Abstract
(2S*,3R*)-1-(biphenyl-4-carbonyl)piperazine-2,3-dicarboxylic acid (PBPD) is a moderate affinity, competitive N-methyl-D-aspartate (NMDA) receptor antagonist with an atypical pattern of selectivity among NMDA receptor 2 subunit (NR2) subunits. We now describe the activity of several derivatives of PBPD tested at both rat brain NMDA receptors using L-[3H]-glutamate binding assays and at recombinant receptors expressed in Xenopus oocytes.
Substituting various branched ring structures for the biphenyl group of PBPD reduced NMDA receptor activity. However, substituting linearly arranged ring structures – fluorenone or phenanthrene groups – retained or enhanced activity.
Relative to PBPD, the phenanthrene derivative (2S*,3R*)-1-(phenanthrene-2-carbonyl)piperazine-2,3-dicarboxylic acid (PPDA) displayed a 30- to 78-fold increase in affinity for native NMDA receptors. At recombinant receptors, PPDA displayed a 16-fold (NR2B) to 94-fold (NR2C) increase in affinity over PBPD.
Replacement of the biphenyl group of PBPD with a 9-oxofluorene ring system resulted in small changes in receptor affinity and subtype selectivity.
2′-Bromo substitution on the biphenyl group of PBPD reduced antagonist affinity 3- to 5-fold at NR2A-, NR2B- and NR2D-containing receptors, but had little effect on NR2C-containing receptors. In contrast, 4′-fluoro substitution of the biphenyl ring of PBPD selectively increased NR2A affinity.
The aromatic rings of PBPD and PPDA increase antagonist affinity and appear to interact with a region of the NMDA receptor displaying subunit heterogeneity. PPDA is the most potent and selective NR2C/NR2D-preferring antagonist yet reported and thus may be useful in defining NR2C/NR2D function and developing related antagonists with improved NMDA receptor subtype selectivity.
Keywords: Xenopus oocyte, recombinant, antagonist, NMDA receptor, NR2, NR2C, NR2D, autoradiography
Introduction
N-methyl-D-aspartate (NMDA) receptors are glutamate-gated, ion channel receptors that play important roles in synaptic transmission and neural plasticity; they are also involved in a wide variety of pathological conditions, such as seizure activity and neuronal cell death subsequent to stroke (Choi, 1998). Native NMDA receptors are multimeric complexes composed of subunits from at least two families: NR1a–h and NR2A–D (Monyer et al., 1994; Nakanishi et al., 1998; Dingledine et al., 1999). In addition, some NMDA receptors may contain an NR3 subunit that appears to inhibit channel activity (Dingledine et al., 1999). The NR2 subunits contain the glutamate binding site of the receptor complex (Laube et al., 1997). Accordingly, the different NR2 subunits confer distinct glutamate site pharmacological properties on the NR1/NR2 heterodimer (Ishii et al., 1993; Buller et al., 1994; Laurie & Seeburg, 1994; Buller & Monaghan, 1997), which correspond to the varied NMDA receptor subtypes observed in the rat brain (Buller et al., 1994; Monaghan et al., 1998; Christie et al., 2000).
Although NMDA receptors have been intensively studied in the decade since their cloning, the only selective NMDA receptor subtype-specific antagonists that have been identified are the ifenprodil-like NR2B selective compounds, which bind at a site distinct from that which either glutamate or glycine binds (Dingledine et al., 1999). Antagonists highly selective for NR2A-, NR2C- or NR2D-containing NMDA receptors have not yet been identified despite significant efforts. Recently, however, the antagonist 5-phosphonomethyl-1,4-dihydroquinoxaline-2,3-dione (PEAQX) has been shown to be 100-fold selective for human NR2A-containing receptors over NR2B-containing receptors (Auberson et al., 2002). Subtype-selective NMDA receptor antagonists are likely to have distinctive therapeutic/adverse effect profiles since the NR2 receptor subunits differ significantly in their anatomical and physiological properties (Ishii et al., 1993; Monyer et al., 1994; Vicini et al., 1998). Given the uniquely diencephalic distribution for NR2D subunits and the predominately cerebellar distribution for NR2C (Watanabe et al., 1992; Buller et al., 1994), antagonists selective for these subunits would be expected to display very different in vivo actions than NR2A/NR2B-selective antagonists. A potential therapeutic application is suggested by knockout studies in which a role for NR2D-containing receptors is demonstrated in a model of allodynia (Minami et al., 2001).
Since the L-glutamate binding site is found on NR2 subunits (which are four distinct gene products), there is the potential for identifying NR2-specific L-glutamate binding site antagonists. However, evaluation of a large number of NMDA receptor antagonists revealed only small variations between antagonists in their pattern of selectivity for native NMDA receptor subtypes (Andaloro et al., 1996). Similarly, at recombinant receptors, competitive antagonists generally display little differentiation between NR2 subunits and usually show a similar potency order of NR2A>NR2B>NR2C>NR2D (high to low affinities) (Ikeda et al., 1992; Buller et al., 1994; Laurie & Seeburg, 1994).
The most distinctive antagonist that we have identified in previous studies was the compound (2S*,3R*)-1-(biphenyl-4-carbonyl)piperazine-2,3-dicarboxylic acid (PBPD) (Buller & Monaghan, 1997). PBPD has high affinities for NR2B- and NR2D-subunit-containing NMDA receptors and lowest affinity at NR2A-containing receptors. It may be that the large hydrophobic biphenyl group of PBPD occupies a space in the receptor (a putative hydrophobic pocket) that confers the distinctive pharmacological properties to PBPD. In the present study, a series of PBPD derivatives with structural variations in the biphenyl group were synthesized and tested to probe the shape of the hydrophobic pocket and to determine if such modifications alter subunit specificity. NMDA receptor selectivity was evaluated at native NMDA receptors with the use of quantitative autoradiography and at recombinant NMDA receptors expressed in Xenopus oocytes using two-electrode voltage clamp electrophysiology.
Methods
Compounds
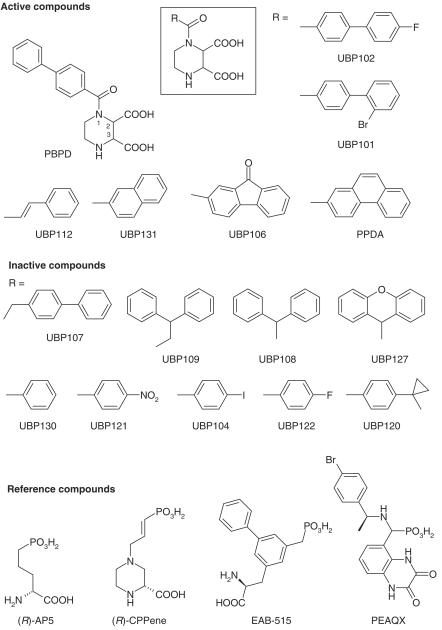
Structures of compounds used in this report are presented in Figure 1. Compounds were synthesized by reaction of the appropriately substituted acid chloride with (2S*,3R*)-piperazine-2,3-dicarboxylic acid (Felder et al., 1960) under modified Schotten–Bauman conditions (full details will be reported elsewhere). Following synthesis and purification, compound structure was verified by 1H NMR and 13C NMR spectroscopy. All compounds had elemental analyses where the determined percentage C, H and N were less than 0.3% different from theoretical values.
