Abstract
To address the question of a possible effect of magnetic fields (MF) at 50 Hz on living systems, gene expression analyses were performed on human primary vascular endothelial cells exposed to MF of various intensities compared to control cells. Exposure protocols included continuous exposure at a single intensity (10 and 700 μT), intermittent exposure at a single intensity (700 μT), and continuous exposure to a variable-intensity field (10–30 μT). The transcriptional response of the cells was investigated using oligonucleotide microarrays containing up to 30 000 unique features. Although in individual experiments genes were identified where the expression appeared to be affected by exposure to MF, none of these genes were regulated in the same manner in subsequent repetition experiments. This is the first report of a transcriptome-wide analysis of the effects of MF exposure on human cells. The lack of a reproducible effect of MF on the expression of any genes in our investigation adds further weight to the evidence that 50-Hz MF are not capable of interacting with biological systems and thus do not represent an endothelial stress factor.
INTRODUCTION
The potential interaction of 50-Hz magnetic fields (MF) produced by domestic electricity supplies with biological systems continues to be a divisive theme in the scientific literature and lay press. Starting with the original paper from Wertheimer and Leeper (1979), a number of epidemiological studies have found a positive association between MF exposure and the presence of various cancers, especially childhood leukemia. Additional studies have found evidence supporting a role of MF in the development of cardiovascular disease (Savitz et al 1999) and Alzheimer's disease (Sobel et al 1996). A majority of studies, however, have failed to show an effect of MF in large populations, leading to a literary quagmire that is difficult to interpret (reviewed in Ahlbom et al 2001). This is not made any clearer by the large number of reports of controlled in vivo and in vitro experiments that also present contradictory findings. Some researchers have found effects of MF exposure including DNA damage, increased proliferation, higher tumor rates, and altered protein expression (Lin et al 1998; Lai and Singh 2004; Wolf et al 2005; Yokus et al 2005). Other reports, including a number of replication studies, have failed to show any evidence of an interaction of MF with the system investigated (Loscher et al 1994; Juutilainen et al 1997; Morehouse and Owen 2000). Our previous studies focussing on MF exposure and heat shock protein expression, both in vitro and in vivo, failed to show any effect (Henderson et al 2003a, 2003b). The lack of convincing evidence for a mechanism of MF interaction with biological systems only adds to this conundrum, although some hypotheses have been proposed, including an MF-responsive DNA promoter sequence and the induction of free radicals (Blank and Goodman 1999; Simko and Mattsson 2004).
With no starting point allowing for targeted investigation of possible pathways or processes that may respond to MF, interrogating a small number of genes has only a negligible chance of finding an effect. Conversely, a technique that is able to simultaneously cover a large number of genes or proteins without requiring specific choices over what they may be is ideally suited to this task. Over recent years, high-throughput transcriptomics and proteomics have provided us with the tools to make this style of experiment not only feasible but also reliable. Microarray analysis allows the investigation of changes in expression patterns across tens of thousands of genes in a single experiment. The main challenge becomes analysis and interpretation of such experiments where a very large data set is created. The disadvantage of using microarray analysis is that an alteration in gene expression may not finally translate into altered protein quantities or activity, while a proteomics approach measures protein levels directly, corresponding to a change in phenotype of the cells.
The first microarray investigations on isolated human cells exposed to MF have recently been published by Luceri et al (2005). This study also looked at yeast cell response to MF by microarray, similar to an earlier study on yeast with a different exposure system (Nakasono et al 2003). Luceri and coworkers found no genes that were significantly affected by MF exposure at intensities up to 100 μT in human lymphocytes or in yeast cells. The scope of these studies was somewhat limited, with only 14 000 genes present on the human array, although the yeast microarray represented the entire genome. Because of the possibility of genome-wide coverage and the ease with which genes of interest can be identified, we have investigated the transcriptional response of human umbilical vein endothelial cells (HUVEC) exposed in vitro to various patterns and intensities of 50-Hz MF. Exposure regimes included continuous exposure to MF at occupational (700 μT) and domestically (10 μT) relevant intensities as well as variable-intensity exposure where the intensity was altered between 10 and 30 μT. To conclude this study, HUVEC were exposed to high-intensity 700-μT MF for 60 min before the field was switched off for 30 min, and this on-off cycle repeated over 24 hours. This report represents the first publication of genome-wide microarray investigations on the response of human cells to MF of various intensities.
