Abstract
The crystal structure of Escherichia coli ribosomal protein L25 bound to an 18-base pair portion of 5S ribosomal RNA, which contains “loop E,” has been determined at 1.8-Å resolution. The protein primarily recognizes a unique RNA shape, although five side chains make direct or water-mediated interactions with bases. Three β-strands lie in the widened minor groove of loop E formed by noncanonical base pairs and cross-strand purine stacks, and an α-helix interacts in an adjacent widened major groove. The structure of loop E is largely the same as that of uncomplexed RNA (rms deviation of 0.4 Å for 11 base pairs), and 3 Mg2+ ions that stabilize the noncanonical base pairs lie in the same or similar locations in both structures. Perhaps surprisingly, those residues interacting with the RNA backbone are the most conserved among known L25 sequences, whereas those interacting with the bases are not.
In Escherichia coli, the 120-nt 5S rRNA binds specifically to three proteins, L25, L18 and L5, forming a separate domain of the ribosome (1). Ribosomal protein L25 binds specifically to a portion of 5S rRNA called loop E, which contains seven non-Watson–Crick base pairs stabilized in the protein-free RNA by several magnesium ions. The structure of the loop E duplex in the absence of protein is significantly distorted from canonical A form RNA (2). Both the hydrogen-bond donors and acceptors that are presented in a widened minor groove and the backbone structure of loop E differ from A form RNA. Its distorted structure is stabilized by a “spine” of Mg2+ bound in the major groove. Further, the major groove of “helix IV,” which lies adjacent to loop E is likewise significantly widened, implying its potential accessibility to sequence-specific protein interactions. Biochemical protection, modification, and interference studies imply that L25 binds to the portion of 5S rRNA including the minor groove side of loop E and the adjacent major groove of helix IV (3–7).
The high-resolution crystal or solution structures of about 16 ribosomal proteins or fragments thereof have been established (8), including the solution NMR structure of protein L25 (9). The structures of these and other ribosomal proteins in the context of the ribosome, where they may all make some interactions with RNA, are beginning to emerge at low resolution (10–12). The solution structure of protein L25 uncomplexed with RNA shows two significantly disordered loops and a β-barrel domain with significant structural similarities to the anti-codon-binding domains of E. coli glutaminyl-tRNA synthetase (13).
Although aminoacyl-tRNA synthetases and sequence-specific DNA-binding proteins must discriminate among RNA and DNA substrates that have largely identical secondary and tertiary structures, ribosomal proteins might be expected to bind to RNA regions that have very distinctive structures as well as varied sequences. Consequently, the structural basis of RNA recognition by this category of proteins may include different features. Indeed, the very recent structure of ribosomal protein L11 bound to a 58-nt fragment of 23S rRNA shows a protein whose structure is complementary to a complicated and unique RNA structure (14, 15).
When L25 complexes with the portion of 5S rRNA containing loop E, its disordered loops assume specific conformations that are observed to interact with an RNA molecule whose structure is largely unchanged from its uncomplexed structure. The major groove of helix IV in the 5S rRNA fragment, which is enlarged relative to A form RNA by cross-strand purine stacks (2), narrows slightly around the α-helix that forms from the disordered protein loop on interaction with the RNA. The protein seems to be recognizing the specific non-A form shape of the RNA backbone as well as three non-Watson–Crick base pairs. Perhaps surprisingly, the most highly conserved amino acid residues among known L25 protein sequences interact with the 5S rRNA backbone. In contrast, those side chains seen interacting with bases are mostly not conserved but covary with the 5S rRNA sequence with which they interact in many cases.
Materials and Methods
Preparation of the L25-RNA Complex.
E. coli ribosomal protein L25 was overexpressed in E. coli BL21 (DE3/pLysS) cells under the control of a T7 promoter (B. Golden and V. Ramakrishnan, personal communication). Cells were disrupted by brief sonication. Cell lysate was clarified by passage over a DEAE-Sepharose column, and the flow-through was purified further by a combination of cation exchange (SP-Sepharose), gel filtration (Sephadex-G50), and Cibacron Blue chromatography. The selenomethionyl version of the L25 protein was prepared by using the procedure of Yang et al. (16).