Figure 1.
Structures of PBPD derivatives and reference compounds. PBPD derivatives were synthesized and tested as inhibitors of L-[3H]-glutamate binding to NMDA receptors in rat brain sections. Compounds that displayed less than 50% inhibition at 100 μM are listed as ‘Inactive Compounds'. The reference compound (R)-AA has the structure of (R)-AP5 in which the phosphonate group is replaced by a carboxy group. (R)-CPP has the structure of (R)-CPPene in which the double bond is reduced.
Receptor autoradiography and quantitative analysis
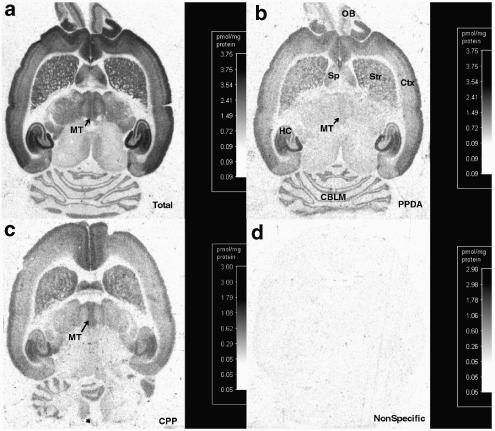
Following halothane anesthesia, brains were removed from adult male Sprague–Dawley rats (200–250 g) and immediately frozen under powdered dry ice. All animal procedures were performed in accordance with institutional and federal animal care guidelines. Horizontal sections (6 μm) were thaw-mounted onto gelatin-subbed slides, and stored at −20°C (<24 h). NMDA receptors were labeled with L-[3H]-glutamate using previously published methods (Monaghan et al., 1988). Briefly, slides were preincubated for 30 min at 0°C in 50 mM Tris-acetate, pH 7.0, followed by two additional preincubations for 10 min at 30°C in fresh buffer. Sections were incubated then with 50 nM L-[3H]-glutamate (40–60 Ci mmol−1; Perkin-Elmer, Boston, MA) in 50 mM Tris-acetate, pH 7.0, containing 5 μM 2-amino-3-hydroxy-5-methylisoxazole-4-propionic acid, 100 μM 4-acetomido-4′-isothiocyanoatostilbene-2,2′-disulfonic acid, 1 μM kainate and 10 μM glycine for 20 min at 0°C. Under these conditions, all anatomically specific binding is displaced by NMDA receptor compounds (Monaghan & Cotman, 1986). Nonspecific binding, defined by including 500 μM NMDA, represented <5% of total binding (see Figure 2). Sections were washed for 20 S in ice-cold buffer and quickly dried by an airstream at room temperature. Slides were then placed against an [3H]-sensitive film (Amersham, Arlington Heights, IL, U.S.A.) with [3H]-Microscale standards (Amersham) and allowed to expose for 3 weeks at 4°C.
Figure 2.
Representative autoradiographs showing compound inhibition of L-[3H]-glutamate binding to NMDA receptors. Autoradiographs are horizontal sections of adult rat brain in (a) absence of inhibitors, total binding, or (b) in the presence of 2 μM PPDA, (c) 6 μM CPP or (d) 500 μM NMDA, nonspecific binding. PPDA preferentially inhibits binding in the medial thalamus (MT) compared to the septum (Sp), striatum (Str), cerebral cortex (Ctx), hippocampus (HC), olfactory bulb (OB) and the cerebellum (CBLM).
[3H]-CGP39653 labels only NR2A subunits in the adult rodent cerebellum and predominately NR2B in the medial striatum (Christie et al., 2000). We used this assay accordingly to further characterize (2S*,3R*)-1-(phenanthren-2-carbonyl)piperazine-2,3-dicarboxylic acid (PPDA) specificity. [3H]-CGP39653 (20 nM; 34 Ci mmol−1; Perkin-Elmer, Boston, M, U.S.A.) was incubated with preincubated tissue sections in 50 mM Tris-acetate, 1 mM MgCl2, pH 7.6, for 30 min at 0°C followed by a 20 S wash in ice-cold buffer.
Computer-assisted image analysis of the autoradiographs (Imagine Research, St Catherines, Ontario, Canada) was used to quantify radioligand binding. Binding densities were calibrated from standard curves of radioactive standards. Using iterative, nonlinear regression analysis (ISI Software, GraphPad, San Diego, CA, U.S.A.) does–response curves were generated for NMDA antagonist inhibition of L-[3H]-glutamate binding. L-[3H]-glutamate Kd values were used to determine antagonist Ki values (Cheng & Prusoff, 1973).
NR subunit expression in Xenopus oocytes
cDNA encoding the NMDAR1a subunit was a generous gift of Dr Shigetada Nakanishi (Kyoto, Japan). cDNA encoding NR2A, NR2C and NR2D were kindly provided by Dr Peter Seeburg (Heidelburg, Germany) and the NR2B [5′UTR]-cDNA was a generous gift of Drs Dolan Pritchett and David Lynch (Philadelphia, PA, U.S.A.). Plasmids were linearized with NotI (NR1a), EcoRI (NR2A, NR2C and NR2D) or SalI (NR2B), and transcribed in vitro with T3 (NR2A, NR2C), SP6 (NR2B) or T7 (NR1a, NR2D) RNA polymerase using the mMessage mMachine Transcription Kits (Ambion, Austin, TX, U.S.A.).
Oocytes were removed from mature female Xenopus laevis (Xenopus One, Ann Arbor, MI, U.S.A.) as previously described (Buller et al., 1994). NMDA receptor subunit RNAs were dissolved in sterile distilled H2O. NR1a and NR2 RNAs were mixed in a molar ratio of 1 : 3. In all, 50 nl of the final RNA mixture was microinjected (15–30 ng total) into the oocyte cytoplasm. Oocytes were incubated in ND-96 solution at 17°C prior to electrophysiological assay (1–5 days).
