Abstract
We investigated the contribution of the xeroderma pigmentosum group C (XPC) gene to DNA repair. We stably transfected XPC cells (XP4PA-SV-EB) with XPC cDNA and selected a partially corrected (XP4PA-SE1) and a fully corrected (XP4PA-SE2) clone. Cell survival after UVC (254 nm) exposure was low for XP4PA-SV-EB, intermediate for XP4PA-SE1, and normal for XP4PA-SE2 cells. XP4PA-SV-EB cells had undetectable XPC mRNA and protein levels. XP4PA-SE1 cells had 130% of normal mRNA but 25% of normal protein levels, whereas XP4PA-SE2 cells had an 18-fold mRNA overexpression and normal XPC protein levels compared with normal cells. We measured cyclobutane pyrimidine dimers (CPD) and 6-4 photoproducts (6-4PP) by using specific mAbs and the ELISA technique. XP4PA-SV-EB cells had no detectable removal of CPD or 6-4PP from their global genome by 24 h after 30 J/m2 UVC exposure. The partially corrected XP4PA-SE1 cells had normal repair of CPD but minimal repair of 6-4PP by 24 h, whereas the fully corrected XP4PA-SE2 cells regained normal CPD and 6-4PP repair capacities. We also exposed pRSVcat plasmid to UVC (to induce CPD and 6-4PP), to UVC + photolyase (to leave only 6-4PP on the plasmid), or to UVB + acetophenone (to induce only CPD). Host cell reactivation of UVB + acetophenone-, but not of UVC + photolyase-treated plasmids was normal in XP4PA-SE1 cells. Thus, increasing XPC gene expression leads to selective repair of CPD in the global genome. Undetectable XPC protein is associated with no repair of CPD or 6-4PP, detectable but subnormal XPC protein levels reconstitute CPD but not 6-4PP repair, and normal XPC protein levels fully reconstitute both CPD and 6-4PP repair.
Cellular integrity depends on the cells' ability to repair DNA damage. UV irradiation is a well known mutagenic agent, and UV-induced DNA damage, if not repaired properly, may lead to cell death, mutations, or carcinogenic transformation. In fact, UV-induced skin cancers are the most frequent neoplasms in Caucasians (1). Nucleotide excision repair (NER) is one of the most versatile and best-studied DNA repair systems. NER eliminates a wide variety of DNA damage, including UV photoproducts (2–5). The sequence of the NER process consists of two broad steps: (i) lesion recognition, strand incision, and damaged nucleotide displacement, and (ii) gap filling by DNA polymerization and ligation (6). Two NER subpathways have been discerned: “global genome repair” (GGR) and “transcription-coupled repair” (TCR) (7). GGR operates genome-wide and is able to remove DNA lesions from all locations in the genome at any moment in the cell cycle. TCR specifically acts on the transcribed strand of active genes, where it rapidly removes elongation-blocking lesions (8).
Three rare, autosomal recessive inherited human disorders are associated with impaired NER activity: xeroderma pigmentosum (XP), Cockayne syndrome, and trichothiodystrophy (reviewed in ref. 9). XP has been studied most extensively. XP patients exhibit extreme sensitivity to UV light, resulting in a high incidence of skin cancers (≈1,000 times that of the general population) (10, 11). About 20% of the XP patients also develop neurologic abnormalities in addition to their skin problems. These clinical findings are associated with cellular defects, including sensitivity to killing and mutagenic effects of UV and the inability of XP cells to repair UV-induced DNA damage (12). Seven different DNA repair genes that correct seven distinct genetic XP complementation groups (XPA–G) have been identified (9). In addition, another entity, XP variant (XPV), exists. Patients suffering from XPV are defective in DNA polymerase η, which is responsible for error-free bypass of UV-induced DNA damage (13, 14).
XPC is the most prevalent form among North Americans and Europeans, but until recently the function of the XPC protein was not well understood. It has been shown that cells from XPC patients are defective only in GGR and have normal TCR capability (8, 15–17). In 1992, the XPC gene was identified after transfection of XPC cells with a human cDNA expression library, resulting in correction of the severe UV sensitivity of these cells (18). The gene is located on chromosome 3p25 and encodes a 940-aa protein (9). Subsequently, the XPC protein was found to complex with HHR23B (19, 20). This complex shows a high affinity for both single- and double-stranded DNA (19–21). In vitro NER studies revealed that the XPC protein (complexed with HHR23) is involved in DNA damage recognition and acts along with XPA protein during early stages of GGR (6, 7, 22, 23).
We wanted to investigate further the contribution of the XPC gene to DNA repair in human cells. We constructed a partially corrected (XP4PA-SE1) and a fully corrected (XP4PA-SE2) cell line by stable transfection of an XPC cell line, XP4PA-SV-EB, with the plasmid pXPC3, which contains XPC cDNA. The ability to repair UV-induced DNA damage was assessed by UV cell survival (24), a plasmid host cell reactivation assay (25), and directly with a photoproduct removal ELISA and specific mAbs (26, 27). Increased, but still subnormal, XPC protein levels led to a partial functional correction in XP4PA-SE1 by reconstituting cyclobutane pyrimidine dimer (CPD) but not 6-4 photoproduct (6-4PP) repair in the cells' global genome.
Materials and Methods
Cell Lines and Culture Conditions.
The simian virus 40 (SV40) immortalized XPC fibroblast cell line XP4PA-SV-EB (18) was generously provided by R. Legerski (M. D. Anderson Cancer Center, Houston, TX). GM637, a normal SV40-immortalized fibroblast cell line was obtained from the Human Genetic Mutant Cell Repository (Camden, NJ). Cells were grown in DMEM supplemented with 2% glutamine and 10% FCS (GIBCO/BRL) in an 8% CO2 humidified incubator at 37°C. The XP4PA-SE1 and XP4PA-SE2 transfectants were grown under the same conditions with the addition of 0.2 mg/ml hygromycin B (Sigma).
