Abstract
The t(9;22) chromosomal translocation is found in almost all patients with chronic myelogenous leukemia. The resultant Bcr-Abl fusion gene expresses a chimeric fusion protein p210bcr-abl with increased tyrosine kinase activity. Hematopoietic progenitors isolated from chronic myelogenous leukemia patients in the chronic phase contain constitutively tyrosine-phosphorylated p62dok protein. p62dok associates with the Ras GTPase-activating protein (RasGAP), but only when p62dok is tyrosine phosphorylated. Here we have investigated the interaction between p62dok and RasGAP and the consequences of p62dok tyrosine phosphorylation on the activity of RasGAP. We have found that p62dok is directly tyrosine phosphorylated by p210bcr-abl, and the sites of phosphorylation are located in the C-terminal half of the p62dok molecule. We have identified five tyrosine residues that are involved in in vitro RasGAP binding and have found that tyrosine-phosphorylated p62dok inhibits RasGAP activity. Our results suggest that p210bcr-abl might lead to the activation of the Ras signaling pathway by inhibiting a key down-regulator of Ras signaling.
Keywords: chronic myelogenous leukemia, Ras
Chronic myelogenous leukemia is a form of leukemia with distinct clinical and pathological features (1). A well-characterized chromosomal translocation within a single primitive myeloid stem cells produces the chimeric bcr-abl gene, which in turn leads to the generation of a 210 kDa Bcr-Abl (p210bcr-abl) fusion protein. Relative to normal c-Abl, p210bcr-abl has increased tyrosine kinase activity and can transform a variety of fibroblastic and hematopoietic cell lines in culture (2, 3). Although c-Abl, as a nonreceptor tyrosine kinase, functions in both the nucleus and the cytoplasm, p210bcr-abl is exclusively cytoplasmic. Both the elevated kinase activity and the subcellular localization of the oncoprotein are considered essential elements of its transforming abilities (4). Although the Ras pathway has been reported to be activated in cells containing p210bcr-abl (5, 6), the mechanism by which p210bcr-abl perturbs normal hematopoiesis has not yet been established.
One of the major targets of p210bcr-abl in hematopoietic progenitors is p62dok, the RasGAP-associated p62 protein (7). p62dok (Dok-1) was first described as the major tyrosine-phosphorylated protein present within cells transformed by different activated tyrosine kinases (8). In addition, its state of hypertyrosine phosphorylation has consistently correlated with the transformed phenotype in cells expressing transforming oncogenes such as v-fps, v-src, v-abl, Ras, and mos (8, 9). Recently, the p62dok protein was isolated both from a p210bcr-abl transformed myeloid cell line and a v-Abl-transformed B cell line (10, 11). Cloning of the Dok-1 cDNA revealed the presence of a pleckstrin homology (PH) domain in the N-terminal portion of the protein molecule. PH domains are thought to mediate protein interactions with cellular membranes, possibly by binding to polyphosphoinositides (12–14). Dok-1 also contains numerous tyrosines in its C-terminal region that are potential targets of cytoplasmic tyrosine kinases. If phosphorylated, these tyrosines could serve as docking sites for proteins that contain SH2 domains. These structural features are reminiscent of the features found in such adaptor molecules as IRS-1 and IRS-2 (15), Gab1 (16–18), and Dos (19). Thus, the overall structure of Dok-1 suggests that it might serve as a docking molecule for different signaling molecules downstream of growth factor receptors.
Although at present the biological function of Dok-1 is unclear, several lines of evidence suggest that it does indeed play a role in distinct signal transduction networks, most likely as a component of a signaling cascade initiated by receptor or membrane-associated tyrosine kinases. The rapid tyrosine phosphorylation of a RasGAP-associated p62 in Rat-1 fibroblasts on epidermal growth factor (EGF) stimulation and in Rat-2 fibroblasts on platelet-derived growth factor stimulation was reported (8). Using a different system, Filvaroff et al. demonstrated the tyrosine phosphorylation of a RasGAP-associated p62 in primary mouse keratinocytes as one of the initial events in calcium-induced terminal differentiation (20). Other agents, such as EGF, type β transforming growth factor, and phorbol ester, did not induce tyrosine phosphorylation of the p62 within keratinocytes, although they did result in the tyrosine phosphorylation of the signaling molecules PLCγ1 and PI3 kinase. In yet a different biological context, application of the mitogen vascular endothelial cell growth factor (VEGF) to bovine endothelial cells resulted in tyrosine phosphorylation and RasGAP association of a p62 protein, most likely Dok-1 (21). The VEGF-stimulated tyrosine phosphorylation of p62 and the VEGF-promoted proliferation of endothelial cells were both inhibited by the tyrosine kinase inhibitor genistein. Thus, Dok-1 may function in signaling pathways downstream of a variety of receptors, in both transformed and nontransformed cells. However, the functional consequences of constitutive tyrosine phosphorylation of Dok-1 are still unclear.
Early studies revealed that when tyrosine phosphorylated, Dok-1 forms an in vivo protein–protein complex with the Ras regulatory protein p120 RasGAP (GAP). GAP interacts with p21ras and stimulates the latter's intrinsic GTPase activity (22). Its catalytic activity is located in the C-terminal portion of the molecule. In cells transformed by tyrosine kinases, GAP can be found to be associated with both Dok-1 and a 190-kDa protein known as p190 RhoGAP. Both proteins interact with the N-terminal SH2-SH3-SH2 region of GAP (23). Unlike the C-terminal region of GAP, the N-terminal region of GAP has no known biochemical activity. However, the N-terminal noncatalytic region of GAP (but not full-length GAP) has been shown to be active in several different biological assays, including cytoskeletal reorganization, adhesion to extracellular matrix (24), transcriptional activation (25), and inhibition of transformation induced by G protein-coupled receptors (26). The biological significance of the RasGAP-p62dok complex is unknown. In this report, we analyze the interaction between GAP and Dok-1, and we demonstrate that the activity of GAP toward Ras is inhibited by the binding of tyrosine-phosphorylated Dok-1 to GAP.
