Abstract
Asthma is an inflammatory disease of the lungs and the transcription factor NF-κB regulates the production of numerous inflammatory mediators that may have a role in the pathogenesis of asthma. Hence, the signalling pathways leading to NF-κB activation are considered prime targets for novel anti-inflammatory therapies. The prevention of NF-κB activity in mice, through the knockout of IKKβ or p65, causes fatal liver degeneration in utero making it difficult to determine the full implications of inhibiting NF-κB activity in tissues physiologically relevant to human diseases.
This study used adenovirus delivery of a dominant inhibitor of NF-κB (IκBαΔN) and dominant-negative IKKα (IKKα(KM)) and IKKβ (IKKβ(KA)) to investigate the role of the individual IKKs in NF-κB activation and inflammatory gene transcription by human pulmonary A549 cells.
Overexpression of IKKβ(KA) or IκBαΔN prevented NF-κB-dependent transcription and DNA binding. IKKβ(KA) also prevented IκBα kinase activity. Similarly, IKKβ(KA) and IκBαΔN overexpression also inhibited IL-1β- and TNFα-dependent increases in ICAM-1, IL-8 and GM-CSF in addition to IL-1β-mediated increases in cyclooxygenase-2 expression, whereas IKKα(KM) overexpression had little effect on these outputs.
IKKβ(KA) also reduced cell viability and induced caspase-3 and PARP cleavage regardless of the stimuli, indicating the induction of apoptosis. This effect seemed to be directly related to IKKβ kinase activity since IκBαΔN only induced PARP cleavage in TNFα-treated cells.
These results demonstrate that inhibition of IKKβ and NF-κB suppresses inflammatory mediator production and reduces A549 cell viability. Thus, novel therapies that target IKKβ could have potent anti-inflammatory effects and may be beneficial in the treatment of certain cancers.
Keywords: NF-kappa B, I kappa B kinase, pulmonary epithelium, inflammation, apoptosis, signal transduction, cytokines, IL-8, GM-CSF, prostaglandin GH synthase
Introduction
Asthma is a chronic inflammatory disease of the lungs, a key characteristic of which is infiltration of the airways by activated immune cells such as macrophages, eosinophils and T-cells (Bousquet et al., 2000). In addition, many structural cells of the airways such as epithelial and smooth muscle cells play an active role in the airway inflammatory process by producing inflammatory mediators including chemokines (IL-8), cytokines (GM-CSF), adhesion molecules (ICAM-1) and enzymes (cyclooxygenase-2 (COX-2) (Bousquet et al., 2000). The transcriptional regulation of such mediators has attracted considerable interest since inhibition of their production could prove beneficial in the treatment of inflammatory diseases (Barnes, 1999). The regulation of many of these genes is thought to be controlled by the ubiquitous transcription factor NF-κB (Ghosh & Karin, 2002), which consists of hetero- or homo-dimers of proteins from the Rel family including RelA (p65), RelB, c-Rel and NF-κB1 (p105/p50) and NF-κB2 (p100/p52) (Ghosh & Karin, 2002). In resting cells, inhibitor of kappa B (IκB) proteins bind NF-κB to prevent activation of NF-κB-dependent transcription. The binding of IL-1β or TNFα to their respective receptors initiates signalling pathways that lead to activation of the IκB kinase (IKK) complex, the degradation of IκBα and the subsequent activation of NF-κB (Ghosh & Karin, 2002). The IKK complex contains a structural protein IKKγ and two kinases, IKKα and IKKβ, both of which are capable of phosphorylating IκBα (Ghosh & Karin, 2002). Phosphorylation of IκBα promotes its ubiquitination, which marks the protein for rapid degradation by the 26S proteasome (Ghosh & Karin, 2002). As the convergence point for numerous stimuli that activate NF-κB, the IKK complex appears to be a logical target for therapeutic strategies aimed at preventing NF-κB activation (Ghosh & Karin, 2002). However, at present, the functional significance of the two IKK kinases in systems relevant to the physiology of human diseases is unclear. Genetic studies in mice indicate that IKKβ is the main IL-1β and TNFα responsive IκB kinase in embryonic fibroblasts (Ghosh & Karin, 2002). Unfortunately, IKKβ knockout mice die in utero due to the increased sensitivity of hepatocytes to TNFα-induced apoptosis, making it difficult to study the role of IKKβ in tissues relevant to specific diseases such as the lung or immune system (Ghosh & Karin, 2002). IKKα knockout mice show abnormal skin and limb development and die shortly after birth (Ghosh & Karin, 2002). Interestingly, replacement of IKKα with a kinase defective IKKα results in a less severe phenotype in which the mice are viable, but demonstrate defects in the activation of NF-κB by receptor activator of NF-kappaB ligand (RANKL) (Ghosh & Karin, 2002). Although these studies suggest that IKKβ is the main cytokine-inducible IκB kinase, it is still uncertain whether this is the case in all cell types or in human cells. Moreover, there is also evidence that both IKKα and IKKβ are required for cytokine-induced NF-κB activation in mouse embryonic fibroblasts (MEFs) (Li et al., 2002).
Since the pulmonary epithelium is the first point of contact for inhaled therapeutic interventions in diseases such as asthma, an understanding of the role of NF-κB and the IKKs in the expression of inflammatory genes in the pulmonary epithelium would provide key information to evaluating the therapeutic potential of inhibiting this pathway. Moreover, elucidating the role of the individual IKKs in NF-κB activation and inflammatory gene transcription may allow targeted inhibition of the IKK responsible for the production of inflammatory mediators relevant to asthma. In the present study, adenoviral-mediated gene transfer of an N-terminal truncated dominant version of IκBα (Krappmann et al., 1996), as well as dominant-negative IKKα and IKKβ (Zandi et al., 1997), was used to examine the role of NF-κB and the IKKs in inflammatory gene expression in pulmonary A549 cells.
