Abstract
ATP-sensitive K+ channels (KATP channels) are tetradimeric complexes of inwardly rectifying K+ channels (Kir6.x) and sulphonylurea receptors (SURs). The SURs SUR2A (cardiac) and SUR2B (smooth muscle) differ only in the last 42 amino acids. In SUR2B, the mutation Y1206S, located at intracellular loop 8, increases the affinity for glibenclamide (GBC) about 10-fold. Here, we examined whether the mutation Y1206S in SUR2A had effects similar to those in SUR2B.
GBC bound to SUR2A with KD=20 nM; the mutation increased affinity ∼5 ×.
In cells, coexpression of SUR2A with Kir6.2 increased the affinity for GBC∼3 ×; with the mutant, the increase was 9 ×.
The mutation did not affect the affinity of SUR2A for openers; coexpression with Kir6.2 reduced opener affinity of wild-type and mutant SUR2A by about 2 ×.
The negative allosteric interaction between the opener, P1075, and GBC at wild-type and mutant SUR2A was markedly affected by the presence of MgATP and by coexpression with Kir6.2.
In inside-out patches, GBC inhibited the wild-type Kir6.2/SUR2A and 2B channels with IC50 values of 27 nM; the mutation shifted the IC50 values to ∼1 nM.
The data show that the mutation Y1206S increased the affinity of SUR2A for GBC and modulated the effects of coexpression. Overall, the changes were similar to those observed with SUR2B(Y1206S), suggesting that the differences in the last 42 carboxy-terminal amino acids of SUR2A and 2B are of limited influence on the binding of GBC and P1075 to the SUR2 isoforms.
Keywords: Sulphonylurea receptor SUR2A; mutation SUR2A(Y1206S), glibenclamide; opener P1075; Kir6.x; effect of coexpression with Kir6.x
Introduction
ATP-sensitive K+ channels (KATP channels) are closed by intracellular ATP and opened by MgADP. They serve important functions in many tissues (Ashcroft & Ashcroft, 1990; Seino & Miki, 2003) and are major drug targets. Sulphonylureas and glinides block these channels in the pancreatic β-cell, thereby promoting insulin secretion, and are used as oral antidiabetics (Sturgess et al., 1985, Gribble & Reimann, 2003). In smooth muscle, KATP channels produce relaxation (Hamilton et al., 1986, Quast, 1993) and openers of these channels have been profiled as antihypertensives, antiasthmatics and against urinary incontinence (Lawson, 1996; Coghlan et al., 2001).
Structurally, KATP channels are tetradimeric complexes of inwardly rectifying K+ channels (Kir6.x) and sulphonylurea receptors (SURs) (Ashcroft & Gribble, 1998; Aguilar-Bryan & Bryan, 1999). The Kir6.x subunits form the pore of the channel. SURs are members of the ATP-binding cassette protein superfamily with two nucleotide binding domains of which, in particular, the second exhibits ATPase activity (Bienengraeber et al., 2000; Matsuo et al., 2000). In addition, SUR carries the binding sites for sulphonylureas and the openers (Aguilar-Bryan et al., 1995; Hambrock et al., 1998; Schwanstecher et al., 1998). The respective binding sites are located on different parts of SUR (Ashfield et al., 1999; Ueda et al., 1999; Uhde et al., 1999; Babenko et al., 2000; Moreau et al., 2000) and are linked by allosteric interactions. Mg2+ salts of adenine nucleotides inhibit glibenclamide (GBC) binding (Hambrock et al., 2002a) and, in a reciprocal manner, GBC releases ATP from SUR (Ueda et al., 1999). High-affinity binding of openers to SUR requires the presence of hydrolysable nucleotides (Schwanstecher et al., 1992; 1998; Quast et al., 1993; Hambrock et al., 1998) and, conversely, openers affect the ATPase activity of SUR (Bienengraeber et al., 2000; Zingman et al., 2001). Sulphonylureas and openers are linked by a negative allosteric interaction of the respective binding sites (Bray & Quast, 1992; Schwanstecher et al., 1992).
SUR is encoded by two genes, which gives rise to two subtypes. The first, SUR1, is mainly found in pancreatic β-cells and in the brain (Aguilar-Bryan et al., 1995; Sakura et al., 1995), and the second, SUR2, in muscle. Alternative splicing of SUR2 leads to two isoforms, which differ in the last C-terminal exon, that is, SUR2A in skeletal and cardiac and SUR2B in smooth muscle (Inagaki et al., 1996; Isomoto et al., 1996). The differences in the last 42 amino acids result in some interesting functional differences between the isoforms. Kir6.2/SUR2A channels have been found to be more resistant to activation by MgADP, nicorandil and diazoxide than their SUR2B-containing counterparts (Shindo et al., 1998; Matsuoka et al., 2000; Matsushita et al., 2002). A stretch of 7 amino acids located in the central part of the last 42 amino acids has been shown to be critical for this difference; this stretch may interact with NBF2, thereby modulating nucleotide binding and hydrolysis in a SUR2-isoform-specific manner (Matsushita et al., 2002). Furthermore, MgATP stabilises the complex of channel openers (P1075 and pinacidil) with the Kir6.2/SUR2B channel more strongly than with Kir6.2/SUR2A (Reimann et al., 2000), and MgATP (in the presence of a ATP-regenerating system) depressed [3H]GBC binding to SUR2B more strongly than that to SUR2A (Hambrock et al., 2002a).
Despite intense effort, the binding of sulphonylureas and openers to SUR and the transduction of this binding into changes of channel activity are not yet completely understood. As an example, recent studies using sulphonylurea [3H]GBC and opener [3H]P1075 as radioligands have revealed unexpected complexities in the binding of these prototypical KATP channel ligands to SUR2B, and it was proposed that SUR2B, expressed alone, may form tetramers (Löffler-Walz et al., 2002). In view of the aforementioned differences between the SUR2 isoforms, it was the aim of this study to examine the ligand binding properties of SUR2A, in particular, the inter-relationship between sulphonylureas and openers. Eventual differences to the results with SUR2B (Hambrock et al., 2001; Löffler-Walz et al., 2002) must be caused by the differences in the carboxy-terminal amino acids of the SUR2 isoforms; hence, this comparison allows one to assess the importance of these amino acids in these phenomena.
The study of [3H]GBC binding to the SUR2 subtypes is hampered by the relatively low affinity of this interaction (∼20 nM for SUR2B) and the considerable binding of GBC to proteins other than SUR2 in intact human embryonic kidney (HEK) cells and in HEK cell membrane preparations (for SUR2B, see Russ et al., 1999; Löffler-Walz et al., 2002). Advantage was therefore taken of the identification of Ser 1237 in SUR1 as a residue important for GBC binding; in SUR2, this residue is replaced by Tyr leading to a marked reduction in the potency of sulphonylureas in blocking the Kir6.2/SUR1 channel (Ashfield et al., 1999). Conversely, the mutation Y1206S in SUR2B increased the affinity of SUR2B for GBC by 25 (Toman et al., 2000) to 10-fold (Hambrock et al., 2001). Therefore, in the [3H]GBC binding experiments, use was often made of the SURY2A(1206S) mutant, and the characterization of this mutant as compared to wild-type SUR2A was the second aim of this study. In addition, the effect of coexpression with Kir6.2 on the binding properties of wild-type and mutant SUR2A was investigated as well as the relationship between binding affinity of GBC and potency in blocking the wild-type and mutant Kir6.2/SUR2A channels.
Methods
Cell culture and transfection
The mutant SUR2 subtypes, SUR2A(Y1206S) and SUR2B-(Y1206S), were constructed from the corresponding murine SUR2 clones (SUR2A: GenBank D86037 and SUR2B: GenBank D86038; Isomoto et al., 1996) using the QuikChange Site-Directed Mutagenesis System (Stratagene, Amsterdam, The Netherlands) as described (Hambrock et al., 2001). HEK 293 cells were cultured in minimum essential medium containing glutamine and supplemented with 10% foetal bovine serum and 20 μg ml−1 gentamycin (Hambrock et al., 1998). Cells were transfected with the mammalian expression vector pcDNA 3.1 (Invitrogen, Karlsruhe, Germany) containing the coding sequences of the murine SUR2 clones mentioned above using LipofectAMINE and Optimem (Invitrogen), and cell lines stably expressing these proteins were generated as described (Hambrock et al., 1998). Cells transiently coexpressing SUR2 and murine Kir6.2 (D50581; Inagaki et al., 1996) or Kir6.1 (D88159; Yamada et al., 1997) were transfected with the plasmids at a molar ratio of 1 : 1. In cotransfections prepared for electrophysiological experiments, the pEGFP-C1 vector (Clontech, Palo Alto, CA, U.S.A.), encoding for green fluorescent protein, was added for identification of transfected cells. At 2–4 days after transfection, cells were used for binding studies and electrophysiological experiments.
