Abstract
Induction of wild-type p53 in mouse fibroblasts causes cell cycle arrest at the G1 phase, whereas coexpression of p53 and the protooncogene c-myc induces apoptosis. Although p53 transcriptional activity generally is required for both pathways, the molecular components mediating p53-dependent apoptosis are not well understood. To identify factors that could mediate p53-induced cell death, we used a comparative RNA differential display procedure. We have identified Pw1/Peg3 as a gene product induced during p53/c-myc-mediated apoptosis. Pw1/Peg3 is not induced during p53-mediated G1 growth arrest nor by c-myc alone. Although it is not clear whether the induction of Pw1/Peg3 depends on p53 activity, we show that Pw1/Peg3 interacts with a p53-inducible gene product Siah1a. We demonstrate that coexpression of Pw1/Peg3 with Siah1a induces apoptosis independently of p53 whereas expression of Pw1/Peg3 or Siah1a separately has no effect on cell death. These data suggest that Siah1a and Pw1/Peg3 cooperate in the p53-mediated cell death pathway. Furthermore, we show that inhibiting Pw1/Peg3 activity blocks p53-induced apoptosis. The observation that Pw1/Peg3 is necessary for the p53 apoptotic response suggests a pivotal role for this gene in determining cell death versus survival.
The molecular processes governing the choice between p53-induced growth arrest or cell death have not been elucidated. One approach to unravel how p53 mediates cell death is to identify genes that are specifically induced under conditions that result in apoptosis. We developed mammalian cell lines that undergo either p53-mediated G1 arrest or apoptosis (1, 2). The VHD cell is a p53-deficient immortalized mouse embryo fibroblast cell line containing a temperature-sensitive mutant p53 (Ala to Val mutation at codon 135, tsp53), which expresses a nonfunctional p53 protein at 37°C to 39°C and a fully functional p53 protein at 32°C. VHD cells undergo G1 growth arrest in a p53-dependent manner at 32°C (1). The VM10 cells express both tsp53 and c-myc and undergo p53-mediated apoptosis at 32°C but are viable at 37–39°C (2). Blocking transcriptional activity of p53 by mdm-2 blocks cell death in VM10 cells, indicating that p53-mediated transcriptional activity is required for apoptosis and that apoptotic specific genes may be induced (2). In this study, we used an RNA differential display method to identify apoptotic-specific genes. This approach resulted in the identification of a cDNA fragment that corresponds to Pw1/Peg3. Pw1/Peg3 recently was identified in mice as a gene expressed throughout postgastrulation development (3) and independently isolated as a maternally imprinted gene called Peg3 (4, 5) (referred to hereafter as Pw1/Peg3). Pw1/Peg3 encodes a large multidomain protein that contains 12 zinc-finger motifs as well as two proline-rich periodic repeat motifs (3). We report here that Pw1/Peg3 is specifically activated during p53/c-myc-mediated apoptosis but not during p53-dependent G1 arrest nor by c-myc alone. The expression of Pw1/Peg3 also is induced in p53/E2F-1-mediated apoptosis. Furthermore, we identified members of the Siah family as Pw1/Peg3 interacting proteins, including Siah1a. This finding is striking because Siah1a also has been implicated in the p53-mediated cell death pathway. The murine Siah1 gene was identified, via differential cDNA analysis, as a p53-induced gene in a leukemia cell line that conditionally undergoes growth arrest and apoptosis (6, 7). It also has been observed that overexpression of human Siah1a results in apoptosis in U937 cell but only induces cellular growth arrest in two different human cell lines (8, 9). BAG1, a ubiquitin-like protein, can suppress p53-mediated growth arrest and apoptosis and has been demonstrated to interact with Siah1a (9). We tested the involvement of Pw1/Peg3 in p53-mediated apoptosis. Although expression of Pw1/Peg3 alone does not induce apoptosis, Pw1/Peg3 is able to cooperate with p53 to induce cell death. More importantly, coexpression of Siah1a and Pw1/Peg3 induces apoptosis independently of p53. Inhibition of Pw1/Peg3 by either antisense mRNA or a dominant negative mutant blocks p53/c-myc-mediated cell death. These data indicate that Siah1a and Pw1/Peg3 are downstream mediators of p53-induced cell death process. Because expression of Siah1a alone induces growth arrest, whereas the expression of Pw1/Peg3 and Siah1a results in apoptosis, we propose that Pw1/Peg3 carries out a pivotal role in determining the choice between cell death or survival.
Materials and Methods
Cell Lines and Transfection.
All cells were maintained in DMEM, supplemented with 10% FBS. They were routinely grown in incubators (Forma Scientific, Marietta, OH) under 5% CO2 at 38°C and shifted to 32°C for 24–48 hr for growth arrest or apoptosis assays. The full-length Pw1/Peg3 cDNA in pBS7 (10) was subcloned into a pMTN mammalian expression vector containing the simian virus 40 promoter and a 9-aa hemagglutinin (HA) tag was in-frame with the amino terminus of Pw1/Peg3 to generate a Pw1/Peg3 expression vector (pMTHAPW). The Pw1/Peg3 cDNA was subcloned into pcDNA3 vector (Invitrogen) in a reverse orientation to create the Pw1/Peg3 antisense construct. The dominant negative Pw1/Peg3, ΔPW1, was reported earlier (10). The Siah expression plasmid is described elsewhere (11). The p53, p21, and c-myc expression plasmids have been described (2, 12). The DNA was transfected into cells by using Fugene transfection reagent according to the manufacturer's protocol (Boehringer Mannheim). For all transient transfection experiments, cells were harvested 48–72 hr after transfection.
