Abstract
The DNA dodecamer CATGGGCCCATG in a crystal structure of resolution 1.3 Å has a conformation intermediate between A and B DNA. This trapping of a stable intermediate suggests that the A and B DNA families are not discrete, as previously believed. The structure supports a base-centered rather than a backbone-centered mechanism for the A ↔ B transition mediated by guanine tracts. Interconversion between A and B DNA provides another means for regulating protein–DNA recognition.
The right-handed B helix is the dominant biological conformation of DNA, but crystal structures of protein–DNA complexes have revealed the duplex to be surprisingly deformable (1). For example, catabolite activator protein bends its operon by 90° (2), and the TATA-binding protein produces severe bending in its cognate site and induces a conformation with some features resembling A DNA (1, 3–6). Indeed, the deformability of DNA is an important determinant of its ability to interact with proteins (7). In protein–DNA complexes, A- tracts, defined as runs of four or more AT base pairs without a disruptive pyrimidine-purine TA step, are nearly always straight and unbent (8, 9). This resistance to bending is a structural feature that proteins can recognize, as is the readiness to bend shown by pyrimidine-purine steps (TA, CG, CA/TG; refs. 1 and 7). Fiber diffraction and solution studies have shown that G- tracts, or runs of four of more GC base pairs, also have special structural properties. G-tracts favor the A DNA helix conformation and, depending on the local water activity, can induce a B-to-A DNA transition (10–13). Sequences with long G tracts usually crystallize as A DNA (14, 15). The decamer CATGGCCATG with a short 4-bp G-tract crystallizes as B DNA (16). In contrast, the present dodecamer CATGGGCCCATG, with a 6-bp G tract has more of an A DNA character and, in fact, is the first DNA oligomer to crystallize in a conformation intermediate to A and B DNA. This suggests that the A ↔ B transition is readily accessible to certain DNA sequences and can be another mode for governing protein–DNA interactions.
Methods
Deoxyoligonucleotides were synthesized by conventional solid-phase phosphoramidite methods. Dodecamers were purified by anion exchange liquid chromatography with a KCl gradient and desalted by reverse-phase liquid chromatography. Crystals were grown at 4°C by vapor diffusion in sitting drops containing 2.3 mM duplex DNA, 120 mM Ca acetate, 4.8 mM spermine, 60 mM Na cacodylate buffer at pH 7.0, and 9.5% (vol/vol) 2-methyl-2,4-pentanediol against a reservoir solution of 25% (vol/vol) 2-methyl-2,4-pentanediol. Crystals were frozen in liquid nitrogen during data collection. Diffraction data for the native crystal were collected at the National Synchrotron Light Source at Brookhaven National Laboratory with 0.978-Å radiation. Diffraction data for the iodinated crystal were collected on a Rigaku R axis IV imaging plate detector with CuKα radiation. The crystals possess P41212 symmetry with one duplex in the asymmetric unit. Data were processed with denzo and scalepack (17). The iodine sites were found by solve and by manual inspection of the Patterson maps (18). Phasing with single isomorphous replacement and anomalous scattering was done by mlphare (19). Density modification and phase extension were performed by the program dm (20). Rigid-body refinement followed by simulated annealing with maximum likelihood treatment of experimental phases was done with cns (21). After R = 27.0%, refinement continued with shelx-97 (Sheldrick, G. M.) with restrained anisotropic treatment of atomic thermal factors as recommended by parvati (22). Water molecules were added to peaks above 3σ in the Fo-Fc map or 1.5σ in the 2Fo-Fc map, which showed reasonable geometry to other potential hydrogen bond donor/acceptors and which decreased Rfree on addition. Helical parameters were calculated with freehelix (1).
Results and Discussion
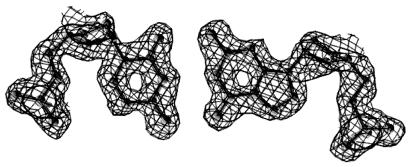
The dodecamer CATGGGCCCATG, which for brevity will be termed the G3C3 helix, was crystallized under mild conditions typical of B DNA rather than the dehydrating conditions that favor A DNA. Crystals diffract to 1.3 Å, and an iodinated derivative plus anomalous scattering measurements allowed direct experimental determination of phases. This process eliminated any prior assumptions as to helix type, which are needed in the molecular replacement method. Fig. 1 shows the high quality of the experimentally phased map before fitting and refinement of the model. Data on the fully refined structure are listed in Table 1.