Membrane preparation and radioligand binding experiments
For cells stably expressing SUR alone, the antibiotic was withdrawn from the culture medium 3–4 days prior to membrane preparation. A crude membrane preparation was obtained by lysing the cells in hypotonic buffer, centrifugation of the lysate at 105 g for 60 min at 4°C and resuspension of the pellet in a Mg2+-free buffer as described (Hambrock et al., 1998). These membranes contain SUR in a concentration similar to that in intact cells. Protein concentration was determined according to Lowry et al. (1951) using bovine serum albumin as the standard.
For radioligand binding competition experiments in membranes in the absence/presence of MgATP, membranes were added to an incubation buffer containing (in mM): HEPES, 5; NaCl, 139; KCl, 5; MgCl2, 0/2.2; EDTA, 1/0 and Na2ATP, 0/1, titrated with NaOH to pH 7.4 and supplemented with the radioligand ([3H]P1075 ∼3 nM or [3H]GBC 2–4 nM) and (unlabelled) GBC, P1075 or levcromakalim. Nonspecific binding (BNS) of [3H]P1075/[3H]GBC was determined in the presence of 10/100 μM P1075 (Hambrock et al., 2001). For [3H]GBC saturation experiments, the radioligand was added in concentrations from 0.2 to 5 × KD. In binding experiments using intact cells, cells were suspended by rinsing with a HEPES-buffered physiological salt solution (PSS) containing (in mM): HEPES, 5; NaCl, 139; KCl, 5; MgCl2, 1.2; CaCl2, 1.25; D(+)-glucose, 11; the buffer was gassed with 95% O2 and 5% CO2 and titrated to pH 7.4 with NaOH at 37°C. Cells were centrifuged two times at 500 × g for 5 min and resuspended in PSS. Incubation was started by the addition of cells (final concentration (1.5–3) × 106 cells ml−1, corresponding to 0.35–0.70 mg protein ml−1) to PSS containing the radioligand in a total volume of 1 ml at pH 7.4 and 37°C.
After equilibrium was reached (15–30 min), incubation was stopped by diluting 0.3 ml aliquots (in triplicate) in 8 ml of ice-cold quench solution (50 mM Tris-(hydroxymethyl)-aminomethane, 154 mM NaCl, pH 7.4). Bound and free ligands were separated by rapid filtration over Whatman GF/B filters (membranes) or GF/C filters (cells). Filters were washed twice with quench solution and counted for [3H] in the presence of 4.5 ml of scintillant (Ultima Gold: Packard, Meriden, CT, U.S.A.).
Electrophysiological experiments
The patch-clamp technique was used in the inside-out configuration as described by Hamill et al. (1981). Patch pipettes were drawn from borosilicate glass capillaries (GC 150 T, Harvard Apparatus, Edenbridge, U.K.) and heat polished using a horizontal microelectrode puller (Zeitz, Augsburg, Germany). Bath and pipette were filled with a high K+-Ringer solution containing (in mM) KCl, 142; NaCl, 2.8; MgCl2, 1; CaCl2, 1; D(+)-glucose, 11; HEPES, 10; titrated to pH 7.4 with NaOH at 23°C. Filled pipettes had a resistance of 1–1.5 MΩ. After excision of the patch, the pipette was moved in front of a pipe with a high K+-Ringer solution containing (in mM) KCl, 143; CaCl2, 1; D(+)-glucose, 11; HEPES, 10; EGTA, 5; the buffer was titrated to pH 7.2 with NaOH at 23°C. MgCl2 was added such that [Mg2+]free was 0.7 mM; ATP, prepared as 100 mM stock solution, was added as indicated. GBC was dissolved as described below and added to the pipe solution. Patches were clamped to −50 mV.
Data were recorded with an EPC9 amplifier (HEKA, Lambrecht, Germany) using the ‘Pulse' software (HEKA). Signals were filtered at 200 Hz using the four-pole Bessel filter of the EPC9 amplifier and sampled with 1 kHz.
Data analysis
Inhibition curves were analysed using the logarithmic form of the Hill equation
Here, A denotes the extent (amplitude) of inhibition, n (=nH) the Hill coefficient and IC50 the midpoint of the curve with pIC50=−log IC50; x is the concentration of the compound under study with px=−log x. If two-component analysis was required, the sum of two logistic terms was used with nH=1 and A2=100−A1. IC50 values were converted into inhibition constants, Ki, by correcting for the presence of the radioligand, L, according to the Cheng & Prusoff (1973) equation
where KD is the equilibrium dissociation constant of the radioligand. In case of homologous competition experiments, the inhibition constant Ki is identical to the KD value. The correction was always <2.0. The fraction of receptors occupied at a given radioligand concentration, L, was calculated according to the Law of Mass Action:
Saturation experiments were analysed by calculating total binding (BTOT) as the sum of specific (BS) and nonspecific binding (BNS). BS was assumed to follow the Law of Mass Action so that
BMAX (fmol mg−1 protein) denotes the concentration of specific binding sites in the preparation, KD is the equilibrium dissociation constant and L is the concentration of the radioligand. In membranes, BNS was proportional to L,
with a as the proportionality constant.
Data are shown as means±s.e.m. Fits of the equations to the data were performed according to the method of least squares using the programme SigmaPlot 6.1 (SPSS Science, Chicago, IL, U.S.A.). The concentration dependence of channel inhibition by GBC was analysed taking all individual data points into account. For binding data, individual experiments were analysed and the parameters averaged assuming that amplitudes and pIC50 values are normally distributed (Christopoulos, 1998). In the text, KD/Ki values are given, followed by the 95% confidence interval in parentheses. When the Hill coefficient was <0.8, analysis of the data according to the two sites model with nH=1 was performed. In calculations involving two mean values with standard errors, propagation of errors was taken into account according to Bevington (1969). Significance of differences between two normally distributed parameters with equal variance was assessed using the two-tailed unpaired Student's t-test.
Materials and solutions
[3H]P1075 (specific activity 4.5 TBq (117 Ci) mmol−1) was purchased from Amersham Buchler (Braunschweig, Germany) and [3H]GBC (specific activity 1.85 TBq (50 Ci) mmol−1) from Perkin-Elmer Life Sciences (Bad Homburg, Germany). The reagents and media used for cell culture and transfection were from Invitrogen. Levcromakalim was a kind gift from GlaxoSmithKline (Harlow, U.K.) and P1075 from Leo Pharmaceuticals (Ballerup, Denmark). GBC was purchased from Sigma (Deisenhofen, Germany) and Na2ATP was from Roche Diagnostics (Mannheim, Germany). KATP channel modulators were dissolved in dimethyl sulphoxide/ethanol (50%/50% (v/v)) to give stock solutions of 0.1 M. These were further diluted with the same solvent or with incubation buffer to give final solvent concentrations <1%. Mg2+-free solutions (no Mg2+ added, contaminating Mg2+⩽10 μM (Forestier & Vivaudou, 1993), EDTA=1 or 5 mM) contained ⩽10 nM [Mg2+]free.
Results
SUR2A and SUR2A(YS): [3H]GBC binding
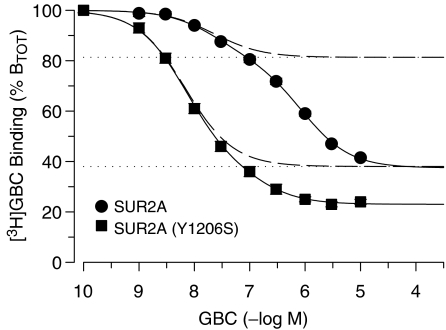
Figure 1 shows the inhibition of [3H]GBC binding by (unlabelled) GBC in membranes from HEK cells stably expressing SUR2A. Experiments were performed in the absence of MgATP, a condition that has been shown to favour GBC binding (Hambrock et al., 2002a; see also below). The inhibition curve extended over three orders of magnitude with a slight hump, indicating the presence of more than one binding component. Two-component analysis gave for the high-affinity component an amplitude of 19% of total binding and a Ki (=KD) value of 20 nM (Table 1). Such high-affinity binding was absent in membranes from HEK cells transfected with the pcDNA 3.1 control vector not encoding SUR2A cells (Hambrock et al., 2002b). In addition, the cyanoguanidine opener P1075, at the saturating concentration of 100 μM, displaced also 19% of total [3H]GBC binding in this series of experiments (Figure 1 and see below). Collectively, this is strong evidence that the high-affinity component represents GBC binding to SUR2A, whereas the low-affinity component represents binding to other GBC sites intrinsic to HEK cell membranes.