RNA Differential Display and Northern Blotting.
Total RNA was extracted from cells by using RNAzol (Cinna/Biotecx Laboratories, Friendswood, TX). A total of 500 ng of total RNA was subjected to the reverse-transcription reaction using the oligonucleotide ATGWACCAKGAICCIYKMKYIG (R = A, G; S = G, C; Y = C, T; K = G, T; M = A, C; W =A, T; I = deoxyinosine) as a primer, and one-fourth of the reverse-transcription products were added to a PCR mix containing 1× PCR buffer (Perkin–Elmer), dNTP (0.25 mM each), 35S-dATP (1 μCi), primers (1.5 nM each), and Ampli-taq (0.5 unit, Perkin–Elmer). A 40-cycle reaction was performed at 94°C for 30 sec, 40°C for 1 min., 72°C for 30 sec as described (13). The PCR primers used were: CIYCYTCIWCACCATGIGWIAIRAIYRC and IYICGSWCTGGWRSIRAIGTIG (R = A, G; S = G, C; Y = C, T; K = G, T; M = A, C; W = A, T; I = deoxyinosine). PCR products were resolved on a 6% polyacrylamide DNA sequencing gel. Differentially expressed bands were excised from the gel, eluted, and reamplified. PCR products then were subcloned by using the TA-cloning system (Invitrogen). Three recombinant clones were sequenced by the dideoxy chain termination method using a sequencing kit (United States Biochemical).
For Northern analysis, equal quantities of total RNA isolated by RNAzol at 10–20 μg per sample were subjected to electrophoresis on a denaturing 1% formaldehyde agarose gel. The RNA was transferred to a nylon membrane and hybridized with 32P-dCTP-labeled probes by using a rapid hybridization system (Amersham Pharmacia).
Immunofluorescence Analysis.
To visualize Pw1/Peg3 protein in situ, cells were plated on glass coverslips in 24-well plates and transfected with Pw1/Peg3 and/or other expressing plasmid. Forty eight to 72 hours after transfection, cells on glass coverslips were fixed in paraformaldehyde (1% in PBS) for 10 min and permeabalized in methanol for another 10 min. The fixed cells were washed twice with PBS, and the coverslips were blocked in 5% of nonfat milk in PBS with 0.05% Tween-20 and 10% goat serum. The coverslips were incubated with anti-HA antibody (12CA5) for 2 hr and rinsed with PBS three times. After incubation with FITC-conjugated secondary antibody (GIBCO-BRL) for 30 min, the coverslips were washed with PBS, and the DNA was stained for 15 min with 4′,6-diamidino-2-phenylindole (5 μg/ml in PBS). The coverslips were mounted and viewed by a Nikon fluorescence microscopy.
Flow Cytometry Analysis.
Cells grown in 100-mm plates were transfected with 2 μg of green fluorescence protein (GFP) expressing plasmid pEGFP-N (CLONTECH) and 10.0 μg final concentration of either control DNA (empty vector), or Pw1/Peg3, or Siah-expressing plasmid, or in combinations (molar ratio is about 1:3). Transfected cells were harvested 48–72 hr later and washed twice in PBS. The cells were fixed in paraformaldehye in PBS for 10 min and permeabalized in 70% ethanol for 16 hr at 4°C. Cells were resuspended in PBS containing 5 μg of propidium iodide and RNase A (5 μg/ml). Flow cytometric analysis was carried out in a fluorescence-activated cell sorting analyzer (Coulter). GFP-positive cells were gated and analyzed for DNA content. On average 10,000 GFP-positive cells were analyzed in each experiment.
Yeast Two-Hybrid Screening and Immunoprecipitation Assays.
Pw1/Peg3 (amino acids 72–1379) was cloned into the pGBT9 vector (CLONTECH) and used to screen an E17d library (CLONTECH) following the Matchmaker Two-Hybrid System protocols (CLONTECH) as described (11). The yeast strain CG-1945 was used in the presence of 25 or 55 mM 3-amino-triazole on selection media for library screening and retransformation experiments. Among the 35 clones that were able to grow, we identified a Siah2 clone missing the ring finger domain (106–326). Full-length Siah1a and Siah2 were cloned in pGAD424 or pGBT9 for two-hybrid experiments using PCR.
To perform protein analysis, the transfected cells in 10-cm dishes were harvested and lysed in 1 ml of lysis buffer (50 mM Tris⋅HCl, pH 8.0/5 mM EDTA/150 mM NaCl/0.5% NP-40/1 mM PMSF). The HA-tagged Pw1/Peg3 protein was immunoprecipitated by using anti-HA antibody (12CA5) and protein A Sepharose (Sigma). Flag-tagged Siah proteins were precipitated by using anti-Flag antibody (Sigma). The p53 protein was detected by using mAb pAB421. The complex then was washed twice with lysis buffer and subjected to electrophoresis on a 7% SDS-polyacrylamide gel. The protein was transferred to a poly(vinylidene difluoride) membrane and developed by using an ECL chemoluminescence kit (Amersham Pharmacia).
Results
Identification of Pw1/Peg3 as a Gene Specifically Induced During p53-Mediated Apoptosis.