MATERIALS AND METHODS
Cell culture conditions and general reagents
The HUVEC were isolated from umbilical cords kindly donated by the Gynecology and Obstetrics Department, University Clinic of Innsbruck. The cells were isolated by enzymatic detachment using collagenase as described elsewhere (Henderson et al 2003b). The cells were routinely passaged in 0.2% gelatin-coated (2% stock solution; Sigma, Steinheim, Germany) polystyrene culture flasks (Becton Dickinson, Meylan Cedex, France) in Endothelial Cell Basal Medium (CC-3121; BioWhittaker Inc, Walkersville, MD, USA) supplemented with EGM SingleQuots Supplements and Growth Factors (CC-4133; BioWhittaker) in a humidified atmosphere containing 5% CO2. Experiments were then performed with 1 × 106 HUVEC seeded onto fibronectin (1 mg/mL; Sigma)-coated cell culture Petri dishes (100 mm; Becton Dickinson) and grown to confluence under the same conditions used for routine cell culture. Medium was changed 30 hours before the cells were harvested. Chemical reagents were purchased from Merck (Darmstadt, Germany) unless otherwise stated and were of analytical grade quality.
Exposure conditions
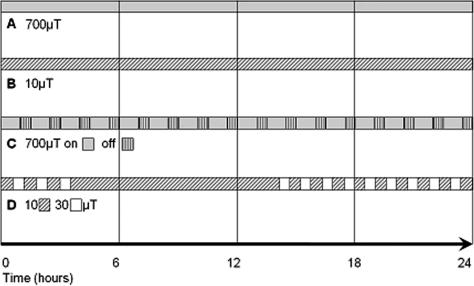
Exposure of HUVEC to defined intensity 50-Hz MF (Fig 1) was achieved by construction of a Helmholtz coil, as described and depicted earlier (Henderson et al 2003a, 2003b). The intensity of the fields used was controlled using a C.A 40 Gaussmeter (Chauvin Arnoux, Paris, France) by measurement before and after each experimental exposure. The earth static magnetic field in Innsbruck is 20 μT. In the first set of experiments (700 μT, 24 hours), control cells were placed in the opposite corner of the same incubator to exclude environmental differences. As such, the “near control” was exposed to an MF of 1% (5 μT) of that of the MF-exposed cells (Henderson et al 2003b). An alternative to the negative control used in later experiments was a sham-exposed control, where cells were placed in a “reversed” Helmholtz coil with a current running through it to control for heating due to the current itself (Henderson et al 2003a). The sham coil is constructed with the upper and lower wire coils wound in the opposite direction, which cancels the magnetic field produced by the current. Sham-exposed cells are exposed to low magnetic field intensity: 0.3 μT at 10 μT exposure (Fig 1B); 70 μT at 700 μT exposure (Fig 1C), 0.8 μT at 30 μT exposure (Fig 1D).
Fig 1.
The exposure regimes followed during the course of this investigation. Magnetic field exposure experiments were performed on human umbilical vein endothelial cells under the following conditions: (A) 700 μT, 24 hours; (B) 10 μT, 24 hours; (C) 700 μT, 60 minutes on, 30 minutes off, over 24 hours; (D) variable intensity, 10– 30 μT, changed every 30 minutes for 3 hours, left at the same intensity for 12 hours, then changed every 30 minutes for a further 9 hours
RNA isolation and microarray analysis
After MF or sham exposure, cells were collected using a plastic cell scraper and washed twice in ice-cold phosphate-buffered saline (pH 7.2) before total RNA was isolated using TriReagent according to the manufacturer's instructions (Sigma, Vienna, Austria). To ensure total removal of contaminants, RNA was further purified using RNeasy spin columns and the “RNA cleanup” protocol (Qiagen, Hilden, Germany).