Electrophysiology
Electrophysiological responses were measured using a standard two-microelectrode voltage clamp as previously described (Buller et al., 1994). The voltage clamp used was a Warner Instruments (Hamden, CT, U.S.A.) model OC-725B Oocyte Clamp, designed to provide fast clamp of large cells. This uses a high impedance voltage probe to measure membrane potential, a current sensing electrode to measure membrane current and to clamp the bath to ground, and a high-voltage amplifier to provide clamping current. The recording buffer contained 116 mM NaCl, 2 mM KCl, 2 mM BaCl2 and 5 mM HEPES, pH 7.4. Response magnitude was determined by the steady plateau response elicited by bath application of 10 μM L-glutamate plus 10 μM glycine at a holding potential of −60 mV. PEAQX was tested against 100 μM NMDA/10 μM glycine-evoked responses. Response amplitudes for the four heteromers were generally between 30 and 100 nA. Attempts were made to keep response magnitudes within this range to minimize activation of the endogenous Cl− current. The presence of a plateau response was taken as an indication of the lack of significant activation of the endogenous Cl− current by Ba2+ in these cells. Antagonist inhibition curves were fit (GraphPad Prism, ISI Software, San Diego, CA, U.S.A.) according to the equation I=Imax−Imax/[1+(IC50/A)n], where Imax is the current response in the absence of antagonist, A is the antagonist concentration and IC50 is the antagonist concentration producing half-maximal inhibition. Apparent Ki values were determined by correcting for agonist affinity according to the equation IC50=IC50 (obs)/1+([agonist]/EC50) as described (Durand et al., 1992). ANOVA with a Newman–Keuls multiple comparison test was used for statistical analysis.
Results
Activity at native NMDA receptor subtypes
Potential antagonists (Figure 1) were tested for the ability to inhibit the binding of L-[3H]-glutamate to NMDA receptors in the rat brain. As shown in Figure 2, essentially all of the L-[3H]-glutamate binding is specifically displaced by NMDA (Figure 2d), and qualitative differences between antagonists are readily observed. For example, PPDA (Figure 2b) preferentially inhibits binding to the medial thalamus whereas the antagonist CPP (Figure 2c) does not. Using quantitative receptor autoradiography, it is possible to detect the binding of L-[3H]-glutamate to each of the four NR2 NMDA receptor subunits. NMDA-sensitive L-[3H]-glutamate binding in the striatum predominately represents binding to NR2B subunits (Buller et al., 1994; Christie et al., 2000). Binding in the midline thalamus represents approximately 50% binding to NR2D (Beaton et al., 1992; Buller et al., 1994) while the remainder is probably to the NR2B subunit (Watanabe et al., 1993; Buller et al., 1994). NMDA-sensitive L-[3H]-glutamate binding in the cerebellar granule cell layer represents approximately 50% to NR2A and 50% to NR2C (Beaton et al., 1992; Buller et al., 1994; Christie et al., 2000). NMDA-sensitive L-[3H]-glutamate binding in the lateral thalamus appears to primarily represent NR2B subunits coexpressed with NR1 subunits that contain exon 5 (Buller et al., 1994). Consistent with the evidence that the NR1 subunit contains the glycine binding domain, and not the glutamate binding site, NR1 subunit composition does not significantly alter glutamate site antagonist pharmacology. However, NMDA receptors containing NR1 subunits possessing exon 5 have a modestly higher antagonist affinity and a lowered agonist affinity (Hollmann et al., 1993).
Several compounds displayed negligible activity at 100 μM (UBP107, UBP108, UBP109, UBP127, UBP121, UBP104, UBP120), or weak activity (UBP122, UBP130; IC50 >100 μM) in displacing NMDA-sensitive L-[3H]-glutamate binding (‘Inactive Compounds' shown in Figure 1). Extending the biphenyl group by one methylene group eliminated activity (UBP107). Changing the linear biphenyl structure of PBPD into a T-shaped biphenyl structure (UBP108, UBP109, UBP127) also eliminated NMDA receptor activity. Replacing the biphenyl group of PBPD with a benzene and a cyclopropyl group (UBP120) also eliminated activity. Removing the distal benzene of PBPD (UBP130) significantly reduced affinity and adding back an NO2 group (UBP121), or a halogen (UBP104 and UBP122), did not restore activity.
For those compounds that inhibited NMDA-sensitive L-[3H]-glutamate binding (‘Active Compounds', except UBP131), dose–response curves were generated and the corresponding Ki values are shown in Table 1. While the mono-substituted benzene derivatives (UBP121, UBP104, UBP122) were weak or inactive, extending the link between the benzene group and the carbonyl with an ethylene group (UBP112) largely restored activity. Interestingly, this compound displayed an altered NMDA receptor specificity compared to PBPD. UBP112 had a more than two-fold higher affinity than PBPD for cerebellar NMDA receptors, while having a lower affinity than PBPD in the other brain regions tested (Table 1). These results suggest that this compound has an enhanced affinity for native NR2A and/or NR2C subunits and a reduced affinity at NR2B-containing receptors.
Table 1.
Compound Ki values (μM) for inhibiting NMDA-sensitive L-[3H]-glutamate or [3H]-CGP-39653 binding in select brain regions
| [3H]-ligand | Medial striatum | Medial thalamus | Lateral thalamus | Cerebellum | |
|---|---|---|---|---|---|
| (R)-α-AA | L-Glutamate | 29±13 | 29±14 | 19±9 | 43±18 |
| CPP | L-Glutamate | 2.9±0.4 | 3.2±0.5 | 1.0±0.1 | 12.3±2.8 |
| PPDA | L-Glutamate | 0.56±0.11 | 0.32±0.06 | 0.17±0.06 | 0.52±0.11a |
| 13.9±2.1 | |||||
| PPDA | CGP-39653 | 0.21±0.03 | 0.17±0.02 | ND | 0.89±0.14 |
| PBPD | L-Glutamate | 24.6±5.0 | 9.79±1.61 | 9.0±1.9 | 125±43 |
| UBP112 | L-Glutamate | 34.47±5.73 | 20.50±1.66 | 13.51±1.26 | 53.23±6.91 |
| UBP106 | L-Glutamate | 16.23±1.78 | 7.19±1.05 | 5.87±1.12 | 61.03±7.61 |
| UBP101 | L-Glutamate | 39.9±6.0 | 17.3±3.2 | 17.4±8.3 | 268±30 |
| UBP102 | L-Glutamate | 35.8±3.3 | 16.0±2.0 | 18.0±5.5 | 100±16 |
PPDA displacement of L-[3H]glutamate binding in the cerebellum was best fit by a two-affinity site model with 55.8±0.03% to the high-affinity site.
For comparison, the reference compounds (R)-α-aminoadipate ((R)-AA) and CPP are also listed in Table 1. For these antagonist, as with other ‘typical' NMDA receptor antagonists (Andaloro et al., 1996), affinity in the medial thalamus was similar or lower than that found for the medial striatum. In contrast, the PBPD derivatives examined here displayed higher affinities (low Ki values) in the NR2D-rich medial thalamus. All compounds displayed lowest affinity in the cerebellum.