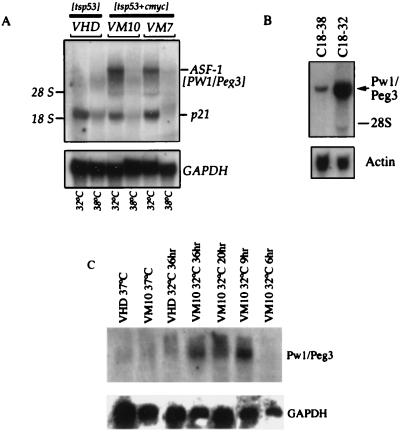
To identify genes that are specifically up-regulated during p53-mediated apoptosis, total RNA was prepared from VM10 and VHD cells plated at both 32°C and 38°C and subjected to RNA differential display analysis. A fragment was identified that was induced specifically during apoptosis in VM10 cells at 32°C (data not shown). The gene fragment initially was named ASF-1 (apoptosis-specific fragment 1). To confirm that ASF-1 is induced during apoptosis, Northern blot analysis was performed on the total RNA isolated from VHD and VM10 cells. In addition, RNA from VM7 cells, an independently derived cell line that expresses tsp53 and c-myc, also was tested to control for clonal variation. We observed that ASF-1 hybridizes to a large transcript with an estimated size of 7–9 kb (Fig. 1A). More importantly, transcripts are readily detected at 32°C in both VM10 and VM7 cells, whereas no or very low levels of transcripts are detected in VHD cells at both temperatures and in VM10 and VM7 cells growing at 38°C (Fig. 1A). To verify that p53 functions in these cells, the expression of a well-characterized p53-induced transcript, p21WAF1/CIP1, was analyzed. The expression of p21WAF1/CIP1 is detected readily in all three cell lines at 32°C, indicating that the p53 protein functions like a wild type at the permissive temperature (Fig. 1A). This result indicates that ASF-1 is specifically induced during p53 and c-myc-mediated apoptosis.
Figure 1.
Identification of ASF-1 (Pw1/Peg3) as a gene specifically induced during p53-mediated apoptosis. (A) Northern blot analysis of Pw1/Peg3 (ASF-1) during p53-mediated cell cycle arrest and apoptosis. Total RNA was prepared from VHD, VM10, and VM7 cells growing at 38°C and cells shifted to 32°C for 24 and 48 hr. Total RNA was extracted and RNA samples of the two time points at 32°C were pooled together. Fifteen micrograms of total RNA from each sample was subjected to electrophoresis on a 1% formaldehyde agarose gel, transferred to a nylon membrane, and hybridized with 32P-labeled ASF-1 probe and a mouse p21 probe. The positions of 28S and 18S ribosomal RNA are indicated. RNA loading was normalized by hybridization with glyceraldehyde-3-phosphate dehydrogenase (GAPDH). (B) Induction of Pw1/Peg3 during p53/E2F-1 mediated apoptosis. The C18 cell is a murine fibroblast cell line expressing tsp53 and E2F-1 and undergoes apoptosis at 32°C as described (1). Total RNA were extracted and analyzed by Northern blotting using ASF-1 as a probe as described in A. A 5-fold increase in Pw1/Peg3 expression was observed in C18 cells at 32°C. (C) Time course of Pw1/Peg3 induction in VM10 cells at 32°C. The VM10 cell and control VHD cells were shifted to 32°C at the time point indicated. Total RNA were extracted and blotted with ASF-1 probe. The control of RNA loading was measured by probing with GAPDH.
A 462-bp segment of ASF-1 was sequenced. Sequence comparison indicated that ASF-1 is identical to Pw1/Peg3 (3, 5). The full-length transcript of Pw1/Peg3 is 8.5 kb, consistent with the observed size of ASF-1 as recognized on a Northern blot. A nonoverlapping probe to Pw1/Peg3 also recognizes a band on a Northern blot of identical size and expression in response to p53 and c-myc as seen for ASF-1 (data not shown). We next examined whether Pw1/Peg3 is induced in other cell systems in which apoptosis can be induced. It has been demonstrated that p53 also can cooperate with E2F-1 to induce apoptosis (1). As shown in Fig. 1B, Northern analysis reveals that Pw1/Peg3 is similarly induced (5-fold induction), suggesting that Pw1/Peg3 is part of a general p53-mediated cell death response and does not depend on the expression of c-myc. Because Pw1/Peg3–1 is not induced by p53 alone in VHD cells (Fig. 1A), our data suggest that the induction of Pw1/Peg3 is apoptosis specific.
The kinetics of Pw1/Peg3 induction in VM10 cells at 32°C was investigated. VM10 cells were transferred to 32°C, total RNA were extracted at different time points, and Pw1/Peg3 expression was examined by Northern blotting. Pw1/Peg3 expression is readily detected at 9 hr after temperature shift (Fig. 1C), which coincides with the complete transition of tsp53 from mutant confirmation to the wild type (14). This finding suggests that induction of Pw1/Peg3 is likely at the transcriptional level.
Coexpression of Pw1/Peg3 and p53 Induces Apoptosis.