Stable Transfection of XP4PA-SV-EB Cells.
The plasmid pXPC3 (18), which contains the cDNA for XPC, together with a hygromycin B resistance gene was generously provided by R. Legerski. A total of 0.25 μg of CsCl-purified pXPC3 was transfected into 0.15 × 106 fibroblasts by using 3 μl of Lipofectamine (GIBCO/BRL) in a total volume of 1 ml OPTI-MEM medium (GIBCO/BRL) for 5 h. Hygromycin B (Sigma) at a concentration of 0.2 mg/ml was added immediately after transfection for selection. After 2–4 wk, single clones were picked and further expanded in hygromycin B-containing medium.
Post-UVC Cell Survival.
Cell survival was determined by assessing cell growth in 35-mm dishes after UVC irradiation (24). A total of 2 × 104 cells were seeded per dish and irradiated with 254-nm UVC at a fluency of 0.16 J/m2 per s, as detected by a calibrated UVC radiometer (International Light, Newburyport, MA; model 12770A with a PT171C detector). Duplicate dishes were exposed to 0, 3, 6, 9, and 12 J/m2 UVC. After 4 days, the cells per dish were counted with a hemocytometer, and cell survival was calculated as the ratio of cell numbers in irradiated vs. unirradiated dishes.
Northern Blot Analysis.
Total cytoplasmic RNA was isolated from cells (RNAqueous-Midi Kit; Ambion, Austin, TX), and Northern blotting was performed following standard procedures as suggested by the manufacturer (NorthernMax Kit; Ambion). For probing, XPC cDNA was released from pXPC3 by SfiI enzyme digestion, gel purified, and 32P-labeled (Strip-EZ DNA; Ambion). Autoradiographic band intensities were measured with a laser densitometer (Molecular Dynamics). After stripping off the XPC probe (Strip-EZ DNA; Ambion), the same nylon membrane was reprobed with β-actin cDNA (CLONTECH) to compensate for variations in RNA transfer.
Western Blot Analysis.
A total of 7 × 106 exponentially growing cells were lysed in 120 μl of SDS sample buffer. The total protein concentration of cell extracts was measured (Bio-Rad protein assay), equal protein amounts (75 μg) were resolved by SDS/8% PAGE, and transferred to a poly(vinylidene difluoride) (PVDF) membrane (Perkin–Elmer). Either a mouse monoclonal anti-XPC antibody (generous gift from E. Lee, San Antonio, TX; working dilution 1:50) or a rabbit polyclonal anti-XPC antibody (generous gift from J. Hoeijmakers, Rotterdam, The Netherlands; working dilution 1:250) was used as a primary antibody. In addition, two different chemiluminescent detection systems, either employing a biotinylated (Western-Light Plus, Perkin–Elmer) or a horseradish peroxidase-labeled secondary antibody (ECL, Amersham Pharmacia), were used according to the manufacturers. Autoradiographic band intensities were measured with the laser densitometer.
ELISA with Photoproduct-Specific Monoclonal Antibodies.
This procedure was described previously (26, 27). Briefly, 5 × 106 cells were seeded on 15-cm culture dishes and irradiated with 0, 15, 22.5, and 30 J/m2 UVC as described above. Cells were harvested immediately, 6 h, or 24 h after irradiation, and genomic DNA was isolated (QIAamp Blood Kit; Qiagen). ELISA (in quadruplicate) was performed in 96-well poly(vinyl chloride) microtiter plates (Dynatech) precoated with 1% protamine sulfate (Sigma). For CPD detection with TDM-2 antibodies, 15 ng of DNA was used, for 6-4PP detection with 64 M-2 antibodies, 150 ng of DNA, respectively (TDM-2 and 64 M-2 were generously provided by T. Mori, Nara, Japan). After adding biotinylated F(ab′)2 goat anti-mouse IgG fragments and streptavidin-peroxidase (Zymed), the absorbance (OD) of products from o-phenylenediamine (Sigma) was measured at 492 nm (SN 02055 Spectra MAX 250; Molecular Devices). The photoproduct removal frequency was calculated as the ratio of OD-related UV equivalents (obtained from the induction curve) from DNA 6 h or 24 h repaired and from immediately harvested DNA after 30 J/m2 UVC.
Plasmid Host Cell Reactivation.
To generate only CPD lesions, the nonreplicating pRSVcat reporter gene plasmid (24, 25) was irradiated with 313-nm monochromatic UVB light (2 J/m2 per s) in the presence of 10−2 M acetophenone (Sigma) for 1 h under anoxic conditions (28, 29). To induce only 6-4PP lesions, the plasmid was irradiated with 1,000 J/m2 UVC (1.6 J/m2 per s) and subsequently treated with photolyase (PharMingen; 1 ml of 0.05 mg/ml pRSVcat + 1 μl of 5 mg/ml photolyase) and 405-nm monochromatic blue light (4.5 J/m2 per s) for 1 h to remove UVC-induced CPD lesions (30, 31). Photolesions after treatment were measured with the ELISA technique as described above, except that for CPD detection 1.5 ng of plasmid DNA and for 6-4PP detection 15 ng of plasmid DNA was used. A total of 0.25 μg of CsCl-purified treated or untreated pRSVcat was transfected into 0.15 × 106 fibroblasts by using 3 μl of Lipofectamine (GIBCO/BRL) in a total volume of 1 ml of OPTI-MEM medium (GIBCO/BRL) for 5 h. After 48 h, the chloramphenicol acetyltransferase (CAT) activity and the total protein amount were measured in the cell lysates (24). Relative CAT activity is expressed as percentage activity obtained from treated plasmids compared with the corresponding untreated control plasmids.