RNA oligonucleotides were chemically synthesized by the Keck Oligonucleotide Synthesis Facility at Yale University and purified according to Correll et al. (17). Single-stranded RNAs were annealed at 65°C before they were combined with the L25 protein in a 1:1 ratio at 4°C (2, 18).
Crystallization and Data Collection.
Crystals were obtained at 19°C by using the sitting drop vapor diffusion method. The reservoir solution contained 20% (vol/vol) 2-methyl-2,4-pentandiol and 0.4 M unbuffered ammonium sulfate. The drops were composed of 6 μl of L25-RNA complex at 10 mg/ml and 4 μl of 20% (vol/vol) 2-methyl-2,4-pentandiol, 50 mM Na cacodylate (pH 6.0), 100 mM KCl, and 10 mM MgCl2. Cocrystals with protein containing selenomethionine were prepared by using the streak-seeding method.
For x-ray diffraction analysis, crystals were flash frozen in liquid propane. Both native and selenomethionine-containing crystals yielded data to 1.8-Å resolution and belonged to space group C2221. Native and multiwavelength anomalous dispersion (MAD) data sets were collected at beamlines X12C and X8C, respectively, of Brookhaven National Laboratory (Upton, NY). Data were reduced and scaled with the computer programs denzo and scalepack (19).
Structure Determination.
Crystal structure of the L25-RNA complex was solved by using the MAD method. The positions of three selenium sites were determined with solve (20). Density modification and phase extension to 2.3-Å resolution with solomon (21) produced an electron density map in which most of the protein and nucleic acid residues could be identified unambiguously with the program o (22). Native data from 20 Å to 1.8 Å (I/σ > 2) were used for structure refinement. Rounds of manual rebuilding, interspersed with torsion-angle simulated annealing and individual B factor refinement with the crystallography and NMR system (23), generated a model with a free R value of 22.5%. The model contains 1,572 nonhydrogen atoms, in addition to 242 waters and 5 magnesium ions (see Table 1). All figures were generated with ribbons (24), except for Fig. 1D, which was prepared with grasp (25).
Table 1.
Crystallographic data
| Data set | Wavelength, Å | Resolution, Å | Unique reflections | Completeness (last shell), % | I/σ (last shell) | Rsym (last shell), % |
|---|---|---|---|---|---|---|
| Native | 0.9806 | 50 −1.80 | 24,727 | 95.6 (77.2) | 32.2 (2.9) | 4.8 (31.3) |
| Edge | 0.9790 | 40 −1.80 | 48,921 | 99.8 (100) | 25.1 (4.1) | 5.3 (31.9) |
| MAD peak | 0.9786 | 40 −1.80 | 48,776 | 99.5 (100) | 21.1 (2.9) | 6.2 (38.7) |
| Remote 1 | 0.9770 | 40 −1.80 | 25,043 | 97.2 (99.1) | 18.4 (2.7) | 6.0 (38.7) |
| Remote 2 | 0.9160 | 40 −1.80 | 24,860 | 96.6 (98.9) | 27.5 (3.0) | 4.8 (26.2) |
| Overall MAD figure of merit: 0.64 (20–2.5Å) R factor: 20.7%; R free: 22.5% (20 − 1.8Å) | ||||||
| Ramachandran analysis | Most favored regions, % | Allowed regions, % | Generously allowed regions, % | Disallowed regions, % |
| (Procheck) | 95.2 | 2.4 | 1.2 | 1.2 |
Rsym = ∑|I − <I>|/∑I, where I is the intensity of each reflection. Figure of merit is defined as the cosine of estimated phase error. R factor = ∑|Fo − Fc|/∑Fo, where Fo and Fc are the observed and calculated structural factors, respectively. R free is the same as R factor but calculated with 10% of the reflections excluded from structure refinement.
Figure 1.