Materials and Methods
Antibodies.
A monoclonal antibody against the C-terminal tag (RYIRS) was kindly provided by S. H. Lin (27) (Univ. of Texas, M. D. Anderson Cancer Center, Houston). Anti-p62dok rabbit polyclonal antibody was described previously (10), as was the use of anti-pTyr monoclonal 4G10 (Upstate Biotechnology). Monoclonal anti-Bcr antibody and antihemagglutinin antibody (12CA5) were from OSI (Uniondale, NY), and anti-GAP (B4F8) was from Santa Cruz Biotechnology.
Cell Culture.
Human 293 cells and monkey COS cells, obtained from Cold Spring Harbor Laboratory Tissue Culture Facility, were maintained in DMEM containing antibiotics 22 units/ml penicillin (GIBCO/BRL), 100 μg/ml streptomycin (Sigma), and 10% calf serum (GIBCO/BRL) at 37°C and 5% CO2. All experiments utilized 293 cells between passages 1 and 20. Mammalian expression constructs were electroporated into cells at 200 V, 960 μF capacitance.
Immunoprecipitation, Immunoblotting, and RasGAP-Binding Assays.
Immunoprecipitation and blotting were as described (10). GST-GAP proteins (constructs kindly provided by J. Schlessinger, New York University, New York) were incubated with lysate for 30 min at 4°C, washed extensively with lysis buffer, and bound proteins separated by SDS/PAGE.
In Vitro Kinase Reactions.
Purified Dok-1 (250 ng), generated by overexpression in Escherichia coli, was incubated with 100 units Abl protein tyrosine kinase (NEB, Beverly, MA) in a buffer recommended by the manufacturer. The reaction was allowed to proceed at 30°C for the indicated time, after which it was stopped by the addition of SDS/PAGE sample buffer. For the reaction depicted in Fig. 1b, anti-Bcr antibodies were used to precipitate proteins from lysates of Mo7 or Mo7/p210 cells (10). Protein A-Sepharose beads (10 μl), to which the precipitated complexes were bound, were incubated with in vitro-translated p62dok according to instructions of the manufacturer. After a 30-min incubation at 30°C, the kinase reaction was stopped by addition of 500 μl precipitation buffer (10), and Dok-1 was precipitated by addition of anti-p62dok antibodies or antiphosphotyrosine antibody (4G10, Upstate Biotechnology).
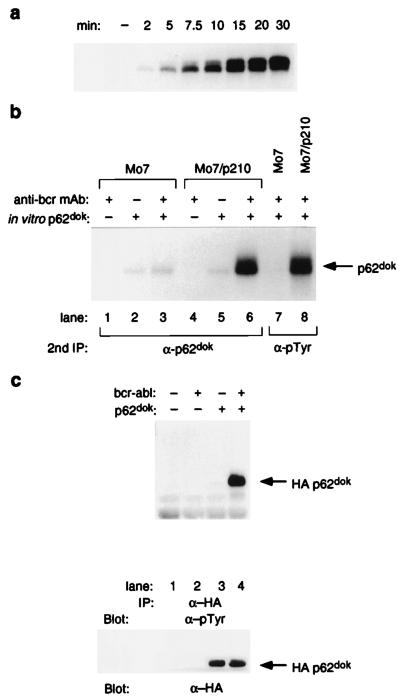
Figure 1.
Tyrosine phosphorylation of p62dok by p210bcr-abl. (a) Time course, in vitro abl kinase reaction utilizing purified recombinant p62dok as substrate. p62dok is phosphorylated in a time-dependent manner by Abl. (b) In vitro p210bcr-abl kinase reaction, utilizing in vitro-translated p62dok as substrate. Monoclonal anti-bcr antibody was used to isolate p210bcr-abl. Immune complex was incubated with in vitro-translated p62dok in the presence of γ-32P-ATP. Arrow indicates phosphorylated p62dok. (c) p210bcr-abl-dependent tyrosine phosphorylation of p62dok, in vivo. Hemagglutinin-tagged Dok-1 and p210bcr-abl were expressed in COS cells. Tyrosine phosphorylation of p62dok was analyzed by precipitating the molecule from lysates derived from transfected cells, resolving the immune complexes by SDS/PAGE and transfer to nitrocellulose, and immunoblotting with anti-pTyr antibody (Top) or antihemagglutinin antibody (Bottom). Arrow indicates p62dok.
Truncations.
The PCR was used to create the tagged Dok-1 truncation products illustrated in Fig. 3a. Each construct was sequenced and subcloned into the pMT2 mammalian expression vector.
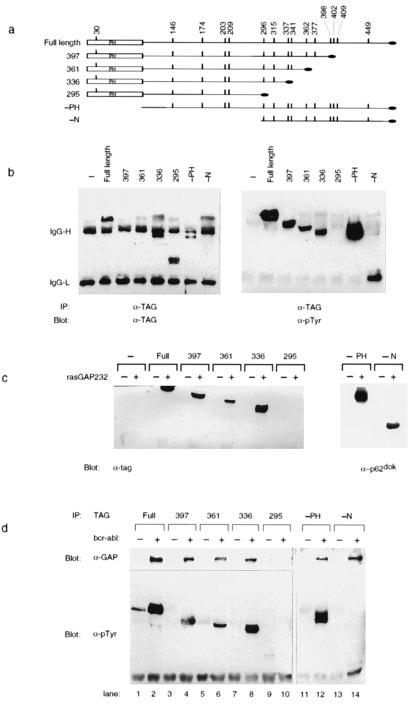
Figure 3.