Figure 1.
Inhibition of [3H]GBC binding to wild-type and mutant SUR2A by GBC in membranes at 37°C in the absence of MgATP (1 mM EDTA, 0 ATP). Data are expressed as % of total binding (BTOT) to show the contributions of GBC binding to SUR2A and to endogenous GBC sites in HEK cell membranes. Individual inhibition curves for GBC (n=4–6) were analysed according to the two-component model with nH=1 and the resulting parameters were averaged as described in Methods. The parameters of the high-affinity components are listed in Table 1; the parameters of the low-affinity component were (wild-type/mutant SUR2A) A2=44±1/15±1% BTOT and KD,2=900 (780, 1100)/130 (90, 200) nM. The broken curves represent the high-affinity component (i.e. binding to SUR2A and SUR2A(YS), respectively), and the dotted horizontal lines the amount of [3H]GBC displaced by 100 μM P1075. Experiments with wild-type were conducted at 3.5 to 3.8 nM [3H]GBC and BTOT was 200±16 fmol mg−1 (n=16); for SUR2A(YS), [3H]GBC was 2.3 nM and BTOT 310±38 fmol mg−1 (n=6).
Table 1.
[3H]GBC binding competition experiments in membranes and cells
| Receptor | GBC | P1075 | |||
|---|---|---|---|---|---|
| Mg ATP (mM) | KD (nM) | A (%BTOT) | Ki (nM) | A (%BTOT/%BS) | |
| SUR2A | 0 | 20 (14, 28) | 19±2 | 1500 (1100, 2000) | 22±2/100 |
| SUR2A(YS) | 0 | 4.3 (3.0, 6.2) | 62±3 | 2200 (1700, 2800) | 56±4/100 |
| 1 | 8.3 (6.3, 11) | 58±3 | 19 (14, 26) | 30±1/63±2 | |
| 1500 (800, 3000) | 17±1/37±2 | ||||
| Cells | 4.9 (3.4, 7.1) | 68±2 | ND | ||
| Kir6.2/SUR2A | 0 | ∼20a | ND | ||
| Cells | 6.2 (5.6, 6.8) | 39±1 | |||
| Kir6.2/SUR2A(YS) | 1 | c.f. Saturation experiments (Table 2) | 55 (39, 78) | 50±5/62±7 | |
| 950 (550, 1600) | 30±3/38±7 | ||||
Parameters are means from the analysis of individual inhibition curves (n=4–6) as described in Methods and are followed by the 95% confidence interval or s.e.m. Pooled inhibition curves are shown in Figures 1 and 3. A=amplitude.
Owing to low expression of Kir6.2+SUR2A this value is only approximate (see text).
For SUR2A(YS), the homologous competition curve was also biphasic. In this case, however, the high-affinity component (i.e. binding to SUR2A(YS) comprised 62±3% of total binding with a Ki value of 4.3 nM (Figure 1, Table 1). First, this showed that the mutation increased the affinity of SUR2A for GBC by ∼5 times. Second, the amplitude of the high-affinity component was ∼3 × higher than that seen with wild-type SUR2A. This is a direct consequence of the increased affinity of mutant SUR2A for GBC: using similar radioligand concentrations in the two cases (2.3 and ∼3.5 nM, respectively), the increase in affinity leads to a higher fractional receptor occupancy by the radioligand, that is, to a larger amplitude of the high-affinity component (cf. equation (3) in Methods).
Effect of MgATP
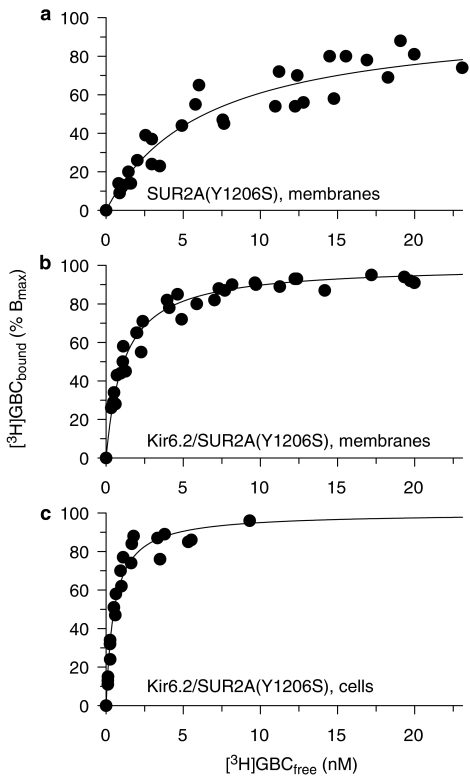
We have shown previously that MgATP reduces GBC binding to all SUR subtypes (Hambrock et al., 2002a). In the presence of MgATP (1 mM), specific [3H]GBC binding to wild-type SUR2A was too small to be reliably measured. With the mutant, the inhibition curve was biphasic and analysis gave a KD of 8.3 nM for the high-affinity component, which represents binding to SUR2A(YS) (Table 1). More detailed information was obtained in [3H]GBC saturation experiments (Figure 2a). Table 2 shows that the KD values obtained in these experiments were in good agreement with those from the competition experiments (Table 1). More importantly, the saturation experiments showed that inhibitory action of MgATP was due to a reduction of the number of GBC binding sites to 55±11%, whereas the KD value remained unchanged.
Figure 2.
Saturation of [3H]GBC binding to SUR2A(YS) in membranes (a, b) and cells (c). Data represent normalised specific binding pooled from three to four experiments. Binding is to SUR2A(YS) expressed alone (a) and to Kir6.2/SUR2A(YS) (b, c). Experiments in membranes were performed in the presence of MgATP (1 mM). Individual experiments were evaluated according to equations (4) and (5) in Methods section; mean parameters of specific binding are listed in Table 2. Nonspecific binding was given by equation (5) with a=39±2, 32±2 and 67±2 fmol mg−1 nM−1 for SUR2A(AS) and Kir6.2/SUR2A(YS) in membranes and cells, respectively.
Table 2.
[3H]GBC binding to SUR2A(YS) as determined in saturation binding experiments
| Receptor | MgATP (mM) | KD (nM) | BMAX (fmol mg−1) |
|---|---|---|---|
| SUR2A(YS) | 0 | 5.6 (3.9, 8.1) | 1144±128 |
| 1 | 6.6 (4.5, 10) | 626±107a | |
| Kir6.2/SUR2A(YS) | 0 | 1.9 (1.1, 2.7) | 772±102 |
| 1 | 1.2 (0.9, 1.5) | 569±48a | |
| 0b | 0.88 (0.76, 1.0)b | 463±12b | |
| Cells | 0.53 (0.40, 0.69) | 310±23 | |
| Kir6.1/SUR2A(YS) | Cells | 0.75 (0.66, 0.87)c | 180±24 |
The respective experiments in the absence and presence of MgATP were carried out side by side using the same membrane preparation(s). Parameters were determined from experiments as shown in Figure 2.
Different from the value in the absence of MgATP (P<0.01 and <0.05, respectively).
Experiments at 23°C; incubation time was 45 min.
Different from Kir6.2/SUR2A(YS) (P<0.05).
Effect of coexpression with Kir6.2
Coexpression with Kir6.x increases the affinity of SUR2B and SUR2B(YS) for GBC in intact cells but not in membranes (Russ et al., 1999; Hambrock et al., 2001). Here, we have examined the effect of coexpression on wild-type and mutant SUR2A. Experiments with wild-type SUR2A proved difficult since coexpression with Kir6.2 at a molar ratio of 1 : 1 reduced [3H]GBC binding to SUR2A to ∼35% (see also Russ et al., 1999) and specific binding was low. In membranes and in the absence of MgATP, GBC inhibition curves were performed and a KD value around 20 nM was estimated for GBC binding (Table 1). This is similar to the value obtained with SUR2A alone (Table 1) and shows that under these conditions, coexpression with Kir6.2 did not increase the affinity of SUR2A for GBC.
