Abstract
We have identified a gene at 11q23, telomeric to MLL, that encodes a guanine nucleotide exchange factor (GEF). This gene is transcribed into a 9.5-kb mRNA containing a 4.6-kb ORF. By Northern analysis, it was found to be expressed in all human tissues examined including peripheral blood leukocytes, spleen, prostate, testis, ovary, small intestine, colon, and minimally in thymus. Analysis of the predicted protein sequence indicates that it has strong homology to several members of the family of Rho GEFs that includes such oncogenes as Dbl, Vav, Tiam, and Bcr. A patient with primary acute myeloid leukemia (AML) and a karyotype of 51,XY,+8,+19,+3mar was found to have the 5′ end of MLL at exon 6 fused in-frame with the 3′ end of almost the entire ORF of this gene, which we named LARG for leukemia-associated Rho GEF. Transcriptional orientation of both genes at 11q23 is from centromere to telomere, consistent with other data that suggest the MLL-LARG fusion resulted from an interstitial deletion rather than a balanced translocation. LARG does not appear to have any homology with other MLL partner genes reported thus far. Thus, LARG represents an additional member of the GEF family and a novel MLL fusion partner in acute myeloid leukemia.
Keywords: LARG, Dbl protein, gene rearrangements
Leukemia is a heterogeneous disease at the molecular level resulting from a variety of alterations in numerous genes important for cell growth, differentiation, and cell death (1, 2). Identification and characterization of these genetic rearrangements has proved invaluable for appropriate diagnosis and prognosis, especially in acute leukemia. Rearrangements of the MLL gene (ALL1, HRX, and Hrtx) located at chromosome band 11q23 are commonly involved in acute leukemia. These rearrangements have been associated with 5–10% of adult and pediatric cases of primary acute leukemias (3–6) and also are found in the majority of patients with secondary leukemias after prior treatment with DNA topoisomerase II inhibitors (e.g., etoposide) (7, 8). MLL consists of at least 36 exons encoding an estimated 430-kDa protein that is thought to function as a positive regulator of gene expression in early embryonic development and hematopoiesis (9, 10). MLL translocation breakpoints cluster within an 8.3-kb region spanning exons 5–11 (11). At least 20 different partner genes with MLL have been cloned (12–23) from the more than 30 different chromosomal translocation partners identified to date (3, 24, 25). The mechanisms by which these rearrangements result in leukemia remain largely unknown.
In addition to chromosomal translocations, other mechanisms of MLL rearrangement have been demonstrated in patients with acute leukemia. The partial tandem duplication (PTD) of MLL is present in approximately 10% of patients with acute myeloid leukemia (AML) and normal cytogenetics and in the majority of patients with AML and trisomy 11 as the sole cytogenetic abnormality (26, 27). This rearrangement is characterized by an internal duplication of MLL spanning exons 2–6 or 2–8 (28). Cytogenetic deletions of 11q23 are also frequently found in acute leukemia. Although they are less likely than 11q23 translocations to involve MLL (29–33), some cases of cytogenetic deletions have been shown to represent cryptic translocations of MLL (34).
In the present study, we identify a novel gene at 11q23, which was named LARG for leukemia-associated Rho guanine nucleotide exchange factor (GEF). LARG has strong sequence homology to several members of the Rho family of GEFs. Further, LARG was found to be fused with MLL in a patient with primary AML. Cytogenetic and molecular evidence suggests that this fusion resulted from an interstitial deletion at 11q23.
Materials and Methods
Patient.
A 38-year-old male (patient 76) with a history of occupational exposure to herbicides was diagnosed with primary AML (FAB-M4). After receiving standard induction chemotherapy, he achieved complete remission by morphological bone marrow analysis and received a successful allogeneic bone marrow transplantation from an HLA identical sibling. He died 6 months later from interstitial pneumonia. There were no signs of relapse at autopsy.
Cytogenetic Analysis and Comparative Genomic Hybridization.
Cytogenetic analysis was performed by using standard techniques on diagnostic bone marrow (35). The criteria used to define a cytogenetic clone and the description of karyotypes followed the recommendations of the International System for Human Cytogenetic Nomenclature (36). Metaphase whole chromosome painting was performed by using standard techniques with a chromosome 11-specific probe (Oncor) (37, 38). Comparative genomic hybridization was performed according to Kallioniemi et al. (39) with modifications (40).
Southern Analysis.
Genomic DNA was isolated by a standard procedure from diagnostic bone marrow (41). Six micrograms of DNA was digested to completion with BamHI and electrophoresed on a 0.8% agarose gel. The gel was Southern-blotted and hybridized with B859, a cDNA probe spanning the 11q23 breakpoint cluster region containing exons 5–11 of the MLL gene (42). Southern analysis, probe radiolabeling, and hybridization were performed following standard procedures (43).
Genomic DNA Phage Library Screening.
Genomic DNA from the diagnostic patient sample was digested with BamHI and ligated to the Lambda DASH II phage vector following the manufacturer's protocol (Stratagene). The ligation mixture was packaged with Gigapack II Gold packaging extract (Stratagene), titered, and plated on XL1-Blue MRA (P2) cells (Stratagene). One million recombinant phage plaques were screened with the B859 probe following standard procedures (43). Positive clones were isolated, amplified, and subcloned into pBluescript II plasmid vector (Stratagene) for further analysis.