Results
Selection of Partially and Fully Corrected XPC Clones.
To investigate the role of the XPC gene in DNA repair in vivo, we stably transfected SV40-transformed XPC fibroblasts with the XPC cDNA containing plasmid pXPC3 (18). After selection with Hygromycin B, single stably transfected clones were screened for their ability to repair UV-induced DNA damage by means of their post-UVC cell survival. Cells were exposed to 0–12 J/m2 UVC, and after 4 days survival was measured. GM637 cells showed normal survival and XP4PA-SV-EB cells showed low survival typical of XPC cells (24) (Fig. 1A). The low cell survival of XPC cells was partially restored in the XP4PA-SE1 transfectant and fully restored in the XP4PA-SE2 transfectant. At 6 J/m2 XP4PA-SE1 had a 6.5-fold (P < 0.0001, Student's t test) higher survival rate than the parental XPC cells. XP4PA-SE2, like normal cells, exhibited a 24-fold (P < 0.0001, Student's t test) higher survival rate compared with XPC. In addition, transfected clones were screened for their ability to reactivate UV- damaged plasmid DNA. UVC-irradiated pRSVcat plasmid was transfected into the cells, and the CAT activity was measured 48 h later in the cell extracts. CAT activity expressed as the relative activity compared with unirradiated control plasmids reflects the DNA repair capacity of the host cells (25). Transfection of 1,000 J/m2 UVC-treated pRSVcat plasmid resulted in 3% reporter gene expression in XP4PA-SV-EB cells and 45% in repair proficient GM637 cells. Stable expression of the XPC gene in XP4PA-SV-EB cells enabled them to enhance their DNA repair capacity of UV-treated plasmids. CAT expression in the XP4PA-SE1 and XP4PA-SE2 clones increased to 10% and 23%, respectively, 3.1-fold and 7.7-fold higher (P < 0.0001, Student's t test) than in the original XPC cell line. Thus, the extent of increased cell survival closely correlated with the extent of increased host cell reactivation in the two partially and fully corrected XPC clones.
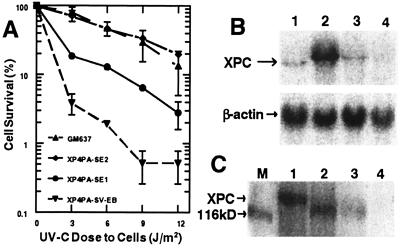
Figure 1.
Selection of partially and fully corrected XPC clones stably transfected with XPC cDNA. (A) Post-UVC cell survival was measured 4 days after exposure to 3–12 J/m2 UVC. The markedly reduced survival of XPC cells (XP4PA-SV-EB) is partially restored (XP4PA-SE1) or fully restored (XP4PA-SE2) in two stable transfectants. Each data point represents the mean of at least two independent measurements (±SEM). (B) Northern blot analysis of XPC-mRNA expression was performed with extracts from normal GM637 cells (lane 1), XP4PA-SE2 (lane 2), XP4PA-SE1 (lane 3), and XP4PA-SV-EB (lane 4). A total of 25 μg of total RNA was loaded per lane, separated by electrophoresis, transferred to a nylon membrane, and probed with a 3.5-kb XPC cDNA fragment. The relative amount of RNA transferred was monitored by probing with β-actin cDNA. (C) Western blot analysis of XPC protein (125 kDa) expression was performed on total cell protein extracts (75 μg) from the same cell lines (lanes 1–4, respectively; M, size marker) with an anti-XPC mouse mAb.
The Partially and Fully Corrected XPC Clones Regained Different Levels of XPC Gene Expression.
To assess the correlation between the functional correction and the recovery of XPC gene expression, we compared XPC mRNA levels from the XP4PA-SE1 and XP4PA-SE2 clones to those from the original XPC cell line and normal GM637 cells by Northern blot hybridization (Fig. 1B). XPC cDNA was used as a probe and a single band about 3.8 kb in size was detected in the normal controls (lane 1) but was undetectable in the XPC cell line XP4PA-SV-EB (lane 4). This is consistent with previously reported descriptions of XPC mRNA expression in normal and XPC cells (24). XP4PA-SE1 cells, which showed only partial correction in the previous assays, were found to express 1.3 times as many XPC transcripts as did the normal cells (lane 3), and the fully corrected XP4PA-SE2 cells exhibited a strong 18-fold elevated XPC mRNA expression compared with normal cells (lane 2). Hybridization of the same membrane with the β-actin cDNA probe (Fig. 1B) revealed similarly high β-actin mRNA levels in all four samples, indicating that the differences in XPC transcription were not because of degradation of RNA samples isolated from the cell lines. We also measured the XPC protein expression levels in our transfectants by Western blot analysis (Fig. 1C). Equal amounts of total cell protein were loaded per lane, separated by SDS/PAGE, and probed with an anti-XPC mouse mAb. A single band about 125 kDa in size was detected in the normal controls (lane 1) but was undetectable in the XPC cells (lane 4). XP4PA-SE1 cells, which showed only partial functional correction, expressed 25% of normal XPC protein (lane 3). The fully corrected XP4PA-SE2 cells showed 90% of normal protein expression (lane 2). Similar signal patterns were obtained by using an anti-XPC rabbit polyclonal antibody (data not shown). The pXPC3 plasmid codes for an XPC cDNA that is truncated by 160 bp from the N terminus compared with the wild-type XPC cDNA (18, 19). The expression vector cloning site contains an in-frame AUG start signal so that the resulting protein is truncated by 53 aa, resulting in a faster migrating band in the transfectants (lanes 2 and 3).
The Partially Corrected Clone XP4PA-SE1 Can Repair CPD but Not 6-4PP.
