Abstract
The crystal structure of Bacillus subtilis orotidine 5′-monophosphate (OMP) decarboxylase with bound uridine 5′-monophosphate has been determined by multiple wavelength anomalous diffraction phasing techniques and refined to an R-factor of 19.3% at 2.4 Å resolution. OMP decarboxylase is a dimer of two identical subunits. Each monomer consists of a triosephosphate isomerase barrel and contains an active site that is located across one end of the barrel and near the dimer interface. For each active site, most of the residues are contributed by one monomer with a few residues contributed from the adjacent monomer. The most highly conserved residues are located in the active site and suggest a novel catalytic mechanism for decarboxylation that is different from any previously proposed OMP decarboxylase mechanism. The uridine 5′-monophosphate molecule is bound to the active site such that the phosphate group is most exposed and the C5-C6 edge of the pyrimidine base is most buried. In the proposed catalytic mechanism, the ground state of the substrate is destabilized by electrostatic repulsion between the carboxylate of the substrate and the carboxylate of Asp60. This repulsion is reduced in the transition state by shifting negative charge from the carboxylate to C6 of the pyrimidine, which is close to the protonated amine of Lys62. We propose that the decarboxylation of OMP proceeds by an electrophilic substitution mechanism in which decarboxylation and carbon-carbon bond protonation by Lys62 occur in a concerted reaction.
Orotidine monophosphate (OMP, 1) decarboxylase catalyzes the final step in the de novo biosynthesis of uridine monophosphate (UMP, 2) (Eq. 1). ![]()
In most prokaryotes, OMP decarboxylase is a dimer of identical subunits whereas in higher organisms, it is part of a bifunctional enzyme that also catalyzes the formation of OMP. Amino acid sequence comparisons suggest that monomeric and bifunctional OMP decarboxylases are structurally homologous with about a dozen residues conserved throughout all species. The enzyme accelerates this decarboxylation reaction by 1017 and is the most proficient enzyme identified so far (1). OMP decarboxylase does not use any cofactors (2). Its mechanism is novel because the carbanion generated by carbon dioxide loss is localized in an sp2 orbital perpendicular to the π system of the pyrimidine. In all other decarboxylases, the carbanion is delocalized either into an adjacent carbonyl or into a covalently bound thiamin, pyridoxal, or pyruvoyl cofactor (3). Although several hypotheses have been advanced to explain how the enzyme stabilizes the carbanion intermediate, the mechanistic details of this reaction are currently unclear.
Three mechanisms have been proposed for OMP decarboxylase (Scheme S1). In the first mechanism (zwitterion mechanism), protonation of the C2 carbonyl group would generate the zwitterion 3, in which the positive charge at N1 could stabilize the negative charge accumulating at C6 during the decarboxylation. This proposal was supported by a model study that demonstrated that the rate of decarboxylation of 1,3-dimethylorotate was 1010× slower than the decarboxylation of 1-methyl-2,4,-dimethoxypyrimidinium-6-carboxylate (4).
Scheme 1.
In the second proposal, a concerted protonation of the C4 carbonyl group and decarboxylation would generate the stabilized carbene 6 rather than the high energy carbanion. This intriguing mechanism was suggested based on theoretical calculations (5–7). However, this mechanism does not explain why the replacement of the C4 carbonyl with a thiocarbonyl has only a small effect on the reaction rate (50% reduction in kcat) whereas replacement of the C2 carbonyl group with a thiocarbonyl has a large effect on the rate (kcat reduced by >10,000-fold) (8).
In the third proposal, also supported by a model study, an active site nucleophile adds to C5 of the pyrimidine to give 7. Decarboxylation then results in the expulsion of this nucleophile (9). The absence of a secondary deuterium isotope effect at C5 and the failure to detect the addition of a nucleophile to C5 of phosphoribofuranosyl barbituric acid have been used to argue against this proposal (10). However, these arguments do not rigorously exclude this mechanism because the possibility remains that the secondary deuterium isotope effect may have been too small to detect, and nucleophilic addition to phosphoribofuranosyl barbituric acid is very unlikely because C5 is an electron rich center in this substrate analog.