Removal of N- and C-terminal residues from p62dok does not prevent its in vitro and in vivo association with RasGAP. (a) Schematic diagram of p62dok truncations. Four C-terminal Dok-1 truncations were constructed. In each case, a different tyrosine (residues 398, 362, 337, and 296) and all amino acids C terminal to the selected tyrosine were replaced with a C-terminal antigenic tag (RYIRS, black oval). Additionally, two N-terminal truncations were constructed, lacking either the PH domain (−PH) or the entire N-terminal region (−N). The vertical bars mark the location of the tyrosines within p62dok. The PH domain is indicated (PH). (b) Tyrosine phosphorylation of p62dok truncations. The indicated Dok-1 truncations and p210bcr-abl were expressed in 293 cells. Antibody directed against the C-terminal tag was used to precipitate Dok-1 proteins. Immune complexes were analyzed by immunoblotting by using antibodies against the C-terminal tag (TAG, Left) or phosphotyrosine (pTyr, Right). Bands derived from IgG are indicated as IgG-H and IgG-L. The only Dok-1 mutant to lack detectable tyrosine phosphorylation in the presence of p210bcr-abl was that which lacked all 10 C-terminal tyrosines (295). (c) Removal of N-terminal and C-terminal residues from p62dok does not prevent its in vitro association with RasGAP. The indicated Dok-1 truncations and Bcr-Abl were expressed in 293 cells. Cell lysates were prepared and utilized in an in vitro binding reaction to glutathione-Sepharose beads to which were bound GST (−) or GST-GAP SH2-SH3-SH2 region. Bound proteins were separated by SDS/PAGE, blotted to nitrocellulose, and detected by immunoblotting with antibodies to the C-terminal tag (Left) or to p62dok (Right). The only Dok-1 mutant unable to associate with GAP was that which lacked all 10 C-terminal tyrosines (295). (d) Removal of N-terminal and C-terminal residues from p62dok does not prevent its in vivo association with RasGAP. Dok-1 truncations and p210bcr-abl were expressed in 293 cells as described above.
Site-Directed Mutagenesis.
All Dok-1 Y→F point mutants were generated by site-directed mutagenesis of double-stranded DNA by using the QuickChange site-directed mutagenesis kit (Stratagene) according to manufacturer's specifications. Mutagenesis was confirmed by sequence analysis.
RasGAP Activity Assay.
Immunoprecipitated RasGAP activity was measured by analyzing the ratio of GTP and GDP bound to Ras as previously described (28), with some modification. Recombinant human H-Ras (Calbiochem) was loaded with [α-32P]GTP at 25°C for 10 min in 20 mM Tris⋅HCl, pH 7.5, containing 5 mM EDTA. RasGAP was produced by transient transfection in 293 cells and obtained by precipitation with anti-RasGAP (10 μg) agarose conjugate (Santa Cruz Biotechnology). Enzyme activity was assayed by incubating bound RasGAP with 60 ng [α-32P]GTP-loaded Ras for 60 min or 90 min. In vitro-phosphorylated Dok-1 (0, 0.05, 0.1, 0.5, and 1 μg) recombinant wild-type or 1 μg of mutant Dok-1 was added to the reaction. Reactions were terminated by adding ice-cold stop buffer containing anti-H-Ras antibody agarose conjugate (Oncogene Science), as described (28). After rocking this mixture for 45 min at 4°C, the resin was washed four times with 0.5 ml of 0.5% (vol/vol) Nonidet P-40 in stop buffer and then once with stop buffer alone. The nucleotide was eluted off resin with 20 μl of 2 mM EDTA/2 mM DTT/0.2% SDS/0.5 mM GDP/0.5 mM GTP by heating to 65°C for 5 min and then separated on polyethyleneimine-cellulose TLC plate (J. T. Baker, Inc., Phil- lipsburg, NJ) developed with 0.75 M KH2PO4 (pH 3.4). The positions of GDP and GTP on the plates were visualized under UV light by using standard GDP and GTP. Ras-bound GDP and GTP were visualized by autoradiography and quantified by using NIH image 1.62 to calculate the GTP/GDP ratio.
Results
Interaction Between p210bcr-abl, p62dok, and RasGAP.
The presence of tyrosine phosphorylation on p62dok in cells containing p210bcr-abl suggested that p62dok is a direct substrate of p210bcr-abl. Nonetheless, it is also possible that p210bcr-abl might activate other cytoplasmic tyrosine kinases that directly phosphorylate p62dok. The ability of Dok-1 to be phosphorylated by p210bcr-abl was tested in three separate assays. First, recombinant Dok-1 generated by overexpression in bacteria was tested as a substrate of the isolated kinase domain of Abl. Fig. 1a illustrates that Dok-1 was phosphorylated in a time-dependent manner by the kinase domain of Abl. This suggested that Dok-1 contains tyrosines whose surrounding amino acid residues comprise a motif that can be recognized by the Abl kinase. Because it is possible that p210bcr-abl might interact with possible substrates differently than the isolated Abl kinase domain, a second assay was used to test the ability of Dok-1 to be phosphorylated by p210bcr-abl. Fig. 1b illustrates that Dok-1, generated by in vitro translation of the Dok-1 cDNA in a rabbit reticulocyte lysate system, could be phosphorylated by p210bcr-abl isolated by immunoprecipitation from lysates of Mo7/p210 cells with anti-Bcr antibody (lane 6). That the incorporated phosphate was bonded to tyrosine was indicated by the ability of antiphosphotyrosine antibodies to precipitate Dok-1 (lane 8). Finally, an in vivo transfection assay was used to test whether p210bcr-abl could phosphorylate Dok-1. Dok-1 cDNA, cloned into a mammalian expression vector downstream of sequences encoding the hemagglutinin Tag, was cotransfected with the cDNA encoding p210bcr-abl into COS cells. Fig. 1c illustrates that Dok-1 was tyrosine phosphorylated in a p210bcr-abl -dependent fashion when overexpressed in COS cells. These results indicate that p62dok can be phosphorylated by p210bcr-abl, and that p62dok might be a substrate of p210bcr-abl within primitive blasts of patients with chronic myelogenous leukemia.