Initial experiments with continuous 700 μT MF were analyzed using commercial Pan Human 10k Arrays (MWG-Biotech, Ebersberg, Germany) containing 9850 unique genes. All subsequent experiments were performed using arrays printed using the Human Oligo 30k Set (A+B+C; MWG-Biotech) on Nexterion epoxysilane– coated glass slides (PEQLAB, Erlangen, Germany) using a MicroGrid II spotting robot (BioRobotics, Cambridge, UK). These genome-wide arrays contained ca. 36 000 features representing 29 952 known and predicted genes.
Labeling and hybridization of the Pan Human 10k arrays was completed by MWG-Biotech. A single round of T7 RNA polymerase directed linear amplification with direct incorporation of Cy5 or Cy3 after cDNA synthesis was performed before overnight hybridization at 42°C. After washing, arrays were scanned using an Affymetrix 428 array scanner (Affymetrix, Santa Clara, CA) driven by Imagene and GeneSight software (BioDiscovery, El Segundo, CA).
Sample preparation for experiments using the Human 30k arrays was also performed with 1 round of linear amplification before postlabeling the RNA with Cy3 or Cy5. Total RNA (2 μg) was taken for cDNA synthesis using a MessageAmp aRNA Kit (Ambion, Austin, TX, USA), before in vitro transcription using T7 RNA polymerase according to the manufacturer's directions. After determination of the RNA concentration, up to 40 μg were fragmented in Affymetrix fragmentation buffer by heating at 94°C for 15 minutes and subsequently stored on ice. Samples were postlabeled with Cy3 and Cy5 dyes, respectively, (CyScribe Direct mRNA labeling kit; Amersham Pharmacia Biotech, Uppsala, Sweden) according to the manufacturer's instructions. For each of the control and exposed (MF) samples, 2 μg of the amplified RNA were labeled. Labeled RNA was ethanol precipitated before being resuspended in hybridization buffer (50% formamide, 5× SSC, 0.1% SDS). Hybridization of the labeled RNA to the microarray was carried out for 16 hours at 42°C in a moistened CMT-Hybridization chamber (Corning, Acton, MA, USA). Nonspecifically bound RNA was removed by washing for 5 minutes each with buffers 1–3 at 30°C, respectively (wash buffer 1: 2× SSC, 0.1% SDS; wash buffer 2: 1× SSC, 0.1% SDS; wash buffer 3: 0.5× SSC). Microarrays were scanned using a GenePix 4000B array scanner and GenePix Pro software (Axon, Union City, CA, USA). Initial analysis of the arrays was performed using GenePix Pro software before normalization using LimmaGUI software (Wettenhall and Smyth 2004). The normalization process used was a print-tip group Loess method based on median values with background correction. The LimmaGUI algorithm normalizes the features within each block on the array, based on the assumption that the global expression within each block is unchanged (Fig 2). Further data analysis was performed using the Excel program from Microsoft.
Fig 2.