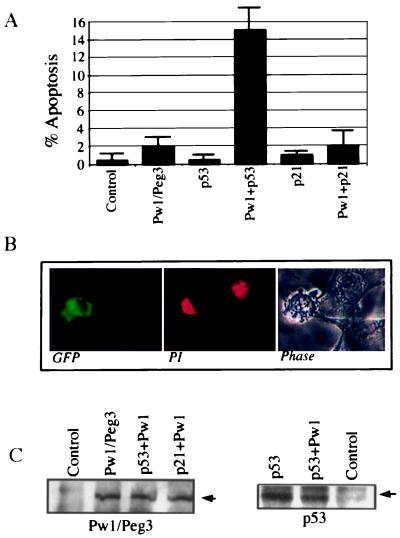
To investigate the function of Pw1/Peg3 during apoptosis, a Pw1/Peg3 expression vector was transfected into BALB/c 3T3 10.1 cells (p53 deficient). The cells were cotransfected with a plasmid carrying the GFP to track transfected cells and analyzed by FACS 48 hr after transfection. Apoptosis was measured by the presence of sub-G1 cells. We observe that expression of Pw1/Peg3 alone does not induce apoptosis (Fig. 2A), and coexpression of Pw1/Peg3 and c-myc induces a moderate increase in apoptosis (data not shown); however, coexpression of Pw1/Peg3 and p53 results in pronounced cell death as measured by cell counts of sub-G1 population (Fig. 2A). Although fewer than 1% of control cells (transfected with empty vector and GFP) contain sub-G1 cells, expression of p53 and Pw1/Peg3 together results in 15% of cells displaying sub-G1 DNA, indicating a pronounced induction of apoptosis (Fig. 2A). The GFP-positive cells display morphological characteristics of apoptosis (Fig. 2B), with condensed nuclei, uneven distribution of DNA, and cell membrane blebbing. The cell death caused by p53 and Pw1/Peg3 is not caused by variations in protein expressions in the experiments as levels of Pw1/Peg3 and p53 proteins remain unchanged (Fig. 2C).
Figure 2.
Coexpression of p53 and Pw1/Peg3 induces apoptosis. (A) 10.1 cells were transfected with empty vector, Pw1/Peg3, p53 and Pw1/Peg3, p21 and Pw1/Peg3. A GFP expressing plasmids pEGEP was cotransfected at molar ratio of 3 to 1. The transfected cells were fixed, stained with propidium iodide, and subjected to FACS analyses. The transfected cells represented by GFP-positive population were gated and analyzed for their DNA content. The level of apoptosis was measured by cells in the sub-G1 population. The level (percent) of apoptotic cells was averaged from three independent experiments. SDs are given. The coexpression of p53 and Pw1/Peg3 results in 15% apoptotic cells, whereas 0.5% of control cells shows cell death. Coexpression of p21WAF1/CIP1 and Pw1/Peg3 does not induce cell death. (B) GFP-positive cells display apoptotic morphology. The 10.1 cells transfected with GFP, p53, and Pw1/Peg3 were fixed on coverslips, stained with propidium iodide, and viewed under a fluorescent microscope. The GFP-positive cells show condensed chromatin and cell membrane blebbing that are characteristics of apoptosis. (C) Expression of Pw1/Peg3 and p53 proteins in transfected cells. The 10.1 cells were transfected with DNA as in the FACS experiments. The Pw1/Peg3 and p53 proteins indicated by arrows were detected by immunoprecipitation and blotting with either anti-HA antibody or pAB421, respectively.
One major cellular response to p53 expression is the induction of p21WAF1/CIP1 expression, which leads to growth arrest. To test whether p21WAF1/CIP1 participates with Pw1/Peg3 to mediate apoptosis, human p21WAF1/CIP1 was cotransfected with Pw1/Peg3 and GFP. FACS analysis of transfected cells showed no significant increase in sub-G1 cells compared to that of cells transfected with Pw1/Peg3 only (Fig. 2A). Thus, p21WAF1/CIP1 expression is not sufficient to cooperate with Pw1/Peg3 to induce cell death in 10.1 cells.
Pw1/Peg3 Interacts with Siah Proteins.
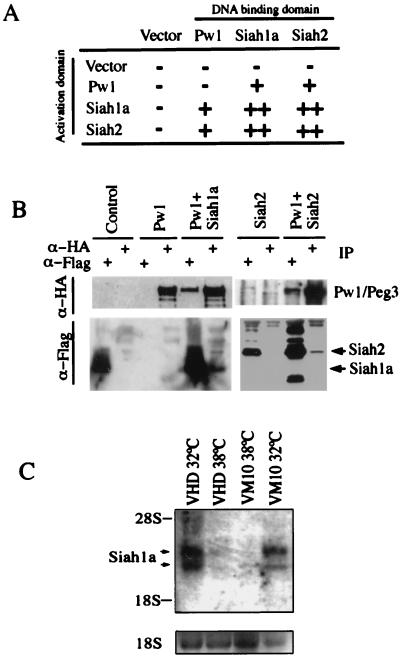
Using a yeast two-hybrid screen to identify Pw1/Peg3 interacting proteins (10), we isolated a fragment (amino acids 106–326) of a previously identified protein, Siah2 (11). Siah2 is one of three functional homologues of the Drosophila gene seven in absentia (sina). Siah2 is highly homologous to Siah1a and Siah1b in mice and huSiah1 in humans (11). Given the strong homology between Siah family members, we tested all murine Siah proteins for interaction with Pw1/Peg3 by using a yeast two-hybrid interaction assay. The cDNAs corresponding to Pw1/Peg3, Siah1a, and Siah2 were fused to the Gal4 DNA binding domain and activation domain separately and tested for interaction. We observed that Pw1/Peg3 interacts with Siah1a and Siah2 and that Siah proteins are capable of homo- and heterotypic (Fig. 3A). The interaction of Pw1/Peg3 with Siah proteins was further confirmed by coexpressing epitope-tagged cDNAs in mammalian cells. The protein complex was detected by immunoprecipitation followed by Western blotting (Fig. 3B). Consistent with the yeast two-hybrid data, we detected stable association between Pw1/Peg3 and Siah1a as well as Siah2 proteins (Fig. 3B). We noted that only a small proportion of Siah proteins and Pw1/Peg3 can be found in a stable complex after coexpression in mammalian cells.