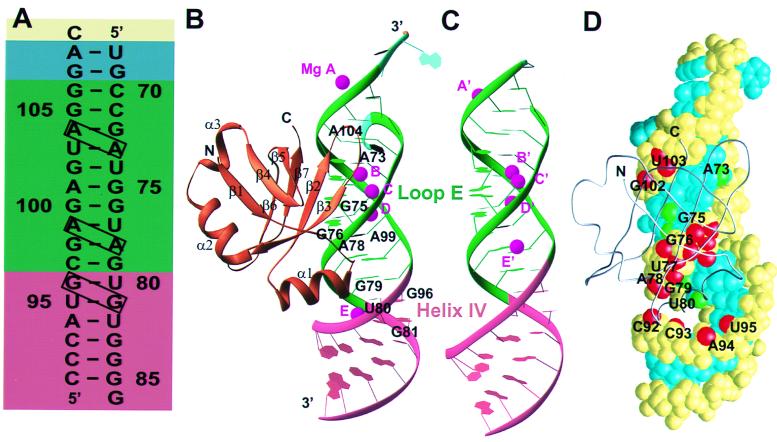
(A) The sequence of the 5S rRNA fragment that was cocrystallized with L25 with the cross-strand purine stacks boxed. (A–C) Helix I of 5S rRNA is drawn in yellow; linker in cyan; loop E in green; helix IV in magenta; magnesium ions in purple; and protein L25 in brown. (B) A ribbon representation of the L25-RNA complex. The β-strands are numbered β1 to β6 and the α-helices are α1 to α3. The five metal ions are MgA to E. (C) Ribbon diagram of the unbound 5S rRNA fragment whose structure is a composite of the loop E dodecamer structure and the structure of helix IV from fragment I (2). (D) A van der Waals surface representation of the 5S rRNA fragment complexed with a backbone representation of L25 protein (gray). Base atoms interacting with the protein are green; backbone atoms contacting protein are red; and the rest of the bases and backbone atoms are blue and yellow, respectively.
Results and Discussion
Overall Structure of the L25-RNA Complex.
Crystallographically suitable crystals of E. coli ribosomal protein L25 complexed with a fragment of 5S rRNA were obtained by complexing L25 with a number of chemically synthesized duplex RNA molecules, all of which contained the loop E sequences but whose lengths varied, and screening these complexes for their crystallization properties. An 18-base pair RNA duplex containing a single nucleotide 3′ overhang on both strands (Fig. 1A) produced cocrystals that diffract to better than 1.8-Å resolution. Although the two single nucleotides at each end of the duplex are self-complementary, they were not base paired in the crystal, as often happens in protein–DNA complexes. The cocrystals are orthorhombic space group C2221, with unit cell dimensions of 75.5, 76.6, and 95.1 Å; the cocrystals have a solvent content of 60%. The crystal structure was solved from an electron density map that was calculated by using data collected from cocrystals of RNA and L25 protein containing selenomethionines and phased by the four-wavelength MAD method. The free R factor of the refined structure to 1.8-Å resolution was 22.5%, whereas the working R factor was 20.7% (Table 1).
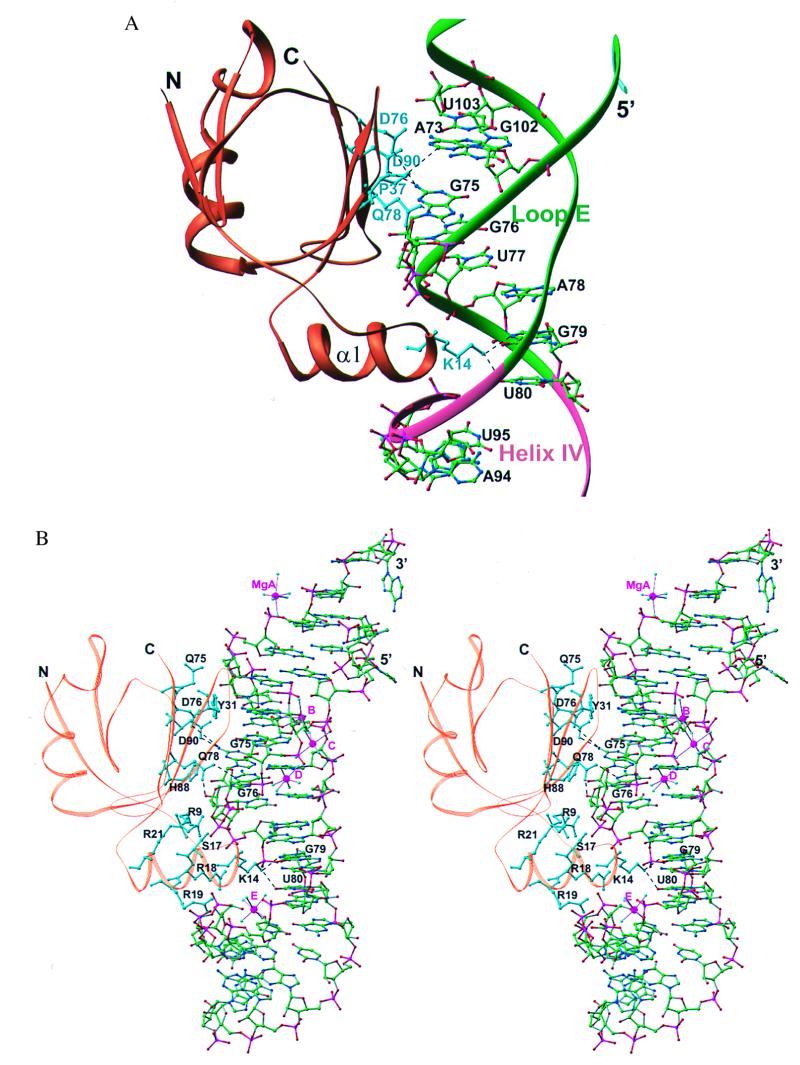
The L25 protein interacts with the minor groove side of loop E, inserting a pair of two-stranded antiparallel β-ribbons (β2, β3, β6, and β7) into the minor groove and the amino end of an α-helix into an adjacent major groove (Figs. 1B and 2; see also Fig. 6A). L25 consists of a seven-stranded β-barrel and three α-helices. The protein makes interactions with 10 of the 18 base pairs and buries ≈1,740 Å2 of RNA and protein from solvent accessibility as calculated by grasp (25). The nucleotides of 5S rRNA interacting with L25 as well as the protein residues seen interacting with RNA agree with those predicted by footprinting and modification interference experiments as well as with NMR studies (4, 9); thus, it seems likely that the interactions observed in this cocrystal structure closely resemble those occurring in solution.
Figure 2.
(A) A ribbon representation of the L25 complex with RNA (color coding as in Fig. 1B). The five amino acid residues interacting with bases are drawn in cyan, and those nucleotides that interact with protein are shown in all-atom representation. (B) A stereo representation of the complex with all atoms of the RNA shown along with the hydrated Mg2+ ions, protein backbone (brown), and those amino acid residues (cyan) making polar interactions with the RNA.
Figure 6.
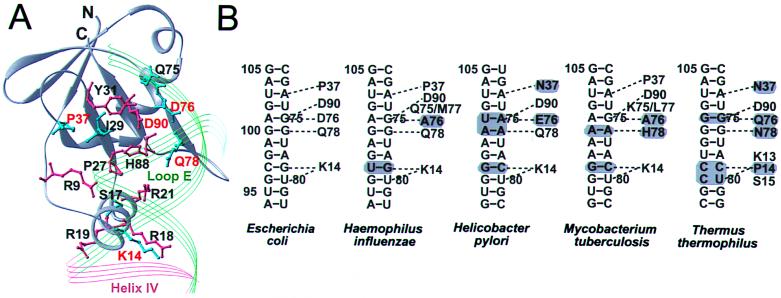
(A) The residues of L25 (gray ribbon) that are conserved among L25 sequences are magenta, and those that are not are cyan. The position of the RNA backbone is also indicated. (B) Sequences of loop E regions from various species and the corresponding residues from L25 that interact; gray shades indicate variations.
Changes in RNA and Protein Structures On Complex Formation.
In contrast with the HIV rev response element RNA and HIV TAR RNA, whose structures change extensively on binding peptides of rev and tat (26, 27), the structure of loop E remains largely unaltered on binding protein L25 (Fig. 1 B and C); 11 base pairs containing 516 atoms of uncomplexed loop E RNA superimpose on the corresponding atoms of the complexed RNA with an rms deviation of 0.42 Å. As is observed in the unbound 5S rRNA fragments (2), the overall shape of the complexed RNA is changed from A form by a pair of 3-base pair motifs in loop E. Each motif forms a cross-strand adenine stack, which distorts the sugar-phosphate backbone and narrows the major groove between the motifs. In the complexed RNA, the major groove of loop E is about 2 Å narrower than that of A form RNA, whereas the minor groove is expanded by 2 Å. In addition, the combination of a cross-strand guanine stack located in helix IV adjacent to a cross-strand adenine-stack widens the major groove by about 5 Å, less than in the uncomplexed RNA. On binding of the protein, there is a small change in helix IV whose helical axis becomes almost perpendicular to that of loop E. This structural change is accompanied by a narrowing of the major groove into which an α-helix has been inserted.
