken before and after the addition of 20 mM DTT. The Inset shows the far-UV CD spectrum of HGD and R14C and is an empirical measure of the secondary structure. The two proteins are nearly identical within the limits of experimental error.
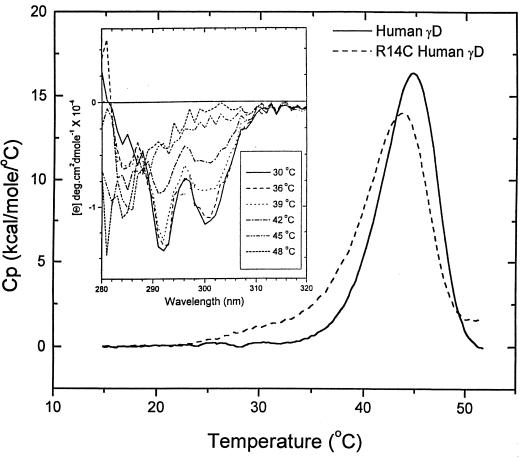
To determine the stabilities of HGD and R14C, we conducted DSC experiments. Data were taken at pH 2 for comparison with previously published work on bovine γB crystallin (28, 29). The increased sensitivity of our instrument enabled us to use 10-fold lower protein concentrations (0.1–0.2 mg/ml) than those used by Kono et al. (28) and Rudolf et al. (29). Under these solution conditions, cysteine oxidation and the consequent protein aggregation and precipitation were found to be minimal. Our DSC data for HGD and R14C (Fig. 5) show a strong endothermic transition centered at 43°C (HGD) and 42°C (R14C) and a much weaker transition centered around 60°C (not shown). We find that as a result of protein aggregation, even the main transition is only partially (50–60%) reversible. This aggregation is also confirmed by the 100-fold increase in the hydrodynamic radii (Rh) of HGD and R14C before (Rh ≈ 2.3 nm) and after (Rh > 230 nm) the DSC experiments, as determined by quasielastic light scattering (data not shown). Therefore, because of the partial reversibility of the unfolding transition, the DSC data cannot be used to calculate thermodynamic quantities such as enthalpies, entropies, and free energies of unfolding.
Figure 5.
DSC scans of HGD and R14C in pH 2 buffer (0.1 M NaCl⋅HCl) at the scan rate of 1°C/min. These are the conditions used for bovine γB (28, 29). The Inset shows the change in the near-UV CD of HGD as the protein unfolds in the range of the thermal transition observed in DSC.
To verify whether the main endothermic transition (with an apparent midpoint, Tm ≅ 43°C) was in fact caused by the unfolding of the protein, we measured the near-UV CD spectra as a function of temperature (Fig. 5 Inset). In general, ellipticities in the near-UV region arise from the tertiary structure of proteins. For the γ-crystallins, this CD region is dominated by the four invariant tryptophan residues (27). In Fig. 5 Inset, it is evident that the tryptophan signals at 292 and 300 nm are eliminated during the thermal unfolding of HGD, with a midpoint comparable to that observed by DSC. Therefore, we attribute the main transition in our DSC data to the overall unfolding of the tertiary structure of the protein. Furthermore, the DSC data show that R14C with an apparent Tm of 42°C is only marginally less stable relative to native HGD.
Thus, our structure and stability studies show that even as the mutant protein aggregates at physiological pH, it maintains its secondary and tertiary structure. These results contradict a common view of cataract formation, according to which protein unfolding or significant destabilization of the protein structure is a prerequisite for protein aggregation and cataract formation (30–34). An example of this view is the hereditary Coppock-like cataract, in which a truncated form of γE crystallin was believed to be overexpressed in the lens (3, 33). Recent studies (30) now link this cataract to a mutation in the γC crystallin gene. However, regardless of the identity of the gene product, it is still presumed that the cataract is formed because of an unstable γ-crystallin (30). The progressive juvenile-onset cataract discussed herein presents an example where the destabilization paradigm does not apply.