Figure 1.
A 1.55-Å electron-density map of a typical base pair after phasing by single isomorphous replacement plus anomalous scattering and density modification but before model building and refinement. The skeleton of the final refined model has been superimposed for reference.
Table 1.
Crystal structure analysis of the CATGGGCCCATG dodecamer
| Parameter | CATGGGCCCATG | CATGGGCC(IC)ATG |
|---|---|---|
| Space group | P41212 | P41212 |
| Unit cell dimensions, Å | a = b = 40.2, c = 77.3 | a = b = 40.3, c = 78.1 |
| Resolution range, Å | 20.0–1.3 | 20.0–2.3 |
| Observations | 128,129 | 75,985 |
| Unique reflections | 16,111 | 5,461 |
| Completeness, % | 98.6, 90.0* | 100, 100* |
| I/σ | 29.5, 7.6* | 50.0, 13.3* |
| Rsym† | 7.0, 17.3* | 6.4, 20.1* |
| Rcullis, centric‡ | 0.47 | |
| Phasing power, centric/acentric§ | 2.07/2.39 | |
| Rcrys¶ | 17.2 | |
| Rfree¶ | 20.0 | |
| Bond lengths rms deviation from ideal, Å | 0.007 | |
| Bond angles rms deviation from ideal, Å | 0.016 |
*Values for highest resolution shell of data: 1.35–1.30 Å for native and 2.38–2.30 Å for the derivative.
†Rsym = Σ|I − 〈I〉|/ΣI.
‡Rcullis, centric = Σcentric∥Fph| − |Fp∥/Σcentric|Fph − Fp|.
§Phasing power = [ΣFh(calc)2/(ΣFph(obs) − Fph(calc))2]1/2.
¶Rcrys = 100 (Σ|Fobs − Fcalc|)/(ΣFobs). Rfree is calculated like Rcrys for a random 5% of the reflections.
As Table 2 indicates, the G3C3 helix shares archetypal traits of both A DNA and B DNA, with which this helix is compared in Fig. 2. Typically, A and B DNA differ in the inclination of base pairs to the helical axis, lateral displacement of base pairs away from the helical axis, width and depth of the minor groove, deoxyribose ring conformation, and crystal packing (9, 14, 15, 19, 23).
Table 2.
Structural features of CATGGGCCCATG and their relationships to A and B DNA
| Feature | B DNA | G3C3 helix | A DNA | Greatest similarity |
|---|---|---|---|---|
| Normal vector plot | Around origin | Around origin | Writhed curve | B |
| Mean parameters* | ||||
| Roll, degree | +1.6 | +2.5 | +10.0 | B |
| Inclination, degree | +3.4 | +5.5 | +20.1 | B |
| X displacement, Å | −0.1 | −2.9 | −4.5 | A/B |
| Minor groove width, ņ | 6.2 | 9.5 | 10.0 | A |
| Helical twist, degree | 35.6 | 31.8 | 30.4 | A |
| Slide, Å | +0.4 | −1.6 | −1.7 | A |
| Sugar puckering | Broad distribution around C2′-endo | C3′-endo | C3′-endo | A |
| Crystal packing | Parallel stacks of helices | A-like | Terminal base pairs against wall of neighboring minor groove | A |
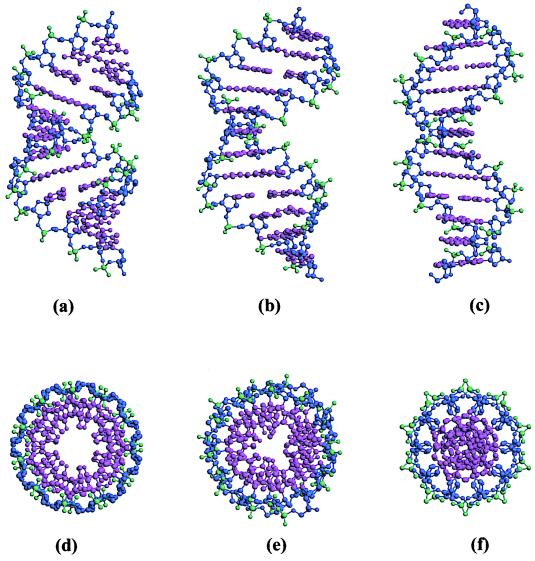
Figure 2.