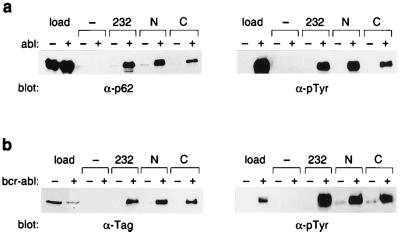
Numerous investigators working with a variety of transformed cell lines have observed the tyrosine phosphorylation of p62dok protein and its association with GAP (see for example refs. 8 and 29). The portion of the GAP molecule responsible for the interaction with p62dok has been mapped to the N-terminal region of GAP, comprising the SH2-SH3-SH2 domain (30). Therefore, the need for p62dok to be tyrosine phosphorylated to associate directly with GAP was tested. When recombinant Dok-1 was tested for the ability to bind in vitro to the N-terminal regions of GAP, only tyrosine-phosphorylated Dok-1 associated in vitro with the N-terminal region of GAP (see Fig. 2a). Similarly, when Dok-1 was overexpressed in 293 cells in the presence or absence of p210bcr-abl it was observed that only tyrosine-phosphorylated Dok-1 bound to GAP (see Fig. 2b). This confirms the long-held suspicion that tyrosine phosphorylation of Dok-1 is necessary for the interaction between Dok-1 and GAP.
Figure 2.
Interaction between p62dok and RasGAP. (a) Tagged p62dok was expressed in E. coli, purified, and incubated in a kinase reaction in the presence of Abl kinase (+) or buffer only (−). After the kinase reaction, p62dok was incubated in an in vitro-binding reaction with glutathione-Sepharose beads to which were bound GST(−), GST-GAP SH2-SH3-SH2 (232), GST-GAP N-SH2 (N), or GST-GAP C-SH2 (C). Proteins bound to GST fusion proteins were separated by SDS/PAGE, blotted to nitrocellulose, and detected by immunoblotting with antibodies to p62dok (α-p62dok) or phosphotyrosine (α-pTyr). (b) Mammalian-expressed p62dok associates with RasGAP in a phosphorylation-dependent manner. (Fig. 1b). C-terminally tagged Dok-1 cDNA was electroporated into 293 cell in the presence (+) or absence (−) of Bcr-Abl. After a 24-h incubation, cells were lysed, and lysates were utilized in an in vitro- binding reaction with glutathione-Sepharose beads to which were bound GST(−), GST-GAP SH2-SH3-SH2 (232), GST-GAP N-SH2 (N), or GST-GAP C-SH2 (C). Proteins bound to GST fusion proteins were separated by SDS/PAGE, blotted to nitrocellulose, and detected by immunoblotting with antibodies to the C-terminal tag (α-tag) or phosphotyrosine (α-pTyr).
p62dok Truncations and RasGAP Binding.
To assess the region of p62dok, in particular the tyrosine(s), involved in the interaction with GAP, a series of truncation mutants of Dok-1 was constructed. The structures of the Dok-1 mutants are illustrated in Fig. 3a. Four different C-terminal truncations were constructed. In each case, in the resulting Dok-1 protein a different tyrosine and all amino acids C terminal to the selected tyrosine were substituted with a C-terminal antigenic tag (27). The tyrosines at which the different constructs were terminated were tyrosines 398, 362, 337, and 296. In addition to the C-terminal truncations, two N-terminal truncations were constructed by replacing sequences encoding N-terminal amino acids with a starting ATG. The resulting Dok-1 protein in one case lacked the PH domain and in the other case lacked all amino acids N terminal to Ala-294 (see Fig. 3a). When coexpressed with p210bcr-abl in 293 cells, each mutant Dok-1 protein became tyrosine phosphorylated, with the exception of the 295 Dok-1 mutant (see Fig. 3b).
Mutant Dok-1 proteins were also assessed for their ability to bind in vitro to the SH2-SH3-SH2 region of GAP. As illustrated in Fig. 3c, all mutant Dok-1 proteins with the exception of the 295 Dok-1 mutant were able to bind in vitro to the SH2-SH3-SH2 region of GAP. Thus, removal of all amino acid residues C terminal to Tyr-337 or N terminal to Ala-294 did not prevent the remaining portion of Dok-1 from binding in vitro to the SH2 region(s) of GAP. The mutant Dok-1 proteins were also assessed for their ability to associate with GAP in vivo. Mutant Dok-1 proteins were expressed in 293 cells in the presence or absence of p210bcr-abl. Then, antibodies directed against the C-terminal antigenic tag were used to precipitate the mutant Dok-1 proteins from extracts prepared from the transfected cells. As illustrated in Fig. 3d, GAP coprecipitated with each mutant Dok-1 protein in a p210bcr-abl-dependent fashion, with the exception of the 295 Dok-1 truncation mutant. These in vivo results complement the results obtained in the in vitro binding assays; specifically, the in vivo results point to the region between Dok-1 residues Ala-294 and Tyr-337 as involved in the association of Dok-1 with the SH2 domain(s) of GAP. Within the primary amino acid sequence of this region of Dok-1 are two tyrosine residues spaced 18 amino acids apart, Tyr-296 and Tyr-315. These tyrosines are implicated as involved in the association of p62dok with GAP. However, our data do not exclude the possibility that other tyrosines within the C-terminal region of Dok-1 are tyrosine phosphorylated and involved in GAP binding. In fact, Tyr-337, Tyr-362, and Tyr-398 of p62dok protein purified from p210bcr-abl expressing Mo7 cells were found phosphorylated by mass spectrometry and protein sequencing (data not shown).