In the case of SUR2A(YS), the higher affinity for GBC allowed saturation experiments of [3H]GBC binding to be performed. In the absence of MgATP, coexpression with Kir6.2 decreased KD from 5.6 to 1.9 nM (Table 2), that is, the affinity for GBC was increased by a factor of 2.9 (1.4, 5.9). The experiments in the presence of MgATP are shown in Figure 2. The presence of Kir6.2 decreased the KD value of GBC binding from 6.6 to 1.2 nM (Table 2), that is, the affinity was increased 5.5 (3.3, 8.9) times. Hence, the coexpression-induced increase in the affinity of SUR2A(YS) for GBC was significantly larger in the presence of MgATP than in its absence (two-tailed t-test performed on the pKD values, P<0.05). In addition, coexpression with Kir6.2 significantly reduced the depressing effect of MgATP on the BMAX values; these were 55% for SUR2A(YS) alone and 74% for coexpression (Table 2).
The effect of coexpression on the affinity of SUR2A(YS) for [3H]GBC was also determined in intact cells. For SUR2A(YS) alone, the KD value was 4.9 nM (Table 1); coexpression with Kir6.2 shifted the value to 0.53 nM and with Kir6.1 to 0.75 nM (Table 2). The two values were different, P<0.05. Hence, affinity was increased by ∼9 and 7 times, respectively, showing a slight dependence of this effect on the Kir6.x subtype.
Opener binding
The effect of the Y1206S mutation on the opener binding properties of SUR2A was studied using the opener [3H]P1075 as the radioligand in the presence of MgATP (1 mM). Inhibition curves with unlabelled P1075 and levcromakalim were monophasic with Hill coefficient 1 and gave KD/Ki values of 21 and 910 nM, respectively (Table 3). Comparison with the corresponding values for SUR2A wild-type (Table 3) showed that the mutation left the affinity of SUR2A for P1075 essentially unchanged but increased the Ki value of levcromakalim by a factor of 2.
Table 3.
[3H]P1075 competition experiments in membranes in the presence of 1 mM MgATP
| Receptor | P1075 | Levcromakalim | GBC | |
|---|---|---|---|---|
| KD (nM) | Ki (nM) | Ki (nM) | A (%Bs) | |
| SUR2A | 17 (15, 20) | 430 (360, 510) | 500 (400, 630) | 100 |
| SUR2A(YS) | 21 (18, 26) | 910 (660, 1260) | 130 (110, 150) | 100 |
| Kir6.2/SUR2A | 28 (26, 29) | ND | 15 (10, 23) | 44±5 |
| 310 (230, 410) | 56±5 | |||
| Kir6.2/SUR2A(YS) | 35 (29, 42) | ND | 1.3 (0.93, 1.8) | 76±3 |
| 21 (14, 32) | 24±3 | |||
Parameters are means derived from three to eight experiments; the pooled inhibition curves for GBC are shown in Figure 4. ND=not determined.
Coexpression of wild-type SUR2A with Kir6.2 shifted KD value of P1075 from 17 to 28 nM (Table 3), that is, by a factor of 1.6 (1.4, 1.9). With mutant SUR2A, there was a similar shift from 21 to 35 nM, that is, by a factor of 1.7 (1.3, 2.2) (Table 3). In both cases, the shifts were significant (P<0.05). This showed that coexpression with Kir6.2 slightly but significantly decreased the affinity of wild-type and mutant SUR2A for P1075.
The allosteric interaction between GBC and P1075
KATP channel openers inhibit [3H]GBC binding to SUR by a negative allosteric interaction and in a MgATP-dependent manner (SUR1: Schwanstecher et al., 1992; SUR2B: Hambrock et al., 2001; Löffler-Walz et al., 2002; Russ et al., 2003). Here, we have examined the inhibition of [3H]GBC binding to (wild-type and mutant) SUR2A by P1075. In the absence of MgATP, P1075 inhibited 22% of total [3H]GBC binding (BTOT) to wild-type SUR2A in a monophasic manner (nH=0.95±0.10) and with Ki=1.5 μM (Table 1; in another series of experiments, maximum inhibition was 19% BTOT (Figure 1)). For SUR2A(YS), the inhibition curve was again monophasic (nH=0.94±0.06) and of low affinity (Ki=2.2 μM); in this case, maximum inhibition was 56±4% BTOT (Table 1; the inhibition of specific binding is shown in Figure 3). The two Ki values agree well with one another, suggesting that the mutation does not much affect the interaction of P1075 with SUR2A in the absence of MgATP.
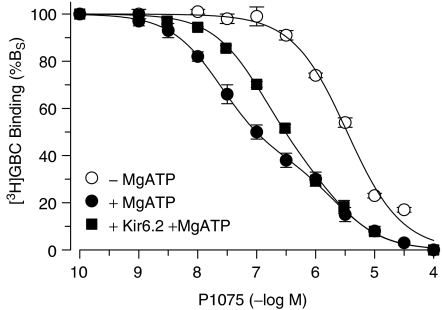
Figure 3.
Inhibition of [3H]GBC binding to SUR2A(YS) by P1075: effect of MgATP (1 mM) and of coexpression with Kir6.2. Data are means±s.e.m. from four to five independent experiments and are expressed as % specific binding (%BS). Mean fitting parameters obtained from the analysis of the individual inhibition curves are listed in Table 1. BTOT values, determined at [3H]GBC concentrations of 2.5–1.5 nM, ranged from 440 to 200 fmol mg−1 and nonspecific binding from 50 to 20% of BTOT.
In the presence of MgATP, P1075 inhibited [3H]GBC binding to SUR2A(YS) in a markedly biphasic manner (Figure 3, Table 1). The high-affinity component comprised 63% of specific binding (BS) with a Ki value of 19 nM. The Ki value agrees very well with that determined with [3H]P1075 as the radioligand (21 nM; Table 3); hence, this component gives the true affinity of SUR2A(YS) for the opener. The low-affinity component (Ki=1.5 μM) agrees well with that seen in the absence of MgATP. Coexpression with Kir6.2 reduced the biphasic nature of the inhibition curve (Figure 3). Inspection of Table 1 shows that this was essentially due to an increase in the Ki value of the high-affinity component by almost three times. Thereby, the Ki ratio of the two components decreased from 79 to 17; the ratio of the amplitudes (%BS) remained unchanged.
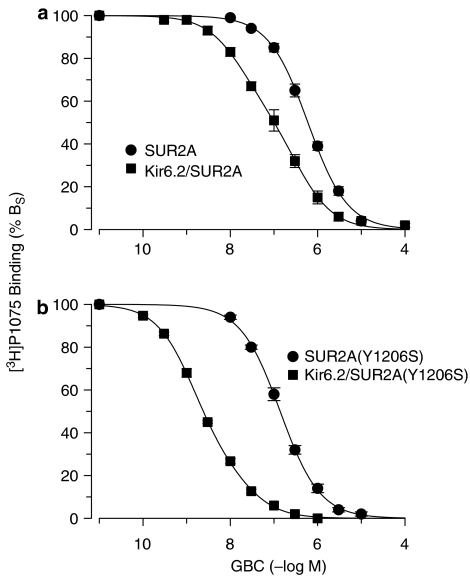
The negative allosteric interaction between GBC and P1075 was also probed using [3H]P1075. Figure 4 shows that GBC inhibited [3H]P1075 binding to SUR2A and SUR2A(YS) with Ki values of 500 and 130 nM, respectively (Table 3). This showed that this type of assay underestimates the true affinity of GBC by about 20 times and the mutation increased the potency of GBC to allosterically interfere with P1075 binding by ∼4 times. Coexpression with Kir6.2 induced a leftward shift and a flattening of the [3H]P1075–GBC inhibition curves (Figure 4). For wild-type SUR2A, coexpression shifted the inhibition curve approximately 5 × to the left (IC50 ∼100 nM) with Hill coefficient nH=0.71±0.02 (Figure 4a). The latter suggested the presence of two classes of GBC sites. The fit of the two component model with nH=1 gave a high-affinity component comprising 44% of BS with Ki=15 nM and a second component with A2=56% BS and Ki,2=310 nM (Table 3). In case of SUR2A(YS), coexpression induced a 54-fold leftward shift (from Ki=130 to 2.4 nM) and a slight flattening (nH=0.81±0.02) (Figure 4b). The fit of the two-component model gave a dominant high-affinity component comprising 76% of the total inhibition with a Ki value of 1.3 nM; the Ki of the low-affinity component was 21 nM and these parameters are listed in Table 3. Owing to the only minute deviation from homogeneity, the parameters of the two sites fit must be considered with caution; however, it is obvious that the effect of coexpression with Kir6.2 was much stronger for mutant than for wild-type SUR2A (Figure 4).
Figure 4.