Methods
Cell culture, reporter cell lines and virus infection
The culture of A549 cells and A549 cells harboring the NF-κB-dependent luciferase reporter, 6κBtk.luc has been described (Bergmann et al., 2000). Dominant negative of IKKα(KM), IKKβ (KA), and dominant IκBα (IκBαΔN) have previously been described (Krappmann et al., 1996; Zandi et al., 1997), an empty adenovirus vector (null) was also used. Cells were infected with viruses at the indicated multiplicity of infection (MOI) overnight and subsequently changed to fresh medium. Prior to stimulation, cells were incubated in serum-free medium for 24 h and then at either 24 or 48 h postinfection, the cells were treated with IL-1β (R&D systems, Abingdon, U.K.) (1 ng ml−1) or TNFα (R&D systems) (10 ng ml−1) for the indicated times.
PAGE and Western blotting
Western blot analysis of proteins was carried out using the NuPAGE system (Invitrogen, Paisley, U.K.) as per the manufacturers' instructions. All samples were run on 4–12% gradient gels in MOPS buffer supplemented with antioxidant (Invitrogen). Proteins were electro-transferred onto nitrocellulose membranes using the NuPAGE transfer buffer. Detection of proteins was performed using ECL (Amersham Pharmacia Biotech, Little Chalfont, Bucks, U.K.) according to the manufacturers' instructions. Antisera used were: anti-IKKα (66781A) (BD Pharminogen Oxford U.K.), anti-IKKβ (#05-535) and anti-IKKγ (#05-632) (Upstate Biotechnology, Milton Keynes, U.K.), antiactive caspase-3 (#9662) and anti-PARP (#9542) (New England Biolabs (U.K.) Ltd, Hitchin, U.K.), COX-2 (sc-1745) (Santa Cruz, CA, U.S.A.), ICAM-1 (sc-8439) (Santa Cruz) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (#4699–9555) (Biogenesis Ltd, Poole, U.K.).
Kinase assay and electrophoretic mobility shift assay
Kinase assays were performed as previously described with the following modifications (Nasuhara et al., 1999). IKK complex was immunoprecipitated with an anti-IKKγ antibody (sc-8330AC) (Santa Cruz). Western blot analysis for IKKγ (#05-632) (Upstate biotechnology) was used to confirm loading. Electrophoretic mobility shift assay (EMSA) was performed as previously described (Nasuhara et al., 1999).
ELISA and RIA
ELISAs for IL-8 and GM-CSF were performed using Duoset ELISA kits (R&D Systems) and carried out according to the manufacturers' instructions. Radioimmunoassay (RIA) for PGE2 was performed using a commercially available PGE2 antibody (Sigma) as previously described (Mitchell et al., 1994).
Cell viability assay
Cell viability was assessed by 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) assay.
RNA extraction and Northern blot analysis
Total cellular RNA was prepared using the Qiagen RNA extraction kit according to the manufacturers' instructions (Qiagen, Crawley, West Sussex, U.K.). Northern blot analysis of COX-2 and GAPDH were performed as previously described (Catley et al., 2003). The probe for IL-8 was generated by reverse transcription–polymerase chain reaction (RT–PCR) amplification using the indicated primers prior to cloning and sequence verification. Probes were 32P-labeled using the random primed method (Ready Prime kit) (Amersham Pharmacia Biotech). Primer pairs (5′>3′) for IL-8 were: CTA GCA CAA GAG CCA GGA AGA (forward), AAC CCT CTG CAC CCA GTT TTC (reverse).
Reverse transcriptase–polymerase chain reaction
Semiquantitative RT–PCR for ICAM-1 and GAPDH was performed as previously described (Bergmann et al., 2000). Primers for GAPDH were as described (Bergmann et al., 2000) and primers for ICAM-1 (5′>3′) were: ACC GGA AGG TGT ATG AAC TG (forward), TTG GCT CCA TGG TGA TCT CT (reverse). For each experiment the exponential phase of the PCR, where starting material is proportional to product formation, was determined as described (Bergmann et al., 2000). Reaction products were analysed by agarose gel electrophoresis and gel images subject to densitometric analysis using Totallab version 1 (NonLinear Dynamics Ltd).
Antisense methods
In total, 15 potential IKKα and IKKβ antisense oligonucleotides were identified by screening the mRNA sequence of IKKα and IKKβ for areas which were likely to be accessible to antisense oligonucletides. These oligonucleotides were then manufactured as chimeric methylphosphonate/phosphorothioate oligonucleotides and transfected into A549 cells using previously described methods (McKay et al., 1999). Antisense-induced reduction of mRNA levels was determined by Taqman real time RT–PCR according to the manufacturer's protocols (PE Biosystems, Foster City, U.S.A.) (primers IKKα 5′-TGT GCA CAC TGT GCA GAG TCA-3′ (forward), 5′-GCT TAC AGC CCA ACA ACT TGC T-3′ (reverse); IKKβ 5′-CCG GAA GTA CCT GAA CCA GTT T-3′ (forward) 5′-GGA CGA TGT TTT CTG GCT TTA GA-3′ (reverse); IKKα probe: 5′-TGA CCA AAC AGC TCC TTG AGG-3′; IKKβ probe: 5′-AGT GAC ATT GCC TCT GCG CTT AGA TAC CTT C-3′). This screening process identified one potent antisense for IKKα (antisense 5′-CAU GCC AGG TAA ATG GCU GC-3′, scrambled control 5′-CAU CGC GAT GAT GAT CGC GA-3′) and one for IKKβ (antisense 5′-CCA GTA GTC GAC GGT CAC TG-3′, scrambled control 5′-CCG AGA TAG CCT TGG CCA TG-3′), which were manufactured as chimeric 2′-O-methoxy/phosphorothioate oligonucleotides (McKay et al., 1999). Concentration–response experiments were analysed by Taqman real time RT–PCR and determined the optimum oligonucleotide concentration required for reduction of IKKα or IKKβ mRNA to be 200 nM. A scrambled control oligonucleotide was used to control for nonantisense effects related to the oligonucleotide base composition.
Data analysis
Data are presented as means±s.e.m. Comparison between groups was performed using one-way analysis of variance with a Bonferroni post-test. Significance was taken where P<0.05 (*), P<0.01 (**) and P<0.001 (***).