Effect of p62dok Point Mutants on RasGAP Binding.
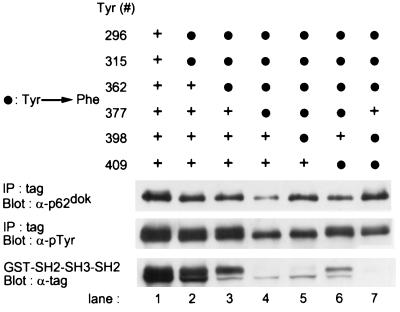
The truncation experiment identified Tyr-296 and Tyr-315 as least involved in the association of Dok-1 to GAP. Indeed, it appeared likely that Tyr-296 and Tyr-315, when phosphorylated, could interact with the two SH2 domains of GAP, similar to the manner in which Tyr-1087 and Tyr-1105 of p190 RhoGAP interacted with GAP (31). To test the role of Tyr-296 and Tyr-315 in GAP binding, we constructed a double Tyr→Phe point mutant Dok-1, in which both tyrosines were altered to phenylalanine. The tagged double mutant Dok-1 protein was expressed in bacteria and phosphorylated in vitro by Abl kinase. Then it was tested for its ability to bind the GAP SH2-SH3-SH2 domain, as described. Interestingly, this double-point mutant retained the ability to become tyrosine phosphorylated and bind to GAP (Fig. 4, lane2). The GAP-SH2 domains bind preferentially to phosphorylated tyrosine residues within the context of pYxxP (11, 32). There are six tyrosine residues in p62dok that have proline in the +3 position: Tyr-296, Tyr-315, Tyr-362, Tyr-377, Tyr-398, and Tyr-409. Therefore, additional mutant proteins in which combinations of tyrosines were changed to phenylalanine were tested for their ability to bind GAP. The Y296F, Y315F, Y362F triple point mutant had reduced ability to bind GAP as compared with the Y296, 315F double point mutant (see Fig. 4, lane 3). Furthermore, when we mutated Tyr-377 to phenylalanine in the context of the Y296, 315, 362F triple mutant, the association of phosphorylated Dok-1 with GAP was significantly reduced (Fig. 4, lane 4). However, even when either Tyr-398 or Tyr-409 was additionally mutated to phenylalanine in the context of the Y296, 315, 362, 377F quadruple mutant, we still observed in vitro association of the mutant protein with GAP (Fig. 4, lanes 5 and 6). Finally, when we mutated five specific tyrosines to phenylalanine (Y296, 315, 362, 398, 409F), we observed that Dok-1 was no longer able to bind in vitro to the SH2-SH3-SH2 domain of GAP (Fig. 4, lane 7). This suggests that these five tyrosines contribute to the in vitro association of Dok-1 with GAP.
Figure 4.
Only when five specific tyrosines are mutated to phenylalanine is the binding of p62dok to the RasGAP SH2-SH3-SH2 region abrogated. The indicated tyrosine-to-phenylalanine mutants of Dok-1 were coexpressed in 293 cells with p210bcr-abl. Cell lysates were prepared and divided into two. Mutant Dok-1 was precipitated from one portion and detected after SDS/PAGE by antibodies to Dok-1 (Top) or antibody to phosphotyrosine (Middle), whereas the other portion was utilized in an in vitro binding reaction to GST-GAP SH2-SH3-SH2 (Bottom). Nonmutated tyrosine residue is indicated (+). Lysates were prepared, and antibodies to the C-terminal tag were used to precipitate Dok-1 proteins. The resulting immunoblot was probed with antibody directed against GAP (Top) or against phosphotyrosine (Bottom).
The Effect of Tyrosine-Phosphorylated p62dok on RasGAP Activity.
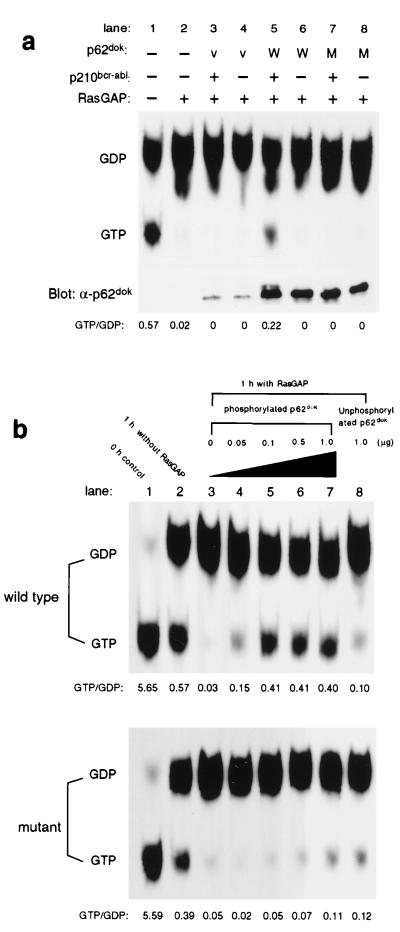
To test whether binding of Dok-1 to GAP had any effect on the activity of GAP, we used an in vitro GAP activity assay (28). This assay measures the ability of GAP to increase the intrinsic GTPase rate of GTP-Ras. We analyzed the in vitro activity of GAP in the presence of tyrosine-phosphorylated wild-type Dok-1 or in the presence of a five Tyr→Phe point mutant unable to associate with GAP (see Fig. 4, lane 7). The assays were carried out with Dok-1 proteins phosphorylated both in vivo and in vitro. In vivo tyrosine-phosphorylated wild-type or mutant Dok-1 (Fig. 4, lane 7) was prepared by immunoprecipitation of Dok-1 from lysates of 293 cells coexpressing p62dok and p210bcr-abl. We observed a significant (approximately 35%) diminution of GAP activity when the assay was conducted in the presence of wild-type tyrosine-phosphorylated Dok-1 (see Fig. 5a, lane 5). In contrast, we observed no diminution of GAP activity when the assay was conducted in the presence of nontyrosine-phosphorylated Dok-1 (Fig. 5a, lane 6) or in the presence of a five Tyr→Phe point mutant Dok-1 unable to bind GAP (see Fig. 5a, lanes 7 and 8). In vitro tyrosine-phosphorylated wild-type or mutant Dok-1 was prepared by phosphorylation of bacterially expressed Dok-1 by c-Abl kinase. Again, we observed a significant diminution of GAP activity when the assay was conducted in the presence of phosphorylated wild-type Dok-1 as compared with GAP activity in the presence of nontyrosine-phosphorylated wild-type Dok-1 or a five Tyr→Phe point mutant Dok-1 unable to bind GAP (compare with Fig. 5b Top and Bottom). These results indicate that the association of tyrosine-phosphorylated Dok-1 with GAP can inhibit the GTPase-activating activity of GAP.