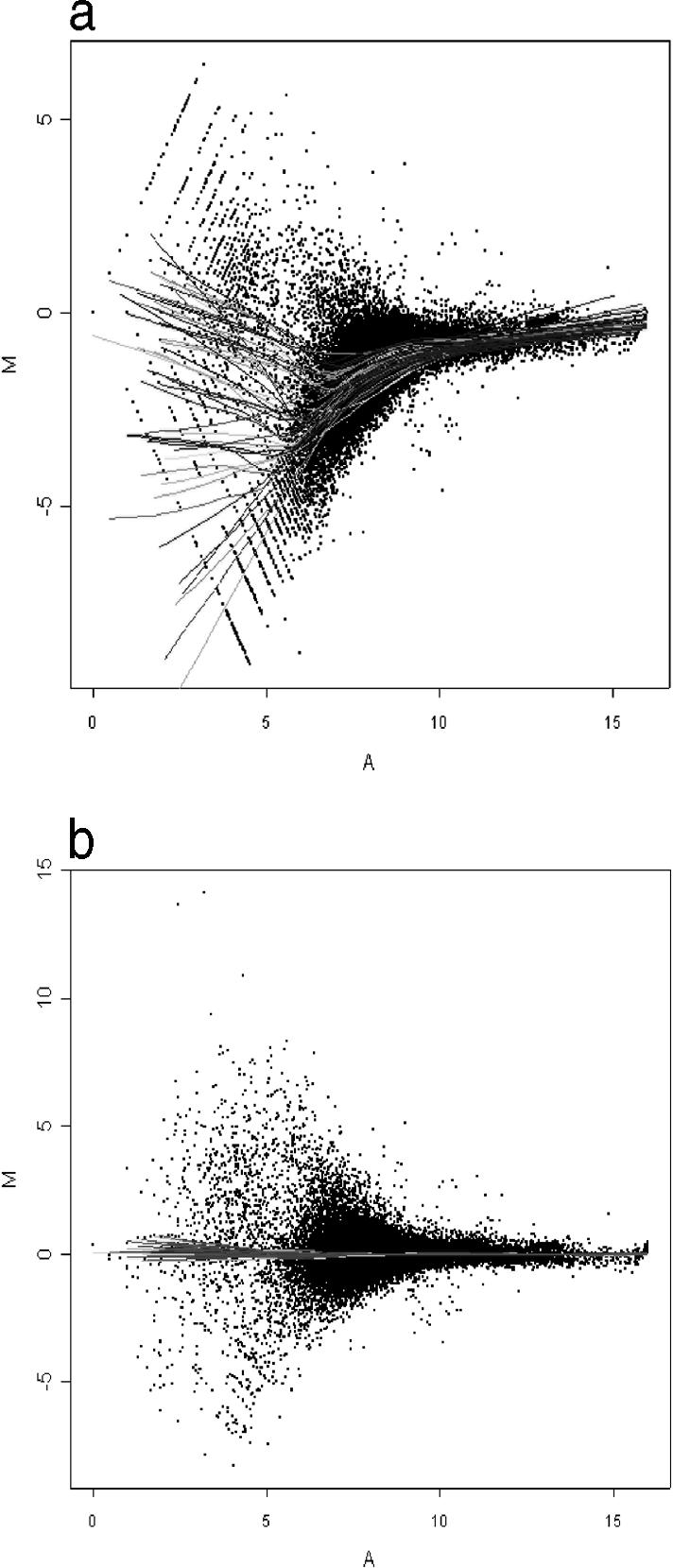
The effect of the normalization procedure on a human 30k microarray using LimmaGUI software. The ratio of Cy5/Cy3 values (log2; M, y-axis) of each feature is plotted against the sum of the intensities in both channels (log2; A, x-axis) before (A) and after (B) normalization. Each of the trend lines depicted represents a line of best fit for the features of 1 of the 24 blocks on the array. The LimmaGUI algorithm normalizes the features within each block, based on the assumption that the global expression change within each block is 0 (log2, equal to no change). Cells were exposed to variable-intensity (10–30 μT) 50-Hz MF for 24 hours and the transcriptome (labeled with Cy3) was compared with a sham-exposed control (Cy5)
To control for bias due to unequal dye incorporation or quantum efficiency, samples from the second experiment were labeled with the opposite dye to the first experiment. These “dye swap” experiments are not technical replicates but provide the opportunity to identify those genes that appear regulated in 1 dye channel only (Quackenbush 2002).