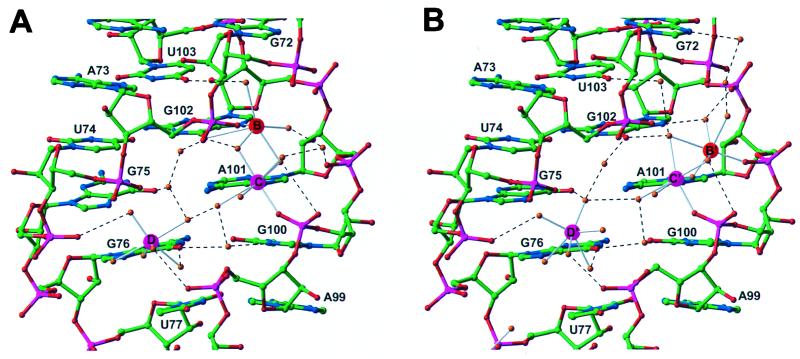
Although both the complexed and uncomplexed RNA bind five magnesium ions in the major groove, only two of these (C and D) are identically and a third (B) similarly positioned in the two RNAs (Figs. 1 B and C and 3). Metal ion A makes an intermolecular crystal contact, which differs in the two crystals, and the position of metal ion E is apparently influenced by the L25 α-helix binding in the major groove. Metal ion B moves slightly, making different inner sphere interactions with the RNA, possibly because of small differences in the RNA backbone conformation resulting from protein binding.
Figure 3.
The three Mg2+ ions bound in the minor groove of loop E are shown as they occur in the L25 complex in A and in the uncomplexed RNA in B. The positions of Mg2+ ions C and D are identical in both, and the position of B is similar.
Although a quantitative comparison between the L25 protein structure complexed to the RNA and its uncomplexed structure cannot be done, because the coordinates of the uncomplexed structure (9) are unavailable, differences exist in the regions that interact with the RNA. The major difference occurs in α-helix α1 (Fig. 1B), which interacts in the major groove of the RNA. This region (residues 14–23) is unstructured in the absence of 5S rRNA. Thus, the RNA provides part of the template that stabilizes the formation of the helical structure.
RNA Recognition by L25 Protein.
The noncanonical base pairs in loop E, which are stabilized by divalent metal ions, provide a nonstandard shape and a complex, nonstandard hydrogen-bonding surface in the minor groove, features that are recognized by the L25 protein. Protein side chains (n = 10) interact with the phosphoribose backbone and are complementary in shape to the considerably widened minor groove of loop E and the substantially wider adjacent major groove. Unlike proteins recognizing duplex DNA or proteins discriminating among similarly shaped tRNA molecules, ribosomal proteins such as L25 can and do identify their target binding sites by the unique shape that they assume.