Substitution with a bromo group at the 2′ position of the biphenyl ring of PBPD (UBP101) reduced antagonist affinity roughly two-fold in each region tested. In contrast, substitution with a fluoro group at the 4′ position (UBP102) reduced affinity nearly two-fold in the forebrain regions, but had no effect in the cerebellum. This may be indicating that relative to PBPD, UBP102 has a reduced affinity specifically for NR2B-containing NMDA receptors.
Compounds PPDA and UBP106 provide an additional link between the two phenyl groups of PBPD, thus generating a rigid, planar structure. Relative to PBPD, inclusion of a carbonyl group in UBP106 modestly increases antagonist affinity, with greater enhancement seen at cerebellar NMDA receptors. Linking the biphenyl groups with another benzene ring, PPDA, caused a strong increase in affinity with an average 40-fold increase in affinity in the forebrain. PPDA affinity in the cerebellum was especially enhanced: PPDA displayed a 78-fold increase in affinity compared to PBPD when using the single affinity site values (125 μM, PBPD; 1.6 μM, PPDA). However, statistical analysis indicates that a two-site fit was better for PPDA than a single-site fit. These two populations were of similar density and had affinities of 0.5 and 14 μM. In contrast, PPDA inhibition of [3H]-CGP39653 binding only displayed a single affinity site in the cerebellum of 0.9 μM. Under the conditions used, [3H]-CGP39653 binding in the cerebellum appears to only represent NR2A subunits (Christie et al., 2000). Overall, for both UBP106 and PPDA, these modifications preferentially increase antagonist affinity for the cerebellum. Since NR2 subunits in the adult rat cerebellum are predominately NR2A- and NR2C-containing, these compounds may have enhanced affinity at native NR2A and/or NR2C subunits relative to NR2B subunits.
Activity at recombinant NMDA receptors
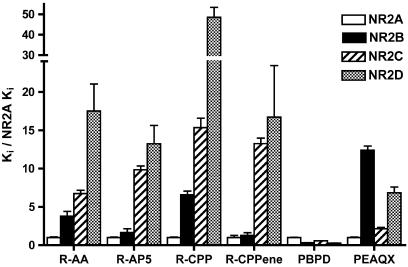
Compounds were tested for their ability to block glutamate/glycine-evoked currents in Xenopus oocytes injected with NMDA receptor subunit cRNA. An example is shown in Figure 3. For reference purposes, we evaluated the well-established NMDA receptor antagonists (R)-AA and (R)-4-(3-phosphonopropyl)piperazine-2-carboxylic acid ((R)-CPP), and the recently described NR2A-selective antagonist PEAQX (Auberson et al., 2002) for their ability to block agonist-evoked responses at recombinant NMDA receptors expressed in Xenopus oocytes. As shown in Figure 4, each of the well-characterized antagonists (R)-AA, (R)-2-amino-5-phosphonopentanoate ((R)-AP5), (R,E)-4-(3-phosphonoprop-2-enyl)piperazine-2-carboxylic acid ((R)-CPPene) and (R)-CPP display the typical competitive antagonist profile with highest affinity for NR2A followed in order by NR2B, NR2C, and then NR2D. (R)-CPP displayed the greatest selectivity for a ‘typical' profile antagonist, with a nearly 50-fold higher affinity for NR2A than NR2D. Unlike the other antagonists, PEAQX displayed a higher affinity for NR2A- and NR2C-containing receptors than for NR2B- and NR2D-containing receptors. In contrast to all of these antagonists, PBPD has its lowest affinity at NR2A-containing receptors.
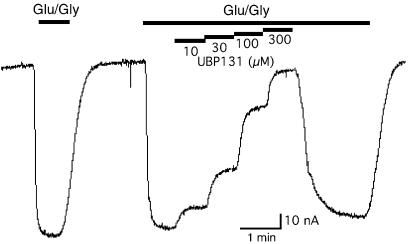
Figure 3.
A representative recording of antagonist blockade of NMDA receptor-mediated responses in Xenopus oocytes. NR1/NR2C RNA-injected oocytes were voltage clamped at −60 mV and inward currents were evoked by bath application of 10 μM L-glutamate plus 10 μM glycine (indicated by the heavy bar). Increasing concentrations of bath applied antagonist (UBP131) reduced the inward currents.
Figure 4.
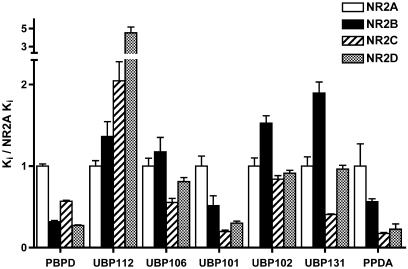
NMDA receptor subunit selectivity of PBPD compared to other competitive antagonists. Each compound was evaluated for the ability to block agonist-induced responses of NR1a/NR2 NMDA receptors expressed in Xenopus oocytes. IC50 values were generated from dose–response curves and the corresponding Ki value is shown above. To compare the subunit selectivity profiles of drugs with various affinities, Ki values were normalized to the drug's NR1/NR2A affinity. The selectivities of PBPD and the novel antagonist PEAQX are shown in comparison with the well-established NMDA receptor antagonists (R)-AA, (R)-CPP, (R)-AP5 and (R)-CPPene. Data for (R)-AP5, (R)-CPPene and PBPD are from Buller & Monaghan (1997) and are presented for comparison. Absolute values and statistical differences are listed in Table 2.
Each of the putative NMDA receptor antagonists was able to block recombinant NMDA receptor responses (Table 2, Figure 5). Each of the PBPD analogs that inhibited NMDA-sensitive L-[3H]-glutamate binding also displayed NMDA receptor antagonist activity. UBP112 differed from each of the other PBPD analogs in having the potency order NR2A>NR2B>NR2C>NR2D, thus generating the typical antagonist profile seen in Figure 2. UBP112's lower affinity than PBPD in the forebrain is consistent with UBP112's lower affinity at NR2B-containing receptors. The higher affinity found for UBP112 than PBPD at cerebellar NMDA receptors is consistent with UBP112's slightly higher relative affinity for NR2A, but is inconsistent with UBP112's low relative affinity for NR2C subunits. UBP112 also displays unexpectedly high affinity in the NR2B/NR2D-enriched medial thalamus. Given UBP112's reduced affinity for NR2D, it would be expected to have a lower affinity, not a higher affinity, in the medial thalamus (NR2B/NR2D) than in the medial striatum (NR2B).
Table 2.