To provide information that might differentiate among the proposed mechanisms for OMP decarboxylase, we undertook structural studies of the enzyme. Here we report the x-ray crystal structure of the Bacillus subtilis OMP decarboxylase complexed with the reaction product, UMP, and analyze the active site interactions in the context of these proposals.
Methods
Overexpression and Purification.
The B. subtilis OMP decarboxylase gene was PCR amplified from genomic DNA and was cloned into the pET-16b expression vector (Novagen) by using standard molecular cloning techniques. This expression construct, which codes for an amino-terminal polyhistidine tag with amino acid sequence MG(H)10SSGHIEGRH-natural N terminus, was subsequently transformed into Escherichia coli BL21(DE3) cells (Novagen). Cells were grown at 37°C in 1 liter of LB medium containing 200 μg/ml of ampicillin. When the culture reached an OD600 of 0.6, the temperature was reduced to 25°C, and the cells were induced with 1 mM IPTG and incubated for 12 hours. Cells were harvested by centrifugation and stored at −80°C until use.
All subsequent protein purification steps were carried out at 4°C or on ice. Cells were lysed with a French Press. After a high-speed centrifugation step to remove insoluble material, the polyhistidine-tagged OMP decarboxylase was purified using Ni-NTA resin (Qiagen). The protein was eluted from the Ni-column in 250 mM imidazole, 300 mM NaCl, 50 mM sodium phosphate buffer (pH 8.0), 2 mM 2-mercaptoethanol (β-ME), 10% vol/vol glycerol, and 50 mM UMP. The purified protein was buffer-exchanged into 20 mM Tris⋅HCl (pH 7.8), 2 mM β-ME, and 10 mM UMP using an ultra-filtration device. The presence of UMP during the chromatography and buffer exchange steps was required to preserve enzyme activity and solubility. The selenomethionine (SeMet) incorporated protein used for multiple wavelength anomalous diffraction phasing was expressed using the methionine auxotrophic E. coli strain, B834(DE3), in minimal medium. The purification procedure for the SeMet protein was identical except that 5 mM β-ME was added to the final buffer to prevent oxidation of the SeMet residues. The resulting SeMetOMP decarboxylase contained eight SeMet residues per monomer.
The purified material was stored at −80°C. Protein concentration was determined by the Bradford method (11) using BSA as a standard. Purity was verified by running samples on 12% SDS polyacrylamide gels followed by Coomassie staining (gels not shown). Spectrophotometric OMP decarboxylase assays following the method of Turnbough (12) were performed to verify enzyme activity for both the polyhistidine-tagged OMP decarboxylase and the polyhistidine-tagged SeMetOMP decarboxylase. Both forms of OMP decarboxylase were fully active.
X-Ray Data Collection.
All x-ray intensity data were measured at the Structural Biology Center undulator beamline (ID19) of the Advanced Photon Source using a mosaic charge-coupled devise-based x-ray detector (13). Data collection statistics are summarized in Table 1. The multiple wavelength anomalous diffraction data sets were collected at cryogenic temperatures to 2.5 Å resolution. Three wavelengths were selected for data collection corresponding to the maximum f" (peak), the minimum f′ (edge), and a reference wavelength (remote). A total of 100° of data plus an additional 100° of inverse beam data were measured at each wavelength by using a 1° oscillation angle measured for 15 sec. The data for the native protein were collected at cryogenic temperature to 2.4 Å resolution. A total of 180° of data were measured by using the oscillation method. Individual frames consisting of a 1° oscillation angle were measured for 30 sec. The final data set was 99.4% complete to 2.4 Å resolution with an overall Rsym of 8.8%. The integration and data scaling were performed by using the program denzo/scalepack (14).
Table 1.
Summary of x-ray diffraction data
| Native | Edge | Peak | Remote | |
|---|---|---|---|---|
| Wavelength, Å | 1.03321 | 0.97949 | 0.97935 | 0.93928 |
| Resolution, Å | 2.4 | 2.5 | 2.5 | 2.5 |
| No. of measurements | 177,412 | 180,489 | 180,208 | 175,717 |
| No. of unique reflections | 29,779 | 26,490 | 25,454 | 26,411 |
| Completeness | ||||
| Overall, % | 99.4 | 97.1 | 97.3 | 97.6 |
| Outermost shell, % | 98.5 | 75.5 | 78.4 | 82.5 |
| R-sym | ||||
| Overall | 0.088 | 0.097 | 0.105 | 0.114 |
| Outermost shell | 0.234 | 0.310 | 0.273 | 0.472 |
Structure Determination and Refinement.