We used an ELISA assay and specific mAbs (26, 27) to further assess the repair kinetics of CPD and 6-4PP in the two transfectants. As shown in Fig. 2, normal cells were able to repair more than 90% of 6-4PP and 37% of CPD within 24 h after irradiation with 30 J/m2 UVC. In contrast, XPC cells had no detectable removal of either 6-4PP or CPD photoproducts from the global genome within 24 h. These results are in good agreement with previously published data (8). The partially corrected clone XP4PA-SE1 had barely detectable removal of 6-4PP, but showed a normal repair capacity for CPD (Fig. 2). In the fully corrected clone XP4PA-SE2, both 6-4PP and CPD repair were reconstituted to normal levels. Thus, increased but subnormal XPC protein levels resulted in a selective recovery of CPD but not 6-4PP repair in XP4PA-SE1.
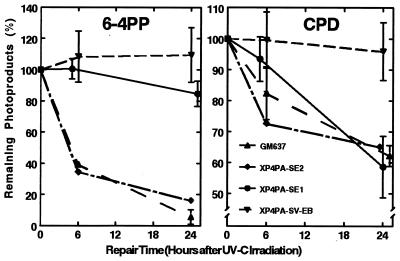
Figure 2.
Direct characterization of the photoproduct repair capacity in the two stably corrected XPC clones. Repair kinetics of 6-4PP (Left) and CPD (Right) from genomic DNA in normal cells (GM637), XPC cells (XP4PA-SV-EB), the partially corrected XP4PA-SE1 clone, and the fully corrected XP4PA-SE2 clone were assessed. Cells were irradiated with 30 J/m2 UVC, and 0, 6, or 24 h after UV treatment, the genomic DNA was isolated from the cells. Remaining photolesions in the DNA were quantitated with an ELISA procedure and specific mAbs (27) against 6-4PP or CPD. Each data point represents the mean of at least two experiments with three ELISA assays performed in quadruplicate (±SEM). The partially corrected XP4PA-SE1 clone showed normal CPD repair but did not repair 6-4PP.
Host Cell Reactivation Demonstrates Selective Recovery of CPD Repair in the Partially Corrected Clone XP4PA-SE1.
To confirm the photoproduct repair results obtained with the ELISA assay, we also conducted in vivo plasmid DNA repair studies. We used pRSVcat plasmid either containing only CPD or 6-4PP lesions. The recovery of CAT activity indirectly reflects the ability of the transfected cells to repair the induced plasmid DNA damage. To selectively induce CPD or 6-4PP lesions, the plasmid was treated with UVB + acetophenone or photoreactivated with blue light in the presence of Escherichia coli photolyase after treatment with 1,000 J/m2 UVC light, respectively. Photolesions were verified and quantitated with specific mAbs and the ELISA technique as described above. We treated the plasmid in a way to induce the same ratio of CPD to 6-4PP as induced by UVC exposure (Fig. 3). One-hour UVB irradiation in the presence of acetophenone generated the same amount of CPD as did irradiation with 1,000 J/m2 UVC, but 6-4PP were only generated in minimally detectable amounts (Fig. 3A). In contrast, with photoreactivation of UVC-treated plasmids, the numbers of UVC-induced 6-4PP remained unchanged, but all CPD lesions were removed (Fig. 3B). Transfection of only CPD-containing plasmids resulted in a significantly reduced CAT activity in XPC cells (32 ± 3%, P = 0.005; Student's t test) compared with normal cells (48 ± 3%), whereas the activities expressed from the two stably corrected clones XP4PA-SE2 (58 ± 5%) and XP4PA-SE1 (46 ± 4%) were not significantly different from normal (Fig. 3C). Transfection of only 6-4PP-containing plasmids also resulted in normal reporter gene expression in the fully corrected clone (90 ± 20%). However, CAT expression in the partially corrected clone (33 ± 5%, P = 0.048; Student's t test; compared with normal cells) was markedly reduced down to levels of XPC cells. We also transfected 6-4PP-containing plasmids into the four cell lines that were irradiated with 500 J/m2 UVC and subsequently photoreactivated. As shown in Fig. 3B, these plasmids carry only about half as many 6-4PP lesions as do the 1,000 J/m2 irradiated and photoreactivated plasmids. CAT expression in the partially corrected clone (51 ± 6%, P = 0.002; Student's t test; compared with normal cells) was also markedly reduced down to levels of XPC cells (51 ± 8%), whereas the fully corrected clone (93 ± 9%) had normal (94 ± 6%) CAT expression. These values of CAT expression are similar to those seen at the higher dose level (1,000 J/m2; Fig. 3C) and thus indicate that host cell reactivation experiments were performed at saturating lesion concentrations. These host cell reactivation data confirm the inability of the partially corrected XP4PA-SE1 cells to repair 6-4PP in whole cell genomic DNA.
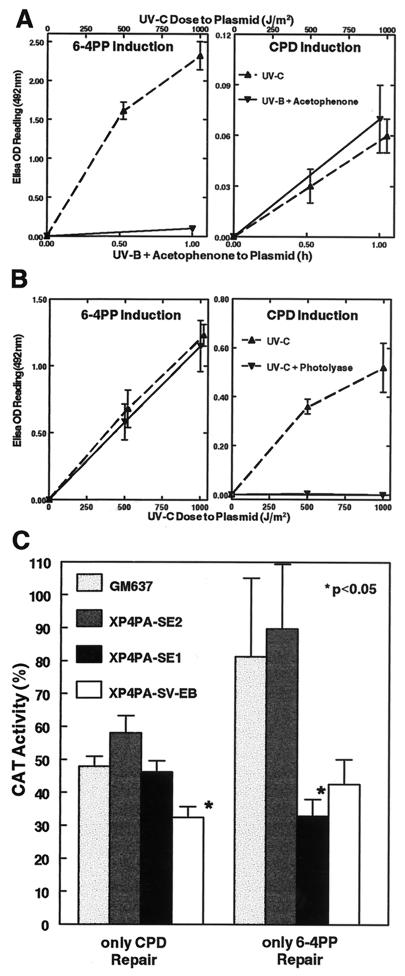
Figure 3.