As a positive control for both the cells used and the microarray analysis process, HUVEC were exposed to an aqueous cigarette smoke extract (Bernhard et al 2004) before the transcriptional profile was interrogated. This led to the identification of a number of cigarette smoke extract–responsive genes, including a wide range of stress proteins many of which have been confirmed by real-time PCR analysis (Henderson et al, unpublished data).
RESULTS
Genome-wide microarray experiments were performed investigating a variety of exposure conditions in addition to the 700-μT high-intensity magnetic field experiments undertaken with the lower-resolution 10 000-gene MWG arrays. The experiments have each been performed for 24 hours at a low domestically relevant intensity (10 μT), at high intensity with a repeated intermittent on-off switching (700 μT; 60 minutes on and 30 minutes off), and at field intensities varying between 10 and 30 μT, respectively (Fig 1).
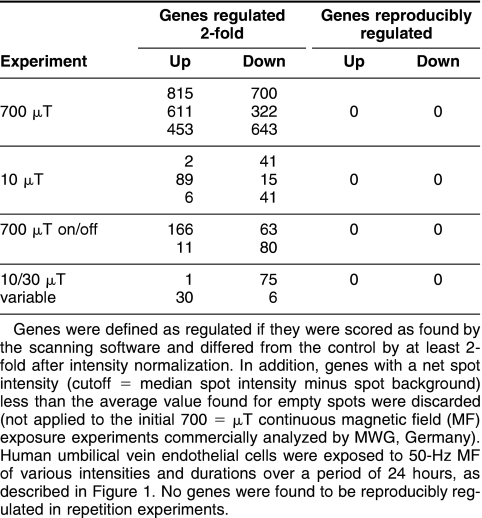
Initial experiments based on an occupational exposure of 700 μT were performed with a constant intensity over 24 hours (Fig 1A). Microarray analysis was completed investigating 10 000 genes, including most of the known and annotated genes. Three independent experiments were performed and the genes found regulated in each experiment compared (Table 1). There was no agreement in the genes regulated on each microarray, leading to the conclusion that the expression of none of the genes was reproducibly affected by exposure to MF in the system used.
Table 1.
Numbers of genes found expressed and regulated at least 2-fold in each array experiment
To better mimic the domestic situation encountered in everyday life, experiments were performed at 10 μT for a time period of 24 hours (Fig 1B). Generally, people are exposed to an average magnetic field intensity under 1 μT, with frequent spikes of intensity as people move nearer to functioning appliances. While 10 μT is an overestimate of the “average” situation, it is relevant to the fields that people are exposed to. In the first experiment performed at these conditions, 2 genes were found to be up-regulated and 41 down-regulated in response to MF exposure (Table 1). In 2 subsequent repetition experiments, in both cases with ‘dye-swap’ labeling, this result could not be confirmed. Thus, no genes were found to be consistently regulated by exposure to a 10-μT magnetic field compared to the sham-exposed control cells.
Some suggestion has been made that a change in field intensity, such as turning on and off an appliance or movement into an altered field, could cause a biological response to MF in tissues (Ivancsits et al 2002). To investigate this hypothesis further, HUVEC were exposed to a high-intensity 700-μT magnetic field, reflecting occupational exposure, which was on for 60 minutes before being switched off for 30 minutes, with the process repeated over 24 hours (Fig 1C). The timing was designed to mirror the situation where a worker is exposed to a high-intensity field for some time before moving away to perform other work or to take a break and then returning to the original task. Although this would not usually occur over a full 24-hour time period, this condition was kept for consistency to allow comparison between all the exposure regimes investigated and to more accurately reflect possible long-term-exposure effects. Comparison of the results of 2 microarray experiments (the second labeled in a dye-swap manner) showed that the expression of no gene was reproducibly influenced by MF exposure (Table 1).