Antagonist Ki values (μM) for inhibiting the responses of recombinant NMDA receptors expressed in Xenopus oocytes (means±s.e.m. (n))
| NR1a/NR2A | NR1a/NR2B | NR1a/NR2C | NR1a/NR2D | |
|---|---|---|---|---|
| (R)-α-AA | 6.5±0.6D (5) | 24.7±4.1D,c (7) | 43.9±2.8D (5) | 114±23 (5) |
| (R)-CPP | 0.041±0.003D,c* (6) | 0.27±0.02d* (5) | 0.63±0.05D (5) | 1.99±0.20 (5) |
| PBPDa | 15.79±0.43b,c,d | 5.01±0.25c | 8.98±0.18d | 4.29±0.11 |
| PEAQX | 0.0054±0.0004B,D (5) | 0.067±0.003C,D (5) | 0.0116±0.0009C (5) | 0.037±0.004 (4) |
| PPDA | 0.55±0.15b,C,D (3) | 0.31±0.02c,d (5) | 0.096±0.006 (4) | 0.125±0.035 (5) |
| UBP112 | 13.2±0.89D (5) | 18.0±2.4D (4) | 27.0±3.1D (5) | 59.7±8.5 (6) |
| UBP106 | 7.97±0.77c (4) | 9.36±1.41c* (6) | 4.42±0.40 (6) | 6.45±0.40 (5) |
| UBP101 | 42.9±5.3b*,C,D (4) | 22.1±5.2 (5) | 8.49±0.72 (4) | 12.9±1.1 (4) |
| UBP131 | 9.6±1.1B,C (5) | 18.2±1.3C,D (5) | 3.89±0.09D (5) | 9.26±0.43 (6) |
| UBP102 | 8.78±0.88B (5) | 13.4±0.8C,D (5) | 7.38±0.38 (5) | 8.02±0.31 (5) |
b, c, d, significantly different from NR2B, NR2C and NR2D, respectively; P<0.05.
b*, c*, d*, significantly different from NR2B, NR2C and NR2D, respectively; P<0.01.
B, C, D, significantly different from NR2B, NR2C and NR2D, respectively; P<0.001.
n=4–6; PBPD results from Buller et al. (1997).
Figure 5.
NMDA receptor subunit selectivity of PBPD derivatives. Each compound was evaluated for the ability to block responses to 10 μM L-glutamate plus 10 μM glycine at each of the NR1a/NR2 NMDA receptors expressed in Xenopus oocytes. To compare the subunit selectivity profiles of drugs with various affinities, Ki values were normalized to the drug's NR1/NR2A affinity. Absolute values and statistical differences are listed in Table 2.
Halogenation of PBPD in different positions resulted in differential effects on NMDA receptor subunit selectivity. Adding a fluoro group to the 4′ position of the biphenyl ring of PBPD (UBP102) doubled the compound's affinity for NR2A, reduced the affinity for NR2B three-fold, reduced NR2D affinity two-fold and had no effect on NR2C affinity. 2'-Bromo substitution of the biphenyl ring of PBPD (UBP101) led to a 3- to 4-fold decrease in affinity at NR2A-, NR2B- and NR2D-containing receptors, while having no effect on NR2C-containing receptors. Thus, halogenation at these two sites had opposite effects on NR2A affinity, similar effects on NR2B (reduced affinity) and minimal effects on NR2C affinity.
Placing a carbonyl group between the two phenyl groups of PBPD to generate the fluorenone derivative UBP106 increased antagonist affinity two-fold for NR2A and NR2C, decreased NR2B affinity to one-half and did not change NR2D affinity. Replacing the biphenyl group with a naphthyl group (UBP131) had generally similar effects – a two-fold increase in NR2A and NR2C affinities, a nearly four-fold reduction in NR2B affinity and a smaller two-fold reduction in NR2D affinity.
The most striking compound found was obtained by linking the two phenyl groups of PBPD through a benzene ring to generate the phenanthrene derivative PPDA. This modification significantly increased antagonist affinity for all subunits, and also altered subunit selectivity. Relative to PBPD, PPDA affinity was higher at NR1/NR2A receptors by 29-fold, NR1/NR2B receptors by 16-fold, NR1/NR2C by 94-fold and NR1/NR2D by 34-fold. Thus, PPDA is a high-affinity antagonist and represents the most selective NR2C and NR2D antagonist reported to date.
Discussion and conclusions
Presently, there are few pharmacological tools available for distinguishing NMDA receptor subtypes. Most competitive NMDA receptor antagonists have been designed as modifications of the glutamate or (R)-AP5 backbone, and these are frequently incorporated into a ring structure such as in CGS-19755 or CPP. These antagonists usually generate a subunit selectivity pattern of NR2A>NR2B>NR2C>NR2D, with highest affinity for NR2A-containing NMDA receptors (Ikeda et al., 1992; Ishii et al., 1993; Buller et al., 1994; Laurie & Seeburg, 1994; Kendrick et al., 1996; Buller & Monaghan, 1997). In the present study, we extend these observations with (R)-AA and (R)-CPP, which also display the NR2A>NR2B>NR2C>NR2D affinity profile. (R)-CPP displayed even greater NR2A>NR2D selectivity than previously reported compounds, with a nearly 50-fold selectivity for NR2A-containing receptors. Together with the relatively high affinity of L-glutamate for NR2D, CPP should be roughly 100-fold selective for NR2A over NR2D. The weak activity of CPP for NR2D may account for observations in the literature that CPP does not block all NMDA receptor-mediated responses (e.g. Harris & Davies, 1992; Binns et al., 1999).
Interestingly, a few atypical antagonists have now been identified that alter this pattern of subunit selectivity. Specifically, the bicyclic compound (3S,4aR,6S,8aR)-6-(1H-tetrazol-5-ylmethyl)decahydroisoquinoline-3-carboxylic acid (LY233536), and the biphenyl-containing compounds (S)-2-amino-3-(5-phosphonomethylbiphenyl-3-yl)propionic acid (EAB-515) and PBPD each display a higher affinity for NR2B than for NR2A-containing receptors and have an improved relative affinity for NR2C- and NR2D-containing receptors (Buller & Monaghan, 1997). The compounds examined in this study extend this general finding – large, bulky substitutions alter the subunit specificity of NMDA receptor antagonists. Thus, these large multiring NMDA receptor antagonists are capable of probing a differing part of the binding pocket which alters the subunit selectivity of the antagonist. Based upon structure–activity analysis of EAB-515, it has been proposed that the biphenyl group of EAB-515 binds to a hydrophobic binding pocket (Bigge, 1993; Cheung et al., 1996). This presumably corresponds to the ‘allowed' region described in various antagonist modeling studies (for a review see Jane et al., 1994). As previously reported (Auberson et al., 2002), PEAQX shows improved NR2A>NR2B selectivity. In contrast to the typical AP5-like antagonists, we further find that PEAQX is distinctive in having a higher affinity for NR2C than NR2B (or NR2D) subunits (Figure 4). As with other ‘atypical' antagonists, PEAQX has a bulky structure potentially allowing interactions with distal regions of the glutamate binding pocket.