Results
Dominant-negative IKKβ prevents NF-κB dependent transcription
In a previous study, we showed that an MOI of 10 is sufficient to infect >90% of A549 cells with an Ad5-based adenoviral vector (Catley et al., 2003). This study also demonstrated inhibition of NF-κB DNA binding and transcriptional activity by overexpression of IκBαΔN (Catley et al., 2003). In the present study, adenoviral-mediated overexpression of dominant-negative versions of IKKα and IKKβ was used to elucidate the role of these two kinases in the activation of NF-κB. Infection with dominant-negative IKKα(KM) had no significant inhibitory effect on NF-κB reporter activation following treatment with either IL-1β or TNFα (Figure 1a and b). However, a small activation of the reporter is seen with MOI 3 of this virus. This is thought to be due to nonspecific effects of the virus on the reporter, since a similar effect on the reporter is seen with the null virus (data not shown). In contrast, at MOI of 0.1–30 dominant-negative IKKβ(KA) significantly reduced reporter activation by both IL-1β and TNFα to or below the basal level, and in each case this effect was proportional to the MOI used (Figure 1c and d). This data therefore suggest an important role for IKKβ, but not IKKα, in the transcriptional activation of NF-κB.
Figure 1.
Effect of IKKα(KM) and IKKβ(KA) on NF-κB-dependent transcription. A549 cells stably transfected with the NF-κB-dependent reporter 6κBtk.luc were infected with various MOIs of either IKKα(KM) or IKKβ(KA) expressing adenovirus. After 48 h, cells were then either not stimulated or stimulated with IL-1β (1 ng ml−1) (a and c) or TNFα (10 ng ml−1) (b and d) for 6 h prior to harvesting for luciferase assay and Western blot analysis. Western blots (upper panels) are representative of two such experiments using anti-HA (a and b) or anti-IKKβ protein (c and d) antibodies. Luciferase data (lower panels) are representative of four such experiments and is expressed as a percentage of stimulation as means±s.e.m.
IKK activity and NF-κB DNA binding activation depends on a functional IKKβ
Analysis of kinase activity of the IKK complex and NF-κB DNA binding by EMSA was undertaken to investigate the effects of dominant-negative IKKs and the dominant IκBαΔN (Figure 2). Following cell stimulation and immunoprecipitation, IKK kinase activity was revealed by the ability to phosphorylate a GST-IκBα (1–54) substrate. In each case, both IL-1β and TNFα strongly induced kinase activity (Figure 2a). However, whereas the dominant-negative IKKα virus had no effect on kinase activity, the dominant-negative IKKβ virus completely ablated the kinase activity induced by both IL-1β and TNFα (Figure 2a). As expected, IκBαΔN showed no effect on kinase activity as it blocks NF-κB activation downstream of the IKK complex. Kinase specificity was confirmed by the inability of immunoprecipitates to phosphorylate a mutated GST-IκBα (1–54: S32S, S36A) in which the two phospho-acceptor serines are replaced by alanine. Specificity of the immunoprecipitation was shown by the lack of activity in lanes using pre-immune antisera in place of the IKKγ antisera for immunoprecipitation. Purified IKK complex from TNFα stimulated HeLa cells was used as a positive control.
Figure 2.
Effect of IKKα(KM) and IKKβ(KA) on IKK activity and NF-κB DNA binding. A549 cells were infected with the indicated viruses (MOI 10) for 48 h prior to stimulation with IL-1β (1 ng ml−1) or TNFα (10 ng ml−1). (a) Cells were harvested at 2-min poststimulation and IKK complex immunoprecipitated using anti-IKKγ antibody. Immunoprecipitates were then divided into two and subjected to either IKK kinase assay using GST-IαBα (1–54) as substrate (upper panel) or Western blot analysis for IKKγ (lower panel). Blots representative of two such experiments are shown. Immunoprecipitates from cells treated with TNFα and IL-1β were also subjected to kinase assay using a mutant substrate, GST-IκBα-(1–54; S32A, S36A), as indicated (Mutant). In addition, immunoprecipitates using preimmune antisera were subject to Western blot analysis and kinase assay as indicated (PI). Purified IKK complex from TNFα-stimulated HeLa cells was used as a positive control (IKK). (b) Cells were treated as in (a) and harvested at 1 h poststimulation and EMSA performed using a probe for NF-κB. Autoradiographs representative of three such experiments are shown.
Analysis of NF-κB nuclear translocation and DNA binding by EMSA revealed three complexes that were competed out by cold competition and therefore represent specific NF-κB species (Figure 2b). Previous studies have demonstrated that these complexes consist of both p50 and p65 by supershift (Newton et al., 1996). The upper bands were most induced in response to IL-1β and TNFα, whereas a middle band appeared to be constitutive. Virus overexpressing the dominant-negative IKKβ and the dominant IκBαΔN completely ablated the inducible DNA binding complexes (Figure 2b). By contrast, neither infection with the dominant-negative IKKα virus nor the null virus resulted in any obvious effect on DNA binding complexes. Taken together with the reporter and the kinase assay data, this indicates that, in A549 cells, IKKβ, and not IKKα, is primarily responsible for the IL-1β- and TNFα-induced activity of the IKK complex and that this is required for NF-κB nuclear translocation, DNA binding and subsequent transcriptional activation. Cytoplasmic fractions from the EMSA were analysed by Western blotting to confirm the expression of FLAG-IKKα(KM), HA-IKKβ(KA) and IκBαΔN protein expression (Figure 2b).
Expression of inflammatory genes depends on NF-κB and requires IKKβ
Having established that IKKβ is the main IκB kinase responsible for NF-κB-dependent transcription, experiments were undertaken to establish the functional consequences of inhibiting this cascade. Northern blot analysis demonstrated significantly increased expression of both COX-2 and IL-8 following stimulation with IL-1β (Figure 3a). However, the TNFα-mediated increase in mRNA was smaller than that for IL-1β and, in the case of COX-2, was barely above background levels. Prior infection with the dominant-negative IKKβ and the dominant IκBαΔN viruses significantly reversed this increase, whereas dominant-negative IKKα and the null virus were without effect. Semiquantitative RT–PCR analysis of ICAM-1 mRNA revealed strong induction by both TNFα and IL-1β and this was dramatically reduced by the dominant-negative IKKβ and the dominant IκBαΔN viruses (Figure 3b). Again the dominant-negative and IKKα and the null viruses had no effect.