Characterization of the photoproduct repair capacity in the two stably corrected XPC clones. Host cell reactivation of a reporter gene plasmid (pRSVcat) was used to assess the 6-4PP or CPD repair capability of the fully and partially corrected transfectants. (A) To induce only CPD lesions on the plasmid, pRSVcat was treated with acetophenone (10−2 M) + 313 nm UVB light (see Methods). 6-4PP and CPD lesions in the double-stranded DNA plasmid after treatment were measured with specific mAbs (as in Fig. 2) and compared with UVC-treated plasmid. The UVB + acetophenone treatment generated minimally detectable amounts of 6-4PP (Left), but induced about the same numbers of CPD as did a 1,000 J/m2 UVC treatment (Right). (B) To induce only 6-4PP lesions on the plasmid, pRSVcat was irradiated with UVC and subsequently photoreactivated with E. coli photolyase and 405 nm blue light (29). After this treatment, the number of UVC-induced 6-4PP was unchanged (Left), but all CPD lesions were removed from the plasmid (Right) as assessed with the ELISA assay. Data points are means of four determinations performed in quadruplicate (±SEM). (C) Either UVB + acetophenone-treated pRSVcat plasmid (only CPD Repair) or 1,000 J/m2 UVC + photolyase-treated pRSVcat plasmid (only 6-4PP Repair) was transfected into normal cells (GM637), the fully corrected XPC clone (XP4PA-SE2), the partially corrected XPC clone (XP4PA-SE1), or XPC cells (XP4PA-SV-EB). Relative CAT activity was measured 48 h after transfection compared with the corresponding untreated control plasmids. Bars represent means of at least triplicate experiments (±SEM). Specific activity with the untreated plasmids ranged from 1.59 to 22.38 nmol per min per mg protein. The partially corrected XP4PA-SE1 clone showed normal CPD repair but did not repair 6-4PP.
Discussion
We wanted to investigate the role of the XPC gene product in DNA nucleotide excision repair in vivo. For that purpose, a plasmid containing XPC cDNA (pXPC3) was stably transfected into an SV40-immortalized XPC cell line (XP4PA-SV-EB). XP4PA-SV-EB has a deletion of two nucleotides T and G at positions 1483 and 1484, which results in a frameshift mutation that leads to a 430-aa truncated XPC protein (about half the size of the wild-type protein), abolishing its function, and leading to undetectable XPC mRNA and protein levels (32). Because this cell line was established in culture from amniotic fluid (33), no information about the severity of the clinical phenotype is available. However, the results of our molecular studies on the cell line (Figs. 1–3) are within the range of results previously reported from other severely affected XPC patients (8, 24).
The pXPC3 plasmid contains XPC cDNA that is missing 160 bp from the N terminus (18, 19). However, pXPC3 fully complemented the repair deficiency in several XPC cell lines, as determined by the restoration of unscheduled DNA synthesis after somatic cell microinjection experiments (18) and by restoration of reporter gene expression in a transient complementation assay (34) (Fig. 3C). We also replaced this cDNA with full-length XPC cDNA in pXPC3 and found a similar extent of host cell reactivation of transiently complemented XPC cells (S.E. and K.H.K., unpublished data). This and the repair results obtained with the fully corrected XPC cell line XP4PA-SE2 (Figs. 1A, 2, and 3C) indicate that the XPC mRNA product from pXPC3 is representative of the XPC mRNA produced in normal cells.
We were able to isolate two XPC clones, XP4PA-SE1 and XP4PA-SE2, stably transfected with pXPC3 that exhibited different levels of restoration. Functional restoration was determined either directly by the cells' sensitivity to killing by UV light (9, 24) or indirectly by the cells' ability to reactivate UV-irradiated plasmids (35). With both assays, XP4PA-SE1 showed a partial and XP4PA-SE2 a full functional correction compared with normal cells (Figs. 1A and 3C).
These results also closely correlated with the clones' restoration of XPC gene expression. XP4PA-SE1 expressed 1.3 times as many XPC transcripts as normal cells and 25% XPC protein compared with normal cells, whereas XP4PA-SE2 expressed 18 times as many XPC transcripts and 90% XPC protein compared with normal cells (Fig. 1 B and C). This suggests a strong XPC gene regulation either on the mRNA and/or the protein level that is different from XPA (36) and XPD (37) genes. XPA or XPD transfectants, which had normal or slightly higher than normal XPA or XPD protein expression, did not show full functional correction. A 4.8-fold XPD protein overexpression (compared with normal cells) in an XPD transfectant resulted in only partial functional correction (37). In the case of XPC, we found that recovery of XPC protein expression to nearly normal levels was sufficient to fully correct the transfected XPC clone (XP4PA-SE2).
We extended the study to see how our findings are reflected by repair of specific DNA lesions. UV irradiation induces two major forms of DNA damage, 6-4PP and CPD (38). Various estimates suggest that the 6-4PP is formed at 10–50% of the frequency of CPD by 254 nm UVC light (9). The shape of the action spectrum for induction of CPD and 6-4PP is similar over many wavelengths, including UVC and UVB (39). An ELISA assay with specific mAbs against 6-4PP or CPD enabled us to separately quantitate the extent of removal of each photoproduct from the global genome in XP4PA-SE1 and XP4PA-SE2 (26, 27). We determined that the partial functional recovery in XP4PA-SE1 was solely attributed to the correction of CPD repair, whereas 6-4PP were minimally repaired in this clone (Fig. 2). XP4PA-SE2 cells had normal removal of both types of photoproducts from their global genome.