Side and top views to the same scale of ideal A DNA (a and d), G3C3 or CATGGGCCCATG (b and e), and ideal B DNA (c and f). Ideal structures are taken from fiber diffraction data (10).
In B DNA, the base pairs are roughly perpendicular to the helical axis, and normal vectors to these base pairs cluster tightly around the axis (Figs. 2c and 3). In contrast, base pairs in A DNA are inclined 15–20° to the overall helical axis and writhe around that axis as shown by sweeping ellipses in the normal vector plot (Figs. 2a and 3). A side view of the G3C3 helix (Fig. 2b) and its normal vector plot (Fig. 3) show that, in this respect, the structure is B-like. However, the view down the helical axis (Fig. 2e) shows an A-like hollow through the center of the column produced by displacement of base pairs off-axis. Fig. 4 shows that the X displacement, or shift of base pairs along their short axis in the direction of the minor groove, averages ≈−3 Å, intermediate between the X displacement expected for B DNA (≈0 Å) and for A DNA (−4 to −6 Å). Fig. 2 d–f also shows that the central “hole” in G3C3 is larger than that for B but smaller than that for A.
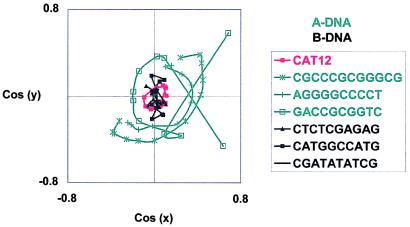
Figure 3.
Normal vector plots of G3C3 (red, CAT12) as well as several typical A DNA (green) and B DNA (black) oligomers. These oligomers were selected from high-quality crystal structures in the Nucleic Acid Database (http://ndbserver.rutgers.edu/). Note the clustering of black and red curves around the origin, which indicates straight helices with base pairs perpendicular to the helical axis. Note also the green arcs, which show the writhe typical of A DNA.
Figure 4.
X displacement of base pairs of G3C3 (red, CAT12) as well as representative A DNA (green) and B DNA (black) helices. Positive X displacement is the shift of a base pair along its minor axis in the direction of the major groove (39). Note that the X displacement of G3C3 is intermediate between the near zero displacement of B DNA and the large negative displacement of A DNA.
The offset of base pairs produces changes in dimensions of the major and minor grooves. Because base pairs are displaced toward the minor groove, A DNA has a wider but shallower minor groove but a deeper but narrower major groove than B DNA. Changes in major and minor groove dimensions provide another means for regulating the accessibility of DNA to proteins. Most proteins bind DNA via the major groove, and the narrower major groove of A DNA suggests that that its conformation is less biologically active. Crystal structures of A and B DNA oligomers show considerable variation from canonical, fiber diffraction-derived geometry. Nevertheless, groove widths of G3C3 are intermediate to the width in ideal A and B DNA.
With respect to sugar conformation, the verdict, “A DNA: C3′-endo; B DNA: C2′-endo” has become a mantra analogous to “Four legs good; two legs bad” from George Orwell's Animal Farm. But like that slogan, the sugar conformation mantra needs revision, because it ignores what we see in the crystallographic data. Whereas A DNA crystal structures show little deviation from C3′-endo sugar puckering (14, 15), B DNA structures have a broad distribution of sugar puckers centered on C2′-endo but extending from C4′-exo to C3′-endo and beyond (9). Indeed, this greater flexibility of the B helix over A may make the B helix a better candidate for recognition by other macromolecules. At 1.3-Å resolution, the sugar puckering of all rings in G3C3 is clear. Except for one sugar of a terminal nucleotide of the duplex (terminal nucleotides enjoy a greater range of motion), which has a C2′-endo pucker, all sugars of G3C3 cluster tightly in the C3′-endo region. In this regard, G3C3 resembles A DNA.
Crystals of G3C3 are not isomorphous with any previous oligonucleotide structure, whether A, B, or Z DNA. Crystalline B DNA dodecamers typically form parallel stacks of helices with overlapping ends. In A DNA crystals, the terminal base pairs of one helix pack against one wall of the broad minor groove of neighboring helices, and G3C3 shares this packing mode. This arrangement leaves large solvent channels between duplexes, and the solvent content of the G3C3 crystals is a high 60%. Much of this solvent is disordered and not localized. Only 93 water molecules and no ions or spermine molecules were located in the 1.3-Å electron density map, which is atypical for diffraction data of such high resolution and quality.