In the present study, we have generated and evaluated a variety of PBPD analogs to probe the hydrophobic pocket/allowed region. Our results indicate that the geometry of this space is quite specific. Each of the antagonists with either a bent or branched arrangement of aromatic rings displayed no activity. However, linear multiring systems were tolerated, and in the case of phenanthrene substitution was preferred.
Of the active compounds evaluated, UBP112 was the only compound displaying the typical D-AP5-like pattern of subunit selectivity (NR2A>NR2B>NR2C>NR2D; high-to-low affinity). UBP112 was also the least bulky of the active PBPD derivatives, further suggesting that bulky side groups alter subunit selectivity. UBP112 and the naphthyl derivative UBP131 are the only two active compounds examined that do not have a benzene ring in the position of the third benzene of PPDA (or the second benzene of PBPD). Since these two compounds have a higher affinity for NR2A than NR2B, while PBPD and PPDA have a higher affinity for NR2B than for NR2A, it appears that the distal benzene ring contributes to the higher affinity for NR2B than NR2A in the PPDA and PBPD structures.
The increased affinity of PPDA for NR2C and NR2D appears to be mediated by a combined effect of each of the three benzene rings. If one compares UBP112 and UBP131, the effect of adding the first benzene is a preferential increase in NR2C and NR2D affinity. Likewise, adding the second ring (comparing PBPD to PPDA) preferentially increases NR2C and NR2D binding affinity and the third ring (comparing UBP131 to PPDA) preferentially increases affinity at NR2C, NR2D and NR2B subunits. Thus, the additional benzene rings may be providing additional interactions with the receptor and/or they may be restricting the molecule to a coplanar structure, which has improved interactions with NR2C and NR2D.
The hydrophobic pocket confers multiple subunit-specific points of antagonist interaction. While the addition of the multiple ring groups confers a general higher relative affinity for NR2B, NR2C and NR2D subunits, minor modifications made to the biphenyl structure of PBPD lead to various other changes in NMDA receptor subunit selectivity. For example, 4′-fluoro substitution (UBP102) reversed the NR2A–NR2B selectivity of PBPD. 2′-Bromo substitution (UBP101) significantly reduced NR2A and NR2B affinity but did not change NR2C affinity. Linking the two phenyl groups of PBPD, as in UBP106 and PPDA, preferentially increased NR2C selectivity. Thus, the region of the NMDA receptor binding site occupied by the biphenyl group of PBPD shows pharmacologically relevant NR2 heterogeneity. The effect of a fluoro group at the 4′-position of the biphenyl ring of PBPD (UBP102) may be analogous to the effect of bromo substitution at the 4 position on the distal benzene ring on the recently described antagonist PEAQX (Auberson et al., 2002), which also leads to an increase in NR2A affinity relative to NR2B affinity.
Recent receptor molecular modeling studies in our laboratory (based upon the X-ray crystallography of the structurally related GluR2 receptor) suggest that the hypothetical hydrophobic pocket/allowed area represents the S1/S2 cleft (Kinarsky et al., in preparation). More specifically, for PPDA, the hydrophobic pocket, may include the groove found in S2 at the base of the ‘H' helix (Armstrong et al., 1998) that leads directly away from the glutamate binding site – S1/S2 hinge region and out toward the surface of the receptor. Since subunit-specific amino-acid residues near the glutamate binding pocket were mostly found near the surface of the S1/S2 domain, only large antagonists such as PBPD or PPDA (or EAB515 and PEAQX) are long enough to come within reach of the subunit-specific amino-acid residues. Thus, further modifications of PBPD or PPDA in the distal benzene ring might be able to further increase subunit-specific interactions.
The observed patterns in NR2 selectivity displayed by the different compounds can account for many, but not all, of the regional variations seen in antagonist displacement of NMDA-sensitive L-[3H]-glutamate binding. PBPD has higher affinity than UBP112 in the forebrain; likewise PBPD has a four-fold higher affinity than UBP112 for NR2B (predominant forebrain site labeled by L-[3H]-glutamate). The reduced NR2B affinity of UBP102 can account for the reduced affinity of UBP102 in forebrain regions and the small affinity increase in the cerebellum can be accounted for by the presence of NR2A subunits. UBP106 binds with twice the affinity of PBPD in the cerebellum while it has affinities more similar to PBPD in the forebrain regions. This is consistent with UBP106's higher affinity than PBPD for both NR2A and NR2C, whereas PBPD has higher affinity than UBP106 at NR2B subunits. In contrast to the typical antagonists R-α-AA and CPP, PBPD and associated derivatives have higher affinity in the medial thalamus than in the medial striatum – an observation consistent with their higher affinities for NR2D-containing than for NR2B-containing NMDA receptors. UBP112, however, has a higher affinity for NR2B than for NR2D and yet has reduced affinity in the medial striatum (but note that it is less NR2B>NR2D selective than the typical antagonists R-α-AA and CPP). Also, UBP112 has a two-fold higher affinity than PBPD for the cerebellum, while UBP112 has similar (NR2A) and three-fold lower affinity (NR2C) than PBPD in its affinity for NR2A and NR2C receptors (which predominate in the cerebellum). Likewise, UBP102 has higher affinity for NR2A and NR2C receptors than NR2B-containing receptors; however, UBP102 has a lower affinity for the NR2A/NR2C-containing cerebellum than the primarily NR2B-containing forebrain. UBP112 also displays an unexpectedly higher affinity in the medial thalamus given UBP112's lower affinity for NR2D subunits. Overall, there appears to be a general trend for antagonists to have slightly lower than predicted affinities in the cerebellum and higher than predicted affinities in the medial thalamus. It is possible that other associated proteins (e.g. NR3?) or possible heterodimers (NR2B/NR2D in the medial thalamus and NR2A/NR2C in the cerebellum) can cause modest changes in the binding site conformation causing a small change in antagonist affinities.
The compounds studied in the present report represent the most selective compounds yet described for NR2A/NR2B selection over NR2C/NR2D (CPP) and for selecting NR2C/NR2D over NR2A/NR2B (PPDA). The degree of selectivity, for example, 50-fold for CPP at NR2A compared to NR2C and six-fold for PPDA at NR2C compared to NR2A, may be adequate for separating subunit-specific NMDA receptor actions in some cases (Hrabetova et al., 2000). The present findings indicate that small changes in antagonist structure in the biphenyl side group of PBPD lead to changes in subunit selectivity. Thus, PPDA with its high affinity and distinct subunit selectivity may represent an important lead compound for the development of NMDA receptor antagonists of yet greater selectivity.