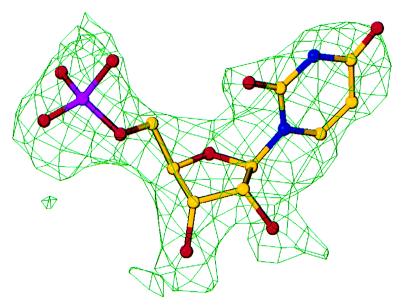
The structure was determined at 2.5 Å resolution by using multiple wavelength anomalous diffraction phasing (15). The anomalous differences for the peak wavelength were used to locate the Se atom positions. DiffE values were generated from the anomalous difference by using the drear package (16). A total of 18 of the 24 possible Se atom sites were found by direct methods using the Shake-and-Bake procedure (17) and the computer program snb (18). The missing six Se atoms, one at the normal N terminus and one in the His tag for each of the three monomers, were later shown to be disordered. The 18 Se atom positions were refined by using the mlphare program (19), and the resulting phases were improved by solvent flattening using the program dm (20). The correct hand for the Se atom model was determined from inspection of the electron density maps. The figure of merit was 0.53 initially and increased to 0.79 after density modification. The final unaveraged experimental map was of excellent quality showing clear main chain and side chain density as well as the active site UMP (Fig. 1). Using this map and the computer program o (21), a continuous C-α carbon tracing was generated for one monomer, starting with Met1 and ending with Gly236. All main chain and side chains atoms were then added, and the second and third monomers were generated by using noncrystallographic symmetry transformations. Before refinement, the individual side chains were adjusted for each monomer.
Figure 1.
Experimental electron density at 2.5 Å resolution (unaveraged) for UMP of monomer C using solvent flattened multiple wavelength anomalous diffraction phases. The contour level is 1σ.
The model was further improved by using alternate cycles of simulated annealing refinement and model building. The refinement was performed by using the torsion angle refinement of the program cns (22). Water molecules were added by using difference electron density maps and were retained in the model if they formed good hydrogen bonds and refined with reasonable B-factors. The progress of the refinement was monitored by using the free R-factor for a sample of data. The final R-factor was 19.3% for 29,779 unique reflections to 2.4 Å resolution, and the free R-factor was 22.8%.
Results and Discussion
Quality of Model.
The asymmetric unit of the OMP decarboxylase crystals contains one-and-a-half OMP decarboxylase dimers (monomers A, B, and C). Monomers A and B form a dimer using a noncrystallographic twofold axis, and monomers C and C′ form a second dimer using a crystallographic twofold axis. All three monomers contain one molecule of the reaction product, UMP, which was added during purification. The entire N-terminal polyhistidine tag and a few residues at the C terminus are disordered in each monomer. The final model of OMP decarboxylase contains residues 1–237 for monomer A, 3–236 for monomer B, and 3–236 for monomer C. The UMP molecule was clearly defined for each monomer, and its refined temperature factors are among the lowest in the entire model. The final model also included 226 well ordered water molecules. The standard deviation of the atomic positions, as estimated from a Luzzatti plot, was 0.31 Å (23). Analysis of the model using procheck (24) showed that 99.4% of the residues are in allowed regions with only one residue, Gln26 of monomer B, in a disallowed region.
Structure of the Monomer.