The G3C3 structure is compared with ideal, fiber diffraction-derived A and B DNA in Table 2. This structure is B-like in the inclination of base pairs, which are effectively perpendicular to the helical axis, and in the small mean roll from one base pair to the next. The structure is A-like in twist, slide, sugar pucker, and minor groove width, and intermediate with respect to X displacement. Because of its wider minor groove, G3C3 packs like A DNA in crystals. It is pointless to contend whether crystal or solution studies identify the “true” structural features of a given DNA sequence; these studies provide different snapshots of the range of conformations adaptable by a selectively flexible molecule. Indeed, the vaunted “tyranny of the lattice” should be appreciated as a source of information about molecular deformability (24).
The G3C3 crystal structure implies that an intermediate state between A and B DNA is readily accessible to short G-tracts. Independent theoretical studies have predicted that the thermodynamic barrier between A and B forms for A-favoring G- tracts in solution is no more than 0.2 kcal/mol (13, 25). Electrostatic repulsion between stacked GC base pairs in the B DNA form creates a destabilizing tension that increases cooperatively with G-tract length (26). In the present case, addition of just two more GC base pairs to the B DNA decamer CATGGCCATG led to a dodecamer that crystallized as an A/B DNA intermediate. Still longer G-tracts clearly favor an A helix (Fig. 3). The trapping of an A/B DNA intermediate in the G3C3 crystal challenges results from CD spectroscopic experiments that suggest that the A ↔ B transition occurs abruptly between two discrete states (11, 12). However, CD absorbance may be insensitive to the presence of AB intermediates containing base-pair inclination angles similar to those of B DNA, because a major component of the CD signal depends on base-pair inclination (27). CD measurements may be detecting only one aspect of the B-to-A transition.
Our results corroborate ideas that Calladine and Drew proposed in 1984 in a seminal paper entitled, “A base-centered explanation of the B-to-A transition in DNA” (ref. 28; see also ref. 29). They proposed that the transition is driven by base-pair stacking and that changes in sugar pucker were only a consequence rather than a cause of the transition. Using wooden models, they demonstrated that a B-to-A transition could be induced by changing only two base-stacking variables: (i) increasing the roll (contrarotation of adjacent base pairs about their long axes) from near 0 to +12° and (ii) changing the slide (motion of a base pair relative to another along their long axes) from near 0 to −1.5 Å, as schematized in Fig. 5. As they expressed it,
Figure 5.
Calladine–Drew base-centered mechanism for the B-to-A helix transition. (a) B DNA with neither slide nor roll. (b) B DNA with addition of a −1.5-Å slide. (c) B DNA with addition of a +12° roll. (d) A DNA, resulting from addition of both slide and roll to B DNA. The G3C3 helix corresponds to the helix in b and can be regarded as a B DNA helix to which slide has been added but not roll. Conversely, it can be thought of as A DNA from which roll has been removed. Figures were created with nustar (X.-J. Lu and W. K. Olson, unpublished work).
… neither roll nor slide by itself is sufficient to account for the observed change in helical structure; but roll and slide in combination do a good job of describing the effect, without the need to invoke a change in twist. All of the outward appearances so commonly noted during the B-to-A transition: the shift of base pairs away from an imaginary central axis, the tilt of base pairs from horizontal, and an overall shortening of the assembly follow primarily from changes in just two of the base-step parameters, roll and slide. (28)
Modern molecular dynamics simulations support the Calladine–Drew theory, suggesting that the transition involves a generalized change in many parameters (30–32) and that “… one parameter alone, such as sugar repuckering… does not necessarily drive the transition” (31).
The G3C3 helix bears out the Calladine–Drew ideas precisely. Mean values of roll and slide for G3C3, after deleting the helix-terminal steps, are +2.5°/−1.6 Å. Corresponding values for the typical B DNA helix are +1.6/+0.4 Å and for A DNA are +10.0/−1.7 Å. Hence, the G3C3 structure essentially corresponds to a halfway point in the Calladine–Drew B-to-A transition, created by using slide but not roll, as shown in Fig. 5b.