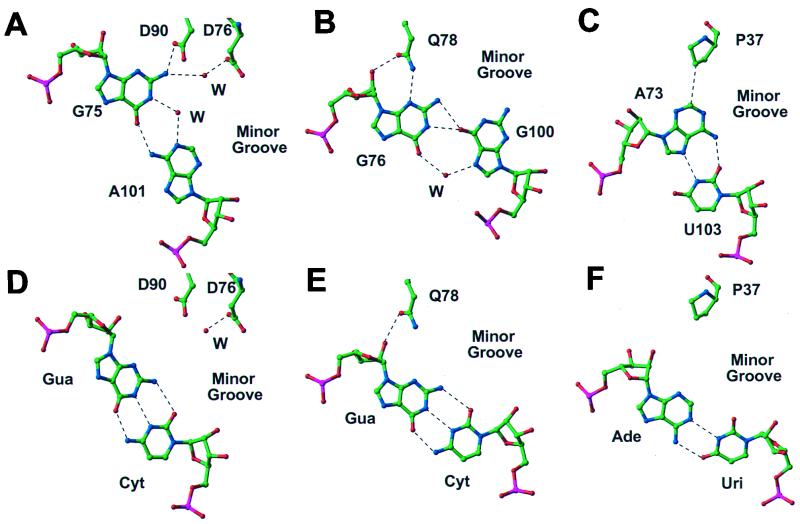
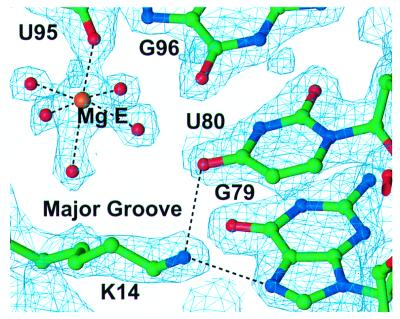
There are five protein side chains making direct or water-mediated contacts with base pairs, one in the major groove and four in the minor groove, and those in the minor groove require the presence of non-Watson–Crick base pairs (Figs. 4 and 5). Although D90 and D76 interact with the N2 of G75, which is base paired with A101, the presence of a standard Watson–Crick or wobble (GU) base pair would put the N2 in a different position, thereby preventing the interaction with the recognition faction (Fig. 4 A and D). Similarly, Q78 interacts with the N3 of G76 and its ribose hydroxyl (Fig. 4B). Once again, G76, which is base paired with G100, if base paired in a standard Watson–Crick or wobble (GU) base pair would be incorrectly positioned to make this recognition interaction with N3 (Fig. 4E). The same is true for A73 whose C2 atom contacts P37 through van der Waals interactions (Fig. 4 C and F). In contrast, K14 interacts with adjacent U80 and G79, which are involved in Watson–Crick base pairing (Fig. 5). However, the major groove is accessible to the α-helix on which K14 resides, only because it has been widened by cross-strand purine stacks.
Figure 4.
Base-specific interactions in the minor groove. The observed direct or water-mediated interactions between the protein and three base pairs are shown in A–C, whereas the lack of interaction expected if GC or AU base pairs were substituted is shown in D–F.
Figure 5.
Electron density for a specific protein interaction with a base in the major groove along with a hydrated Mg2+ ion. A simulated-annealed (2Fo–Fc) composite omit map calculated at 1.8-Å resolution and contoured at 1.5 σ is superimposed on the final model and shows Lys-14 interacting with two RNA bases in the major groove of helix IV.
Divalent metal ions play an important role in stabilizing this unique structure of loop E that is being specifically recognized by protein L25. NMR studies have shown that, in the absence of divalent metal ions and in the absence of the protein, loop E assumes a less well structured conformation (28). Thus, it seems that the metal ions and the protein act synergistically to stabilize the same RNA structure, and both may play a coordinated role in forming the RNA architecture that is necessary for building the ribosome structure from RNA in vivo.
Although the structure of L25 protein shows a striking similarity to the anti-codon-binding domains of E. coli glutaminyl-tRNA synthetase (13), as previously observed (9), these two proteins interact with their RNA substrates in completely unrelated ways by using different parts of the structures. Further, the synthetase domain binds and recognizes primarily the splayed-out, unpaired bases of the anticodon loop. L25, on the other hand, recognizes duplex RNA. It remains possible, however, that in the context of the ribosome, L25 may make additional interactions with other parts of 5S rRNA (6) or with 23 S rRNA, and these interactions might be related to those made by the synthetase domains. The 5-Å resolution structure of the 50S ribosomal subunit shows many ribosomal proteins making interactions with multiple RNA segments (10).
Conservation and Variation in L25 and 5S rRNA Sequences.
Examination of homologous L25 protein sequences and the sequences of the 5S rRNA to which they bind shows an unexpected result: the protein side chains that interact with the bases and the 5S rRNA sequences with which they interact are mostly not conserved, whereas the protein residues that bind the phosphoribose backbone are mostly conserved (Fig. 6A). Of the five residues that bind to the bases, only one, D90, is conserved. In contrast, of the 10 side chains interacting with the backbone, only 3, Q75, I29, and S17 (Fig. 6A), are not conserved. This pattern of sequence conservation differs from that of the sequence-specific DNA-binding proteins, such as the E. coli methionine repressor and catabolite gene activator protein (29, 30). In these instances, both the protein side chains that directly recognize the DNA sequence as well as the bases with which they interact are highly conserved among the various bacterial organisms.