Restriction Mapping and Sequencing.
Multiple restriction enzymes were used to map the 17-kb fragment. The Human Cancer Genetics sequencing facility at The Ohio State University performed sequencing of DNA and cDNA clones by using an Applied Biosystems model 377 Stretch DNA sequencing system (Perkin–Elmer). All sequences were compared against the GenBank databases by using the Basic Local Alignment Search Tool (blast).
Reverse Transcription–PCR (RT-PCR).
Total cellular RNA was isolated from diagnostic bone marrow by using RNA STAT-60 following the manufacturer's protocol (Tel-Test, Friendswood, TX). RT was performed by using random hexamers and Superscript II reverse transcriptase (Life Technologies, Grand Island, NY). This cDNA was amplified on a model 9700 thermal cycler (Perkin–Elmer Applied Biosystems) with AmpliTaq DNA polymerase (Perkin–Elmer) by using an upstream primer designed from the B859 probe of MLL (5′-GGAAGTCAAGCAAGCAGGTC-3′) and a downstream primer (5′-CATACTTGCACTGTTGTCAT-3′) designed from a genomic fragment that shared 88% sequence identity with a murine expressed sequence tag (EST) (see Results). The product was analyzed by electrophoresis on an ethidium bromide-stained 1% agarose gel and subcloned by TA cloning (Invitrogen) for sequencing.
Northern Analysis.
Using restriction enzymes HaeIII and BstXI, an 862-bp region of unknown sequence was cut out of the RT-PCR-amplified MLL-LARG fusion. This fragment, named HB862, did not contain any MLL sequence and was used as a probe for analysis of LARG expression in a human multiple tissue Northern blot (CLONTECH) following the manufacturer's instructions.
cDNA Cloning.
A human normal prostate 5′ STRETCH cDNA library (CLONTECH) was screened according to the manufacturer's recommendations. Positive clones were isolated, cloned into pBluescript II SK+, and sequenced. From the first-round consensus sequence, two probes were designed from the 5′ and 3′ ends and amplified by PCR. Screening of the library was performed a second time with the 5′ and 3′ probes. Based on this consensus sequence, rapid amplification of the 5′ and 3′ cDNA ends was performed on normal human jejunum by using a commercial kit (Boehringer Mannheim). The sequence obtained from the positive clones was assembled and compared with EST sequences in GenBank and The Institute for Genomic Research (TIGR) databases by using blast. Sequences found in this search then were added to the consensus by using dnastar sequence analysis software (DNAstar, Madison, Wisconsin). Protein motif analysis was performed by using the expasy prosite database (44).
Fluorescence in Situ Hybridization (FISH).
A 15-kb FISH probe was cut out of the 17-kb subcloned fragment with restriction enzymes and contained only the new gene. The probe was labeled with the BioNickô DNA labeling system (Life Technologies) by using biotin-14-dATP and hybridized to metaphase chromosome preparations obtained from peripheral blood lymphocytes from a normal male according to the manufacturer's instructions (Oncor). The probe was precipitated in the presence of human placental and herring testis DNAs by using a BlocKit (Oncor) before denaturation and overnight hybridization with target metaphases. Probe signals were detected by using FITC-conjugated avidin followed by a single amplification step. The chromosomes were counterstained with propidium iodide and antifade. Slides were examined on a Zeiss Axioskop epifluorescent microscope equipped with dual band pass filters and a filter wheel, and images were obtained by using macprobe software and the PowerGene FISH System (Perceptive Scientific Systems, League City, TX).
Radiation Hybrid Mapping.
Primers from the 5′ end of the 17-kb genomic band were used to amplify a 500-bp product from a Genebridge4 (GB4) radiation hybrid panel (Research Genetics, Huntsville, AL). The PCR amplification was performed by using Taq polymerase (Boeringer Mannheim) in a Perkin–Elmer Cetus 9600 thermal cycler, and the products were visualized by ethidium bromide staining after electrophoresis in a 1.5% agarose gel. The scoring data were submitted to the Whitehead Institute/Massachusetts Institute of Technology Center for Genome Research mapping server (http://www-genome.wi.mit.edu/cgi-bin/contig/rhmapper.pl).
Chromosomal mapping of sequence-tagged site (STS) markers was performed to confirm the FISH data by using the GB4 radiation hybrid map displayed by The National Center of Biotechnology Information (www.ncbi.nlm.nih.gov/genemap) (45). Additional fine mapping was carried out with KIAA0382, a partial sequence containing the 3′ end of the gene (46).
Results
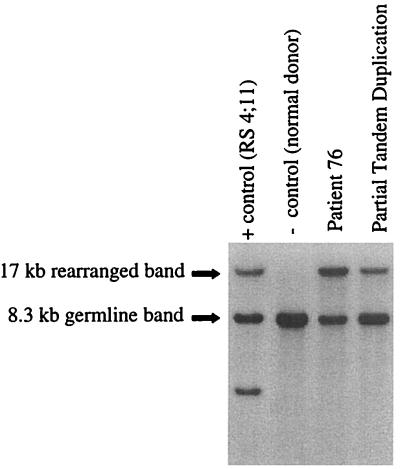
Cytogenetic analysis performed on the case of AML revealed a karyotype of 51,XY,+8,+19,+3 mar1[19]/46,XY[1]. A chromosome 11-specific painting probe hybridized to all three marker chromosomes. Comparative genomic hybridization studies indicated an amplification of the 11q22qter. The MLL gene at 11q23 was rearranged on Southern analysis with a single restriction fragment and with the appearance of amplification (Fig. 1). This single restriction band on Southern analysis can be found with the PTD of MLL (28) or with chromosomal translocations where the reciprocal fusion is lost (47). No PTD could be confirmed on further analysis.