Acknowledgments
We thank Drs Shigetada Nakanishi, David Lynch, and Peter Seeburg for providing NMDA receptor cDNA constructs, and Dr Yves Auberson for providing PEAQX. This work was supported by NIH Grant MH60252. Reprint requests should be sent to Dr Bihua Feng, 986260 Nebraska Medical Center, Omaha, NE 68198-6260 .
Abbreviations
- EAB-515
(S)-2-amino-3-(5-phosphonomethylbiphenyl-3-yl)propionic acid
- LY233536
(3S,4aR,6S,8aR)-6-(1H-tetrazol-5-ylmethyl)decahydroisoquinoline-3-carboxylic acid
- NMDA
N-methyl-D-aspartate
- NR2
NMDA receptor 2 subunit
- PBPD
(2S*,3R*)-1-(biphenyl-4-carbonyl)piperazine-2,3-dicarboxylic acid
- PEAQX
5-phosphonomethyl-1,4-dihydroquinoxaline-2,3-dione
- ∣PPDA
(2S*,3R*)-1-(phenanthren-2-carbonyl)piperazine-2,3-dicarboxylic acid
- (R)-AA
(R)-α-aminoadipate
- (R)-AP5
(R)-2-amino-5-phosphonopentanoate
- (R)-CPP
(R)-4-(3-phosphonopropyl)piperazine-2-carboxylic acid
- (R)-CPPene
(R,E)-4-(3-phosphonoprop-2-enyl)piperazine-2-carboxylic acid
- UBP101
(2S*,3R*)-1-(2′-bromobiphenyl-4-carbonyl)piperazine-2,3-dicarboxylic acid
- UBP102
(2S*,3R*)-1-(4′-fluorobiphenyl-4-carbonyl)piperazine-2,3-dicarboxylic acid
- UBP104
(2S*,3R*)-1-(4-iodobenzoyl)piperazine-2,3-dicarboxylic acid
- UBP106
(2S*,3R*)-1-(9-oxo-9H-fluorene-2-carbonyl)piperazine-2,3-dicarboxylic acid
- UBP107
(2S*,3R*)-1-(2-biphenyl-4-yl-acetyl)piperazine-2,3-dicarboxylic acid
- UBP108
(2S*,3R*)-1-diphenylacetylpiperazine-2,3-dicarboxylic acid
- UBP109
(2S*,3R*)-1-(3,3-diphenylpropionyl)piperazine-2,3-dicarboxylic acid
- UBP112
(2S*,3R*)-1-(3-phenylacryloyl)piperazine-2,3-dicarboxylic acid
- UBP120
(2S*,3R*)-1-(1-phenylcyclopropanecarbonyl)piperazine-2,3-dicarboxylic acid
- UBP121
(2S*,3R*)-1-(4-nitrobenzoyl)piperazine-2,3-dicarboxylic acid
- UBP122
(2S*,3R*)-1-(4-fluorobenzoyl)piperazine-2,3-dicarboxylic acid
- UBP130
(2S*,3R*)-1-benzoylpiperazine-2,3-dicarboxylic acid
- UBP131
(2S*,3R*)-1-(naphthalene-2-carbonyl)piperazine-2,3-dicarboxylic acid
References
- ANDALORO V.J., JANE D.J., TSE H.W., WATKINS J.C., MONAGHAN D.T. Pharmacology of NMDA receptor subtypes. Soc. Neurosci. Abstr. 1996;22:604. [Google Scholar]
- ARMSTRONG N., SUN Y., CHEN G.Q., GOUAUX E. Structure of a glutamate-receptor ligand-binding core in complex with kainate. Nature. 1998;395:913–917. doi: 10.1038/27692. [DOI] [PubMed] [Google Scholar]
- AUBERSON Y.P., ALLGEIER H., BISCHOFF S., LINGENHOEHL K., MORETTI R., SCHMUTZ M. 5-Phosphonomethylquinoxalinediones as competitive NMDA receptor antagonists with a preference for the human 1A/2A, rather than 1A/2B receptor composition. Bioorg. Med. Chem. Lett. 2002;12:1099–1102. doi: 10.1016/s0960-894x(02)00074-4. [DOI] [PubMed] [Google Scholar]
- BEATON J.A., STEMSRUD K., MONAGHAN D.T. Identification of a novel N-methyl-D-aspartate receptor population in the rat medial thalamus. J. Neurochem. 1992;59:754–757. doi: 10.1111/j.1471-4159.1992.tb09433.x. [DOI] [PubMed] [Google Scholar]
- BIGGE C.F. Structural requirements for the development of potent N-methyl-D-aspartic acid (NMDA) receptor antagonists. Biochem. Pharmacol. 1993;45:1547–1561. doi: 10.1016/0006-2952(93)90294-7. [DOI] [PubMed] [Google Scholar]
- BINNS K.E., TURNER J.P., SALT T.E. Visual experience alters the molecular profile of NMDA-receptor-mediated sensory transmission. Eur. J. Neurosci. 1999;11:1101–1104. doi: 10.1046/j.1460-9568.1999.00544.x. [DOI] [PubMed] [Google Scholar]
- BULLER A.L., LARSON H.C., SCHNEIDER B.E., BEATON J.A., MORRISETT R.A., MONAGHAN D.T. The molecular basis of NMDA receptor subtypes: native receptor diversity is predicted by subunit composition. J. Neurosci. 1994;14:5471–5484. doi: 10.1523/JNEUROSCI.14-09-05471.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BULLER A.L., MONAGHAN D.T. Pharmacological heterogeneity of NMDA receptors: characterization of NR1a/NR2D heteromers expressed in Xenopus oocytes. Eur. J. Pharmacol. 1997;320:87–94. doi: 10.1016/s0014-2999(96)00880-1. [DOI] [PubMed] [Google Scholar]
- CHENG Y., PRUSOFF W.H. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- CHEUNG N.S., O'ALLAGHAN D., RYAN M.C., DUTTON R., WONG M.G., BEART P.M. Structure–activity relationships of competitive NMDA receptor antagonists. Eur. J. Pharmacol. 1996;313:159–162. doi: 10.1016/0014-2999(96)00608-5. [DOI] [PubMed] [Google Scholar]
- CHOI D. Antagonizing excitotoxicity: a therapeutic strategy for stroke. Mt. Sinai J. Med. 1998;65:133–138. [PubMed] [Google Scholar]
- CHRISTIE J.M., JANE D.E., MONAGHAN D.T. Native N-methyl-D-aspartate receptors containing NR2A and NR2B subunits have pharmacologically distinct competitive antagonist binding sites. J. Pharmacol. Exp. Ther. 2000;292:1169–1174. [PubMed] [Google Scholar]
- DINGLEDINE R., BORGES K., BOWIE D., TRAYNELIS S.F. The glutamate receptor ion channels. Pharmacol. Rev. 1999;51:7–61. [PubMed] [Google Scholar]