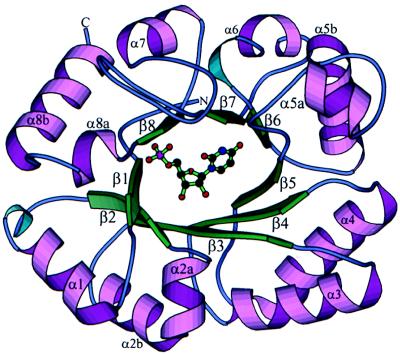
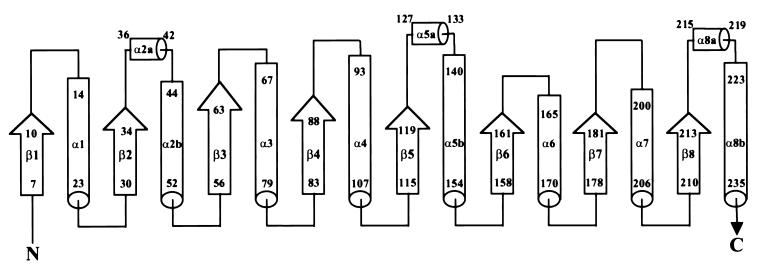
The structure of the monomer uses a triose phosphate isomerase (TIM) barrel fold (25) (Fig. 2). A topology diagram labeled with the first and last residue for each secondary structural element is shown in Fig. 3. Altogether, the fold contains eight β-strands and 11 α-helices. In addition to the alternating structure of eight β-strands and α-helices, there are additional short helices located between strands β2 and β3, between strands β5 and β6, and after strand β8. The α-helices range in length from 6 to 15 residues, and the β-strands range in length from 4 to 8 residues. A search of the structural database by using the program dali (27) showed no striking similarity to other proteins, other than the generic TIM barrel fold.
Figure 2.
Ribbon drawing of the OMP decarboxylase monomer. Helices and strands are labeled, and UMP is shown as a ball-and-stick model. The drawing was prepared with molscript (26).
Figure 3.
Topology diagram showing the OMP decarboxylase fold. The first and last residue numbers are given for each α-helix and β-strand.
Structure of the Dimer.
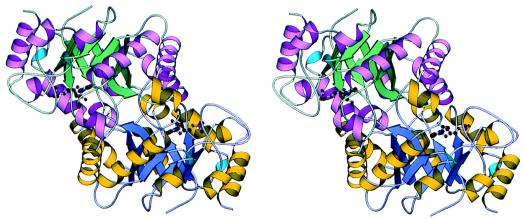
The active form of OMP decarboxylase is a dimer of identical subunits (Fig. 4). The dimer is formed by an extensive set of contacts between twofold-related TIM barrels. This twofold rotation places the top of each barrel (top defined as the face nearest the carboxy-terminal ends of the β-strands) in close contact to each other. The barrel tops are offset such that the N-terminal ends of helices α3 and α4 of one barrel are approximately over the center of the second TIM barrel. The capping of one monomer by the other is required to complete the active site as described below. The dimer interface is formed by many polypetide segments, including the β2-α2A loop, the β3-α3 loop, the β4-α4 loop, the β5-α5A loop, the α5A -α5B loop, β7-α7 loop, the β8-α8 loop, and parts of helices α2A, α3, and α5B. The longest β-strand in the structure, β3, extends from the active site of one monomer to the active site of the other monomer. Four absolutely conserved residues are found in or near this segment; Asp60 and Lys62 are located in the active site of one monomer, and Asp65 and Ile66 are located in the active site of the second monomer. His88, which is conserved in all OMP decarboxylase sequences but makes no direct contacts with the UMP, donates a hydrogen bond to the main chain carbonyl oxygen atom of His64 from an adjacent monomer.
Figure 4.
Stereodiagram showing the OMP decarboxylase dimer. Different colors are used for each subunit, and UMP is shown as a ball-and-stick model. The drawing was prepared with molscript (26).
Active Site.