Figure 3.
Interaction of Pw1/Peg3 with Siah proteins and expression of Siah1a during p53-mediated growth arrest and apoptosis. (A) Interaction between Pw1/Peg3 and the Siah family of proteins in yeast. Pw1/Peg3, Siah1a, and Siah2 were fused to the Gal4 DNA binding domain and activation domain separately and were tested in yeast for interaction. The empty vectors were used as negative controls. + indicates growth and LacZ staining, whereas − indicates no growth on selective media. Siah1a and Siah2 showed strong homo- and heterotypic interactions, as indicated by ++. (B) 293 cells plated in 10-cm dishes were transiently transfected with expression vectors encoding Flag-tagged Siah1a and Siah2 with HA-tagged Pw1/Peg3. Transfection of empty vector was used as a control. The cells were harvested and lysed 48 hr later. Half of the lysate was immunoprecipitated with anti-Flag antibody (Sigma), and the other half was precipitated by using antibody against HA (12CA5). The immunocomplex was subjected to SDS/PAGE, and Western blot was carried out by using either α-HA or α-Flag antibodies as indicated. Pw1/Peg3/Siah complex is readily detected. (C) Induction of Siah1a expression by p53. Total RNA was prepared from VHD and VM10 as described in Fig. 1A. Northern analysis of Siah1a mRNA was carried out by using a 32P-labeled Siah1a probe. The Siah1a mRNA appears as a doublet. The RNA loading is shown below.
Induction of Apoptosis by Coexpression of Pw1/Peg3 and Siah1a.
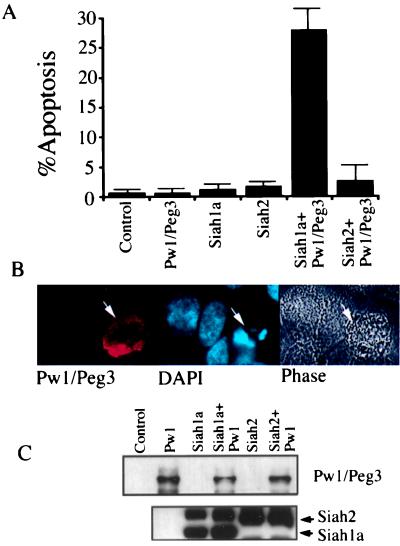
The observation that Pw1 induces cell death in cooperation with p53 suggests that Pw1/Peg3-mediated apoptosis requires additional p53 downstream target genes. Given the observation that Pw1/Peg3 interacts with Siah1a, and that the murine Siah1a has been reported recently to be specifically up-regulated by p53 during apoptosis in a leukemic cell line (6, 7), we speculated that the Pw1/Peg3:Siah1a interaction may reflect the involvement of both proteins in a coordinated p53 response. It recently was reported that the human Siah1 is induced by p53 and that forced expression of Siah1 results in either growth arrest or cell death depending on the cell types (9). As a first step to test whether Siah participates in apoptosis induced by the coexpression of c-myc and p53, we examined Siah expression in the VM10 cell line. Using Northern analysis, we observe that Siah1a is induced by p53 alone as well as by coexpression of p53 and c-myc (Fig. 3C). This finding is in contrast to Pw1/Peg3, which is induced only by expression of both p53 and c-myc (see Fig. 1A). Thus, conditions that induce the expression of Pw1/Peg3 also induce the expression of Siah1a. To investigate the effect of Pw1/Peg3 and Siah coexpression on apoptosis, constructs were transfected into 10.1 cells along with GFP-expressing plasmid. FACS analyses were performed to quantify levels of apoptosis as measured by the percentage of cells in sub-G1. Coexpression of Siah1a and Pw1/Peg3 results in a pronounced increase in cells with sub-G1 DNA content (Fig. 4A). The transfected cells display typical morphological characteristics of apoptosis including chromatin condensation and fragmentation (Fig. 4B). In contrast, expression of either Siah1a or Siah2 alone did not induced apoptosis (Fig. 4A). Siah2 in combination with Pw1/Peg3 showed only a small increase upon cell death (Fig. 4A). The Pw1/Peg3 and Siah protein levels in the experiments were confirmed by immunoprecipitation-Western blot (Fig. 4C). Similar results were obtained by using the human lung carcinoma cell line H1299 (data not shown). Taken together, these results indicate that Pw1/Peg3 and Siah1a cooperatively induce apoptosis in the absence of p53.
Figure 4.
Induction of apoptosis by coexpression of Pw1/Peg3 and Siah1a. (A) 10.1 cells transfected with vector, Pw1/Peg3, Siah1a, Siah2, and GFP-expressing plasmids, as well as in combinations were subject to FACS analysis as described in Fig. 2. The percentages of apoptosis were quantified by sub-G1 cells in three independent experiments. SDs are given. (B) In situ fluorescence staining shows that Pw1/Peg3-expressing cells display condensation and fragmentation of DNA. 10.1 cells grown on coverslips were transfected with Pw1/Peg3 and Siah1a-expressing plasmids. The cells were fixed and stained with antibody against HA epitope tag on Pw1/Peg3 and nuclei by 4′,6-diamidino-2-phenylindole (DAPI). The phase contrast morphology of the cell also is shown. The fragmented nucleus is indicated by an arrow. (C) Expression of Pw1/Peg3 and Siah proteins in transfected cells. The 10.1 cells were transfected with DNA as in the FACS experiments. The Pw1/Peg3 and Siah proteins were detected by immunoprecipitation followed by Western blotting.