Figure 1.
Southern analysis of an AML cell line, a normal donor, and two patient bone marrow samples. This blot was made from BamHI-digested DNA and hybridized with the B859 probe. A germ-line MLL 8.3-kb band is seen in each sample. In the positive control RS4;11 cell line, two rearranged bands are seen in addition to the germ-line band. Each band represents one portion of the translocation. The second lane is a normal donor negative control. In the last two lanes, both patient 76 and the PTD show a single 17-kb rearranged band. However, the 17-kb band in the patient 76 lane shows greater intensity relative to the germ-line band unlike the rearrangement seen with the PTD.
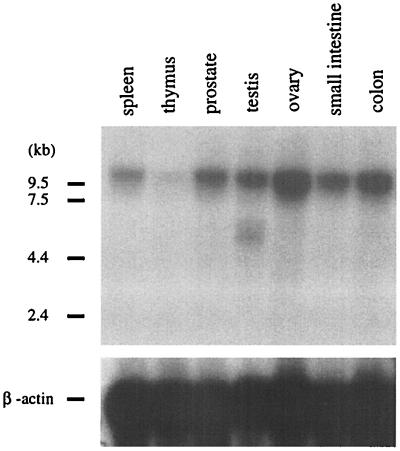
A lambda phage library was constructed with genomic DNA from the case of AML and screened with the B859 probe from MLL. A 17-kb fragment was identified and subcloned. Restriction mapping and partial sequencing of this band revealed a 121-bp segment that shared 88% sequence identity with a murine EST (GenBank accession no. C86803), consistent with exonic sequence. RT-PCR primers were constructed from this sequence and the B859 probe, and RT-PCR was performed by using cDNA from the patient's bone marrow sample. The cDNA amplicon contained 924 bp that had no sequence homology to any known genes in existing databases. A portion of this cDNA was cut into a smaller 862-bp fragment and used as a probe (HB862) to screen a human multiple tissue Northern blot (Fig. 2). A 10-kb transcript was detected in spleen, prostate, testis, ovary, small intestine, and colon, and minimally in the thymus. Peripheral blood leukocytes failed to show expression on the commercial blot; however, subsequent Northern analysis of other normal human leukocytes from normal healthy donors showed expression (data not shown).
Figure 2.
Multiple tissue Northern analysis. This human tissue Northern blot was hybridized with the HB862 cDNA probe. (Upper) A major 10-kb transcript was expressed in the spleen, prostate, testis, ovary, small intestine, colon, and minimally in the thymus. A similar transcript was detected from human leukocytes from normal healthy donors (data not shown). A smaller, approximately 6-kb transcript, seen in the testis RNA, most likely represents a splice variant. (Lower) A β-actin control probe is shown.
A normal prostate cDNA library was screened with the HB862 probe to identify the full-length cDNA sequence of the new gene. Two rounds of library screening yielded a total of 5.1 kb of sequence, which was compared with known sequences in public databases and found to have 2.5 kb of overlap with a 6.2-kb partial cDNA sequence KIAA0382 (GenBank accession no. AB002380) (46). Additional ESTs also were added to the contiguous sequence. 3′ Rapid amplification of cDNA ends (RACE) performed on normal human jejunum RNA yielded a product of 497 bp that confirmed the 3′ end of the gene. 5′ RACE also performed on jejunum RNA gave 60 bp of additional sequence on the 5′ end of the gene and completed the ORF.
The overlapping subclones, ESTs, and PCR products were assembled into one contiguous sequence, resulting in a total cDNA size of 9,501 nt. This sequence was submitted to GenBank (accession no. AF180681). Computer analysis indicated a 4,635-nt ORF encoding a 1,544-aa protein with an estimated molecular mass of 173.2 kDa. From prostate cDNA library-derived subclones, an alternatively spliced exon of 57 bp was observed (nucleotides 150–206) that was not observed in 5′ rapid amplification of cDNA ends products from jejunum or testis tissue.
Motif analysis using the expasy prosite database indicated at least four significant regions of homology to known functional domains. Most interesting were identification of a Dbl homology (DH) and a pleckstrin homology (PH) domain, which function in tandem in proteins in the Dbl family of GEFs (48). The other two domains identified were a PDZ domain and a bipartite nuclear localization signal. Additionally, an independent comparison with the PDZ-Rho GEF protein also identified the presence of a Lsc homology (LH) domain, a motif that was named for sequence identity to a region from the Lsc gene (49). Amino acid sequences for these functional domains were compared with several GEF family members specific for the Rho family of GTPases by using the DNAstar megaalign analysis program. This analysis revealed a strong degree of homology for each domain, suggesting that this novel gene represents an additional member of the Rho GEF family (Fig. 3).
Figure 3.