It is well established that, in cataract, protein condensates are important scattering elements responsible for making the lens opaque (35, 36). Our studies clearly suggest that the progressive juvenile-onset cataract discussed herein results from the accumulation of mutant protein aggregates and precipitates. This aggregation is induced by a reactive cysteine residue at the surface of the R14C mutant. Thus, a pharmacological agent that blocks the reactive thiol should prevent the formation of this cataract. Very recently, several other human and animal cataracts have been identified and linked to mutations in the genes of the lens γ-crystallins (30). Our work on one particular cataract suggests that comparing the solution properties of the normal and mutant proteins is an effective way to understand the molecular basis for the formation of cataractogenic light scattering elements.
Acknowledgments
We thank Drs. Mohan Chellani and Lung-Nan Lin (Microcal) for the DSC data, Profs. Thaddeus Dryja and Felix Villars and Dr. George Thurston for critical comments, and Ms. Kris Bowring for help with manuscript preparation. This work was supported by National Institutes of Health Grants EY05127 to G.B.B., EY10535 to J.P., and GM17980 to J.A.K.
Abbreviation
- HGD
human recombinant γD crystallin
- SE
size exclusion
- DSC
differential scanning calorimetry
- Tph
phase separation temperature
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.040554397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.040554397
References
- 1.Stephan D A, Gillanders E, Vanderveen D, Freas-Lutz D, Wistow G, Baxevanis A D, Robbins C M, VanAuken A, Quesenberry M I, Bailey-Wilson J, et al. Proc Natl Acad Sci USA. 1999;96:1008–1012. doi: 10.1073/pnas.96.3.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Russell P, Meakin S O, Hohman T C, Tsui L C, Breitman M L. Mol Cell Biol. 1987;7:3320–3323. doi: 10.1128/mcb.7.9.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brakenhoff R H, Aarts H J M, Reek F H, Lubsen N H, Schoenmakers J G G. J Mol Biol. 1990;216:519–532. doi: 10.1016/0022-2836(90)90380-5. [DOI] [PubMed] [Google Scholar]
- 4.Siezen R J, Thomson J A, Kaplan E D, Benedek G B. Proc Natl Acad Sci USA. 1987;84:6088–6092. doi: 10.1073/pnas.84.17.6088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meakin S O, Breitman M L, Tsui L C. Mol Cell Biol. 1985;5:1408–1414. doi: 10.1128/mcb.5.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broide M L, Berland C R, Pande J, Ogun O O, Benedek G B. Proc Natl Acad Sci USA. 1991;88:5660–5664. doi: 10.1073/pnas.88.13.5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andley U P, Mathur S, Griest T A, Petrash J M. J Biol Chem. 1996;271:31973–31980. doi: 10.1074/jbc.271.50.31973. [DOI] [PubMed] [Google Scholar]
- 8.Asherie N, Pande J, Lomakin A, Ogun O, Hanson S R A, Smith J B, Benedek G B. Biophys Chem. 1998;75:213–227. doi: 10.1016/s0301-4622(98)00208-7. [DOI] [PubMed] [Google Scholar]
- 9.Hanson S R A, Smith D L, Smith J B. Exp Eye Res. 1998;67:301–312. doi: 10.1006/exer.1998.0530. [DOI] [PubMed] [Google Scholar]
- 10.Braginskaya T G, Dobichin P D, Ivanova M A, Klubin V V, Lomakin A V, Noskin V A, Shmelev G E, Tolpina S P. Physica Scr. 1983;28:73–79. [Google Scholar]
- 11.Pike E R. In: Scattering Techniques Applied to Supramolecular and Nonequilibrium Systems. Chen S H, Chu B, Nossal R, editors. New York: Plenum; 1981. pp. 179–200. [Google Scholar]