Figure 3.
Effect of IKKα(KM), IKKα(KA) and IκBαΔN on IL-1β- and TNFα-induced expression of COX-2, IL-8 and ICAM-1. Cells were either not infected or infected with viral vectors for IKKα(KM), IKKβ(KA), IκBαΔN or a null virus as indicated. At 48 h postinfection the cells were either not stimulated (NS) or treated as indicated with IL-1β (1 ng ml−1) or TNFα (10 ng ml−1) for 4 h and total RNA was prepared. (a) Northern blot analysis was performed using probes for IL-8, COX-2 and GAPDH. Blots representative of at least four such experiments are shown. Autoradiographs were subjected to densitomeric analysis, normalised to GAPDH, expressed as a percentage of the IL-1β stimulated sample and plotted as means±s.e.m. (a, lower panels). (b) RT–PCR was performed using primers specific for ICAM-1 and GAPDH. Gels representative of at least three such experiments are shown. Gels were subjected to densitomeric analysis, normalised to GAPDH and optical densities were plotted as means±s.e.m. (b, lower panel).
To confirm these effects observed at the mRNA level, Western blot analysis for ICAM-1 and COX-2 was performed along with ELISA for IL-8 and GM-CSF and RIA for PGE2 (Figure 4). However, whereas in our previous experiments no effect on cell viability was observed, it was apparent that increasing the incubation time to 18 h following the serum starvation led to noticeable losses in cell viability (see Figure 5), which prevented meaningful analysis using this protocol. In an effort to ameliorate this issue, cells were infected with viruses and then incubated in serum-free medium overnight prior to being either not stimulated or treated with cytokines the following day. Cells were then harvesting after 18 h (i.e. less than a total of 48 h post viral infection). Using this modified protocol, no significant loss of cell viability was observed (data not shown). In the absence of any prior stimulation, infection with all four viruses had no effect on expression of ICAM-1 or COX-2 (Figure 4a). Stimulation with IL-1β produced an increase in both COX-2 and ICAM-1 expression and this was unaffected by infection with either the dominant-negative IKKα or the null virus (Figure 4a). Consistent with the mRNA data, both the dominant IκBαΔN and the dominant-negative IKKβ viruses prevented the IL-1β-mediated increases in COX-2 and ICAM-1 protein (Figure 4a). Stimulation with TNFα also strongly induced expression of ICAM-1 protein, but not COX-2. Again this was unaffected by the dominant-negative IKKα and the null viruses, but was prevented by the IκBαΔN and the dominant-negative IKKβ viruses.
Figure 4.
The effect of IKKα(KM), IKKβ(KA) and IκBαΔN on the expression of inflammatory mediators. Cells were either not infected or infected with the indicated virus (MOI 10) for 24 h prior to stimulation with either IL-1β (1 ng ml−1) or TNFα (10 ng ml−1) for a further 18 h. (a) Cells were harvested for Western blot analysis of ICAM-1, COX-2 and GAPDH. Blots representative of five such experiments are shown. (b and c) Cell culture medium was removed and analysed for GM-CSF and IL-8 release by ELISA and for PGE2 release by RIA. Data (n=4) are plotted as the mean±s.e.m.
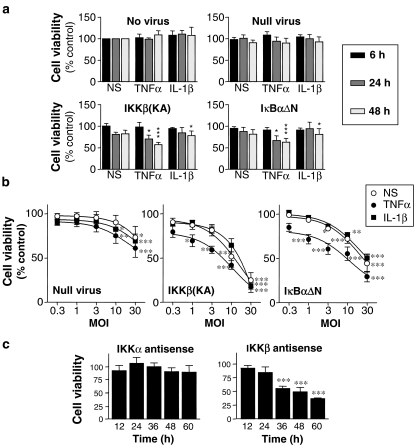
Figure 5.
Cell viability following infection with recombinant adenoviruses and inflammatory cytokines. (a) Cells were either not infected or infected with the indicated virus (MOI 10) for 48 h prior to stimulation with either IL-1β (1 ng ml−1) or TNFα (10 ng ml−1) or no stimulation (NS). Cells were harvested after 6, 24 and 48 h for MTT assay of cell viability. (b) Cells were infected with various MOI of the indicated viruses for 48 h prior to stimulation with IL-1β (1 ng ml−1), TNFα (10 ng ml−1) or no stimulation (NS). After a further 24 h cells were harvested for MTT assay. (c) Cells were transfected with antisense oligonucleotides to either IKKα or IKKβ for the indicated times prior to harvesting for MTT assay. Data in (a) (n=6–7) and (b) (n=6–8) are expressed as either a percentage of uninfected and unstimulated samples harvested at each time point. In (c), data (n=4) are expressed as a percentage of samples transfected with a scrambled control oligonucleotide. All data are plotted as means±s.e.m.
Supernatants from the above experiments were analysed for IL-8 and GM-CSF expression by ELISA. Infection with the dominant-negative IKKα and IKKβ, dominant IκBαΔN or the null viruses again had no effect on the basal release of IL-8 or GM-CSF (data not shown). Stimulation with TNFα or IL-1β strongly induced the release of IL-8 and GM-CSF (Figure 4b). However, the TNFα-induced release of IL-8 and GM-CSF (59 ng ml−1 and 43 pg ml−1 respectively) was considerably lower than that for IL-1β (236 ng ml−1 and 224 pg ml−1 respectively). Infection with the dominant-negative IKKα or the null virus appeared to show some inhibition of release of both IL-8 and GM-CSF. However, this was only partial, whereas infection with the dominant-negative IKKβ or IκBαΔN viruses significantly reduced both IL-8 and GM-CSF release to at or below basal levels.