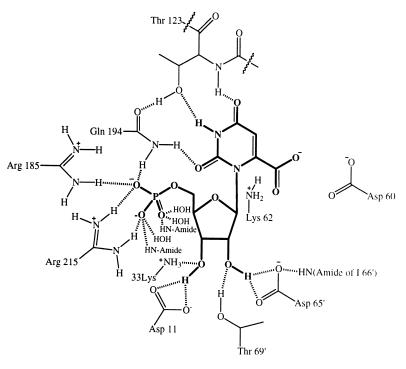
The active site is located at the end of the TIM barrel that corresponds to the carboxy-terminal ends of the β-strands and amino-terminal ends of the α-helices (Fig. 4) and near the interface of two monomers. About 18 residues appear to line the active site cavity. These residues are contributed by the loops after β-strands β1, β2, β3, β5, β6, β7, and β8. Thus, the active site stretches almost completely across one end of the TIM barrel. Of the active site residues, 15 are contributed from one monomer, and three are contributed from β3-α3 loop of the adjacent monomer. Sequence alignments using OMP decarboxylase sequences from diverse species show that 12 residues are absolutely conserved. These include Asp11, Lys33, Asp60, Lys62, Asp65, Ile66, His88, Pro182, Gly183, Arg185, Gln194, and Arg215. In addition, Thr123 in B. subtilis OMP decarboxylase is always Thr or Ser in other OMP decarboxylase sequences, and residues Val119, Leu122, Val160, and Val212 in B. subtilis OMP decarboxylase are always hydrophobic in other OMP decarboxylase sequences. A ball-and-stick model of UMP and several key active site residues is shown in Fig. 5. Extensive interactions are observed between UMP and the enzyme. Based on the crystal structure of OMP decarboxylase with bound UMP, we have assigned roles for amino acid residues in the OMP decarboxylase active site. These interactions are illustrated in Figs. 5 and 6.
Figure 5.
Active site of OMP decarboxylase. The stereoview shows UMP and the key amino acid residues. The red spheres represent water molecules. The drawing was prepared with ribbons (29).
Figure 6.
Schematic representation of the OMP decarboxylase active site. Only charged and hydrophilic residues are included. The carboxylate of OMP is based on modeling studies. Hydrogen bonds are shown as dashed lines.
The active site contains UMP, the product of the OMP decarboxylase reaction. In fact, we were not able to purify and crystallize B. subtilis OMP decarboxylase in the absence of UMP. Instead, the unliganded OMP decarboxylase formed heavy precipitates during purification. In the presence of UMP, OMP decarboxylase remained soluble until crystal growth occurred. The UMP density was clear and consistent in all three monomers. In the OMP decarboxylase active site, UMP exists in the syn comfomation, with a glycosidic torsion angle of 68.0°. The ribosyl group is in the C3′-exo conformation, and the phosphate group is extended out and away from the ribose with torsion angles near 180°. All of the conformational parameters are within the range expected for unbound UMP (28), suggesting that the OMP decarboxylase active site introduces little strain within the pyrimidine nucleotide molecule.
The binding site of the 5′-monophosphate group is located nearest the surface of the protein. Ten hydrogen bonds are provided by Arg185 (two), Gln194 (one), Gly214 (one), Arg215 (three), and one by each of three water molecules. By inference, phosphate acts as acceptor for all 10 hydrogen bonds. For the ribosyl group, the 2′-hydroxyl group contributes a bifurcated hydrogen bond to Asp65, and the 3′-hydroxyl group contributes a bifurcated hydrogen bond to Asp11. The extensive hydrogen bonding between UMP and, by inference, OMP, provides significant binding energy with little conformational flexibility. It is possible that these interactions are responsible for driving the pyrimidine base into the active site pocket, where it is highly shielded from solvent and close to amino acid side chains required for catalysis.
In the pyrimidine binding site, the amino group of Gln194 donates hydrogen bonds to O2 and one of the phosphate oxygen atoms. Thr123 accepts a hydrogen bond from N3 and donates a hydrogen bond to the carbonyl oxygen atom of Gln194. O4 is sandwiched between the methyl group of Thr123 and the side chain of Leu122 and accepts a hydrogen bond from the amide group of Thr123.
In addition, the structure of UMP bound to OMP decarboxylase clearly indicates the location of the carboxylate binding pocket for the substrate, OMP. A model for OMP can be generated by adding a carboxylate to the C6 position, assuming that the conformation of UMP does not change in the active site. This assumption seems justified on the basis that the UMP binding site appears to be rigid and involves many contacts between ligand and protein atoms. The carboxylate pocket is lined by the side chains of Lys33, Asp60, and Lys62 of one monomer and Asp65 of an adjacent monomer. Assuming that these side chains are rigid, the carboxylate has the fewest close contact when it is in the plane of the pyrimidine ring. The OMP model shows that the carboxylate group is pointed directly at Asp60, which in turn is sandwiched between Lys33 and Lys62. Lys62 is located below the bond that joins the carboxylate to the pyrimidine ring (Fig. 5).