Figure 5.
Tyrosine-phosphorylated p62dok inhibits RasGAP activity. (a) In vivo phosphorylated wild-type Dok-1 by p210bcr-abl inhibits GAP activity toward Ras. However, a mutant Dok-1 that does not bind to GAP has no effect on GAP activity. (b) In vitro-phosphorylated Dok-1 by Abl tyrosine kinase also inhibits GAP activity, whereas a mutant Dok-1 has no effect. v, vector only; W, wild-type p62dok; M, five-tyrosine mutant p62dok.
Peptide Competition Analysis.
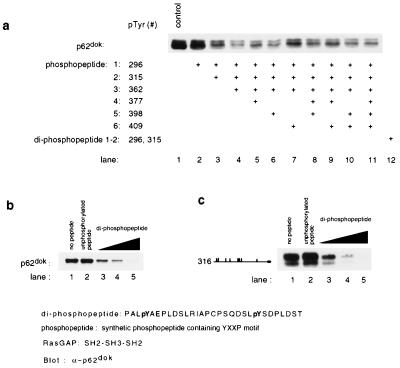
We synthesized tyrosine-phosphorylated peptides (15 mer) corresponding to the region surrounding each of tyrosines in YxxP motif (see Fig. 6 Legend). Then we attempted to inhibit the interaction between Dok-1 and the SH2-SH3-SH2 domain of GAP by including the peptides in the in vitro binding reaction described in Fig. 2. Interestingly, any mixture of the singly tyrosine-phosphorylated peptides containing pYxxP motif, including the five GAP-binding tyrosines (Fig. 4, lane 7), failed to inhibit in vitro association of Dok-1 with the SH2-SH3-SH2 domain of GAP (Fig. 6a, lanes 2–11, and data not shown).
Figure 6.
Peptide competition analysis indicates that Tyr-296 and Tyr-315 play a critical role in the binding of p62dok to RasGAP. The ability of the diphosphopeptide to inhibit binding of Dok-1 to GAP suggests that the proper positioning of pTyr-296 and pTyr-315 in tandem is critical for the interaction of the two molecules. (a) Combinations of phosphopeptides corresponding to the regions surrounding individual tyrosines in p62dok fail to inhibit the binding of p62dok to RasGAP. However, a diphosphopeptide corresponding to residues 293–322 is able to inhibit binding of Dok-1 to the GAP SH2-SH3-SH2 region. Binding analysis was conducted as described. Synthetic phosphopeptides used for this experiment were: 1, SPPALpYAEPLDS (pTyr-296); 2, SQDSLpYSDPLDS (pTyr-315); 3, PKEDPIpYDEPEGL (pTyr-362); 4, VPPQG LpYDLPREPK (pTyr-377); 5, RVKEEGpYELPYNPATDD (pTyr-398); 6, NPATDD pYAVPPPR (pTyr-409); and diphosphopeptide, PALpYAEPLDSLRIAPCPSQDS LpYSDPLDST (pTyr-296 and pTyr-315). For control, unphosphorylated peptides were used. Each phosphopeptide was added in concentration of 50 μM. (b) Dose-dependent inhibition of p62dok binding to RasGAP by diphosphopeptide (Dok-1 aa 293–322). The diphosphopeptide was added in concentration of 0.5, 5, or 50 μM. The unphosphorylated peptide was added in concentration of 50 μM. (c) Dose-dependent inhibition of a truncated Dok-1 (the truncation construct 316: residues 316–481) binding to RasGAP by diphosphopeptide. The diphosphopeptide was added in concentration of 0.5, 5, or 50 μM. The unphosphorylated peptide was added in concentration of 50 μM.
Recently, Hu and Settleman demonstrated that the critical determinants of p190 RhoGAP mediating the association of p190 with GAP are two tyrosines spaced 17 residues apart, Tyr-1087 and Tyr-1105 (31). These tyrosines lie within the motif YxxPxD, and each appears to bind simultaneously to the two SH2 domains of GAP. There are two tyrosines of p62dok that lie within the motif YxxPxD, Tyr-296, and Tyr-315. Similar to the tyrosines within p190 RhoGAP, they are spaced 18 residues apart. We synthesized a diphosphopeptide of 30 amino acids corresponding to the residues surrounding Tyr-296 and Tyr-315 and attempted to inhibit the in vitro interaction between p62dok and RasGAP by including the diphosphopeptide in the binding reaction. The diphosphopeptide completely inhibited the in vitro association of wild-type p62dok to RasGAP [Fig. 6 a (lane 12) and b). Furthermore, the diphosphopeptide was able to inhibit the GAP association of a truncated Dok-1 that lacked all amino acids N terminal to Tyr-315 (see Fig. 6c). These results suggest that although five tyrosines within the C-terminal region of Dok-1 can contribute to GAP binding, Tyr-296 and Tyr-315 double mutant may play the predominant role. Because a mixture of five singly phosphorylated peptides (including Tyr-296 and Tyr-315) was not able to completely inhibit the GAP binding (see Fig. 6a, lane 10), our results also suggest that a configuration that positions the two YxxPxD motifs (Tyr-296 and Tyr-315) is critical for the interaction of p62dok with RasGAP.