Comparison of LARG amino acid sequence to various other proteins. Numbers refer to amino acid position of each protein. White letters in black boxes represent amino acid identity. (A) Predicted amino acid sequence of LARG relative to other PDZ-containing proteins. (B) Predicted LH domain of LARG compared with several Rho GEFs and Lsc. (C) Predicted DH domain compared with several Rho GEFs and Dbl. (D) Predicted PH domain of LARG compared with several Rho GEFs and pleckstrin.
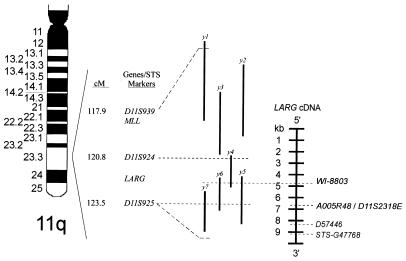
FISH on metaphase preparations from normal peripheral leukocytes was performed with the 15-kb probe from the rearranged genomic fragment. Eight metaphases were analyzed and each demonstrated a single signal on each chromosome 11 in band q23. No signals were present on any other chromosome. Physical mapping with the GB4 radiation hybrid panel placed the unknown gene 0.00 cR from marker AFMB048ZA9 and 4.71 cR from marker WI-7302 in 11q23.3. Additional fine mapping of the gene was determined with KIAA0382. Five STS markers were found on the GB4 radiation hybrid map in 11q23.3: WI-8803, D57446, and stSG47768 constitute different cDNA fragments of the gene whereas D11S2318E and A005R48 overlap most of the same cDNA sequence. MLL is located in 11q23.3 between D11S939 (117.9 cM) and D11S924 (120.8 cM). Our STS marker data place the unknown gene in a 5′ to 3′ orientation into the adjacent, telomeric interval to MLL between D11S924 (120.8 cM) and D11S925 (123.5 cM) (Fig. 4).
Figure 4.
Schema of chromosome 11q shows the telomeric relation of LARG to MLL as determined by radiation hybrid mapping. D11S939, D11S924, and D11S925 anchor markers are shown from the GB4 radiation hybridization map and are depicted immediately upstream and downstream to both genes. WI-8803, A005R48, D11S2318E, D57446, and STS-G47768 STSs were identified from searching the GB4 map with the KIAA0382 partial cDNA sequence. These STSs are shown relative to their position in the cDNA sequence of LARG and support its having a 5′ to 3′, centromeric to telomeric orientation. A yeast artificial chromosome (YAC) map determined from WI-8803 and the Whitehead Institute/Massachusetts Institute of Technology database further supports the orientation of LARG. YAC symbols: y1 = 797-E7, y2 = 822-G-8, y3 = 785-C-6, y4 = 828-G-11, y5 = 969-D-7, y6 = 901-A-11, y7 = 936-D-9.
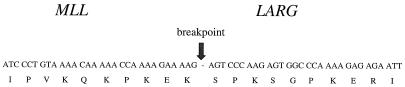
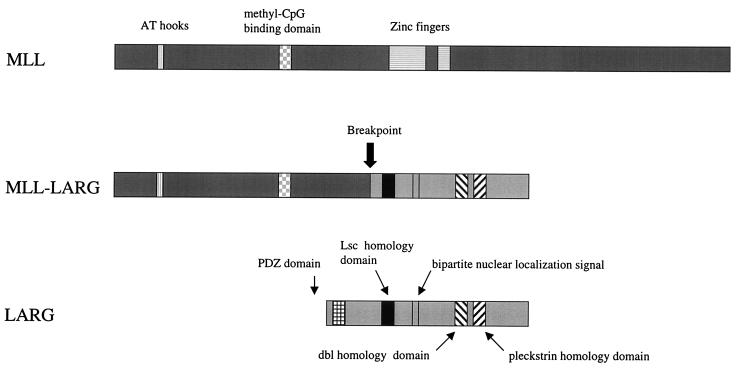
The original genomic hybridization to B859 produced a 17-kb band that included the new gene and MLL. Both genes mapped to 11q23.3. To determine how the new gene was involved with MLL in a case of AML, RT-PCR was performed with a 5′ B859 primer and 3′ primer from the new gene. Sequence analysis of the 1,136-bp product revealed a fusion of exon 6 of MLL and the new gene that maintained the ORF of both genes (Fig. 5). The breakpoint in the new gene was at its 5′ end after nucleotide 931 with 80% of the ORF contained in the in-frame fusion. We therefore named this gene LARG, for leukemia-associated Rho GEF. A schema illustrating the protein domains of both the wild-type and chimeric gene products is shown in Fig. 6.
Figure 5.
Amino acid and cDNA sequence of MLL and LARG at the breakpoint determined by RT-PCR show an in-frame fusion.
Figure 6.
Schematic representation of MLL, MLL-LARG, and LARG proteins. In LARG, the PDZ domain, LH domain, bipartite nuclear localization signal, DH domain, and PH domain are illustrated (Bottom). The predicted translocation product, MLL-LARG, is shown containing the AT hooks and methyl-CpG binding domain of MLL and the bipartite nuclear localization signal, LH, DH domain, and PH domain of LARG.