Analysis of supernatants by RIA for PGE2, the major prostanoid produced by these cells (Mitchell et al., 1994), demonstrated no effect of these four viruses on PGE2 release from unstimulated cells. In agreement with the above mRNA and protein data for COX-2, TNFα showed little effect on PGE2 release (Figure 4c). In contrast, IL-1β induced a large increase in the level of PGE2 release and this was unaffected by the dominant-negative IKKα or the null viruses (Figure 4c). Again the dominant IκBαΔN and the dominant-negative IKKβ viruses reduced PGE2 release to near basal levels (Figure 4c).
Inhibition of NF-κB and IKKβ activity promotes cell death
A number of reports have suggested a role for NF-κB in preventing entry into apoptosis, and indeed the above experiments also suggest that the inhibition of NF-κB may adversely effect A549 cell viability (Karin & Lin, 2002). Initial experiments revealed little or no effect of the null or IKKα(KM) overexpressing virus, whereas both the IKKβ(KA) and the IκBαΔN expressing viruses resulted in significant loss of cell viability at MOIs of up to 30 (data not shown). In each case this cell death was associated with rounding up of cells, cytoplasmic blebbing and nuclear condensation, which are all signs of apoptosis (data not shown). To further explore this effect, cells were treated with the various viruses (MOI 10) for 48 h and then either not stimulated or treated with TNFα or IL-1β. Mitochondrial succinate dehydrogenase activity was assessed at the times indicated using the MTT assay as an indicator of cell viability (Figure 5a). In each case, TNFα, IL-1β or null virus in combination or alone resulted in no significant effect on cell viability at any time point. However, both the IKKβ(KA) and the IκBαΔN expressing viruses resulted in significant reductions in viability when incubated with TNFα or to a lesser extent IL-1β for 24 or 48 h. Importantly, for the previous experiments detailed above, there was no effect of either virus following 48 h of viral incubation plus a 6 h stimulation period. These data therefore indicate that the observed repressive effects on reporter activity and gene expression precede any loss of cell viability due to inhibition of NF-κB activity. Since effects on viability were observed following 48 h of virus plus a further 24 h of stimulation, this time point was selected for analysis of the effect of increasing MOI (Figure 5b). In the presence of the null virus there was little effect on viability at or below an MOI of 10. However, in the presence of TNFα, viability was reduced to 61±10.3% of control values at an MOI of 30. By contrast both the IKKβ(KA) and the IκBαDN overexpressing viruses produced a highly significant concentration-dependent effect on cell viability (Figure 5b). In each case IL-1β showed no additional effect, but the response was enhanced by TNFα.
To further confirm the specificity of this response, a complementary strategy using modified 2′-O-methoxy/phosphorothioate chimeric antisense oligonucleotides directed to IKKα and IKKβ was adopted (Figure 5a). In these experiments, optimised antisense to IKKα resulted in little or no effect on cell viability compared to a control oligonucleotide, whereas the IKKβ antisense again produced a marked time dependent reduction in cell viability (Figure 5b).
To investigate whether the observed loss of cell viability was due to apoptosis or necrosis, Western blotting for active caspase-3 and cleaved (poly(ADP-ribose) polymerase) PARP was performed. Caspase-3 is one of the key executioners of apoptosis and is responsible for the cleavage of many key enzymes, including PARP (Fernandes-Alnemri et al., 1994). The activation of caspase-3 requires the inactive pro-caspase to be cleaved by upstream caspases, such as caspase-8, into p20 and p12 subunits (Fernandes-Alnemri et al., 1996). The p20 and p12 subunits then associate to form active caspase-3, rapid autocatalysis of the p20 subunit produces p19, which is processed by a slow autocatalysis step to produce p17 (Fernandes-Alnemri et al., 1996). Poly(ADP-ribosylation) is a post-translational modification of proteins, which is involved in DNA replication and repair, transcription and cell death (Soldani & Scovassi, 2002). One of the key proteins involved in Poly(ADP-ribosylation) is PARP-1 (Soldani & Scovassi, 2002). PARP-1 is cleaved in vivo by caspase-3 and -7 (Soldani & Scovassi, 2002). However, PARP-1 may also be cleaved in the absence of caspase-3. The caspase-mediated cleavage of PARP is believed to facilitate cellular disassembly and thus serves as a marker for apoptosis (Oliver et al., 1998). Western blot analysis for PARP in untreated or IL-1β-treated cells showed no evidence of PARP cleavage in uninfected cells, cells infected with the null virus or cells infected with either dominant-negative IKKα or IκBαΔN viruses (Figure 6). In contrast, infection with dominant-negative IKKβ induced PARP cleavage to the 89 and 24 kDa products, indicative of cleavage by caspases. However, cells treated with TNFα showed a different profile of PARP cleavage. Uninfected cells and cells infected with the null or the dominant-negative IKKα expressing virus showed no PARP cleavage, whereas expression of both IKKβ(KA) and IκBαΔN induced PARP cleavage. This is consistent with other reports, which demonstrate increased sensitivity to TNFα-mediated apoptosis in cells where NF-κB activation is defective (Li et al., 1999b). To further confirm that expression of IKKβ(KA) induces the apoptotic cascade the membranes were striped and probed for p19/p17 fragments of active caspase-3. In unstimulated, IL-1β- or TNFα-treated cells, active caspase-3 was absent in uninfected cells, cells infected with null virus cell and cells expressing IκBαΔN. A low level of p19 was seen in IL-1β treated IKKα(KM) infected cells indicating a low level of caspase-3 activation. In contrast, cells expressing IKKβ(KA) show bands at 17 and 19 kDa corresponding to the p19/p17 fragments of active caspase-3. This indicates that caspase-3 is active in A549 cells where IKKβ is not active.
Figure 6.
Effect of IKKα(KM), IKKβ(KA), and IκBαΔN on PARP cleavage and caspase-3 activation. Cells were infected with the indicated viruses (MOI 10) for 48 h prior to stimulation with IL-1β (1 ng ml−1) or TNFα (10 ng ml−1) for 18 h. Cell lysates were then harvested and analysed for cleavage of PARP (upper panels) or active caspase-3 (lower panels) by Western blotting. Blots representative of two such experiments are shown.