Inhibition of p53/c-myc-Mediated Apoptosis by Antisense Pw1/Peg3 mRNA and a Dominant Negative Mutant.
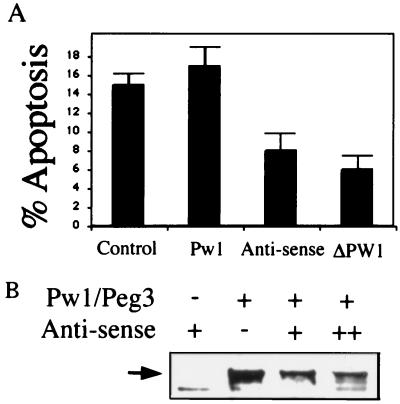
We have demonstrated that Pw1/Peg3 is induced in p53/c-myc-mediated apoptosis and forced expression of Pw1/Peg3 and Siah1a induces cell death. To test whether Pw1/Peg3 is required during p53/c-myc mediated apoptosis, we transfected a vector expressing Pw1/Peg3 antisense mRNA into VM10 cells at 32°C to block PW1 expression. We also tested the effects of a deletion mutant of Pw1/Peg3 containing residues 1–592 (ΔPW1). The ΔPW1 has been shown to act as a dominant negative mutant in Pw1/Peg3-mediated NF-κB activation pathway in several cell types (10). The GFP-expressing plasmid was cotransfected, and FACS analysis was performed. At 32°C, 15% of VM10 cells transfected with control vector and GFP contained sub-G1 DNA, indicating that 15% of control cells undergo apoptosis (Fig. 5A). Expression of Pw1/Peg3 has little effect on cell death. Transfection of either anti-sense Pw1/Peg3 or ΔPW1 reduces apoptosis to half the level of control. The effect of antisense on Pw1/Peg3 protein expression was tested in 293 cells. Transfection of antisense Pw1 reduced levels of the transfected Pw1/Peg3 protein expression in a dose-dependent manner (Fig. 5B). The levels of ΔPW1 and Pw1/Peg3 in these transfection experiments are also similar (data not shown). Our data suggest that blocking the function of Pw1/Peg3 inhibits p53/c-myc-mediated apoptosis.
Figure 5.
Inhibition of p53/c-myc-induced apoptosis by antisense Pw1/Peg3 and a dominant negative mutant. (A) VM10 cells were transfected with vector control, Pw1/Peg3, Pw1/Peg3 antisense mRNA, and a deletion mutant Pw1/Peg3 containing amino acids 1–592 (ΔPw1). The cells were cotransfected with pEGFP as described. The transfected cells were shifted to 32°C for 24 h and analyzed by FACS as in Fig. 2. Percentages of sub-G1 cells (apoptotic) are the averages of three independent experiments. (B) Antisense Pw1 reduces Pw1/Peg3 protein levels. 293 cells were transfected with either 2 μg of Pw1/Peg3, 2 μg of ΔPW1, or the two together in different ratios (1:1 and 1:3). The HA-tagged Pw1/Peg3 was detected by immunoprecipitation-Western blotting. Transfection efficiency was monitored by cotransfecting with 0.5 μg of pEGFP.
Discussion
The tumor suppressor activity of p53 depends on its ability to induce either G1 growth arrest or apoptosis. The contribution of p53-induced gene expression in the apoptotic pathway is still controversial because few genes have been shown to be specific to this process. In this study, we report the identification of Pw1/Peg3 as a gene that is specifically induced during apoptosis. Most importantly, we have demonstrated that Pw1/Peg3 is capable of cooperating with Siah1a, a p53-inducible gene, to induce apoptosis. Furthermore, expression of antisense Pw1/Peg3 or a truncated form of Pw1/Peg3 diminishes the ability of p53/c-myc to induce apoptosis, suggesting that Pw1/Peg3 activity is required for the cell death pathway. The induction of Pw1/Peg3 during apoptosis is unique in that it requires both the activation of p53 and c-myc expression. Whether Pw1/Peg3 is a direct target of p53, c-myc, or both p53 and c-myc has yet to be determined. In addition to p53/c-myc-induced apoptosis, Pw1/Peg3 is induced during p53/E2F-1-induced apoptosis as well as serum deprivation-induced cell death in PC12 cells (data not shown), suggesting that expression of Pw1/Peg3 is involved in several apoptotic pathways. Taken together, our data strongly suggest that Pw1/Peg3 is a critical down-stream effector in the p53-mediated cell death pathway.