Discussion
The GAP-Associated 62-kDa protein, p62dok, was first described as the major tyrosine-phosphorylated protein present within cells transformed by different activated tyrosine kinases. Among the cell types in which Dok-1 has been shown to be constitutively tyrosine phosphorylated are primary hematopoietic progenitors isolated from chronic myelogenous patients in the chronic phase (7). These cells contain the p210bcr-abl oncogene, an aberrant form of the Abl tyrosine kinase produced as a result of the t(9;22) chromosomal translocation (the Philadelphia chromosome). In this study, we have presented evidence that p62dok is a direct target of the p210bcr-abl kinase. In addition, we have demonstrated that tyrosine phosphorylation of Dok-1 is necessary for its association with the N-terminal SH2-SH3-SH2 region of p120 RasGAP. Our data suggest that five phosphotyrosines, pTyr-296, pTyr-315, pTyr-362, pTyr-398, and pTyr-409, of p62dok contribute to GAP association. Because tyrosine-phosphorylated p62dok isolated from cells expressing p210bcr-abl appears as a doublet in one-dimensional gel electrophoresis and as three to four spots in two-dimensional gel electrophoresis (10), multiple species of p62dok clearly exist in cells. Although Tyr-296 and Tyr-315 appear to play a critical role in in vitro GAP association, at present we do not know whether they play a similar role in vivo. We also do not know either the stoichiometry of in vivo phosphorylation or the order in which each tyrosine becomes phosphorylated in vivo. Because there are only two SH2 domains in GAP, it is possible that when two phosphotyrosines of p62dok are binding to GAP, other phosphotyrosines are available to interact with other SH2 domain-containing proteins. We anticipate that the intracellular interaction of p210bcr-abl, p62dok, and GAP is complex. Further investigation is necessary to study tyrosine phosphorylation of p62dok and its interacting proteins.
Finally and most importantly, we have demonstrated that Dok-1 binding to RasGAP negatively affects GAP's activity in vitro toward the GTPase activity of Ras. Previously, Skorski et al. reported (33) that p210bcr-abl negatively regulate the GTPase-promoting activity of GAP in chronic myelogenous leukemia cells. Our results are in agreement with their report. Pendergast et al. reported that Bcr-Abl forms a complex with Grb2 in vivo, and this interaction is required for Bcr-Abl-induced Ras activation (5). It is possible that there are several mechanisms for Bcr-Abl to activate Ras, including the following two scenarios: (i) by modulating the association and activity of the Grb2/Sos exchange factor complex; and (ii) by phosphorylating p62dok, which in turn down-regulates RasGAP activity.
It is currently unknown whether phosphorylation of p62dok affects the MAP kinase-signaling pathway, or whether p62dok plays a role in a signaling pathway parallel to a MAP kinase pathway. Both GAP and Dok-1 are predominantly cytosolic, although a portion of the GAP/Dok-1 complex translocates to the inner surface of the plasma membrane on growth factor stimulation (34).
Although at present the biological function of p62dok is not known, our data suggest that one effect of p62dok constitutive tyrosine phosphorylation is to disrupt the activity of a key down-regulator of the Ras signaling pathway. Thus, constitutive p62dok tyrosine phosphorylation might directly contribute to the phenotypic and behavioral differences observed in chronic myelogenous leukemia hematopoietic progenitors (35).
Acknowledgments
We thank B. Clarkson, S. Swendeman, Y. Lazebnik, and J.T. Kadonaga for critically reading and reviewing the manuscript, L. van Aelst for valuable discussion, and M. Moran for the RasGAP plasmid. This work was supported by a National Institutes of Health grant (CA64593) (N.C. and R.K.) and by the Robertson Fund (N.K.).