The fourth set of experiments involved cells exposed to variable intensities of MF during a period of 24 hours. These experiments were again designed to mimic the domestic situation with constantly changing the field intensities while keeping the overall intensity to a domestically relevant level. The intensity was altered between 10 and 30 μT every 30 minutes for 3 hours and kept constant at 10 μT for 12 hours (to mirror the situation when people are sleeping and not moving into or out of magnetic fields) before resuming cycles of intensity shifting for a further 9 hours (Fig 1D). Once again, microarray analysis of RNA isolated from cells exposed to these conditions did not reveal any genes that were reproducibly regulated (Table 1).
DISCUSSION
The omnipresence of MF from domestic electricity sources in modern society makes investigation of possible deleterious health effects of exposure of vital importance. If 50 Hz MF can be shown to interact with biological systems leading to disease, as has been implied by a number of epidemiological studies, then measures such as improved shielding may be undertaken to prevent this.
In our previous investigations, we found no evidence of 50-Hz MF affecting the expression of heat shock protein 60 or 70 in HUVEC in vitro or in an in vivo mouse restenosis model (Henderson et al 2003a, 2003b). In addition, there were no persistent effects of MF exposure in mice on any of the other parameters investigated, including cell infiltration and neo-intima growth (Henderson et al 2003a).
In this study, HUVEC were exposed to 50-Hz MF encompassing domestic and occupational intensities. Exposure regimes included continuous exposure to a single intensity, which, although less relevant for the real-life situation, enables comparison with earlier published results. A noncontinuous field is better able to mimic the actual exposure that people receive, whether it is switched on and off, or whether the intensity is continually altered. The first scenario is more representative of a professional exposure where a machine is used for some time before being switched off and later switched on again. In domestic life, however, we are continuously exposed to MF, which varies in intensity as we move to or from different sources of MF, circumstances that may be imitated by altering the intensity over the course of the exposure. Regardless of the timing protocol or intensity used to expose the cells to MF, no genes were identified showing a reproducible effect after 24 hours. Genes that were found to be regulated at least 2-fold on each of the arrays were almost exclusively limited to those with a low intensity, which means that any small difference in expression or background is strongly amplified compared to features with a higher intensity. In all these cases, when the experiment was repeated using a dye-swap protocol, these genes were not found to be regulated in the same manner by exposure to MF, if at all.
The lack of change in gene expression profiles shown in these experiments is in good agreement with the only other microarray analysis of human tissue published (Luceri et al 2005). In that study, the effect of exposure of lymphocyte and yeast cells to continuous MF fields ranging from 1 to 100 μT for 18 hours were analyzed on microarrays containing only 14 000 and 6200 genes, respectively. The authors were also unable to identify any candidate genes that were significantly affected in either eukaryotic system. An earlier publication investigating the yeast transcriptional response to very intense MF (10–300 mT) also failed to find any genes affected under these conditions (Nakasono et al 2003). Interestingly, the authors also performed 2-dimensional protein gel analysis of yeast exposed to the same conditions, again failing to find an effect of MF exposure.
Although this study has been comprehensive with a variety of different exposure situations, each of these was investigated at only a single time point more representative of chronic exposure. Thus, it is possible that some genes are regulated at earlier or later time points. The recently published microarray analysis of human lymphocytes exposed to continuous MF of also examined only a single time point, in this case 18 hours. Endothelial cells were used for all experiments, consistent with our previous work looking at the stress response of endothelial cells to high-intensity MF. Again, it cannot be ruled out that other types of tissues may respond to MF exposure where endothelial cells in our system have failed to do so. In the past, publications have generally examined only a small number of genes looking for a response to MF exposure (Kang et al 1998; Bodega et al 2005; De Mattei et al 2005). The recent publication of the first microarray analysis on human tissue was an important first step in high-throughput analysis of the MF effects. This publication represents an important extension of this work to include a variety of continuous and noncontinuous exposure regimes and intensities while using a genome-wide platform of investigation to study whether MF is able to interact with biological systems in vitro.