Discussion
In this report, we describe a gene named LARG whose predicted protein is an additional member of the Dbl family of proteins. Like all Dbl family members, LARG contains a DH domain in tandem with a PH domain. The Dbl family of proteins is generally structurally dissimilar except for the presence of these tandem DH/PH domains (50). Members of the Dbl protein family function as GEFs, most often for the Rho family of GTPases (51). Rho GEFs promote the exchange of GDP for GTP and thereby activate members of the Rho family, which include Rho, Rac, and cdc42 (52). Classically, Rho GTPases are known to regulate the formation of actin cytoskeletal structures; however, it is becoming increasingly evident that they also are involved in transcriptional regulation, membrane trafficking, and control of cell growth (51, 52).
Protein domains similar to LARG's predicted domains have been studied in depth in other Rho GEF proteins. The DH domain has been shown to be responsible for the nucleotide exchange activity of GEFs toward Rho GTPases (53). Positioned adjacent to the DH domain, the PH domain functions in membrane localization through its interaction with membrane lipids and proteins (54). PDZ domains have been identified in well over 75 different proteins and have been implicated in protein–protein interactions often involving transmembrane signaling pathways (55). Nuclear localization signals are responsible for the targeting and translocation of proteins from the cytoplasm to the nucleus through nuclear pores (56, 57). Lastly, the LH domain of PDZ-RhoGEF has been shown to bind activated α subunits of heterotrimeric G proteins of the Gα12 family (49). The corresponding amino-terminal domain of p115 RhoGEF has been shown to mediate direct stimulation of GEF activity by Gα13 (58).
Beyond their capacity as regulators of GTPases, many Rho GEFs are known for their role as oncogenes, for example, Vav, Dbl, Tiam, Lbc, Lsc, and Lfc (59). The oncogenic potential of the Rho GEFs is noteworthy as many initially were identified because of their ability to transform NIH 3T3 cells into a malignant phenotype (52). This oncogenic potential is thought to be mediated by the DH domain through altered expression and activation of Rho GTPases. One Rho GEF, Bcr, has been implicated in leukemia through a recurrent chromosomal translocation. Bcr is fused with Abl in t(9;22) and this gene fusion is termed the Philadelphia chromosome as seen in chronic myeloid leukemia (CML), ALL, and rarely, AML (60). Interestingly, the 5′ portion of Bcr is retained in the Philadelphia chromosome fusion product whereas, in the MLL-LARG fusion, the 3′ portion of the LARG was retained. However, like the MLL-LARG fusion, the DH and PH domains of Bcr are retained in the p210 Bcr-Abl fusion protein of CML, but not in the smaller p190 fusion protein of ALL (50).
Through radiation hybrid mapping and FISH studies, LARG was mapped to 11q23, telomeric to MLL. Like MLL, LARG is oriented in a 5′ to 3′ direction. In this case of AML studied, the mapping information taken together with routine cytogenetics provide data that are most consistent with the interpretation of an interstitial deletion involving MLL and LARG in 11q23. This deleted segment includes at its 5′ end, the 3′ portion of MLL, and at its 3′ end, the 5′ portion of LARG. LARG appears to be the first gene that is fused with MLL as the result of an interstitial deletion at 11q23 although the evidence accumulated thus far does not exclude the possibility of another mechanism. One possibility could be a translocation involving the homologous chromosome 11 with the subsequent deletion of the reciprocal LARG-MLL fusion. Another possible mechanism might involve the phenomenon termed segmental jumping translocation (SJT). SJT is the amplification and translocation of a chromosomal segment to structurally abnormal chromosomes and notably has been observed with an 11q segment containing MLL in secondary AML (61).
Importantly, the deletion-induced MLL-LARG fusion at 11q23 appears to explain the single 17-kb rearranged restriction fragment observed by Southern blotting, which is seen with the PTD of MLL, but not usually with balanced translocations involving MLL. On two separate restriction enzyme digests (HindIII not shown), the rearranged fragment appeared amplified, consistent with the fact that comparative genomic hybridization studies indicated an amplification of 11q22ter, and all three marker chromosomes were hybridized by the chromosome 11-specific painting probe. However, exact quantification of the amplified copy number was not possible, likely because of variation in efficiency of transfer of BamHI- and HindIII-digested DNA fragments for Southern blotting.
The majority of MLL fusion proteins lack consistent sequence homology among each other. Likewise, LARG does not appear to have any homology to other known MLL fusion partners, although like many partner genes, it does encode a nuclear localization signal that is retained in the fusion with MLL. Without exception, gene fusions involving MLL are expressed from the MLL promoter, and in cases of 11q23 translocation, the reciprocal fusion is usually deleted, out of frame, or simply not expressed (62). This finding suggests that the presence of the 5′ end of MLL within the fusion is important for leukemogenesis. The MLL-LARG rearrangement is expressed as an in-frame fusion from the MLL promoter and because this rearrangement likely occurred as the result of an interstitial deletion, a reciprocal fusion does not exist. Therefore, the MLL-LARG fusion protein may be an important contributor to leukemogenesis. The fact that 80% of LARG's ORF is retained as part of the fusion would support this contribution. However, we cannot rule out that an interstitial deletion at 11q23 resulted in the removal of an important tumor suppressor gene.