Considerable attention has been paid to runs of adenine, A- tracts, whose primary characteristics are (i) an optionally narrowed minor groove produced by the large propeller twist AT base pairs, which are held together by only two hydrogen bonds, and (ii) helix rigidity produced by the sawhorse-like stacking of the propellered base pairs. In contrast, the primary characteristics of this G-tract intermediate seem to be (i) a wide minor groove by virtue of lack of propeller twist in GC base pairs, which are kept flatter by their three hydrogen bonds, (ii) absence of roll because of the significant overlap of the stacked base pairs, and (iii) appreciable slide of adjacent flat base pairs along their long axes. The result is a helix essentially blocked halfway through its B ↔ A conversion.
Just as the rigidity of A-tracts has been used extensively for protein–DNA recognition, the A ↔ B DNA deformability of G- tracts could also provide a means of regulating protein activity. Crystallographic studies have shown that DNase I can bind to both an A DNA sequence and a B DNA sequence but can cleave only the B DNA sequence (33–35). Similarly, a run of six guanines in a polypurine tract is the signal for termination of digestion of a DNA/RNA hybrid helix by RNase H of HIV (36). Otwinoski et al. (37) concluded that the capability of a particular sequence to adopt the A DNA-like conformation observed in the crystal structure of the protein–DNA complex determines the binding specificity of trp repressor. More generally, the introduction of an α-helix into the major groove induces broad conformational changes characteristic of a B-to-A conversion (38). A ↔ B helix interconversion is a phenomenon that raises important biological and chemical questions and deserves further study.
Acknowledgments
We thank D. Cascio for collecting the synchrotron data set, K. Goodwill and Thang K. Chiu for help with crystallographic computations, and J. Šponer for stimulating discussions. This work was supported by National Institutes of Health Program Project Grant GM31299.
Footnotes
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1DC0), and in the Nucleic Acid Database (accession code BD0026).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.040571197.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.040571197
References
- 1.Dickerson R E. Nucleic Acids Res. 1998;26:1906–1926. doi: 10.1093/nar/26.8.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schultz S C, Shields G C, Steitz T A. Science. 1991;253:1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- 3.Kim Y, Geiger J H, Hahn S, Sigler P B. Nature (London) 1993;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- 4.Juo Z S, Chiu T K, Leiberman P M, Baikalov I, Berk A J, Dickerson R E. J Mol Biol. 1996;261:239–254. doi: 10.1006/jmbi.1996.0456. [DOI] [PubMed] [Google Scholar]
- 5.Kim J L, Nikolov D B, Burley S K. Nature (London) 1993;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- 6.Guzikevich-Guerstein G, Shakked Z. Nat Struct Biol. 1996;3:32–37. doi: 10.1038/nsb0196-32. [DOI] [PubMed] [Google Scholar]
- 7.Dickerson R E, Chiu T K. Biopolymers. 1998;44:361–403. doi: 10.1002/(SICI)1097-0282(1997)44:4<361::AID-BIP4>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 8.Sprous D, Young M A, Beveridge D L. J Mol Biol. 1999;285:1623–1632. doi: 10.1006/jmbi.1998.2241. [DOI] [PubMed] [Google Scholar]
- 9.Dickerson R E. In: Oxford Handbook of Nucleic Acid Structure. Neidle S, editor. Oxford: Oxford Sci.; 1999. pp. 145–197. [Google Scholar]
- 10.Arnott S. In: Oxford Handbook of Nucleic Acid Structure. Neidle S, editor. Oxford: Oxford Sci.; 1999. pp. 1–38. [Google Scholar]
- 11.Ivanov V I, Minchenkova L E, Schyolkina A K, Poletayev A I. Biopolymers. 1973;12:89–110. doi: 10.1002/bip.1973.360120109. [DOI] [PubMed] [Google Scholar]