Siah1a and Siah2 are the mammalian homologues of the Drosophila sina gene, which is involved in a signaling pathway that leads to the formation of the photoreceptor R7 in the eye (15). Recently, it was shown that sina acts to promote R7 differentiation by targeting the Tramtrak repressor for degradation through a ubiquitin-mediated processes (16). The vertebrate homologues of sina consist of three members in mice: Siah1a, Siah1b, and Siah2. The function of the murine Siah proteins have not been fully characterized; however, Siah2 has been shown to possess a similar ubiquitin-targeting activity as sina to target DCC and N-CoR for proteasome-mediated degradation (17). In contrast, the murine Siah1 gene was identified as a p53-inducible gene by differential display analysis in a leukemic cell line induced for p53-mediated growth arrest and apoptosis (7). It later was shown that expression of Siah1 leads to apoptosis in human U937 cells (8). This observation led to the proposal that Siah1 may participate in either p53-mediated cell death or growth arrest. Matsuzawa et al. (9) further demonstrated that the human Siah1a gene is induced by p53 and genotoxic stress. Moreover, it has been shown that overexpression of Siah1a induces growth arrest in the human embryonic kidney line 293 and in human immortalized GM701 fibroblasts, but no apoptosis is observed in these cells (9). These results show that the involvement of Siah1 in apoptosis is system dependent and requires other proteins. In this report, we show that expression of Siah1a alone does not induce apoptosis in murine fibroblasts. We further identified that Siah proteins interact with Pw1/Peg3 directly and that coexpression of Siah1a and Pw1/Peg3 induces apoptosis. These results suggest an highly regulated mechanism exists at least in our system in which two transcripts are induced by p53 and c-myc, which encode proteins that subsequently interact and cooperate to induce cell death. If Pw1/Peg3 is not induced, as seen in the case of p53 induction in the absence of c-myc, cell death does not occur and growth arrest ensues. In this scenario, the presence or absence of Pw1 will dictate whether cells undergo death or growth arrest. Our results suggests that Pw1/Peg3 induction is a key step in the cell death pathway.
The cooperation between Pw1/Peg3 and Siah1a in inducing apoptosis is further strengthened by the finding that Siah1a also interacts with BAG1, an ubiquitin-like protein that binds to Bcl-2 and enhances the antiapoptotic activity of Bcl-2 (18–20). The BAG1-Siah1a interaction appears to be inhibitory to Siah1a function, because overexpression of BAG1 abrogates p53-mediated growth arrest, likely by inhibiting Siah1a functions (9). It also raises the possibility that BAG1 can interfere with apoptosis through two pathways, one by enhancing Bcl-2 activity and the other by interacting with Siah1a. It has been shown that Siah1a can induce growth arrest of cells (9). Because p21WAF1/CIP1 cannot cooperate with Pw1/Peg3 to induce apoptosis, it suggests that the growth arrest per se is either irrelevant or insufficient for apoptosis or that Siah1a is sufficient for the growth arrest function (9).
Pw1/Peg3 has been identified independently in three distinct contexts, i.e., development (3), imprinting (4), and apoptosis (this study). Pw1/Peg3 is expressed strongly upon gastrulation and is expressed at high levels in skeletal muscle and subregions of the brain in the adult (3). High levels of expression in muscle and nerve may reflect similar molecular strategies in cell death and terminal differentiation. In contrast, other cell types may require a p53-mediated response to induce Pw1/Peg3 expression. It is noteworthy that Pw1/Peg3 is syntenic with human 19q13.1–13.3, which contains the myotonic dystrophy gene, and loss of heterozygosity in this locus is associated with gliomas (21) suggesting that Pw1/Peg3 may act as a tumor suppressor gene. Pw1/Peg3 also was identified as a maternally imprinted gene (4). Imprinted genes often are associated with cellular regulatory mechanisms that control proliferation and differentiation leading to the establishment of the correct size of the fetus by the end of gestation. Mice carrying a null mutation for the Pw1/Peg3 locus have been generated recently (22). These mice develop specific defects in the brain leading to defects in maternal behavior and are smaller possibly because of decreased cellular proliferation (22). It undoubtedly will be of interest to test whether the p53-apoptotic pathway is intact in these animals.
The deduced amino acid sequence of Pw1/Peg3 reveals that it is a relatively large protein containing several identifiable protein motifs, including 12 C2H2 zinc fingers, two acidic and proline-rich periodic repeat domains, as well as other domains containing periodic repeats of amino acids (3, 4). The prevalence of these domains suggested to us that Pw1/Peg3 may interact with other proteins. We previously have reported that the Pw1/Peg3 protein interacts with TRAF2 (10) implicating it in the tumor necrosis factor (TNF)-NF-κB activation pathway and that Pw1/Peg3 expression leads to NF-κB activation (10). Paradoxically, it has been observed that activation of NF-κB can lead to cellular resistance to apoptosis (23, 24). One possibility is that Pw1/Peg3 and Siah1a can generate a cellular response similar to cells responding to TNF-α treatment. TNF-α is known to elicit both strong NF-κB activation and cell death (reviewed in ref. 25). The other possibility is that the dying cells and cells that activate NF-κB fall into two distinct populations. Alternatively, expression of Siah1a abrogates the ability of Pw1/Peg3 to activate NF-κB; however, preliminary investigations revealed that coexpression of Siah1a and Pw1/Peg3 does not block NF-κB activation. The interaction of Pw1/Peg3 with TRAF2 may underlie a mechanism whereby Pw1/Peg3 interactions with Siah1 influence cell death pathways mediated through TRAF2.
Acknowledgments
We thank Dr. Andrew Chan for synthesizing the oligonucleotides used in this study, Drs. Jim Manfredi, Stuart Aaronson, and Giovanna Marazzi for helpful discussion of the experiments, and Drs. M. Goldfarb and R. Krauss for critical readings of the manuscript. The experiments were supported by a start-up fund from the Mount Sinai School of Medicine, a grant from Merck Co. & Inc., and a grant from the National Institutes of Health (X.W.), and a Kenner-Fellow Award from the American Heart Association-New York City (D.A.S.).