As new partner genes with MLL are discovered, more clues are presented for MLL's role in leukemogenesis; however, few conclusions can yet be made. One of the central questions regarding MLL's contribution to leukemogenesis is whether its derivative fusion protein acts in a gain-of-function or in a dominant-negative role. In terms of the MLL-LARG fusion, we cannot rule out that a truncation and/or a conformational change in LARG contributed to leukemogenesis in light of the oncogenic potential of other Dbl family members. Amino-terminal truncation of one closely related Dbl family protein causes transformation of NIH 3T3 cells in expression studies (63), and the elimination of the LH domain specifically was thought to contribute to this transformation in another (49). In the predicted MLL-LARG fusion protein, the extreme amino-terminal end of LARG was truncated; however, the LH domain remained undisturbed.
Cytogenetic deletions of 11q23 are commonly observed in primary AML (64). These breaks often are described as either terminal or interstitial deletions. However, terminal deletions most likely do not exist and rather represent interstitial deletions with breaks close to the telomere of 11q. MLL is not always deleted or rearranged in these cases, suggesting that there are other genes on the long arm of chromosome 11 that are involved in leukemia. LARG rearrangements could provide an additional explanation for these other 11q abnormalities.
Acknowledgments
We thank Dr. Peter Aplan and Dr. Krzysztof Mrózek for their critical review of the manuscript. We thank Lucia Culley for excellent technical assistance in FISH analysis. This work was supported in part by National Cancer Institute Grants P30 CA16058 and CA-09338, the National Research Service Award, Division of Cancer Prevention and Control, Bethesda, MD, and the Coleman Leukemia Research Fund.
Abbreviations
- AML
acute myeloid leukemia
- STS
sequence-tagged site
- GEF
guanine nucleotide exchange factor
- PTD
partial tandem duplication
- RT-PCR
reverse transcription–PCR
- EST
expressed sequence tag
- FISH
fluorescence in situ hybridization
- GB4
Genebridge4
- DH
Dbl homology
- PH
pleckstrin homology
- LH
Lsc homology
- LARG
leukemia-associated Rho GEF
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF180681).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.040569197.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.040569197
References
- 1.Look A T. Science. 1997;278:1059–1064. doi: 10.1126/science.278.5340.1059. [DOI] [PubMed] [Google Scholar]
- 2.Rabbitts T H. Nature (London) 1994;372:143–149. doi: 10.1038/372143a0. [DOI] [PubMed] [Google Scholar]
- 3.Cimino G, Moir D T, Canaani O, Williams K, Crist W M, Katzav S, Cannizzaro L, Lange B, Nowell P C, Croce C M, Canaani E. Cancer Res. 1991;51:6712–6714. [PubMed] [Google Scholar]
- 4.Djabali M, Selleri L, Parry P, Bower M, Young B D, Evans G A. Nat Genet. 1992;2:113–118. doi: 10.1038/ng1092-113. [DOI] [PubMed] [Google Scholar]
- 5.Tkachuk D C, Kohler S, Cleary M L. Cell. 1992;71:691–700. doi: 10.1016/0092-8674(92)90602-9. [DOI] [PubMed] [Google Scholar]
- 6.Ziemin-van der Poel S, McCabe N R, Gill H J, Espinosa R, III, Patel Y, Harden A, Rubinelli P, Smith S D, LeBeau M M, Rowley J D, Diaz M O. Proc Natl Acad Sci USA. 1991;88:10735–10739. doi: 10.1073/pnas.88.23.10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pui C H, Relling M V, Rivera G K, Hancock M L, Raimondi S C, Heslop H E, Santana V M, Ribeiro R C, Sandlund J T, Mahmoud H H, et al. Leukemia. 1995;9:1990–1996. [PubMed] [Google Scholar]
- 8.Smith M A, McCaffrey R P, Karp J E. J Natl Cancer Inst. 1996;88:407–408. doi: 10.1093/jnci/88.7.407. [DOI] [PubMed] [Google Scholar]
- 9.Nilson I, Lochner K, Siegler G, Greil J, Beck J D, Fey G H, Marschalek R. Br J Haematol. 1996;93:966–972. doi: 10.1046/j.1365-2141.1996.d01-1748.x. [DOI] [PubMed] [Google Scholar]
- 10.Rasio D, Schichman S A, Negrini M, Canaani E, Croce C M. Cancer Res. 1996;56:1766–1769. [PubMed] [Google Scholar]
- 11.Gu Y, Alder H, Nakamura T, Schichman S A, Prasad R, Canaani O, Saito H, Croce C M, Canaani E. Cancer Res. 1994;54:2326–2330. [PubMed] [Google Scholar]