This selective repair of CPD over 6-4PP in XP4PA-SE1 was also demonstrated by utilizing a pRSVcat plasmid-based host cell reactivation system (25). To generate 6-4PP, plasmid was treated with UVC and subsequently photoreactivated with E. coli photolyase and 405-nm monochromatic light. This procedure is known to remove >99% of UVC-induced CPD lesions, as shown by agarose gel pattern indicating sensitivity to nicking of supercoiled plasmid form I by treatment with T4 endonuclease V (specifically cleaves at CPD but not 6-4PP sites) (25, 30). However, this detection system loses its sensitivity at UVC doses >30–50 J/m2 to plasmid. We photoreactivated plasmid irradiated with 1,000 J/m2 UVC light. Using the ELISA technique and photoproduct-specific antibodies, we were able to verify the type and quantity of photoproducts present in the same stock of plasmids, which were also used for host cell reactivation. We confirmed that photoreactivation removes all detectable CPD lesions from the UVC-treated plasmid but does not change the numbers of 6-4PP (Fig. 3B). Similarly, we found that plasmid treatment with UVB light and in the presence of acetophenone (28, 29) resulted in the same amount of CPD lesions as did treatment with 1,000 J/m2 UVC. However, 6-4PP were generated only minimally (Fig. 3A). According to previous studies (25), we estimated that these treatments resulted in the formation of about 100 CPD lesions per pRSVcat plasmid. In these host cell reactivation experiments, we used the same ratio of CPD to 6-4PP as was produced by UVC. This ratio also holds for UVB in natural sunlight (39). We did not examine the relative repair at different artificial ratios.
Based on the results from host cell reactivation of pRSVcat plasmids that exclusively contain either CPD or 6-4PP (Fig. 3C), we were also able to estimate the number of remaining CAT expression-inactivating lesions per CAT gene (40). If the number of inactivating lesions follows the Poisson distribution, the CAT inactivation curve is defined by e−m, where m is the number of inactivating lesions. The negative natural logarithm of the CAT activity from plasmids containing either CPD or 6-4PP then represents the mean number of remaining inactivating CPD or 6-4PP lesions per CAT gene, respectively. These calculations may apply only to a single UV dose, in our case 1,000 J/m2, as the negative natural logarithm will vary with the dose in case of nonlinear CAT inactivation curves. We calculated that the partially corrected clone XP4PA-SE1 had a mean value of 0.77 inactivating CPD and 1.11 inactivating 6-4PP lesions per CAT gene. The mean values for the fully corrected XP4PA-SE2 clone were 0.54 inactivating CPD and 0.11 inactivating 6-4PP lesions. The corresponding values for the normal and XPC cells were 0.74 and 1.12 CPD, and 0.21 and 0.85 6-4PP inactivating lesions, respectively. This confirms that the XP4PA-SE1 transfectant regained a normal CPD repair capacity but cannot repair 6-4PP, whereas XP4PA-SE2 regained normal repair capacities for both types of photoproducts. Nearly identical results in terms of 6-4PP repair were obtained with 500 J/m2 UVC-irradiated and photolyase-treated plasmids, indicating that the repair reaction was saturated.
Our results support a competitive damage recognition scheme for CPD and 6-4PP in the global genome NER between XPC and XPA in vivo. In vitro studies showed that the XPC protein (complexed with HHR23A or -B) is essential for a productive GGR (7), but is not involved in the TCR subpathway (8, 17). Evans et al. (41) proposed a model in which TFIIH and XPC may be involved in the initial opening of double-stranded DNA around a lesion. Mu et al. (42) reported that the first detectable NER intermediate contained XPC together with XPA, single-strand DNA-binding factor RPA, and TFIIH. In addition, Li et al. (6) found that XPC is necessary to promote a stable binding of XPA to UV-damaged DNA. Wakasugi and Sancar (23) found that XPC-HHR23 participates in the assembly of the excision nucleases, but is then no longer present in the ultimate dual incision complex. Whether the XPC-HHR23 protein complex (7) or the XPA-RPA complex (22) is the initiator of GGR, in any event, both proteins have to act together to successfully initiate repair of both types of UV-induced DNA damage, CPD and 6-4PP. Previously, XPA was shown to selectively repair and eliminate 6-4PP in vivo in accordance with its greater binding affinity for 6-4PP than for CPD in vitro (43–47). Our in vivo results suggest that the XPC gene product leads to selective repair of CPD in the global genome at the natural ratio of CPD to 6-4PP (Figs. 2 and 3). This may explain the extremely high damage discrimination exhibited by the NER machinery. The idea of two damage recognition proteins each selectively recognizing a major environmentally important form of DNA damage may be also supported by the existence of the recently reported human DNA polymerase η. This polymerase, defective in postreplication-deficient XP variant patients, can specifically bypass CPD lesions but not 6-4PP lesions (13, 14, 48).
However, the molecular mechanism responsible for our observed selective repair of CPD by XPC remains to be elucidated. Wakasugi and Sancar (22) report a high in vitro binding affinity of XPC protein to a DNA fragment containing one 6-4PP, which exceeded the binding affinity of XPA to 6-4PP. Therefore, it is unlikely that a greater direct binding affinity of XPC to CPD than to 6-4PP leads to a selective repair of CPD. Hwang et al. (49) proposed that NER of CPD in the global genome is initiated by the binding of UV-DDB (UV damage-specific DNA-binding protein, XP-E). XPC-HHR23 would then recognize UV-DDB bound to the lesion. This indirect binding of XPC to CPD might exceed the XPC binding affinity to 6-4PP, thus, leading to selective repair of CPD. We also cannot exclude the possibility that the N-terminal truncation of the XPC protein used in this study might account for the selective CPD repair observed. It is possible that the truncation affected the 6-4PP (or a general DNA damage) binding domain of XPC lowering its 6-4PP (or general DNA damage) binding affinity. The lower binding affinity could be compensated by nearly normal XPC protein levels (XP4PA-SE2 transfectant). Restricted XPC protein amounts (XP4PA-SE1 transfectant) would then lead to detectable DNA repair deficiencies, which, in the case of CPD repair, could be compensated by an indirect UV-DDB-mediated binding of XPC to CPD.