The total lack of transcriptional response to various forms of 50-Hz MF exposure found in this study adds strongly to the evidence that this form of nonionizing radiation is incapable of interacting with living systems in a way that alters gene expression, making health effects unlikely. Thus, 50-Hz MF cannot be considered a risk factor for endothelial cell damage leading to incipient atherosclerosis.
Acknowledgments
The authors wish to thank Stefan Schmidt and Reinhard Kofler for invaluable advice on microarray analysis. This work was funded by the Austrian Association of Electricity Companies (VEÖ) and the European Union network of excellence European Vascular Genomics Network (EVGN, LSHM-CT-2003-503254).
REFERENCES
- Ahlbom IC, Cardis E, Green A, Linet M, Savitz D, Swerdlow A. Review of the epidemiologic literature on EMF and Health. Environ Health Perspect. 2001;109 Suppl 6:911–933. doi: 10.1289/ehp.109-1240626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhard D, Huck CW, Jakschitz T, Pfister G, Henderson B, Bonn GK, Wick G. Development and evaluation of an in vitro model for the analysis of cigarette smoke effects on cultured cells and tissues. J Pharmacol Toxicol Methods. 2004;50:45–51. doi: 10.1016/j.vascn.2004.01.003.1056-8719(2004)050[0045:DAEOAI]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Blank M, Goodman R. Electromagnetic fields may act directly on DNA. J Cell Biochem. 1999;75:369–374. doi: 10.1002/(sici)1097-4644(19991201)75:3<369::aid-jcb2>3.3.co;2-1.0730-2312(1999)075[0369:EFMADO]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Bodega G, Forcada I, Suarez I, Fernandez B. Acute and chronic effects of exposure to a 1-mT magnetic field on the cytoskeleton, stress proteins, and proliferation of astroglial cells in culture. Environ Res. 2005;98:355–362. doi: 10.1016/j.envres.2004.12.010.0013-9351(2005)098[0355:AACEOE]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- De Mattei M, Gagliano N, and Moscheni C. et al. 2005 Changes in polyamines, c-myc and c-fos gene expression in osteoblast-like cells exposed to pulsed electromagnetic fields. Bioelectromagnetics. 26:207–214. [DOI] [PubMed] [Google Scholar]
- Henderson B, Tagwerker A, and Mayrl C. et al. 2003a Progression of arteriovenous bypass restenosis in mice exposed to a 50 Hz magnetic field. Cell Stress Chaperones. 8:373–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson BR, Pfister G, Boeck G, Kind M, Wick G. Expression levels of heat shock protein 60 in human endothelial cells in vitro are unaffected by exposure to 50 Hz magnetic fields. Cell Stress Chaperones. 2003b;8:172–182. doi: 10.1379/1466-1268(2003)008<0172:elohsp>2.0.co;2.1466-1268(2003)008[0172:ELOHSP]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivancsits S, Diem E, Pilger A, Rudiger HW, Jahn O. Induction of DNA strand breaks by intermittent exposure to extremely-low-frequency electromagnetic fields in human diploid fibroblasts. Mutat Res. 2002;519:1–13. doi: 10.1016/s1383-5718(02)00109-2.0027-5107(2002)519[0001:IODSBB]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Juutilainen J, Huuskonen H, Komulainen H. Increased resorptions in CBA mice exposed to low-frequency magnetic fields: an attempt to replicate earlier observations. Bioelectromagnetics. 1997;18:410–417. doi: 10.1002/(sici)1521-186x(1997)18:6<410::aid-bem2>3.0.co;2-5.0197-8462(1997)018[0410:IRICME]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Kang KI, Bouhouche I, Fortin D, Baulieu EE, Catelli MG. Luciferase activity and synthesis of Hsp70 and Hsp90 are insensitive to 50Hz electromagnetic fields. Life Sci. 1998;63:489–497. doi: 10.1016/s0024-3205(98)00298-7.0024-3205(1998)063[0489:LAASOH]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Lai H, Singh NP. Magnetic-field-induced DNA strand breaks in brain cells of the rat. Environ Health Perspect. 