Discussion
Numerous studies have investigated the role of the individual IKKs in the activation of NF-κB using knockout mouse strategies (Ghosh & Karin, 2002). Since IKKβ−/− mice die in utero, most of the studies examining transcriptional control have been carried out in MEFs and indicate that, in contrast to IKKα, IKKβ is required for activation of NF-κB by IL-1β and TNFα (Li et al., 1999b; Ghosh & Karin, 2002). The data presented in this present study demonstrate that in pulmonary A549 cells, stimulation of NF-κB-dependent transcription, the phosphorylation of IκBα and the induction of NF-κB DNA binding in response to TNFα or IL-1β is also dependent on IKKβ and not IKKα. Moreover, prevention of IKKβ, but not IKKα, kinase activity inhibited the production of known inflammatory products, including ICAM-1, GM-CSF, IL-8, COX-2 and PGE2. This is consistent with studies in HeLa cells and alveolar macrophages, which demonstrate prevention of NF-κB activity and inflammatory mediator production after removal of IKKβ kinase activity (Mercurio et al., 1997; Conron et al., 2002). Thus, the data presented here further endorses the role of IKKβ as the main IκB kinase in response to inflammatory stimuli in human pulmonary cells. It also demonstrates that novel therapeutic agents, which target IKKβ activation, will prevent the expression of inflammatory mediators and may have potential as anti-inflammatory drugs. However, the specific ablation of IKKβ in the epidermis of mice causes a progressive inflammatory skin disease (Pasparakis et al., 2002), indicating that inhibition of IKKβ may also have detrimental effects. Likewise, the inhibition of IKKβ may also affect signalling through toll receptors and this could possibly have a suppressive effect on innate immunity in the lungs and lead to higher levels of infective episodes (Imler & Hoffmann, 2001).
In contrast, a number of recent studies have indicated that IKKα plays a significant role in the TNFα-mediated activation of NF-κB-dependent transcription, possibly by translocating to the nucleus and phosphorylating either p65 or histone H3 (Li et al., 2002; Anest et al., 2003; Yamamoto et al., 2003). However, the data presented above show that overexpression of IKKα(KM) has little effect on transcription of NF-κB responsive genes after TNFα, or IL-1β, stimulation suggesting that such events do not occur in the present system. Thus, while it is difficult reconcile this role for IKKα in general TNFα-mediated NF-κB activation, it is possible that IKKα activates NF-κB in response to certain stimuli or in specific cell types (Ghosh & Karin, 2002). For example, IKKα is required for p100 processing to p52 and this process appears to be important for B-cell survival and the formation and organisation of lymphoid tissues (Ghosh & Karin, 2002). Moreover, IKKα knockin mice have a specific defect in RANKL signalling in mammary epithelial cells and the NIK-IKKα pathway is thought to function in lymphotoxin-β signalling (Matsushima et al., 2001). Therefore, while IKKβ clearly plays a key role in responses to IL-1β and TNFα and it remains possible that IKKα may mediate responses to a different range of stimuli.
In the present study, expression of kinase defective IKKβ(KA), and to a slightly lesser extent IκBαΔN, reduced the viability of A549 cells. This effect is generally consistent with studies showing fatal TNFα-induced hepatocyte apoptosis in IKKβ, IKKγ and p65 knockout mice (Li et al., 1999a; Ghosh & Karin, 2002). Furthermore, as NF-κB is known to play a central role in the transcriptional induction of various cellular inhibitors of apoptosis (cIAPs), it is likely that in the present study these NF-κB-dependent genes are inhibited along with the classical inflammatory genes COX-2, IL-8 and GM-CSF (Karin & Lin, 2002). Consequently, the successful inhibition of NF-κB may be expected to promote apoptosis. Thus, in IKKβ(KA) overexpressing cells, PARP and caspase-3 cleavage was induced in unstimulated cells and cells treated with IL-1β and TNFα. This may predispose these cells towards an apoptotic phenotype and suggests, as has been previously reported, that constitutive activation of NF-κB may be required to maintain cell viability (Orlowski & Baldwin Jr, 2002). In contrast, overexpression of IκBαΔN only induced PARP cleavage in TNFα-treated cells, and this was without apparent effect on activation of caspase-3. While this is consistent with the greater loss of viability in the presence of TNFα, the lack of detectable caspase-3 activation suggests the involvement of alternative apoptotic pathways such as caspase-7, which is known to cleave PARP in vivo (Soldani & Scovassi, 2002).
The finding that both dominant-negative IKKβ and dominant IκBαΔN overexpression leads to loss of cell viability, yet only dominant-negative IKKβ induces caspase-3 cleavage implies that IKKβ kinase activity may have a specific role in repressing the caspase-3 pathway. Certainly, the fact that IKKβ is a target for caspase-3 mediated cleavage indicates that removal of IKKβ activity may be a critical event in the decision between life and death for the cell (Tang et al., 2001; Karin & Lin, 2002). Thus, it is possible that IKKβ kinase activity targets proteins other than the IκBs. Such an activity may be required to prevent caspase-3 activation and maintain cell viability.
In terms of lung inflammation, the induction of apoptosis by IKKβ inhibitors could aid the clearance of activated inflammatory cells from the lung, and therefore lead to reduced inflammation of the airways (Bousquet et al., 2000; Senftleben et al., 2001). However, a common feature of asthmatic airways is a shedding of the epithelium, which itself correlates with airway hyper-responsiveness (Bousquet et al., 2000). Thus, IKKβ inhibitors could promote epithelial damage and lead to increased airway hyper-responsiveness. Conversely, in the context of COPD, apoptosis may prove beneficial, for example, by preventing goblet cell hyperplasia. Finally, it may be possible to use IKKβ inhibitors to reduce NF-κB activity, rather than completely eliminating it. This may prevent the induction of apoptosis and reduce problems of associated tissue damage. Clearly, these issues require testing in appropriate animal models. Alternatively, inflammation and activation of NF-κB are also associated with various cancers, and it is likely that the antiapoptotic effects of NF-κB are a contributory factor to tumour survival (Karin & Lin, 2002). The data presented here document a profound effect of inhibiting NF-κB on A549 adenocarcinoma cell survival. Furthermore, these data suggest the existence of IKKβ-dependent pathways, which lead to cell death and activation of caspase-3, as well as NF-κB-dependent effects (inhibited by IκBαΔN), which while also leading to cell death do not appear to involve caspase-3 activation. These data therefore suggest the need for detailed investigation of the potential roles of these pathways in mediating cell death in the lung epithelium and suggest the potential utility of small molecule inhibitors of IKKβ in the treatment of lung adenocarcinoma and possibly other cancers.