Inhibition of [3H]P1075 binding to wild-type and mutant SUR2A by GBC: effect of coexpression with Kir6.2. (a) SUR2A and (b) SUR2A(YS). Data are means from three to six experiments; fitting parameters are listed in Table 3. Experiments were performed in membrane in the presence of MgATP (1 mM).
Inhibition of the Kir6.2/SUR2A(YS) channel by GBC
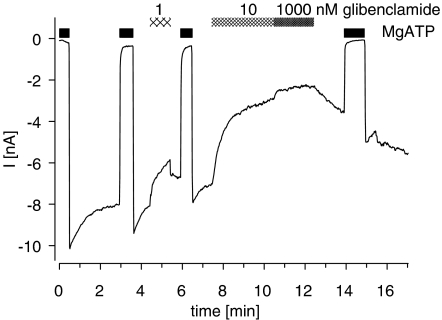
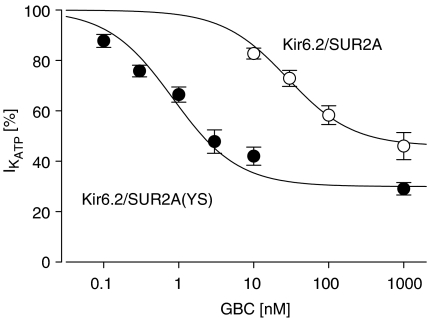
To assess the functional effect of the mutation Y1206S in SUR2A, inhibition of Kir6.2/SUR2A(YS) by GBC was examined in inside-out patches. Figure 5 shows the current from a patch exposed to increasing concentrations of GBC. At 1 μM, the highest concentration used, the channel was not completely blocked and 37% of the ATP-sensitive current remained. The concentration dependence of the inhibition is illustrated in Figure 6, and it gave an IC50 value of 0.83 nM and a maximum inhibition of 70% (Table 4). The corresponding data for the wild-type channel that we reported earlier (Russ et al., 2001) are also shown in Figure 6; IC50 was 26 nM (Table 4). Hence, the mutation increased the potency of GBC by a factor of 31 (21, 45). There was also a difference in the maximum inhibition between wild-type and mutant channel.
Figure 5.
Recording from an inside-out patch showing inhibition of the Kir6.2/SUR2A(YS) channel by GBC. After excision of the patch into nucleotide-free solution, a current was present, which was abolished by superfusion with MgATP (1 mM) and showed some run-down. MgATP was applied repeatedly to induce refreshment of channels from run-down and GBC intermittently as indicated by the hatched bars. Holding potential was −50 mV; experiments were performed in symmetrical high K+ buffer at 23°C. Data were corrected manually for run-down and for weakening of the MgATP block. Note the slow washout of GBC (1000 nM).
Figure 6.
Concentration-dependent inhibition of wild-type and mutant Kir6.2/SUR2A channels by GBC. Data were obtained as shown in Figure 5 and normalised with respect to the ATP-sensitive current ( ) prior to the application of GBC. Concentration dependencies were analysed using the Hill equation with nH=1; the parameters are listed in Table 4. n=4–10 per data point; data for wild type are from Russ et al. (2001).
) prior to the application of GBC. Concentration dependencies were analysed using the Hill equation with nH=1; the parameters are listed in Table 4. n=4–10 per data point; data for wild type are from Russ et al. (2001).
Table 4.
Concentration-dependent inhibition of wild-type and mutant Kir6.2/SUR2 channels by GBC in inside-out patches
| Channel | IC50 (nM) | A (%) |
|---|---|---|
| Kir6.2/SUR2Aa | 26 (23, 29) | 54±3 |
| Kir6.2/SUR2A(YS) | 0.83 (0.58, 1.2) | 70±2 |
| Kir6.2/SUR2Ba | 27 (22, 32) | 63±2 |
| Kit6.2/SUR2B(YS) | 1.0 (0.70, 1.5) | 65±2 |
In addition, we have examined the sensitivity of the Kir6.2/SUR2B(YS) channel to inhibition by GBC. The inhibition curve (not illustrated) was very similar to that of the Kir6.2/SUR2A(YS) channel (see parameters in Table 4). Comparison with the data for the wild-type channel (Russ et al., 2001; here: Table 4) shows that the Kir6.2/SUR2B(YS) channel was 27 (17, 41) times more sensitive to inhibition by GBC than the corresponding wild-type channel (Table 4). Hence, there was no difference between the two SUR2 isoforms in this respect. With SUR2B, there was no difference in the maximum inhibition between wild-type and mutant channel (Table 4).
Discussion
Binding properties of wild-type and mutant SUR2A
The KD value of GBC binding to wild-type SUR2A in membranes and in the absence of MgATP (20 nM) was similar to that determined before for SUR2B (KD=22 nM; Löffler-Walz et al., 2002). Hence, there is no difference between the SUR2 isoforms in their affinity for GBC under these conditions. Owing to high nonspecific binding, [3H]GBC binding experiments were not possible in the presence of MgATP nor in intact cells. The KD value determined here for P1075 binding to SUR2A in the presence of MgATP (Table 3) is the same as that determined earlier (Hambrock et al., 1999) and is 2–5 times lower than that of SUR2B (Hambrock et al., 1999; Felsch et al., 2004). The mutation Y1206S increased the affinity of SUR2A for GBC ∼5 times (Table 1) and enhanced the potency of GBC to inhibit [3H]P1075 binding ∼4 × (Table 3) but it did not affect the affinity of SUR2A for openers (Table 3). These results are in quantitative agreement with the effects of the Y1206S-mutation of SUR2B (Hambrock et al., 2001; Löffler-Walz et al., 2002; Russ et al., 2003), showing that the differences in the carboxy-terminal amino acids of SUR2 do not modulate the effect of the mutation on ligand binding of SUR2 expressed alone.
Effects of coexpression with Kir6.x
Coexpression of wild-type and mutant SUR2A with Kir6.x had two major effects: it increased the affinity for GBC and it decreased the inhibitory effect of MgATP. Before discussing these effects in detail, we note that in the absence of Kir6.2, SUR1 is immature, core glycosylated and retained in the endoplasmic reticulum. Upon coexpression with Kir6.2, Kir6.2/SUR1 complexes assemble to form the tetradimeric channel, which transits through the Golgi apparatus, obtains its mature glycosylation and is incorporated in the membrane (Clement IV et al., 1997; Zerangue et al., 1999; Crane & Aguilar-Bryan, 2004). Therefore, at least three factors may contribute to the observed effects of coexpression (a) differences in the glycosylation of SUR, (b) contacts with the cytoskeleton that SUR alone retained in the endoplasmic reticulum cannot make, and (c) changes in SUR conformation induced by the coupling to Kir.
Turning now to the increase in affinity, we note that this effect was absent with wild-type SUR2A in membranes in the absence of MgATP. In case of the mutant, however, coexpression with Kir6.2 increased the affinity for GBC in membranes, and the effect was enhanced by the presence of MgATP (Table 2). This is in contrast to our earlier results with SUR2B(YS); in this case, coexpression with Kir6.x did not increase the affinity for GBC in membranes, regardless of the presence of MgATP (Hambrock et al., 2001). This difference suggests that the carboxy-terminal tail of SUR2 plays a role in the coupling of Kir6.2 to SUR2 to increase GBC affinity. The results in Tables 1 and 2 further show that the mutation augmented the effect of coexpression on the affinity of SUR2A for GBC. This is also seen in the [3H]P1075–GBC inhibition curves: in the absence of Kir6.2, the mutation increased the potency of GBC ∼4 × (Table 3); after coexpression with Kir6.2, the mutation increased potency ∼12 × (Table 3, high-affinity sites). In cells, coexpression with Kir6.x gave concordant results for the SUR2 isoforms: Kir6.x increased the GBC affinity of wild-type and mutant SUR2 and Kir6.2 was ∼2 × more efficient than Kir6.1 in this respect (SUR2A: Table 2; SUR2B: Hambrock et al., 2001).
Which of the three factors mentioned above may be responsible for these effects? Work with SUR1 suggests that Kir-induced conformation changes in SUR are important. For SUR1, the increase in affinity for sulphonylureas and repaglinide that is induced by coexpression with Kir6.2 critically depends on the first 10 to 14 N-terminal amino acids of Kir6.2 (Hansen et al., 2005; Stephan et al., unpublished observations); similarly, the block of Kir6.2/SUR1 channels by these compounds depends on the presence of these amino acids (Babenko et al., 1999; Reimann et al., 1999, Hansen et al., in press). Assuming that this holds true also for Kir6.2/SUR2A, one might hypothesise that the mutation Y1206S, located in the 8th intracellular loop of SUR2A, changes the binding pocket of SUR2A in a way that coupling with the amino-terminus of Kir6.2 is improved.