Acknowledgments
We thank Dr. M. Bergel for invaluable help with the Western blotting, Dr. R. Legerski for the XPC cell line and the pXPC3 plasmid, Dr. T. Mori for the photoproduct-specific mAbs, Dr. E. Lee and Dr. J. Hoeijmakers for providing the anti-XPC protein antibodies, and Dr. R. Rahn for suggestions regarding the UVB + acetophenone plasmid treatment. S.E. was supported by a grant from the Deutsche Forschungsgemeinschaft.
Abbreviations
- XP
xeroderma pigmentosum
- CPD
cyclobutane pyrimidine dimers
- 6-4PP
pyrimidine-pyrimidone (6-4) photoproducts
- CAT
chloramphenicol acetyltransferase
- NER
nucleotide excision repair
- GGR
global genome repair
- TCR
transcription-coupled repair
- SV40
simian virus 40
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.040559697.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.040559697
References
- 1.Kraemer K H. Proc Natl Acad Sci USA. 1997;94:11–14. doi: 10.1073/pnas.94.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ma L, Hoeijmakers J H, van der Eb A J. Biochim Biophys Acta. 1995;1242:137–163. doi: 10.1016/0304-419x(95)00008-4. [DOI] [PubMed] [Google Scholar]
- 3.Sancar A. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 4.Wood R D. Annu Rev Biochem. 1996;65:135–167. doi: 10.1146/annurev.bi.65.070196.001031. [DOI] [PubMed] [Google Scholar]
- 5.de Laat W L, Jaspers N G, Hoeijmakers J H. Genes Dev. 1999;13:768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- 6.Li R Y, Calsou P, Jones C J, Salles B. J Mol Biol. 1998;281:211–218. doi: 10.1006/jmbi.1998.1949. [DOI] [PubMed] [Google Scholar]
- 7.Sugasawa K, Ng J M, Masutani C, Iwai S, van der Spek P J, Eker A P, Hanaoka F, Bootsma D, Hoeijmakers J H. Mol Cell. 1998;2:223–232. doi: 10.1016/s1097-2765(00)80132-x. [DOI] [PubMed] [Google Scholar]
- 8.van Hoffen A, Venema J, Meschini R, van Zeeland A A, Mullenders L H. EMBO J. 1995;14:360–367. doi: 10.1002/j.1460-2075.1995.tb07010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bootsma D, Kraemer K H, Cleaver J E, Hoeijmakers J H. In: The Genetic Basis of Human Cancer. Vogelstein B, Kinzler K W, editors. New York: McGraw–Hill; 1998. pp. 245–274. [Google Scholar]
- 10.Kraemer K H, Lee M M, Scotto J. Arch Dermatol. 1987;123:241–250. doi: 10.1001/archderm.123.2.241. [DOI] [PubMed] [Google Scholar]
- 11.Kraemer K H, Lee M M, Andrews A D, Lambert W C. Arch Dermatol. 1994;130:1018–1021. [PubMed] [Google Scholar]
- 12.van Steeg H, Kraemer K H. Mol Med Today. 1999;5:86–94. doi: 10.1016/s1357-4310(98)01394-x. [DOI] [PubMed] [Google Scholar]
- 13.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. Nature (London) 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 14.Johnson R E, Kondratick C M, Prakash S, Prakash L. Science. 1999;285:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- 15.Venema J, van Hoffen A, Natarajan A T, van Zeeland A A, Mullenders L H. Nucleic Acids Res. 1990;18:443–448. doi: 10.1093/nar/18.3.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Venema J, van Hoffen A, Karcagi V, Natarajan A T, van Zeeland A A, Mullenders L H. Mol Cell Biol. 1991;11:4128–4134. doi: 10.1128/mcb.11.8.4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mu D, Sancar A. J Biol Chem. 1997;272:7570–7573. doi: 10.1074/jbc.272.12.7570. [DOI] [PubMed] [Google Scholar]
- 18.Legerski R, Peterson C. Nature (London) 1992;359:70–73. doi: 10.1038/359070a0. [DOI] [PubMed] [Google Scholar]
- 19.Masutani C, Sugasawa K, Yanagisawa J, Sonoyama T, Ui M, Enomoto T, Takio K, Tanaka K, van der Spek P J, Bootsma D. EMBO J. 1994;13:1831–1843. doi: 10.1002/j.1460-2075.1994.tb06452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shivji M K, Eker A P, Wood R D. J Biol Chem. 1994;269:22749–22757. [PubMed] [Google Scholar]
- 21.Reardon J T, Mu D, Sancar A. J Biol Chem. 1996;271:19451–19456. doi: 10.1074/jbc.271.32.19451. [DOI] [PubMed] [Google Scholar]
- 22.Wakasugi M, Sancar A. J Biol Chem. 1999;274:18759–18768. doi: 10.1074/jbc.274.26.18759. [DOI] [PubMed] [Google Scholar]
- 23.Wakasugi M, Sancar A. Proc Natl Acad Sci USA. 1998;95:6669–6674. doi: 10.1073/pnas.95.12.6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khan S G, Levy H L, Legerski R, Quackenbush E, Reardon J T, Emmert S, Sancar A, Li L, Schneider T D, Cleaver J E, Kraemer K H. J Invest Dermatol. 1998;111:791–796. doi: 10.1046/j.1523-1747.1998.00391.x. [DOI] [PubMed] [Google Scholar]