The nonconserved protein side chains of L25 involved in base recognition and the bases of 5S rRNA with which they interact, seem to covary among the species that have been examined (Fig. 6B). Approximate model building of the 5S rRNA and L25 molecules of these various bacterial homologues suggests that the changes in the protein side chains can accommodate the changes in the base sequence allowing an altered form of sequence-specific recognition to occur.
Why is it that an aspect of L25 function as seemingly important as direct base-specific recognition is not conserved among the bacterial species? Perhaps L25 protein is a latecomer on the ribosome evolutionary scene. If 5S rRNA sequences within the loop E region had already diverged among the various species before the introduction of L25 into the ribosome, then this protein would have to accommodate to the different sequences in each of the species. It may be relevant to note that an L25 homologue is not present in the archaeal ribosome or in all of the eubacterial species of ribosomes (31). An alternative explanation for the covariation of L25 side chains and the bases with which they interact is the possibility that sequence-specific interaction is not disrupted by a mutation of either the protein or RNA-interacting residues such that function is not lost by changes in either. If that is the case, then the sequence of the protein and the RNA involved in the interaction could simply drift.
Acknowledgments
We thank P. B. Moore for discussions and comments on this paper, N. Ban and J. Berendzen for help in data collection, and V. Ramakrishnan for the clone that overproduces protein L25. We also wish to thank N. Ban, J. Wang, J. Ippolito, P. Nissen, S. Kamtekar, G. Cheetham, B. Golden, J. Warren, S. Basu, J. Pata, and C. Correll for advice and discussions. This work was supported in part by National Institutes of Health Grant GM-22778 to T.A.S.
Abbreviation
- MAD
multiwavelength anomalous dispersion
Note Added in Proof
As this article was being submitted, a paper describing the solution NMR structure of E. coli L25-RNA complex (32) was published. Although the NMR structure is similar to the crystal structure described here, some differences are observed in both the protein and RNA structures, with rms deviations of 2.2 Å for both protein Cα and RNA phosphorus atoms between the equivalent portions of the crystal and solution structures. These differences may arise from there being no divalent metal ions included in the NMR studies and a shorter 5S rRNA fragment sequence containing an artificial tetra-loop being used in the solution structure determination.
Footnotes
Data deposition: The atomic coordinates and the diffraction amplitudes have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1DFU and NDB ID code PR0018).
References
- 1.Chen-Schmeisser U, Garret R A. FEBS Lett. 1977;74:287–291. doi: 10.1016/0014-5793(77)80866-1. [DOI] [PubMed] [Google Scholar]
- 2.Correll C C, Freeborn B, Moore P B, Steitz T A. Cell. 1997;91:705–712. doi: 10.1016/s0092-8674(00)80457-2. [DOI] [PubMed] [Google Scholar]
- 3.Garret R A, Noller H F. J Mol Biol. 1979;132:637–648. doi: 10.1016/0022-2836(79)90379-6. [DOI] [PubMed] [Google Scholar]
- 4.Huber P W, Wool I G. Proc Natl Acad Sci USA. 1984;81:322–326. doi: 10.1073/pnas.81.2.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ciesiolka J, Lorenz S, Erdmann V A. Eur J Biochem. 1992;204:575–581. doi: 10.1111/j.1432-1033.1992.tb16670.x. [DOI] [PubMed] [Google Scholar]