- 12.Ida K, Kitabayashi I, Taki T, Taniwaki M, Noro K, Yamamoto M, Ohki M, Hayashi Y. Blood. 1997;90:4699–4704. [PubMed] [Google Scholar]
- 13.Rubnitz J E, Behm F G, Downing J R. Leukemia. 1996;10:74–82. [PubMed] [Google Scholar]
- 14.Taki T, Sako M, Tsuchida M, Hayashi Y. Blood. 1997;89:3945–3950. [PubMed] [Google Scholar]
- 15.Taki T, Shibuya N, Taniwaki M, Hanada R, Morishita K, Bessho F, Yanagisawa M, Hayashi Y. Blood. 1998;92:1125–1130. [PubMed] [Google Scholar]
- 16.Thirman M J, Levitan D A, Kobayashi H, Simon M C, Rowley J D. Proc Natl Acad Sci USA. 1994;91:12110–12114. doi: 10.1073/pnas.91.25.12110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.So C W, Caldas C, Liu M M, Chen S J, Huang Q H, Gu L J, Sham M H, Wiedemann L M, Chan L C. Proc Natl Acad Sci USA. 1997;94:2563–2568. doi: 10.1073/pnas.94.6.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sobulo O M, Borrow J, Tomek R, Reshmi S, Harden A, Schlegelberger B, Housman D, Doggett N A, Rowley J D, Zeleznik-Le N J. Proc Natl Acad Sci USA. 1997;94:8732–8737. doi: 10.1073/pnas.94.16.8732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Osaka M, Rowley J D, Zeleznik-Le N J. Proc Natl Acad Sci USA. 1999;96:6428–6433. doi: 10.1073/pnas.96.11.6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Megonigal M D, Rappaport E F, Jones D H, Williams T M, Lovett B D, Kelly K M, Lerou P H, Moulton T, Budarf M L, Felix C A. Proc Natl Acad Sci USA. 1998;95:6413–6418. doi: 10.1073/pnas.95.11.6413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hillion J, Le Coniat M, Jonveaux P, Berger R, Bernard O A. Blood. 1997;90:3714–3719. [PubMed] [Google Scholar]
- 22.Taki T, Kano H, Taniwaki M, Sako M, Yanagisawa M, Hayashi Y. Proc Natl Acad Sci USA. 1999;96:14535–14540. doi: 10.1073/pnas.96.25.14535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taki T, Ohnishi H, Shinohara K, Sako M, Bessho F, Yanagisawa M, Hayashi Y. Cancer Res. 1999;59:4261–4265. [PubMed] [Google Scholar]
- 24.Thirman M J, Gill H J, Burnett R C, Mbangkollo D, McCabe N R, Kobayashi H, Ziemin-van der Poel S, Kaneko Y, Morgan R, Sandberg A A, et al. N Engl J Med. 1993;329:909–914. doi: 10.1056/NEJM199309233291302. [DOI] [PubMed] [Google Scholar]
- 25.Secker-Walker L M. Leukemia. 1998;12:776–778. doi: 10.1038/sj.leu.2401011. [DOI] [PubMed] [Google Scholar]
- 26.Caligiuri M A, Strout M P, Schichman S A, Mrózek K, Arthur D C, Herzig G P, Baer M R, Schiffer C A, Heinonen K, Knuutila S, et al. Cancer Res. 1996;56:1418–1425. [PubMed] [Google Scholar]
- 27.Caligiuri M A, Strout M P, Lawrence D, Arthur D C, Baer M R, Yu F, Knuutila S, Mrózek K, Oberkircher A R, Marcucci G, et al. Cancer Res. 1998;58:55–59. [PubMed] [Google Scholar]
- 28.Schichman S A, Caligiuri M A, Gu Y, Strout M P, Canaani E, Bloomfield C D, Croce C M. Proc Natl Acad Sci USA. 1994;91:6236–6239. doi: 10.1073/pnas.91.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kobayashi H, Espinosa R, 3rd, Fernald A A, Begy C, Diaz M O, Le Beau M M, Rowley J D. Genes Chromosomes Cancer. 1993;8:246–252. doi: 10.1002/gcc.2870080407. [DOI] [PubMed] [Google Scholar]
- 30.Harbott J, Mancini M, Verellen-Dumoulin C, Moorman A V, Secker-Walker L M. Leukemia. 1998;12:823–827. doi: 10.1038/sj.leu.2401018. [DOI] [PubMed] [Google Scholar]
- 31.Raimondi S C, Frestedt J L, Pui C H, Downing J R, Head D R, Kersey J H, Behm F G. Blood. 1995;86:1881–1886. [PubMed] [Google Scholar]
- 32.Cimino G, Nakamura T, Gu Y, Canaani O, Prasad R, Crist W M, Carroll A J, Baer M, Bloomfield C D, Nowell P C, et al. Cancer Res. 1992;52:3811–3813. [PubMed] [Google Scholar]
- 33.Cherif D, Bernard O, Paulien S, James M R, Le Paslier D, Berger R. Leukemia. 1994;8:578–586. [PubMed] [Google Scholar]
- 34.Strout M P, Mrozek K, Heinonen K, Sait S N, Shows T B, Aplan P D. Genes Chromosomes Cancer. 1996;16:204–210. doi: 10.1002/(SICI)1098-2264(199607)16:3<204::AID-GCC8>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 35.Caligiuri M A, Schichman S A, Strout M P, Mrózek K, Baer M R, Frankel S R, Barcos M, Herzig G P, Croce C M, Bloomfield C D. Cancer Res. 1994;54:370–373. [PubMed] [Google Scholar]