- 25.Protic-Sabljic M, Kraemer K H. Proc Natl Acad Sci USA. 1985;82:6622–6626. doi: 10.1073/pnas.82.19.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakagawa A, Kobayashi N, Muramatsu T, Yamashina Y, Shirai T, Hashimoto M W, Ikenaga M, Mori T. J Invest Dermatol. 1998;110:143–148. doi: 10.1046/j.1523-1747.1998.00100.x. [DOI] [PubMed] [Google Scholar]
- 27.Mori T, Nakane M, Hattori T, Matsunaga T, Ihara M, Nikaido O. Photochem Photobiol. 1991;54:225–232. doi: 10.1111/j.1751-1097.1991.tb02010.x. [DOI] [PubMed] [Google Scholar]
- 28.Rahn R O, Landry L C, Carrier W L. Photochem Photobiol. 1974;19:75–78. doi: 10.1111/j.1751-1097.1974.tb06476.x. [DOI] [PubMed] [Google Scholar]
- 29.Lamola A A. Photochem Photobiol. 1969;9:291–294. doi: 10.1111/j.1751-1097.1969.tb07292.x. [DOI] [PubMed] [Google Scholar]
- 30.Protic-Sabljic M, Kraemer K H. Photochem Photobiol. 1986;43:509–513. doi: 10.1111/j.1751-1097.1986.tb09528.x. [DOI] [PubMed] [Google Scholar]
- 31.Parris C N, Kraemer K H. Proc Natl Acad Sci USA. 1993;90:7260–7264. doi: 10.1073/pnas.90.15.7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li L, Bales E S, Peterson C A, Legerski R J. Nat Genet. 1993;5:413–417. doi: 10.1038/ng1293-413. [DOI] [PubMed] [Google Scholar]
- 33.Daya-Grosjean L, James M R, Drougard C, Sarasin A. Mutat Res. 1987;183:185–196. doi: 10.1016/0167-8817(87)90061-7. [DOI] [PubMed] [Google Scholar]
- 34.Carreau M, Eveno E, Quilliet X, Chevalier-Lagente O, Benoit A, Tanganelli B, Stefanini M, Vermeulen W, Hoeijmakers J H, Sarasin A. Carcinogenesis. 1995;16:1003–1009. doi: 10.1093/carcin/16.5.1003. [DOI] [PubMed] [Google Scholar]
- 35.Moriwaki S, Stefanini M, Lehmann A R, Hoeijmakers J H, Robbins J H, Rapin I, Botta E, Tanganelli B, Vermeulen W, Broughton B C, Kraemer K H. J Invest Dermatol. 1996;107:647–653. doi: 10.1111/1523-1747.ep12584287. [DOI] [PubMed] [Google Scholar]
- 36.Levy D D, Saijo M, Tanaka K, Kraemer K H. Carcinogenesis. 1995;16:1557–1563. doi: 10.1093/carcin/16.7.1557. [DOI] [PubMed] [Google Scholar]
- 37.Gozukara E M, Parris C N, Weber C A, Salazar E P, Seidman M M, Watkins J F, Prakash L, Kraemer K H. Cancer Res. 1994;54:3837–3844. [PubMed] [Google Scholar]
- 38.Clingen P H, Arlett C F, Cole J, Waugh A P, Lowe J E, Harcourt S A, Hermanova N, Roza L, Mori T, Nikaido O. Photochem Photobiol. 1995;61:163–170. doi: 10.1111/j.1751-1097.1995.tb03955.x. [DOI] [PubMed] [Google Scholar]
- 39.Chan G L, Peak M J, Peak J G, Haseltine W A. Int J Radiat Biol Relat Stud Phys Chem Med. 1986;50:641–648. doi: 10.1080/09553008614551041. [DOI] [PubMed] [Google Scholar]
- 40.Barrett S F, Robbins J H, Tarone R E, Kraemer K H. Mutat Res. 1991;255:281–291. doi: 10.1016/0921-8777(91)90032-k. [DOI] [PubMed] [Google Scholar]
- 41.Evans E, Moggs J G, Hwang J R, Egly J M, Wood R D. EMBO J. 1997;16:6559–6573. doi: 10.1093/emboj/16.21.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mu D, Wakasugi M, Hsu D S, Sancar A. J Biol Chem. 1997;272:28971–28979. doi: 10.1074/jbc.272.46.28971. [DOI] [PubMed] [Google Scholar]
- 43.Jones C J, Wood R D. Biochemistry. 1993;32:12096–12104. doi: 10.1021/bi00096a021. [DOI] [PubMed] [Google Scholar]
- 44.Mitchell D L. Photochem Photobiol. 1988;48:51–57. doi: 10.1111/j.1751-1097.1988.tb02785.x. [DOI] [PubMed] [Google Scholar]
- 45.Cleaver J E. Photochem Photobiol. 1996;63:377–379. doi: 10.1111/j.1751-1097.1996.tb03047.x. [DOI] [PubMed] [Google Scholar]
- 46.Cleaver J E, Charles W C, McDowell M L, Sadinski W J, Mitchell D L. Cancer Res. 1995;55:6152–6160. [PubMed] [Google Scholar]
- 47.Cleaver J E, McDowell M, Jones C, Wood R, Karentz D. Somatic Cell Mol Genet. 1994;20:327–337. doi: 10.1007/BF02254721. [DOI] [PubMed] [Google Scholar]
- 48.Masutani C, Araki M, Yamada A, Kusumoto R, Nogimori T, Maekawa T, Iwai S, Hanaoka F. EMBO J. 1999;18:3491–3501. doi: 10.1093/emboj/18.12.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hwang B J, Ford J M, Hanawalt P C, Chu G. Proc Natl Acad Sci USA. 1999;96:424–428. doi: 10.1073/pnas.96.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]