Turning to the MgATP sensitivity of GBC binding, we note that with SUR2A(YS) expressed alone, MgATP reduced the number of GBC binding sites by ∼50% leaving KD unchanged; upon coexpression with Kir6.2, MgATP reduced BMAX by ∼25% (Table 2). Qualitatively similar results were obtained earlier with SUR2B(YS): in this case, MgATP reduced BMAX by ∼70% and the effect was essentially reversed by coexpression with Kir6.2 (Hambrock et al., 2002a; Löffler-Walz et al., 2002). Mechanistically, these observations may be explained on the basis of the tetramer model that we have proposed recently for SUR2B (Löffler-Walz et al., 2002) and that, based on the present results, can be extended to SUR2A: the model proposes that SUR2, expressed alone, forms tetramers. In the presence of MgATP, the subunits are linked by strong negative cooperativity of GBC binding, allowing high-affinity binding of only one (SUR2B) to two (SUR2A) molecules of GBC per tetramer. Coexpression with Kir6.2 weakens the negative allosteric interactions between the SUR2 subunits, so that more molecules of GBC can bind per channel. Summing up the last points one can say that coexpression with Kir6.x increases the binding of GBC to SUR2 by two distinct mechanisms, an increase in affinity and a (partial) reversal of the MgATP-induced reduction in high-affinity GBC binding sites. The magnitude of these effects depends on the Kir6.x subtypes, the SUR2 isoforms and the preparation (cells vs membranes±MgATP).
Allosteric interactions between GBC and P1075
The allosteric interactions between the SUR binding sites for GBC and P1075 are most prominent in the presence of MgATP, that is, when SUR is in the high-affinity state for openers. Considering the [3H]GBC–P1075 inhibition curve with SUR2A(YS) first, it is recalled that the curve was biphasic. The Ki value of the high-affinity component corresponded to the true affinity of P1075; that of the low-affinity component agreed with that found in the absence of MgATP (Table 1). With SUR2B(YS), very similar observations were made (Hambrock et al., 2001; Löffler-Walz et al., 2002). This pattern, which is shared by a large group of openers (Russ et al., 2003), may again be interpreted in terms of the tetramer model for SUR2. The high-affinity component reflects binding of the opener to the SUR subunits not occupied by [3H]GBC; this weakens binding of the radioligand by a negative allosteric interaction between the subunits. The low-affinity component reflects opener binding to the SUR subunit(s) occupied by [3H]GBC leading to the complete displacement of the radioligand (Löffler-Walz et al., 2002).
Coexpression with Kir6.2 did not change the amplitude ratio of the [3H]GBC–P1075 inhibition curve but modified the Ki values: the high-affinity Ki was shifted ∼3 times to the right in good agreement with the observations that coexpression weakens opener affinity (Table 3). The low-affinity Ki was slightly shifted leftwards so that the heterogeneity of the curve was reduced. Two points may be taken as circumstantial evidence for the tetramer model. First, the biphasic nature of the [3H]GBC–P1075 inhibition curves first observed with SUR2B(YS) extends also to SUR2A(YS); hence, the phenomenon is general for SUR2. Second, coexpression with Kir6.2 does not change the biphasic nature of the curves (for SUR2B, see Hambrock et al., 2001); in the complete channel, however, biochemical evidence shows that SUR is present as a tetramer (Clement IV et al., 1997; Shyng & Nichols, 1997).
Adopting the complementary approach and studying the [3H]P1075–GBC inhibition curves for wild-type and mutant SUR2A, it is noted that these curves were monophasic with Ki values ∼20 times higher than the true KD of GBC binding. Coexpression with Kir6.2 rendered the inhibition curves biphasic and the Ki values of the high-affinity component agreed well with the KD values of GBC binding to wild-type and mutant SUR2A. Analogous observations had been made with wild-type and mutant SUR2B (Hambrock et al., 2001). These results can again be interpreted within the framework of the tetramer model for SUR2A assuming that binding of [3H]P1075 to some subunits of the tetramer shifts all subunits uniformly to a low-affinity state for GBC binding. Coexpression with Kir6.2 weakens the interactions between the subunits and creates an asymmetry: GBC now binds with its normal affinity to the SUR2A subunits not occupied by [3H]P1075 and thereby weakens the binding of the radioligand; the low-affinity component reflects displacement of the radioligand by GBC binding to the other subunits.
GBC binding and channel closing
The potency of GBC in blocking wild-type and mutant Kir6.2/SUR2A channels in inside-out patches at 23°C compared well with the KD values of GBC binding to the channels in membranes in the absence of MgATP at 37°C (Tables 4, 1 and 2). For Kir6.2/SUR2A(YS), [3H]GBC binding experiments were also performed at 23°C; then, the KD value (0.88 nM, Table 2) was in exact agreement with the inhibition constant for channel closing (0.84 nM). For GBC interacting with Kir6.2/SUR2B(YS), there was also a good correspondence between the binding affinity (2.7 nM; value at 37°C, 1 mM MgATP; Hambrock et al., 2001) and the potency for channel block (1 nM; Table 4). The close agreement between affinity and potency of GBC is not predicted by simple models of the tetradimeric channel, in which binding of one or four GBC molecules to independent sites is required for channel closure (Dörschner et al., 1999; Russ et al., 1999), and more complex models may be needed. In addition, if GBC binding to Kir6.x/SUR2 channels in intact cells is compared to channel block in the whole-cell configuration in the presence of nucleotides, the binding curves are at the left of the channel blocking curves by factors ranging from 7 (Kir6.1/SUR2B, Russ et al., 1999) to >20 (Kir6.2/SUR2B and /SUR2B(YS); Hambrock et al., 2001). This shows that in the presence of nucleotides, the signal transduction, which links binding of sulphonylureas to channel closure is even more complex. Furthermore, full occupation of the GBC sites of SUR in the Kir6.2/SUR2 channels reduced channel open probability in isolated patches in the absence of nucleotides only to 46–30% of control (Table 4), an observation that also holds for the Kir6.2/SUR1 channel (review: Proks et al., 2002).
In conclusion, we have shown here that the mutation Y1206S in SUR2A increases the affinity for GBC binding and enhances the effect of coexpression on GBC affinity. Similarly, the mutation increases the potency of GBC in blocking the channels formed with Kir6.2. In addition, coexpression of wild-type and mutant SUR2A with Kir6.2 affects essentially all aspects of ligand binding to SUR, increasing the affinity for GBC, decreasing that for P1075 and modifying the allosteric interactions between GBC and opener, and GBC and nucleotide binding. Subtle differences with SUR2B show that the differences in carboxy-terminal amino acids of SUR2 play only a minor role in these effects.
Acknowledgments
This study was supported by the Deutsche Forschungsgemeinschaft (Qu 100/3-1, U.L., D. S. and U.Q.). We thank Drs Y. Kurachi and Y. Horio (Osaka) for the generous gift of the murine clones of SUR2A, 2B and Kir6.x.