- DURAND G.M., GREGOR P., ZHENG X., BENNETT M.V., UHL G.R., ZUKIN R.S. Cloning of an apparent splice variant of the rat N-methyl-D-aspartate receptor NMDAR1 with altered sensitivity to polyamines and activators of protein kinase C. Proc. Natl. Acad. Sci. U.S.A. 1992;89:9359–9363. doi: 10.1073/pnas.89.19.9359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FELDER E, MAFFEI S., PIETRA S., PITRÈ D. Catalytic hydrogenation of piperazine carboxylic acids. Helv. Chim. Acta. 1960;117:888–896. [Google Scholar]
- HARRIS N.C., DAVIES J. Cortically evoked excitatory synaptic transmission in the cat red nucleus is antagonised by D-AP5 but not by D-AP7. Brain Res. 1992;594:176–180. doi: 10.1016/0006-8993(92)91046-h. [DOI] [PubMed] [Google Scholar]
- HOLLMANN M., BOULTER J., MARON C., BEASLEY L., SULLIVAN J., PECHT G., HEINEMANN S. Zinc potentiates agonist-induced currents at certain splice variants of the NMDA receptor. Neuron. 1993;10:943–954. doi: 10.1016/0896-6273(93)90209-a. [DOI] [PubMed] [Google Scholar]
- HRABETOVA S., SERRANO P., BLACE N., TSE H.W., SKIFTER D.A., JANE D.E., MONAGHAN D.T., SACKTOR T.C. Distinct NMDA receptor subpopulations contribute to long-term potentiation and long-term depression induction. J. Neurosci. (Online) 2000;20:RC81. doi: 10.1523/JNEUROSCI.20-12-j0002.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IKEDA K., NAGASAWA M., MORI H., ARAKI K., SAKIMURA K., WATANABE M., INOUE Y., MISHINA M. Cloning and expression of the epsilon 4 subunit of the NMDA receptor channel. FEBS Lett. 1992;313:34–38. doi: 10.1016/0014-5793(92)81178-o. [DOI] [PubMed] [Google Scholar]
- ISHII T., MORIYOSHI K., SUGIHARA H., SAKURADA K., KADOTANI H., YOKOI M., AKAZAWA C., SHIGEMOTO R., MIZUNO N., MASU M., NAKANISHI S. Molecular characterization of the family of the N-methyl-D-aspartate receptor subunits. J. Biol. Chem. 1993;268:2836–2843. [PubMed] [Google Scholar]
- JANE D.E., OLVERMAN H.J., WATKINS J.C. Agonists and Competitive Antagonists: Structure–Activity and Molecular Modelling Studies. Oxford: Oxford University Press; 1994. [Google Scholar]
- KENDRICK S.J., LYNCH D.R., PRITCHETT D.B. Characterization of glutamate binding sites in receptors assembled from transfected NMDA receptor subunits. J. Neurochem. 1996;67:608–616. doi: 10.1046/j.1471-4159.1996.67020608.x. [DOI] [PubMed] [Google Scholar]
- LAUBE B., HIRAI H., STURGESS M., BETZ H., KUHSE J. Molecular determinants of agonist discrimination by NMDA receptor subunits: analysis of the glutamate binding site on the NR2B subunit. Neuron. 1997;18:493–503. doi: 10.1016/s0896-6273(00)81249-0. [DOI] [PubMed] [Google Scholar]
- LAURIE D.J., SEEBURG P.H. Ligand affinities at recombinant N-methyl-D-aspartate receptors depend on subunit composition. Eur. J. Pharmacol. 1994;268:335–345. doi: 10.1016/0922-4106(94)90058-2. [DOI] [PubMed] [Google Scholar]
- MINAMI T., MATSUMURA S., OKUDA-ASHITAKA E., SHIMAMOTO K., SAKIMURA K., MISHINA M., MORI H., ITO S. Characterization of the glutamatergic system for induction and maintenance of allodynia. Brain Res. 2001;895:178–185. doi: 10.1016/s0006-8993(01)02069-8. [DOI] [PubMed] [Google Scholar]
- MONAGHAN D.T., ANDALORO V.J., SKIFTER D.A. Molecular determinants of NMDA receptor pharmacological diversity. Prog. Brain Res. 1998;116:158–177. doi: 10.1016/s0079-6123(08)60437-9. [DOI] [PubMed] [Google Scholar]
- MONAGHAN D.T., COTMAN C.W. Identification and properties of N-methyl-D-aspartate receptors in rat brain synaptic plasma membranes. Proc. Natl. Acad. Sci. U.S.A. 1986;83:7532–7536. doi: 10.1073/pnas.83.19.7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MONAGHAN D.T., OLVERMAN H.J., NGUYEN L., WATKINS J.C., COTMAN C.W. Two classes of N-methyl-D-aspartate recognition sites: differential distribution and differential regulation by glycine. Proc. Natl. Acad. Sci. U.S.A. 1988;85:9836–9840. doi: 10.1073/pnas.85.24.9836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MONYER H., BURNASHEV N., LAURIE D.J., SAKMANN B., SEEBURG P.H. Developmental and regional expression in the rat brain and functional properties of four NMDA receptors. Neuron. 1994;12:529–540. doi: 10.1016/0896-6273(94)90210-0. [DOI] [PubMed] [Google Scholar]
- NAKANISHI S., NAKAJIMA Y., MASU M., UEDA Y., NAKAHARA K., WATANABE D., YAMAGUCHI S., KAWABATA S., OKADA M. Glutamate receptors: brain function and signal transduction. Brain Res. Brain Res. Rev. 1998;26:230–235. doi: 10.1016/s0165-0173(97)00033-7. [DOI] [PubMed] [Google Scholar]
- VICINI S., WANG J.F., LI J.H., ZHU W.J., WANG Y.H., LUO J.H., WOLFE B.B., GRAYSON D.R. Functional and pharmacological differences between recombinant N-methyl-D-aspartate receptors. J. Neurophysiol. 1998;79:555–566. doi: 10.1152/jn.1998.79.2.555. [DOI] [PubMed] [Google Scholar]
- WATANABE M., INOUE Y., SAKIMURA K., MISHINA M. Developmental changes in distribution of NMDA receptor channel subunit mRNAs. Neuroreport. 1992;3:1138–1140. doi: 10.1097/00001756-199212000-00027. [DOI] [PubMed] [Google Scholar]
- WATANABE M., INOUE Y., SAKIMURA K., MISHINA M. Distinct spatio-temporal distributions of the NMDA receptor channel subunit mRNAs in the brain. Ann. N.Y. Acad. Sci. 1993;707:463–466. doi: 10.1111/j.1749-6632.1993.tb38099.x. [DOI] [PubMed] [Google Scholar]