Abbreviations
- PH
pleckstrin homology
- Dok-1
p62dok
- pTyr or pY
phosphotyrosine
- RasGAP
Ras GTPase-activating protein
- GAP
p120 RasGAP
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.040547997.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.040547997
References
- 1.Clarkson B, Strife A. Leukemia. 1993;7:1683–1721. [PubMed] [Google Scholar]
- 2.Konopka J, Watanabe S M, Witte O N. Cell. 1984;37:1035–1042. doi: 10.1016/0092-8674(84)90438-0. [DOI] [PubMed] [Google Scholar]
- 3.Lugo T G, Pendergast A M, Muller A J, Witte O N. Science. 1990;247:1079–1082. doi: 10.1126/science.2408149. [DOI] [PubMed] [Google Scholar]
- 4.McWhirter J R, Wang J Y J. Mol Cell Biol. 1991;11:1553–1565. doi: 10.1128/mcb.11.3.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pendergast A M, Quilliam L A, Cripe L D, Bassing C H, Dai Z, Li N, Batzer A, Rabun K M, Gishizky M L. Cell. 1993;75:175–185. [PubMed] [Google Scholar]
- 6.Cortez D, Stoica G, Pierce J H, Pendergast A M. Oncogene. 1996;13:2589–2594. [PubMed] [Google Scholar]
- 7.Wisniewski D, Strife A, Wojciechowicz D, Lambek C, Clarkson B. Leukemia. 1994;8:688–693. [PubMed] [Google Scholar]
- 8.Ellis C, Moran M, McCormick F, Pawson T. Nature (London) 1990;343:377–381. doi: 10.1038/343377a0. [DOI] [PubMed] [Google Scholar]
- 9.Zhao J F, Nakano H, Sharma S. Oncogene. 1995;11:161–173. [PubMed] [Google Scholar]
- 10.Carpino N, Wisniewski D, Strife A, Marshak D, Kobayashi R, Stillman B, Clarkson B. Cell. 1997;88:197–204. doi: 10.1016/s0092-8674(00)81840-1. [DOI] [PubMed] [Google Scholar]
- 11.Yamanashi Y, Baltimore D. Cell. 1997;88:205–211. doi: 10.1016/s0092-8674(00)81841-3. [DOI] [PubMed] [Google Scholar]
- 12.Musacchio A, Gibson T, Rice P, Thompson J, Saraste M. Trends Biochem Sci. 1993;18:343–348. doi: 10.1016/0968-0004(93)90071-t. [DOI] [PubMed] [Google Scholar]
- 13.Lemmon M A, Ferguson K M, O'Brien R, Sigler P B, Schlessinger J. Proc Natl Acad Sci USA. 1995;92:10472–10476. doi: 10.1073/pnas.92.23.10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kavran J M, Klein D E, Lee A, Falasca M, Isakoff S J, Skolnik E Y, Lemmon M A. J Biol Chem. 1998;273:30497–30508. doi: 10.1074/jbc.273.46.30497. [DOI] [PubMed] [Google Scholar]
- 15.White M F. Mol Cell Biochem. 1998;182:3–11. [PubMed] [Google Scholar]
- 16.Holgado-Madruga M, Emlet D R, Moscatello D K, Godwin A K, Wong A J. Nature (London) 1996;379:560–564. doi: 10.1038/379560a0. [DOI] [PubMed] [Google Scholar]
- 17.Maroun C R, Holgado-Madruga M, Royal I, Naujokas M A, Fournier T M, Wong A J, Park M. Mol Cell Biol. 1999;19:1784–1799. doi: 10.1128/mcb.19.3.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lecoq-Lafon C, Verdier F, Fichelson S, Chretien S, Gisselbrecht S, Lacombe C, Mayeux P. Blood. 1999;93:2578–2585. [PubMed] [Google Scholar]
- 19.Raabe T, Riesgo-Escovar J, Liu X, Bausenwein B S, Deak P, Maroy P, Hafen E. Cell. 1996;85:911–920. doi: 10.1016/s0092-8674(00)81274-x. [DOI] [PubMed] [Google Scholar]
- 20.Filvaroff E, Calautti E, McCormick F, Dotto G P. Mol Cell Biol. 1992;12:5319–5328. doi: 10.1128/mcb.12.12.5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo D, Jia Q, Song H Y, Warren R S, Donner D B. J Biol Chem. 1995;270:6729–6733. doi: 10.1074/jbc.270.12.6729. [DOI] [PubMed] [Google Scholar]
- 22.Trahey M, McCormick F. Science. 1987;238:542–545. doi: 10.1126/science.2821624. [DOI] [PubMed] [Google Scholar]
- 23.Moran M F, Koch C A, Anderson D A, Ellis C, England L, Martin G S, Pawson T. Proc Natl Acad Sci USA. 1990;87:8622–8626. doi: 10.1073/pnas.87.21.8622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGlade J, Brunkhorst B, Anderson D, Mbamalu G, Settleman J, Dedhar S, Rozakis-Adcock M, Chen L B, Pawson T. EMBO J. 1993;12:3073–3081. doi: 10.1002/j.1460-2075.1993.tb05976.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schweighoffer F, Barlat I, Chevallier-Multon M C, Tocque B. Science. 1992;256:825–827. doi: 10.1126/science.1317056. [DOI] [PubMed] [Google Scholar]
- 26.Xu N, McCormick F, Gutkind J S. Oncogene. 1994;9:597–601. [PubMed] [Google Scholar]
- 27.Luo W, Liang T C, Li J M, Hsieh J T, Lin S H. Arch Biochem Biophys. 1996;329:215–220. doi: 10.1006/abbi.1996.0211. [DOI] [PubMed] [Google Scholar]
- 28.Bollag G, McCormick F. Methods Enzymol. 1995;255:161–170. doi: 10.1016/s0076-6879(95)55020-8. [DOI] [PubMed] [Google Scholar]
- 29.Moran M F, Polakis P, McCormic F, Pawson T, Ellis C. Mol Cell Biol. 1991;11:1804–1812. doi: 10.1128/mcb.11.4.1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marengere L E, Pawson T. J Biol Chem. 1992;267:22779–22786. [PubMed] [Google Scholar]
- 31.Hu K Q, Settleman J. EMBO J. 1997;16:473–483. doi: 10.1093/emboj/16.3.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holland S J, Gale N W, Gish G D, Roth R A, Songyang Z, Cantley L C, Henkemeyer M, Yancopoulos G D, Pawson T. EMBO J. 1997;16:3877–3888. doi: 10.1093/emboj/16.13.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skorski T, Kanakaraj P, Ku D, Nieborowska M, Canaani E, Zon G, Perussia B, Calabretta B. J Exp Med. 1994;179:1855–1865. doi: 10.1084/jem.179.6.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park S, Jove R. J Biol Chem. 1993;268:25728–25734. [PubMed] [Google Scholar]
- 35.Strife A, Perez A, Lambek C, Wisniewski D, Bruno S, Darzynkiewicz Z, Clarkson B. Cancer Res. 1993;53:401–409. [PubMed] [Google Scholar]