2004;112:687–694. doi: 10.1289/ehp.6355.0091-6765(2004)112[0687:MDSBIB]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Head M, Blank M, Han L, Jin M, Goodman R. Myc-mediated transactivation of HSP70 expression following exposure to magnetic fields. J Cell Biochem. 1998;69:181–188. doi: 10.1002/(sici)1097-4644(19980501)69:2<181::aid-jcb8>3.0.co;2-o.0730-2312(1998)069[0181:MTOHEF]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Loscher W, Wahnschaffe U, Mevissen M, Lerchl A, Stamm A. Effects of weak alternating magnetic fields on nocturnal melatonin production and mammary carcinogenesis in rats. Oncology. 1994;51:288–295. doi: 10.1159/000227352.0030-2414(1994)051[0288:EOWAMF]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Luceri C, Filippo CD, and Giovannelli L. et al. 2005 Extremely low-frequency electromagnetic fields do not affect DNA damage and gene expression profiles of yeast and human lymphocytes. Radiat Res. 164:277–285. [DOI] [PubMed] [Google Scholar]
- Morehouse CA, Owen RD. Exposure to low-frequency electromagnetic fields does not alter HSP70 expression or HSF-HSE binding in HL60 cells. Radiat Res. 2000;153:658–662. doi: 10.1667/0033-7587(2000)153[0658:etlfef]2.0.co;2.0033-7587(2000)153[0658:ETLEFD]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Nakasono S, Laramee C, Saiki H, McLeod KJ. Effect of power-frequency magnetic fields on genome-scale gene expression in Saccharomyces cerevisiae. Radiat Res. 2003;160:25–37. doi: 10.1667/rr3006.0033-7587(2003)160[0025:EOPMFO]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Quackenbush J. Microarray data normalization and transformation. Nat Genet. 2002;32:496–501. doi: 10.1038/ng1032.1061-4036(2002)032[0496:MDNAT]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Savitz DA, Liao D, Sastre A, Kleckner RC, Kavet R. Magnetic field exposure and cardiovascular disease mortality among electric utility workers. Am J Epidemiol. 1999;149:135–142. doi: 10.1093/oxfordjournals.aje.a009779.0002-9262(1999)149[0135:MFEACD]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Simko M, Mattsson MO. Extremely low frequency electromagnetic fields as effectors of cellular responses in vitro: possible immune cell activation. J Cell Biochem. 2004;93:83–92. doi: 10.1002/jcb.20198.0730-2312(2004)093[0083:ELFEFA]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Sobel E, Dunn M, Davanipour Z, Qian Z, Chui HC. Elevated risk of Alzheimer's disease among workers with likely electromagnetic field exposure. Neurology. 1996;47:1477–1481. doi: 10.1212/wnl.47.6.1477.0028-3878(1996)047[1477:EROADA]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Wertheimer N, Leeper E. Electrical wiring configurations and childhood cancer. Am J Epidemiol. 1979;109:273–284. doi: 10.1093/oxfordjournals.aje.a112681.0002-9262(1979)109[0273:EWCACC]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Wettenhall JM, Smyth GK. limmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics. 2004;20:3705–3706. doi: 10.1093/bioinformatics/bth449.1367-4803(2004)020[3705:LAGUIF]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Wolf FI, Torsello A, and Tedesco B. et al. 2005 50-Hz extremely low frequency electromagnetic fields enhance cell proliferation and DNA damage: possible involvement of a redox mechanism. Biochim Biophys Acta. 1743:120–129. [DOI] [PubMed] [Google Scholar]
- Yokus B, Cakir DU, Akdag MZ, Sert C, Mete N. Oxidative DNA damage in rats exposed to extremely low frequency electromagnetic fields. Free Radic Res. 2005;39:317–323. doi: 10.1080/10715760500043603.1071-5762(2005)039[0317:ODDIRE]2.0.CO;2 [DOI] [PubMed] [Google Scholar]