- 12.Ivanov V I, Minchenkova L E, Minyat E E, Frank-Kamenetskii M D, Schyolkina A K. J Mol Biol. 1974;87:817–833. doi: 10.1016/0022-2836(74)90086-2. [DOI] [PubMed] [Google Scholar]
- 13.Minchenkova L E, Schyolkina A K, Chernov B K, Ivanov V I. J Biomol Struct Dyn. 1986;4:463–476. doi: 10.1080/07391102.1986.10506362. [DOI] [PubMed] [Google Scholar]
- 14.Wahl M C, Sundaralingam M. In: Oxford Handbook of Nucleic Acid Structure. Neidle S, editor. Oxford: Oxford Sci.; 1999. pp. 117–144. [Google Scholar]
- 15.Wahl M C, Sundaralingam M. Biopolymers. 1997;44:45–63. doi: 10.1002/(SICI)1097-0282(1997)44:1<45::AID-BIP4>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 16.Goodsell D S, Kopka M L, Cascio D C, Dickerson R E. Proc Natl Acad Sci USA. 1993;90:2930–2934. doi: 10.1073/pnas.90.7.2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Otwinowski Z. In: Data Collection and Processing. Sawyer L, Isaacs N, Bailey S, editors. Daresbury, U.K.: Sci. Eng. Res. Counc.; 1993. pp. 56–63. [Google Scholar]
- 18.Terwilliger T C, Berendzen J. Acta Crystallogr D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berman H M. Biopolymers. 1997;44:23–44. doi: 10.1002/(SICI)1097-0282(1997)44:1<23::AID-BIP3>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 20.Cowtan K. Joint CCP 4 ESF-EACBM Newsl Protein Crystallogr. 1994;31:34. [Google Scholar]
- 21.Brunger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 22.Merritt E A. Acta Crystallogr D. 1999;55:1109–1117. doi: 10.1107/s0907444999003789. [DOI] [PubMed] [Google Scholar]
- 23.Lavery R, Zakrzewska K. In: Oxford Handbook of Nucleic Acid Structure. Neidle S, editor. Oxford: Oxford Sci.; 1999. pp. 39–76. [Google Scholar]
- 24.Dickerson R E, Goodsell D S, Neidle S. Proc Natl Acad Sci USA. 1994;91:3579–3583. doi: 10.1073/pnas.91.9.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foloppe N, MacKerell A D., Jr Biophys J. 1999;76:3206–3216. doi: 10.1016/S0006-3495(99)77472-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Šponer J, Gabb H A, Leszczynski J, Hobza P. Biophys J. 1997;73:76–87. doi: 10.1016/S0006-3495(97)78049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gray D G, Ratliff R L, Vaughan M R. Methods Enzymol. 1992;211:389–406. doi: 10.1016/0076-6879(92)11021-a. [DOI] [PubMed] [Google Scholar]
- 28.Calladine C R, Drew H R. J Mol Biol. 1984;178:773–782. doi: 10.1016/0022-2836(84)90251-1. [DOI] [PubMed] [Google Scholar]
- 29.Calladine C R, Drew H R. Understanding DNA: The Molecule and How It Works. San Diego: Academic; 1997. [Google Scholar]
- 30.Yang L, Pettitt B M. J Phys Chem. 1996;100:2564–2566. [Google Scholar]
- 31.Cheatham T E, III, Kollman P A. J Mol Biol. 1996;259:434–444. doi: 10.1006/jmbi.1996.0330. [DOI] [PubMed] [Google Scholar]
- 32.Sprous D, Young M A, Beveridge D L. J Phys Chem B. 1998;102:4658–4667. [Google Scholar]
- 33.Suck D, Lahm A, Oefner C. Nature (London) 1988;332:464–468. doi: 10.1038/332464a0. [DOI] [PubMed] [Google Scholar]
- 34.Lahm A, Suck D. J Mol Biol. 1991;221:645–647. doi: 10.1016/0022-2836(91)90502-w. [DOI] [PubMed] [Google Scholar]
- 35.Weston S A, Lahm A, Suck D. J Mol Biol. 1992;226:1237–1256. doi: 10.1016/0022-2836(92)91064-v. [DOI] [PubMed] [Google Scholar]
- 36.Palaniappan C, Fuentes G M, Rodriguez-Rodriguez L, Fay P J, Bambara R A. J Biol Chem. 1996;271:2063–2070. [PubMed] [Google Scholar]
- 37.Otwinowski Z, Schevitz R W, Zhang R G, Lawson C L, Joachimiak A, Marmorstein R Q, Luisi B F, Sigler P B. Nature (London) 1988;335:321–329. doi: 10.1038/335321a0. [DOI] [PubMed] [Google Scholar]
- 38.Olson W K, Gorin A A, Lu X-J, Hock L M, Zhurkin V B. Proc Natl Acad Sci USA. 1998;95:11163–11168. doi: 10.1073/pnas.95.19.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dickerson R E, Bansal M, Calladine C R, Diekmann S, Hunter W N, Kennard O, von Kitzing E, Lavery R, Nelson H C M, Olson W K, et al. J Mol Biol. 1989;205:787–791. [Google Scholar]