- 12.Chirgadze Y N, Driessen H P C, Wright G, Slingsby C, Hay R E, Lindley P F. Acta Crystallogr D. 1996;52:712–721. doi: 10.1107/S0907444996000352. [DOI] [PubMed] [Google Scholar]
- 13.Najmudin S, Nalini V, Driessen H P C, Slingsby C, Blundell T L, Moss D S, Lindley P F. Acta Crystallogr D. 1993;49:223–233. doi: 10.1107/S0907444992007601. [DOI] [PubMed] [Google Scholar]
- 14.Peitsch M C. Bio/Technology. 1995;13:658–660. [Google Scholar]
- 15.Guex N, Peitsch M C. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 16.Guex N, Diemand A, Peitsch M C. Trends Biochem Sci. 1999;24:364–367. doi: 10.1016/s0968-0004(99)01427-9. [DOI] [PubMed] [Google Scholar]
- 17.Hay R E, Andley U P, Petrash J M. Exp Eye Res. 1994;58:573–584. doi: 10.1006/exer.1994.1052. [DOI] [PubMed] [Google Scholar]
- 18.Bhat S P, Spector A. DNA. 1984;3:287–295. doi: 10.1089/dna.1.1984.3.287. [DOI] [PubMed] [Google Scholar]
- 19.Pande J, Lomakin A, Fine B, Ogun O, Sokolinski I, Benedek G B. Proc Natl Acad Sci USA. 1995;92:1067–1071. doi: 10.1073/pnas.92.4.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clark J I, Benedek G B. Biochem Biophys Res Commun. 1980;95:482–489. doi: 10.1016/0006-291x(80)90763-9. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka T, Ishimoto C, Chylack L T., Jr Science. 1977;197:1010–1012. doi: 10.1126/science.887936. [DOI] [PubMed] [Google Scholar]
- 22.Ishimoto C, Sun S T, Nishio I, Goalwin P, Tanaka T. Proc Natl Acad Sci USA. 1979;76:4414–4419. doi: 10.1073/pnas.76.9.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanaka T, Nishio I, Sun S-T, Rubin S, Tung W, Chylack L T., Jr Invest Ophthalmol Visual Sci. 1983;24:522–525. [PubMed] [Google Scholar]
- 24.Clark J I, Giblin F J, Reddy V N, Benedek G B. Invest Ophthalmol Visual Sci. 1982;22:186–190. [PubMed] [Google Scholar]
- 25.Clark J I, Carper D. Proc Natl Acad Sci USA. 1987;84:122–125. doi: 10.1073/pnas.84.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pande J, Ogun O, Nath C, Benedek G B. Exp Eye Res. 1993;57:257–264. doi: 10.1006/exer.1993.1123. [DOI] [PubMed] [Google Scholar]
- 27.Mandal K, Bose S K, Chakrabarti B, Siezen R J. Biochim Biophys Acta. 1985;832:156–164. doi: 10.1016/0167-4838(85)90327-9. [DOI] [PubMed] [Google Scholar]
- 28.Kono M, Sen A C, Chakrabarti B. Biochemistry. 1990;29:464–470. doi: 10.1021/bi00454a022. [DOI] [PubMed] [Google Scholar]
- 29.Rudolph R, Siebendritt R, Nesslauer G, Sharma A K, Jaenicke R. Proc Natl Acad Sci USA. 1990;87:4625–4629. doi: 10.1073/pnas.87.12.4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heon E, Priston M, Schorderet D F, Billingsley G D, Girard P O, Lubsen N, Munier F L. Am J Hum Genet. 1999;65:1261–1267. doi: 10.1086/302619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heijtmancik J F. Am J Hum Genet. 1998;62:520–525. doi: 10.1086/301774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Francis P J, Berry V, Moore A T, Bhattacharya S. Trends Genet. 1999;15:191–196. doi: 10.1016/s0168-9525(99)01738-2. [DOI] [PubMed] [Google Scholar]
- 33.Brackenhoff R H, Henskens H A M, van Rossum M W P C, Lubsen N, Schoenmakers J G G. Hum Mol Genet. 1994;3:279–283. doi: 10.1093/hmg/3.2.279. [DOI] [PubMed] [Google Scholar]
- 34.Harding J. Cataract: Biochemistry, Epidemiology and Pharmacology. London: Chapman & Hall; 1991. [Google Scholar]
- 35.Benedek G B, Pande J, Thurston G M, Clark J I. Prog Retin Eye Res. 1999;18:391–402. doi: 10.1016/s1350-9462(98)00023-8. [DOI] [PubMed] [Google Scholar]
- 36.Bettleheim F A. In: The Ocular Lens: Structure, Function, and Pathology. Maisel H, editor. New York: Dekker; 1985. pp. 265–300. [Google Scholar]