Abbreviations
- GBC
glibenclamide
- HEK cells
human embryonic kidney 293 cells
- KATP channel
ATP-sensitive K+ channel
- Kir
inwardly rectifying K+ channel
- P1075
N-cyano-N′-(1,1-dimethylpropyl)-N″-3-pyridylguanidine
- SUR
sulphonylurea receptor
- SUR2A(YS)
SUR2A(Y1206S)
References
- AGUILAR-BRYAN L., BRYAN J. Molecular biology of adenosine triphosphate-sensitive potassium channels. Endocrine Rev. 1999;20:101–135. doi: 10.1210/edrv.20.2.0361. [DOI] [PubMed] [Google Scholar]
- AGUILAR-BRYAN L., NICHOLS C.G., WECHSLER S.W., CLEMENT IV J.P., BOYD A.E., III, GONZÁLES G., HERRERA-SOZA H., NGUY K., BRYAN J., NELSON D.A. Cloning of the β cell high-affinity sulfonylurea receptor: a regulator of insulin secretion. Science. 1995;268:423–426. doi: 10.1126/science.7716547. [DOI] [PubMed] [Google Scholar]
- ASHCROFT F.M., GRIBBLE F.M. Correlating structure and function in ATP-sensitive K+ channels. Trends Neurosci. 1998;21:288–294. doi: 10.1016/s0166-2236(98)01225-9. [DOI] [PubMed] [Google Scholar]
- ASHCROFT S.J.H., ASHCROFT F.M. Properties and functions of ATP-sensitive K-channels. Cell. Signal. 1990;2:197–214. doi: 10.1016/0898-6568(90)90048-f. [DOI] [PubMed] [Google Scholar]
- ASHFIELD R., GRIBBLE F.M., ASHCROFT S.J.H., ASHCROFT F.M. Identification of the high-affinity tolbutamide site on the SUR1 subunit of the KATP channel. Diabetes. 1999;48:1341–1347. doi: 10.2337/diabetes.48.6.1341. [DOI] [PubMed] [Google Scholar]
- BABENKO A.P., GONZALEZ G., BRYAN J. The tolbutamide site of SUR1 and a mechanism for its functional coupling to KATP channel closure. FEBS Lett. 1999;459:367–376. doi: 10.1016/s0014-5793(99)01215-6. [DOI] [PubMed] [Google Scholar]
- BABENKO A.P., GONZALEZ G., BRYAN J. Pharmaco-topology of sulfonylurea receptors – separate domains of the regulatory subunits of KATP channel isoforms are required for selective interaction with K+ channel openers. J. Biol. Chem. 2000;275:717–720. doi: 10.1074/jbc.275.2.717. [DOI] [PubMed] [Google Scholar]
- BEVINGTON P.R. Data Reduction and Error Analysis for the Physical Sciences 1969New York: McGraw-Hill; pp. 55–65 and 92–118 [Google Scholar]
- BIENENGRAEBER M., ALEKSEEV A.E., ABRAHAM M.R., CARRASCO A.J., MOREAU C., VIVAUDOU M., DZEJA P.P., TERZIC A. ATPase activity of the sulfonylurea receptor: a catalytic function for the KATP channel complex. FASEB J. 2000;14:1943–1952. doi: 10.1096/fj.00-0027com. [DOI] [PubMed] [Google Scholar]
- BRAY K.M., QUAST U. A specific binding site for K+ channel openers in rat aorta. J. Biol. Chem. 1992;267:11689–11692. [PubMed] [Google Scholar]
- CHENG Y., PRUSOFF W.H. Relationship between the inhibition constant (Ki) and the concentration of inhibitor which causes 50% inhibition (IC50) of an enzymatic reaction. Biochem. Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- CHRISTOPOULOS A. Assessing the distribution of parameters in models of ligand-receptor interaction: to log or not to log. Trends Pharmacol. Sci. 1998;19:351–357. doi: 10.1016/s0165-6147(98)01240-1. [DOI] [PubMed] [Google Scholar]
- CLEMENT IV J.P., KUNJILWAR K., GONZALEZ G., SCHWANSTECHER M., PANTEN U., AGUILAR-BRYAN L., BRYAN J. Association and stoichiometry of KATP channel subunits. Neuron. 1997;18:827–838. doi: 10.1016/s0896-6273(00)80321-9. [DOI] [PubMed] [Google Scholar]
- COGHLAN M.J., CARROLL W.A., GOPALAKRISHNAN M. Recent developments in the biology and medicinal chemistry of potassium channel modulators: update from a decade of progress. J. Med. Chem. 2001;44:1627–1653. doi: 10.1021/jm000484+. [DOI] [PubMed] [Google Scholar]
- CRANE A., AGUILAR-BRYAN L. Assembly, maturation, and turnover of KATP channel subunits. J. Biol. Chem. 2004;279:9080–9090. doi: 10.1074/jbc.M311079200. [DOI] [PubMed] [Google Scholar]
- DÖRSCHNER H., BREKARDIN E., UHDE I., SCHWANSTECHER C., SCHWANSTECHER M. Stoichiometry of sulfonylurea-induced ATP-sensitive potassium channel closure. Mol. Pharmacol. 1999;55:1060–1066. doi: 10.1124/mol.55.6.1060. [DOI] [PubMed] [Google Scholar]
- FELSCH H., LANGE U., HAMBROCK A., LÖFFLER-WALZ C., RUSS U., CARROLL W.A., GOPALAKRISHNAN M., QUAST U. Interaction of a novel dihydropyridine K+ channel opener, A-312110, with recombinant sulphonylurea receptors and KATP channels: comparison with the cyanoguanidine P1075. Br. J. Pharmacol. 2004;141:1098–1105. doi: 10.1038/sj.bjp.0705718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FORESTIER C., VIVAUDOU M. Modulation by Mg2+ and ADP of ATP-sensitive potassium channels in frog skeletal muscle. J. Membr. Biol. 1993;132:87–94. doi: 10.1007/BF00233054. [DOI] [PubMed] [Google Scholar]
- GRIBBLE F.M., REIMANN F. Sulphonylurea action revisited: the post-cloning era. Diabetologia. 2003;46:875–891. doi: 10.1007/s00125-003-1143-3. [DOI] [PubMed] [Google Scholar]
- HAMBROCK A., LÖFFLER-WALZ C., KLOOR D., DELABAR U., HORIO Y., KURACHI Y., QUAST U. ATP-sensitive K+ channel modulator binding to sulfonylurea receptors SUR2A and SUR2B: opposite effects of MgADP. Mol. Pharmacol. 1999;55:832–840. [PubMed] [Google Scholar]
- HAMBROCK A., LÖFFLER-WALZ C., KURACHI Y., QUAST U. Mg2+ and ATP dependence of KATP channel modulator binding to the recombinant sulphonylurea receptor, SUR2B. Br. J. Pharmacol. 1998;125:577–583. doi: 10.1038/sj.bjp.0702109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAMBROCK A., LÖFFLER-WALZ C., QUAST U. Glibenclamide binding to sulphonylurea receptor subtypes: dependence on adenine nucleotides. Br. J. Pharmacol. 2002a;136:995–1004. doi: 10.1038/sj.bjp.0704801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAMBROCK A., LÖFFLER-WALZ C., RUSS U., LANGE U., QUAST U. Characterization of a mutant sulfonylurea receptor SUR2B with high affinity for sulfonylureas and openers: Differences in the coupling to Kir6.x subtypes. Mol. Pharmacol. 2001;60:190–199. doi: 10.1124/mol.60.1.190. [DOI] [PubMed] [Google Scholar]
- HAMBROCK A., PREISIG-MÜLLER R., RUSS U., PIEHL A., HANLEY P.J., RAY J., DAUT J., QUAST U., DERST C. Four novel splice variants of sulfonylurea receptor 1. Am. J. Physiol. (Cell Physiol.) 2002b;283:C587–C598. doi: 10.1152/ajpcell.00083.2002. [DOI] [PubMed] [Google Scholar]
- HAMILL O.P., MARTY A., NEHER E., SAKMANN B., SIGWORTH F.J. Improved patch-clamp techniques for high-resolution current recording from cells and cell-free membrane patches. Pflügers Arch. Eur. J. Physiol. 1981;391:85–100. doi: 10.1007/BF00656997. [DOI] [PubMed] [Google Scholar]
- HAMILTON T.C., WEIR S.W., WESTON A.H. Comparison of the effects of BRL 34915 and verapamil on electrical and mechanical activity in rat portal vein. Br. J. Pharmacol. 1986;88:103–111. doi: 10.1111/j.1476-5381.1986.tb09476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HANSEN A.M.K., HANSEN J.B., CARR R.D., ASHCROFT F.M., WAHL P. Kir6.2-dependent high-affinity repaglinide binding to β-cell KATP channels. Br. J. Pharmacol. 2005;144:551–557. doi: 10.1038/sj.bjp.0706082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- INAGAKI N., GONOI T., CLEMENT IV J.P., WANG C.Z., AGUILAR-BRYAN L., BRYAN J., SEINO S. A family of suphonylurea receptors determines the pharmacological properties of ATP-sensitive K+ channels. Neuron. 1996;16:1011–1017. doi: 10.1016/s0896-6273(00)80124-5. [DOI] [PubMed] [Google Scholar]