An additional pocket, which may accommodate the second product, CO2, is observed near the C5 position of the pyrimidine base. This pocket is lined exclusively with hydrophobic contacts, including the side chains of Val119, Leu122, Pro182, Val160, Val212, and the methylene groups of Lys33 and Lys62. The location, size, and properties of this pocket suggest that a molecule of CO2 could be accommodated until UMP is released, implying ordered product release.
Catalytic Mechanism of OMP Decarboxylase.
The active site interactions identified from the structure are shown schematically in Fig. 6. The most striking feature of the active site is the absence of acidic residues next to the C2 and the C4 carbonyl groups of the substrate. Specifically, Lys62 (Lys93 in yeast OMP decarboxylase), which was previously proposed to function as the carbonyl activating residue (6, 30), is near neither carbonyl group. In addition, there is no nucleophile close to C5 of the pyrimidine. The structure therefore demonstrates that all three mechanistic proposals outlined in the introduction fail to adequately describe the mechanism of OMP decarboxylase. We propose an alternative mechanism, which is outlined in Fig. 7A.
Figure 7.
Proposed reaction mechanism for OMP decarboxylase. (A) Schematic representation of the reaction with the proposed transition state. (B) Stereoview of a model of the enzyme substrate complex. The OMP was generated by modeling a carboxylate group at C6 coplanar to the pyrimidine ring of UMP. The coordinates of UMP, Asp60, and Lys 62 were used without modification from the crystal structure.
Analysis of the structure suggests at least three critical components to the catalytic mechanism. First, the anionic carboxylate of the substrate is positioned in a negatively charged region of the protein close to the carboxylate of Asp 60, and the carbon of the pyrimidine destined to become the carbanion is positioned close to the positively charged ammonium group of Lys 62 (Fig. 7B). This arrangement of charged residues results in the destabilization of the ground state and in the stabilization of negative charge accumulation at C6 in the transition state. This is an elegant example of how an enzyme can accelerate a reaction by binding the transition state more tightly than the ground state (31). Second, the extensive interactions between the substrate and the enzyme provide the binding energy needed to force the two carboxylate groups into close proximity. Finally, as the decarboxylation reaction proceeds, the weakly basic σ bond linking the carboxy group to the pyrimidine becomes progressively more basic, eventually reaching an approximate pKa value of 35 for the conjugate acid of the putative vinyl carbanion intermediate. Because the protonated amine of Lys62 (pKa = 7.5) is positioned close to this bond, it is highly unlikely that a high energy vinyl carbanion will be formed. Instead, when the C-C bond becomes sufficiently basic, proton transfer from Lys62 will occur. We therefore propose that the OMP decarboxylase catalyzed reaction proceeds by a bimolecular electrophilic substitution mechanism (SE2) in which decarboxylation and protonation are concerted (32).
The proposal in Fig. 7A is in agreement with most of the previously published experimental observations on the enzyme. The presence of a positively charged lysine adjacent to the C6 carbon of the pyrimidine was originally suggested based on the high affinity inhibition of the enzyme by phosphoribofuranosyl barbituric acid (Ki = 9 × 10−12 M) (30, 33). This lysine residue (Lys62) is highly conserved, and mutation of the equivalent residue in the yeast OMP decarboxylase (K93C) gave inactive enzyme (30). The phosphate group on the substrate plays an important role in catalysis. If it is removed, kcat is reduced by 105 (34). This is consistent with the use of substrate binding energy to hold the two carboxylate groups in close proximity. OMP decarboxylase shows a V/K vs. pH profile with a maximum at pH = 7 (35). We propose that Lys62 is protonated (pKa = 7.5) and Asp60 is deprotonated (pKa = 6.5) under these conditions. Replacement of the C4 carbonyl group of the substrate with a thiocarbonyl group (4-thioOMP) has only a small effect on kcat (50% reduction). In contrast, the C2 thiocarbonyl containing analog (2-thioOMP) is not a substrate (kcat is reduced by at least 5,000-fold) (8). This suggests that hydrogen bonding to the C2 carbonyl is much more important in stabilizing the transition state for the decarboxylation than hydrogen bonding to the C4 carbonyl. It is unclear why these two substrate analogs show such different reactivities. One possibility is that the enzymatic synthesis of 2-thioOMP did not give the anticipated product. Isotope effect studies suggest that the protonation and the decarboxylation are not concerted (35, 36). This result does not contradict our mechanistic proposal if we assume that a substantial component of the observed solvent isotope effect is attributable to substrate binding or to a protein conformational change. Although the carbon isotope effect suggests that the decarboxylation is rate limiting, the observed solvent isotope effect is relatively small (1.3 ± 0.2). This suggests that the interaction of the NH proton with the CC bond is weak in the transition state for the decarboxylation and that the proton transfer occurs late on the reaction coordinate because of the low basicity of the CC bond.