- 36.Mitelman F. ISCN: An International System for Human Cytogenetic Nomenclature. Basel: Karger; 1995. [Google Scholar]
- 37.Lichter P, Cremer T, Borden J, Manuelidis L, Ward D C. Hum Genet. 1988;80:224–234. doi: 10.1007/BF01790090. [DOI] [PubMed] [Google Scholar]
- 38.Pinkel D, Straume T, Gray J W. Proc Natl Acad Sci USA. 1986;83:2934–2938. doi: 10.1073/pnas.83.9.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kallioniemi A, Kallioniemi O P, Sudar D, Rutovitz D, Gray J W, Waldman F, Pinkel D. Science. 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- 40.Monni O, Joensuu H, Franssila K, Knuutila S. Blood. 1996;87:5269–5278. [PubMed] [Google Scholar]
- 41.Gustincich S, Manfioletti G, Del Sal G, Schneider C. BioTechniques. 1991;11:298–301. [PubMed] [Google Scholar]
- 42.Gu Y, Nakamura T, Alder H, Prasad R, Canaani O, Cimino G, Croce C M, Canaani E. Cell. 1992;71:701–708. doi: 10.1016/0092-8674(92)90603-a. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 44.Hofmann K, Bucher P, Falquet L, Bairoch A. Nucleic Acids Res. 1999;27:215–219. doi: 10.1093/nar/27.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deloukas P, Schuler G D, Gyapay G, Beasley E M, Soderlund C, Rodriguez-Tome P, Hui L, Matise T C, McKusick K B, Beckmann J S, et al. Science. 1998;282:744–746. doi: 10.1126/science.282.5389.744. [DOI] [PubMed] [Google Scholar]
- 46.Nagase T, Ishikawa K, Nakajima D, Ohira M, Seki N, Miyajima N, Tanaka A, Kotani H, Nomura N, Ohara O. DNA Res. 1997;4:141–150. doi: 10.1093/dnares/4.2.141. [DOI] [PubMed] [Google Scholar]
- 47.Super H G, Strissel P L, Sobulo O M, Burian D, Reshmi S C, Roe B, Zeleznik-Le N J, Diaz M O, Rowley J D. Genes Chromosomes Cancer. 1997;20:185–195. [PubMed] [Google Scholar]
- 48.Seasholtz T M, Majumdar M, Brown J H. Mol Pharmacol. 1999;55:949–956. doi: 10.1124/mol.55.6.949. [DOI] [PubMed] [Google Scholar]
- 49.Fukuhara S, Murga C, Zohar M, Igishi T, Gutkind J S. J Biol Chem. 1999;274:5868–5879. doi: 10.1074/jbc.274.9.5868. [DOI] [PubMed] [Google Scholar]
- 50.Whitehead I P, Campbell S, Rossman K L, Der C J. Biochim Biophys Acta. 1997;1332:F1–F23. doi: 10.1016/s0304-419x(96)00040-6. [DOI] [PubMed] [Google Scholar]
- 51.Olson M F. J Mol Med. 1996;74:563–571. doi: 10.1007/s001090050060. [DOI] [PubMed] [Google Scholar]
- 52.Hall A. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 53.Hart M J, Eva A, Zangrilli D, Aaronson S A, Evans T, Cerione R A, Zheng Y. J Biol Chem. 1994;269:62–65. [PubMed] [Google Scholar]
- 54.Shaw G. BioEssays. 1996;18:35–46. doi: 10.1002/bies.950180109. [DOI] [PubMed] [Google Scholar]
- 55.Fanning A S, Anderson J M. Curr Opin Cell Biol. 1999;11:432–439. doi: 10.1016/S0955-0674(99)80062-3. [DOI] [PubMed] [Google Scholar]
- 56.Gorlich D. Curr Opin Cell Biol. 1997;9:412–419. doi: 10.1016/s0955-0674(97)80015-4. [DOI] [PubMed] [Google Scholar]
- 57.Yoneda Y. J Biochem (Tokyo) 1997;121:811–817. doi: 10.1093/oxfordjournals.jbchem.a021657. [DOI] [PubMed] [Google Scholar]
- 58.Hart M J, Jiang X, Kozasa T, Roscoe W, Singer W D, Gilman A G, Sternweis P C, Bollag G. Science. 1998;280:2112–2114. doi: 10.1126/science.280.5372.2112. [DOI] [PubMed] [Google Scholar]
- 59.Van Aelst L, D'Souza-Schorey C. Genes Dev. 1997;11:2295–2322. doi: 10.1101/gad.11.18.2295. [DOI] [PubMed] [Google Scholar]
- 60.Faderl S, Talpaz M, Estrov Z, O'Brien S, Kurzrock R, Kantarjian H M. N Engl J Med. 1999;341:164–172. doi: 10.1056/NEJM199907153410306. [DOI] [PubMed] [Google Scholar]
- 61.Tanaka K, Arif M, Eguchi M, Kyo T, Dohy H, Kamada N. Blood. 1997;89:596–600. [PubMed] [Google Scholar]
- 62.Johansson B, Moorman A V, Secker-Walker L M. Leukemia. 1998;12:828–833. doi: 10.1038/sj.leu.2401019. [DOI] [PubMed] [Google Scholar]
- 63.Hart M J, Sharma S, elMasry N, Qiu R G, McCabe P, Polakis P, Bollag G. J Biol Chem. 1996;271:25452–25458. doi: 10.1074/jbc.271.41.25452. [DOI] [PubMed] [Google Scholar]
- 64.Strout M P, Marcucci G, Caligiuri M A, Bloomfield C D. Ann Hematol. 1999;78:251–264. doi: 10.1007/s002770050511. [DOI] [PubMed] [Google Scholar]