- ISOMOTO S., KONDO C., YAMADA M., MATSUMOTO S., HIGASHIGUCHI O., HORIO Y., MATSUZAWA Y., KURACHI Y. A novel sulfonylurea receptor forms with BIR (KIR6.2) a smooth muscle type ATP-sensitive K+ channel. J. Biol. Chem. 1996;271:24321–24324. doi: 10.1074/jbc.271.40.24321. [DOI] [PubMed] [Google Scholar]
- LAWSON K. Potassium channel activation: a potential therapeutic approach. Pharmacol. Ther. 1996;70:39–63. doi: 10.1016/0163-7258(96)00003-4. [DOI] [PubMed] [Google Scholar]
- LÖFFLER-WALZ C., HAMBROCK A., QUAST U. Interaction of KATP channel modulators with sulfonylurea receptor SUR2B: implication for tetramer formation and allosteric coupling of subunits. Mol. Pharmacol. 2002;61:407–414. doi: 10.1124/mol.61.2.407. [DOI] [PubMed] [Google Scholar]
- LOWRY O.H., ROSEBROUGH N.J., FARR A.L., RANDALL R.J. Protein measurement with the folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- MATSUO M., TANABE K., KIOKA N., AMACHI T., UEDA K. Different binding properties and affinities for ATP and ADP among sulfonylurea receptor subtypes, SUR1, SUR2A, and SUR2B. J. Biol. Chem. 2000;275:28757–28763. doi: 10.1074/jbc.M004818200. [DOI] [PubMed] [Google Scholar]
- MATSUOKA T., MATSUSHITA K., KATAYAMA Y., FUJITA A., INAGEDA K., TANEMOTO M., INANOBE A., YAMASHITA S., MATSUZAWA Y., KURACHI Y. C-terminal tails of sulfonylurea receptors control ADP-induced activation and diazoxide modulation of ATP-sensitive K+ channels. Circ. Res. 2000;87:873–880. doi: 10.1161/01.res.87.10.873. [DOI] [PubMed] [Google Scholar]
- MATSUSHITA K., KINOSHITA K., MATSUOKA T., FUJITA A., FUJIKADO T., TANO Y., NAKAMURA H., KURACHI Y. Intramolecular interaction of SUR2 subtypes for intracellular ADP-induced differential control of KATP channels. Circ. Res. 2002;90:554–561. doi: 10.1161/01.res.0000012666.42782.30. [DOI] [PubMed] [Google Scholar]
- MOREAU C., JACQUET H., PROST A.-L., D'HAHAN N., VIVAUDOU M. The molecular basis of the specificity of action of KATP channel openers. EMBO J. 2000;19:6644–6651. doi: 10.1093/emboj/19.24.6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PROKS P., REIMANN F., GREEN N., GRIBBLE F., ASHCROFT F. Sulfonylurea stimulation of insulin secretion. Diabetes. 2002;51 Suppl 3:S368–S376. doi: 10.2337/diabetes.51.2007.s368. [DOI] [PubMed] [Google Scholar]
- QUAST U. Do the K+ channel openers relax smooth muscle by opening K+ channels. Trends Pharmacol. Sci. 1993;14:332–337. doi: 10.1016/0165-6147(93)90006-6. [DOI] [PubMed] [Google Scholar]
- QUAST U., BRAY K.M., ANDRES H., MANLEY P.W., BAUMLIN Y., DOSOGNE J. Binding of the K+ channel opener [3H]P1075 in rat isolated aorta: relationship to functional effects of openers and blockers. Mol. Pharmacol. 1993;43:474–481. [PubMed] [Google Scholar]
- REIMANN F., GRIBBLE F.M., ASHCROFT F.M. Differential response of KATP channels containing SUR2A or SUR2B subunits to nucleotides and pinacidil. Mol. Pharmacol. 2000;58:1318–1325. doi: 10.1124/mol.58.6.1318. [DOI] [PubMed] [Google Scholar]
- REIMANN F., TUCKER S.J., PROKS P., ASHCROFT F.M. Involvement of the N-terminus of Kir6.2 in coupling to the sulphonylurea receptor. J. Physiol. (London) 1999;518:325–336. doi: 10.1111/j.1469-7793.1999.0325p.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RUSS U., HAMBROCK A., ARTUNC F., LÖFFLER-WALZ C., HORIO Y., KURACHI Y., QUAST U. Coexpression with the inward rectifier K+ channel Kir6.1 increases the affinity of the vascular sulfonylurea receptor SUR2B for glibenclamide. Mol. Pharmacol. 1999;56:955–961. [PubMed] [Google Scholar]
- RUSS U., LANGE U., LÖFFLER-WALZ C., HAMBROCK A., QUAST U. Interaction of the sulfonylthiourea HMR 1883 with sulfonylurea receptors and recombinant ATP-sensitive K+ channels: comparison with glibenclamide. J. Pharmacol. Exp. Ther. 2001;299:1049–1055. [PubMed] [Google Scholar]
- RUSS U., LANGE U., LÖFFLER-WALZ C., HAMBROCK A., QUAST U. Binding and effect of KATP channel openers in the absence of Mg2+ Br. J. Pharmacol. 2003;139:368–380. doi: 10.1038/sj.bjp.0705238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAKURA H., ÄMMÄLÄ C., SMITH P.A., GRIBBLE F.M., ASHCROFT F.M. Cloning and functional expression of the cDNA encoding a novel ATP-sensitive potassium channel subunit expressed in pancreatic β-cells, brain, heart and skeletal muscle. FEBS Lett. 1995;377:338–344. doi: 10.1016/0014-5793(95)01369-5. [DOI] [PubMed] [Google Scholar]
- SCHWANSTECHER M., BRANDT C., BEHRENDS S., SCHAUPP U., PANTEN U. Effect of MgATP on pinacidil-induced displacement of glibenclamide from the sulphonylurea receptor in a pancreatic β-cell line and rat cerebral cortex. Br. J. Pharmacol. 1992;106:295–301. doi: 10.1111/j.1476-5381.1992.tb14331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHWANSTECHER M., SIEVERDING C., DÖRSCHNER H., GROSS I., AGUILAR-BRYAN L., SCHWANSTECHER C., BRYAN J. Potassium channel openers require ATP to bind to and act through sulfonylurea receptors. EMBO J. 1998;17:5529–5535. doi: 10.1093/emboj/17.19.5529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SEINO S., MIKI T. Physiological and pathophysiological roles of ATP-sensitive K+ channels. Prog. Biophys. Mol. Biol. 2003;81:133–176. doi: 10.1016/s0079-6107(02)00053-6. [DOI] [PubMed] [Google Scholar]
- SHINDO T., YAMADA M., ISOMOTO S., HORIO Y., KURACHI Y. SUR2 subtype (A and B)-dependent differential activation of the cloned ATP-sensitive K+ channels by pinacidil and nicorandil. Br. J. Pharmacol. 1998;124:985–991. doi: 10.1038/sj.bjp.0701927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHYNG S.-L., NICHOLS C.G. Octameric stoichiometry of the KATP channel complex. J. Gen. Physiol. 1997;110:655–664. doi: 10.1085/jgp.110.6.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STURGESS N.C., ASHFORD M.L., COOK D.L., HALES C.N. The sulphonylurea receptor may be an ATP-sensitive potassium channel. Lancet. 1985;2:474–475. doi: 10.1016/s0140-6736(85)90403-9. [DOI] [PubMed] [Google Scholar]
- TOMAN A., UHDE I., SCHWANSTECHER M.A single residue in SURs is essential for sulphonylurea binding Naunyn-Schmiedeberg's Arch.Pharmacol. 2000361SupplR75(Abstract) [Google Scholar]
- UEDA K., KOMINE J., MATSUO M., SEINO S., AMACHI T. Cooperative binding of ATP and MgADP in the sulfonylurea receptor is modulated by glibenclamide. Proc. Natl. Acad. Sci. U.S.A. 1999;96:1268–1272. doi: 10.1073/pnas.96.4.1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UHDE I., TOMAN A., GROSS I., SCHWANSTECHER C., SCHWANSTECHER M. Identification of the potassium channel opener site on sulfonylurea receptors. J. Biol. Chem. 1999;274:28079–28082. doi: 10.1074/jbc.274.40.28079. [DOI] [PubMed] [Google Scholar]
- YAMADA M., ISOMOTO S., MATSUMOTO S., KONDO C., SHINDO T., HORIO Y., KURACHI Y. Sulphonylurea receptor 2B and Kir6.1 form a sulphonylurea-sensitive but ATP-insensitive K+ channel. J. Physiol. (London) 1997;499:715–720. doi: 10.1113/jphysiol.1997.sp021963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZERANGUE N., SCHWAPPACH B., JAN Y.N., JAN L.Y. A new ER trafficking signal regulates the subunit stoichiometry of plasma membrane KATP channels. Neuron. 1999;22:537–548. doi: 10.1016/s0896-6273(00)80708-4. [DOI] [PubMed] [Google Scholar]
- ZINGMAN L.V., ALEKSEEV A.E., BIENENGRAEBER M., HODGSON D., KARGER A.B., DZEJA P.P., TERZIC A. Signaling in channel/enzyme multimers: ATPase transitions in SUR module gate ATP-sensitive K+ conductance. Neuron. 2001;31:233–245. doi: 10.1016/s0896-6273(01)00356-7. [DOI] [PubMed] [Google Scholar]