Although decarboxylation by an SE2 mechanism is unprecedented, related reactions involving the protonation of high energy anionic intermediates have been identified. The protonation of the carbon mercury bond catalyzed by organomercurial lyase occurs by an SE2 mechanism (37), and the remarkable carbon hydrogen bond protonation observed in the metal ion free hydrogenase is also likely to proceed by an SE2 mechanism (38). Therefore, we conclude that the SE2 mechanism for OMP decarboxylase described above provides the best explanation of the available structural and kinetic data.
Table 2.
Crystallographic data and refinement statistics
| Space group cell dimensions, Å | P21212 |
| a | 78.41 |
| b | 89.76 |
| c | 105.90 |
| Z, monomers | 12 |
| Protein fraction | 0.57 |
| Vm, Å3/Da | 2.157 |
| Resolution limits, Å | 20–2.4 |
| No. of protein atoms | 5,371 |
| No. of ligand atoms | 63 |
| No. of water molecules | 226 |
| R-factor, % | 19.3 |
| Free R-factor, % | 22.8 |
| rms bond, Å | 0.013 |
| rms angle, ° Average B-factors, Å2 | 1.62 |
| Protein main chain | 27.3 |
| Protein side chain | 29.3 |
| Ligand | 18.5 |
| Water molecules | 27.1 |
Acknowledgments
Diffraction data for this study were collected at Argonne National Laboratory at the Structural Biology Center beamline ID-19 of the Advanced Photon Source, which is supported by the United States Department of Energy. This work was supported by the National Institutes of Health Grants RR-01646 (to S.E.E.) and DK44083 (to T.P.B.), the W. M. Keck Foundation, and the Lucille P. Markey Charitable Trust.
Abbreviations
- OMP
orotidine 5′-monophosphate
- UMP
uridine 5′-monophosphate
- SeMet
selenomethionine
- TIM
triose phosphate isomerase
- SE2
bimolecular electrophilic substitution
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1dbt).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.259441296.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.259441296
References
- 1.Radzicka A, Wolfenden R. Science. 1995;267:90–93. doi: 10.1126/science.7809611. [DOI] [PubMed] [Google Scholar]
- 2.Cui W, DeWitt J G, Miller S M, Wu W. Biochem Biophys Res Commun. 1999;259:133–135. doi: 10.1006/bbrc.1999.0737. [DOI] [PubMed] [Google Scholar]
- 3.O'Leary M H. In: The Enzymes. Sigman D S, editor. Vol. 20. San Diego: Academic; 1992. pp. 235–269. [Google Scholar]
- 4.Beak P, Siegel B. J Am Chem Soc. 1976;98:3601–3606. doi: 10.1021/ja00428a035. [DOI] [PubMed] [Google Scholar]
- 5.Lee J K, Houk K N. Science. 1997;276:942–945. doi: 10.1126/science.276.5314.942. [DOI] [PubMed] [Google Scholar]
- 6.Chen J, McAllister M A, Lee J K, Houk K N. J Org Chem. 1998;63:4611–4619. [Google Scholar]
- 7.Nakanishi M P, Wu W. Tetrahedron Lett. 1998;39:6271–6272. [Google Scholar]
- 8.Shostak K, Jones M E. Biochemistry. 1992;31:12155–12161. doi: 10.1021/bi00163a026. [DOI] [PubMed] [Google Scholar]
- 9.Silverman R B, Groziak M P. J Am Chem Soc. 1982;104:6434–6439. [Google Scholar]
- 10.Acheson S A, Bell J B, Jones M E, Wolfenden R. Biochemistry. 1990;29:3198–3202. doi: 10.1021/bi00465a007. [DOI] [PubMed] [Google Scholar]