Abbreviations
- HA
hemagglutinin
- GFP
green fluorescence protein
- ASF-1
apoptosis-specific fragment 1
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.040378897.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.040378897
References
- 1.Wu X, Levine A J. Proc Natl Acad Sci USA. 1994;91:3602–3606. doi: 10.1073/pnas.91.9.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen J, Wu X, Lin J, Levine A J. Mol Cell Biol. 1996;16:2445–2452. doi: 10.1128/mcb.16.5.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Relaix F, Weng X, Marazzi G, Yang E, Copeland N, Jenkins N, Spence S E, Sassoon D. Dev Biol. 1996;177:383–396. doi: 10.1006/dbio.1996.0172. [DOI] [PubMed] [Google Scholar]
- 4.Kuroiwa Y, Kaneko-Ishino T, Kagitani F, Kohda T, Li L L, Tada M, Suzuki R, Yokoyama M, Shiroishi T, Wakana S, et al. Nat Genet. 1996;12:186–190. doi: 10.1038/ng0296-186. [DOI] [PubMed] [Google Scholar]
- 5.Kim J, Ashworth L, Branscomb E, Stubbs L. Genome Res. 1997;7:532–540. doi: 10.1101/gr.7.5.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nemani M, Linares-Cruz G, Bruzzoni-Giovanelli H, Roperch J P, Tuynder M, Bougueleret L, Cherif D, Medhioub M, Pasturaud P, Alvaro V, et al. Proc Natl Acad Sci USA. 1996;93:9039–9042. doi: 10.1073/pnas.93.17.9039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amson R B, Nemani M, Roperch J P, Israeli D, Bougueleret L, Le Gall I, Medhioub M, Linares-Cruz G, Lethrosne F, Pasturaud P, et al. Proc Natl Acad Sci USA. 1996;93:3953–3957. doi: 10.1073/pnas.93.9.3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roperch J P, Lethrone F, Prieur S, Piouffre L, Israeli D, Tuynder M, Nemani M, Pasturaud P, Gendron M C, Dausset J, et al. Proc Natl Acad Sci USA. 1999;96:8070–8073. doi: 10.1073/pnas.96.14.8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsuzawa S, Takayama S, Froesch B A, Zapata J M, Reed J C. EMBO J. 1998;17:2736–2747. doi: 10.1093/emboj/17.10.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Relaix F, Wei X J, Wu X, Sassoon D A. Nat Genet. 1998;18:287–291. doi: 10.1038/ng0398-287. [DOI] [PubMed] [Google Scholar]
- 11.Della N G, Senior P V, Bowtell D D. Development (Cambridge, UK) 1993;117:1333–1343. doi: 10.1242/dev.117.4.1333. [DOI] [PubMed] [Google Scholar]
- 12.Lin J, Reichner C, Wu X, Levine A J. Mol Cell Biol. 1996;16:1786–1793. doi: 10.1128/mcb.16.4.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang P, Pardee A B. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 14.Gannon J V, Lane D P. Nature (London) 1991;349:802–806. doi: 10.1038/349802a0. [DOI] [PubMed] [Google Scholar]
- 15.Tang A H, Neufeld T P, Kwan E, Rubin G M. Cell. 1997;90:459–467. doi: 10.1016/s0092-8674(00)80506-1. [DOI] [PubMed] [Google Scholar]
- 16.Li S, Li Y, Carthew R W, Lai Z C. Cell. 1997;90:469–478. doi: 10.1016/s0092-8674(00)80507-3. [DOI] [PubMed] [Google Scholar]
- 17.Hu G, Zhang S, Vidal M, Baer J L, Xu T, Fearon E R. Genes Dev. 1997;11:2701–2714. doi: 10.1101/gad.11.20.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takayama S, Sato T, Krajewski S, Kochel K, Irie S, Millan J A, Reed J C. Cell. 1995;80:279–284. doi: 10.1016/0092-8674(95)90410-7. [DOI] [PubMed] [Google Scholar]
- 19.Takayama S, Kochel K, Irie S, Inazawa J, Abe T, Sato T, Druck T, Huebner K, Reed J C. Genomics. 1996;35:494–498. doi: 10.1006/geno.1996.0389. [DOI] [PubMed] [Google Scholar]
- 20.Takayama K, Ogata K, Nakanishi Y, Yatsunami J, Kawasaki M, Hara N. Cancer J Sci Am. 1996;2:212–215. [PubMed] [Google Scholar]
- 21.Rubio M P, Correa K M, Ueki K, Mohrenweiser H W, Gusella J F, von Deimling A, Louis D N. Cancer Res. 1994;54:4760–4763. [PubMed] [Google Scholar]
- 22.Li L, Keverne E B, Aparicio S A, Ishino F, Barton S C, Surani M A. Science. 1999;284:330–333. doi: 10.1126/science.284.5412.330. [DOI] [PubMed] [Google Scholar]
- 23.Wu M, Lee H, Bellas R E, Schauer S L, Arsura M, Katz D, FitzGerald M J, Rothstein T L, Sherr D H, Sonenshein G E. EMBO J. 1996;15:4682–4690. [PMC free article] [PubMed] [Google Scholar]
- 24.Sonenshein G E. Semin Cancer Biol. 1997;8:113–119. doi: 10.1006/scbi.1997.0062. [DOI] [PubMed] [Google Scholar]
- 25.Wallach D. Trends Biochem Sci. 1997;22:107–109. doi: 10.1016/s0968-0004(97)01015-3. [DOI] [PubMed] [Google Scholar]