In conclusion, this report shows that activation of NF-κB relies on the kinase activity of IKKβ and is independent of IKKα. In addition, removal of IKKβ activity prevented the expression of a number of known inflammatory mediators indicating that targeted disruption of IKKβ activity may have potent anti-inflammatory effects and could provide a therapeutically useful antitumour activity.
Acknowledgments
IKKα(KM), IKKβ(KA) and the null virus were a gift from Professor Michael Karin. The IκBαΔN virus was a gift from Dr Peter Loser. M.C.C. and J.E.C. are collaborative students supported by Aventis Pharmaceuticals and the MRC and BBSRC respectively. N.S.H. is an MRC collaborative student funded by Novartis Pharmaceuticals. R.N. is a CIHR New Investigator.
Abbreviations
- COPD
chronic obstructive pulmonary disease
- COX-2
cyclooxygenase-2
- ECL
enhanced chemi-luminesence
- HA
heamaglutinin
- IKK
I kappa B kinase
- MEF
mouse embryonic fibroblast
- MOI
multiplicity of infection
- NIK
NF-κB inducing kinase
- PARP
poly(ADP-ribose) polymerase
- RANKL
receptor activator of NF-kappaB ligand
- TNFR-1
TNF receptor 1
References
- ANEST V., HANSON J.L., COGSWELL P.C., STEINBRECHER K.A., STRAHL B.D., BALDWIN A.S. A nucleosomal function for IkappaB kinase-alpha in NF-kappaB-dependent gene expression. Nature. 2003;423:659–663. doi: 10.1038/nature01648. [DOI] [PubMed] [Google Scholar]
- BARNES P.J. Therapeutic strategies for allergic diseases. Nature. 1999;402 suppl:B31–B38. doi: 10.1038/35037026. [DOI] [PubMed] [Google Scholar]
- BERGMANN M., BARNES P.J., NEWTON R. Molecular regulation of granulocyte-macrophage colony-stimulating factor in human lung epithelial cells by IL-1, IL-4 and IL-13 involves both transcriptional and post-transcriptional mechanisms. Am. J. Respir. Cell Mol. Biol. 2000;22:582–589. doi: 10.1165/ajrcmb.22.5.3889. [DOI] [PubMed] [Google Scholar]
- BOUSQUET J., JEFFERY P.K., BUSSE W.W., JOHNSON M., VIGNOLA A.M. Asthma. From bronchoconstriction to airways inflammation and remodeling. Am. J. Respir. Crit. Care Med. 2000;161:1720–1745. doi: 10.1164/ajrccm.161.5.9903102. [DOI] [PubMed] [Google Scholar]
- CATLEY M.C., CHIVERS J.E., CHIVERS J.E., CAMBRIDGE L.M., HOLDEN N., SLATER D.M., STAPLES K.J., BERGMANN M.W., LOSER P., BARNES P.J., NEWTON R. IL-1beta-dependent activation of NF-kappaB mediates PGE2 release via the expression of cyclooxygenase-2 and microsomal prostaglandin E synthase. FEBS Lett. 2003;547:75–79. doi: 10.1016/s0014-5793(03)00672-0. [DOI] [PubMed] [Google Scholar]
- CONRON M., ANDREAKOS E., PANTELIDIS P., SMITH C., BEYNON H.L., DUBOIS R.M., FOXWELL B.M. Nuclear factor-kappaB activation in alveolar macrophages requires IkappaB kinase-beta, but not nuclear factor-kappaB inducing kinase. Am. J. Respir. Crit. Care Med. 2002;165:996–1004. doi: 10.1164/ajrccm.165.7.2107058. [DOI] [PubMed] [Google Scholar]
- FERNANDES-ALNEMRI T., ARMSTRONG R.C., KREBS J., SRINIVASULA S.M., WANG L., BULLRICH F., FRITZ L.C., TRAPANI J.A., TOMASELLI K.J., LITWACK G., ALNEMRI E.S. In vitro activation of CPP32 and Mch3 by Mch4, a novel human apoptotic cysteine protease containing two FADD-like domains. Proc. Natl. Acad. Sci. U.S.A. 1996;93:7464–7469. doi: 10.1073/pnas.93.15.7464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FERNANDES-ALNEMRI T., LITWACK G., ALNEMRI E.S. CPP32, a novel human apoptotic protein with homology to Caenorhabditis elegans cell death protein Ced-3 and mammalian interleukin-1 beta-converting enzyme. J. Biol. Chem. 1994;269:30761–30764. [PubMed] [Google Scholar]
- GHOSH S., KARIN M. Missing pieces in the NF-kappaB puzzle. Cell. 2002;109 Suppl:S81–S96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- IMLER J.L., HOFFMANN J.A. Toll receptors in innate immunity. Trends Cell Biol. 2001;11:304–311. doi: 10.1016/s0962-8924(01)02004-9. [DOI] [PubMed] [Google Scholar]
- KARIN M., LIN A. NF-kappaB at the crossroads of life and death. Nat. Immunol. 2002;3:221–227. doi: 10.1038/ni0302-221. [DOI] [PubMed] [Google Scholar]
- KRAPPMANN D., WULCZYN F.G., SCHEIDEREIT C. Different mechanisms control signal-induced degradation and basal turnover of the NF-kappaB inhibitor IkappaB alpha in vivo. EMBO J. 1996;15:6716–6726. [PMC free article] [PubMed] [Google Scholar]
- LI Q., VAN ANTWERP D., MERCURIO F., LEE K.F., VERMA I.M. Severe liver degeneration in mice lacking the IkappaB kinase 2 gene. Science. 1999a;284:321–325. doi: 10.1126/science.284.5412.321. [DOI] [PubMed] [Google Scholar]