- 6.Shpanchenko O V, Zvereva M L, Dontsova O A, Nierhaus K H, Bogdanov A A. FEBS Lett. 1996;394:71–75. doi: 10.1016/0014-5793(96)00872-1. [DOI] [PubMed] [Google Scholar]
- 7.Leontis N B, Westhof E. RNA. 1998;4:1134–1153. doi: 10.1017/s1355838298980566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramakrishnan V, White S. Trends Biochem Sci. 1998;23:208–212. doi: 10.1016/s0968-0004(98)01214-6. [DOI] [PubMed] [Google Scholar]
- 9.Stoldt M, Wohnert J, Gorlach M, Brown L. EMBO J. 1998;17:6377–6384. doi: 10.1093/emboj/17.21.6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ban N, Nissen P, Hansen J, Capel M, Moore P B, Steitz T A. Nature (London) 1999;400:841–847. doi: 10.1038/23641. [DOI] [PubMed] [Google Scholar]
- 11.Cate J H, Yusupov M M, Yusupova G Z, Earnest T N, Noller H F. Science. 1999;285:2095–2104. doi: 10.1126/science.285.5436.2095. [DOI] [PubMed] [Google Scholar]
- 12.Clemons W M, Jr, May J L C, Wimberly B T, McCutcheon J P, Capel M S, Ramakrishnan V. Nature (London) 1999;400:833–840. doi: 10.1038/23631. [DOI] [PubMed] [Google Scholar]
- 13.Rould M A, Perona J J, Soll D, Steitz T A. Science. 1989;246:1135–1142. doi: 10.1126/science.2479982. [DOI] [PubMed] [Google Scholar]
- 14.Wimberly B T, Guymon R, McChutcheon J P, White S W, Ramakrishnan V. Cell. 1999;97:491–502. doi: 10.1016/s0092-8674(00)80759-x. [DOI] [PubMed] [Google Scholar]
- 15.Conn G L, Draper D E, Lattman E E, Gittis A G. Science. 1999;284:1171–1174. doi: 10.1126/science.284.5417.1171. [DOI] [PubMed] [Google Scholar]
- 16.Yang W, Hendrickson W A, Kalman E T, Crouch R J. J Biol Chem. 1990;265:13553–13559. [PubMed] [Google Scholar]
- 17.Correll C C, Munishkin A, Chan Y-L, Ren Z, Wool I G, Steitz T A. Proc Natl Acad Sci USA. 1998;95:13436–13441. doi: 10.1073/pnas.95.23.13436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abdel-Meguid S S, Moore P B, Steitz T A. J Mol Biol. 1983;171:207–215. doi: 10.1016/s0022-2836(83)80353-2. [DOI] [PubMed] [Google Scholar]
- 19.Otwinowski Z, Minor W. In: Methods in Enzymology. Carter C W Jr, Sweet R M, editors. Vol. 276. San Diego: Academic; 1997. pp. 307–325. [DOI] [PubMed] [Google Scholar]
- 20.Terwilliger T C, Berendzen J. Acta Crystallogr D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abrahams J P, Leslie A G W. Acta Crystallogr D. 1998;54:905–921. [Google Scholar]
- 22.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 23.Brünger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 24.Carson M. J Appl Crystallogr. 1991;24:958–961. [Google Scholar]
- 25.Nicolls A, Bharadwaj R, Honig B. Biophys J. 1993;64:A166. (abstr.). [Google Scholar]
- 26.Peterson R D, Feigon J. J Mol Biol. 1996;264:863–877. doi: 10.1006/jmbi.1996.0683. [DOI] [PubMed] [Google Scholar]
- 27.Puglisi J D, Chen L, Blanchard S, Frankel A D. Science. 1995;270:1200–1203. doi: 10.1126/science.270.5239.1200. [DOI] [PubMed] [Google Scholar]
- 28.Leontis N B, Ghosh P, Moore P B. Biochemistry. 1986;25:7389–7392. doi: 10.1021/bi00371a021. [DOI] [PubMed] [Google Scholar]
- 29.Somers W S, Phillips S E V. Nature (London) 1992;359:387–393. doi: 10.1038/359387a0. [DOI] [PubMed] [Google Scholar]
- 30.Schultz S C, Shields G C, Steitz T A. Science. 1991;253:1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- 31.Moore P B. In: Ribosomal RNA, Structure, Evolution, Processing, and Function in Protein Synthesis. Zimmermann R A, Dahlberg A E, editors. Boca Raton, FL: CRC; 1996. pp. 199–236. [Google Scholar]
- 32.Stoldt M, Wöhnert J, Ohlenschläger O, Görlach M, Brown L. EMBO J. 1999;18:6508–6521. doi: 10.1093/emboj/18.22.6508. [DOI] [PMC free article] [PubMed] [Google Scholar]