- 11.Bradford M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 12.Turnbough C L, Jr, Kerr K H, Funderburg W R, Donahue J P, Powell F E. J Biol Chem. 1987;262:10239–10245. [PubMed] [Google Scholar]
- 13.Stanton M, Philips W C, O'Mara D, Naday I, Westbrook E. Nucl Instrum Methods Phys Res Sect A. 1993;325:558–567. [Google Scholar]
- 14.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 15.Hendrickson W A. Science. 1991;254:51–58. doi: 10.1126/science.1925561. [DOI] [PubMed] [Google Scholar]
- 16.Blessing R H, Guo D Y, Langs D A. In: Direct Methods for Solving Macromolecular Structures. Fortier S, editor. Dordrecht, The Netherlands: Kluwer; 1997. [Google Scholar]
- 17.Hauptman H A. In: Crystallographic Computing 5: From Chemistry to Biology. Moras D, Podnarny A D, Thierry J C, editors. Oxford: Oxford Univ. Press; 1991. pp. 324–332. [Google Scholar]
- 18.Miller R, Gallo S M, Khalak H G, Weeks C M. J Appl Crystallogr. 1994;27:613–621. [Google Scholar]
- 19.Otwinowski Z. In: Isomorphous Replacement and Anomalous Scattering. Wolf W, Evans P R, Leslie A G W, editors. Daresbury Laboratories, Warrington, U.K.: SERC Proceedings; 1991. pp. 80–88. [Google Scholar]
- 20.Cowtan K. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography. 1994;31:34–38. [Google Scholar]
- 21.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 22.Brünger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J-S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 23.Luzzati P V. Acta Crystallogr. 1952;5:802–810. [Google Scholar]
- 24.Laskowski R A, MacAuthur M W, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 25.Banner D W, Bloomer A C, Petsko G A, Phillips D C, Wilson I A. Biochem Biophys Res Commun. 1976;72:146–155. doi: 10.1016/0006-291x(76)90972-4. [DOI] [PubMed] [Google Scholar]
- 26.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 27.Holm L, Sander C. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 28.Saenger W. In: Principles of Nucleic Acid Structure. Cantor C R, editor. New York: Springer; 1984. pp. 51–104. [Google Scholar]
- 29.Carson M. J Appl Crystallogr. 1991;24:958–961. [Google Scholar]
- 30.Smiley J A, Jones M E. Biochemistry. 1992;31:12162–12168. doi: 10.1021/bi00163a027. [DOI] [PubMed] [Google Scholar]
- 31.Wolfenden R, Snider M, Ridgway C, Miller B. J Am Chem Soc. 1999;121:7419–7420. [Google Scholar]
- 32.Fukuto J M, Jensen F R. Acc Chem Res. 1983;16:177–184. [Google Scholar]
- 33.Levine H L, Brody R S, Westheimer F H. Biochemistry. 1980;19:4993–4999. doi: 10.1021/bi00563a010. [DOI] [PubMed] [Google Scholar]
- 34.Miller B G, Traut T W, Wolfenden R. Bioorg Chem. 1998;26:283–288. [Google Scholar]
- 35.Smiley J A, Paneth P, O'leary M H, Bell J B, Jones M E. Biochemistry. 1991;30:6216–6223. doi: 10.1021/bi00239a020. [DOI] [PubMed] [Google Scholar]
- 36.Ehrlich J I, Hwang C-C, Cook P F, Blanchard J S. J Am Chem Soc. 1999;121:6966–6967. [Google Scholar]
- 37.Begley T P, Walts A E, Walsh C T. Biochemistry. 1986;25:7192–7200. doi: 10.1021/bi00370a064. [DOI] [PubMed] [Google Scholar]
- 38.Thauer R K, Klein A R, Hartmann G C. Chem Rev. 1996;96:3031–3042. doi: 10.1021/cr9500601. [DOI] [PubMed] [Google Scholar]