- LI X., MASSA P.E., HANIDU A., PEET G.W., ARO P., SAVITT A., MISCHE S., LI J., MARCU K.B. IKKalpha, IKKbeta, and NEMO/IKKgamma are each required for the NF-kappa B-mediated inflammatory response program. J. Biol. Chem. 2002;277:45129–45140. doi: 10.1074/jbc.M205165200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI Z.W., CHU W., HU Y., DELHASE M., DEERINCK T., ELLISMAN M., JOHNSON R., KARIN M. The IKKbeta subunit of IkappaB kinase (IKK) is essential for nuclear factor kappaB activation and prevention of apoptosis. J. Exp. Med. 1999b;189:1839–1845. doi: 10.1084/jem.189.11.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MATSUSHIMA A., KAISHO T., RENNERT P.D., NAKANO H., KUROSAWA K., UCHIDA D., TAKEDA K., AKIRA S., MATSUMOTO M. Essential role of nuclear factor (NF)-kappaB-inducing kinase and inhibitor of kappaB (IkappaB) kinase alpha in NF-kappaB activation through lymphotoxin beta receptor, but not through tumor necrosis factor receptor I. J. Exp. Med. 2001;193:631–636. doi: 10.1084/jem.193.5.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MCKAY R.A., MIRAGLIA L.J., CUMMINS L.L., OWENS S.R., SASMOR H., DEAN N.M. Characterization of a potent and specific class of antisense oligonucleotide inhibitor of human protein kinase C-alpha expression. J. Biol. Chem. 1999;274:1715–1722. doi: 10.1074/jbc.274.3.1715. [DOI] [PubMed] [Google Scholar]
- MERCURIO F., ZHU H., MURRAY B.W., SHEVCHENKO A., BENNETT B.L., LI J., YOUNG D.B., BARBOSA M., MANN M., MANNING A., RAO A. IKK-1 and IKK-2: cytokine-activated IkappaB kinases essential for NF-kappaB activation. Science. 1997;278:860–866. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- MITCHELL J.A., BELVISI M.G., AKARASEREENONT P., ROBBINS R.A., KWON O.J., CROXTALL J., BARNES P.J., VANE J.R. Induction of cyclo-oxygenase-2 by cytokines in human pulmonary epithelial cells: regulation by dexamethasone. Br. J. Pharmacol. 1994;113:1008–1014. doi: 10.1111/j.1476-5381.1994.tb17093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NASUHARA Y., ADCOCK I.M., CATLEY M., BARNES P.J., NEWTON R. Differential IKK activation and IkappaBalpha degradation by interleukin-1beta and tumor necrosis factor-alpha in human U937 monocytic cells: evidence for additional regulatory steps in kappaB-dependent transcription. J. Biol. Chem. 1999;274:19965–19972. doi: 10.1074/jbc.274.28.19965. [DOI] [PubMed] [Google Scholar]
- NEWTON R., ADCOCK I.M., BARNES P.J. Superinduction of NF-kappa B by actinomycin D and cycloheximide in epithelial cells. Biochem. Biophys. Res. Commun. 1996;218:518–523. doi: 10.1006/bbrc.1996.0093. [DOI] [PubMed] [Google Scholar]
- OLIVER F.J., DE LA R.G., ROLLI V., RUIZ-RUIZ M.C., DE MURCIA G., MURCIA J.M. Importance of poly(ADP-ribose) polymerase and its cleavage in apoptosis. Lesson from an uncleavable mutant. J. Biol. Chem. 1998;273:33533–33539. doi: 10.1074/jbc.273.50.33533. [DOI] [PubMed] [Google Scholar]
- ORLOWSKI R.Z., BALDWIN A.S., Jr NF-kappaB as a therapeutic target in cancer. Trends Mol. Med. 2002;8:385–389. doi: 10.1016/s1471-4914(02)02375-4. [DOI] [PubMed] [Google Scholar]
- PASPARAKIS M., COURTOIS G., HAFNER M., SCHMIDT-SUPPRIAN M., NENCI A., TOKSOY A., KRAMPERT M., GOEBELER M., GILLITZER R., ISRAEL A., KRIEG T., RAJEWSKY K., HAASE I. TNF-mediated inflammatory skin disease in mice with epidermis-specific deletion of IKK2. Nature. 2002;417:861–866. doi: 10.1038/nature00820. [DOI] [PubMed] [Google Scholar]
- SENFTLEBEN U., LI Z.W., BAUD V., KARIN M. IKKbeta is essential for protecting T cells from TNFalpha-induced apoptosis. Immunity. 2001;14:217–230. doi: 10.1016/s1074-7613(01)00104-2. [DOI] [PubMed] [Google Scholar]
- SOLDANI C., SCOVASSI A.I. Poly(ADP-ribose) polymerase-1 cleavage during apoptosis: an update. Apoptosis. 2002;7:321–328. doi: 10.1023/a:1016119328968. [DOI] [PubMed] [Google Scholar]
- TANG G., YANG J., MINEMOTO Y., LIN A. Blocking caspase-3-mediated proteolysis of IKKbeta suppresses TNF-alpha-induced apoptosis. Mol. Cell. 2001;8:1005–1016. doi: 10.1016/s1097-2765(01)00380-x. [DOI] [PubMed] [Google Scholar]
- YAMAMOTO Y., VERMA U.N., PRAJAPATI S., KWAK Y.T., GAYNOR R.B. Histone H3 phosphorylation by IKK-alpha is critical for cytokine-induced gene expression. Nature. 2003;423:655–659. doi: 10.1038/nature01576. [DOI] [PubMed] [Google Scholar]
- ZANDI E., ROTHWARF D.M., DELHASE M., HAYAKAWA M., KARIN M. The IkappaB kinase complex (IKK) contains two kinase subunits, IKKalpha and IKKbeta, necessary for IkappaB phosphorylation and NF-kappaB activation. Cell. 1997;91:243–252. doi: 10.1016/s0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]