Abstract
The ability of the Minos transposable element to function as a transformation vector in anopheline mosquitoes was assessed. Two recently established Anopheles gambiae cell lines were stably transformed by using marked Minos transposons in the presence of a helper plasmid expressing transposase. The markers were either the green fluorescent protein or the hygromycin B phosphotransferase gene driven by the Drosophila Hsp70 promoter. Cloning and sequencing of the integration sites demonstrated that insertions in the cell genome occurred through the action of Minos transposase. Furthermore, an interplasmid transposition assay established that Minos transposase is active in the cytoplasmic environment of Anopheles stephensi embryos: interplasmid transposition events isolated from injected preblastoderm embryos were identified as Minos transposase-mediated integrations, and no events were recorded in the absence of an active transposase. These results demonstrate that Minos vectors are suitable candidates for germ-line transformation of anopheline mosquitoes.
Germ-line transformation provides an important link between gene identification and gene function. This approach is especially necessary for organisms in which genetic analysis is limited. Characteristic example is anopheline mosquitoes, the obligatory vectors of a devastating infectious disease, malaria. Although intensifying molecular studies in Anopheles are identifying new genes at an increasing rate, functional gene characterization is hindered by limitations of genetic analysis and the lack of germ-line transformation technology.
Efforts to transform anophelines have been pursued since the first and only reported case of foreign DNA introduction into the Anopheles gambiae genome 12 years ago (1). These efforts were intensified after the successful development of routine transformation techniques using the Minos transposable element in the Mediterranean fruitfly Ceratitis capitata (2) and the mariner and Hermes elements in the yellow fever mosquito Aedes aegypti (3, 4).
Since then, both transgenesis and transposon mobility assays have been used to establish that several transposable elements are efficient gene transfer vectors with a broad host range (5–14). However, anopheline eggs are notoriously difficult to inject, because of their surrounding tough chorion and vitelline membrane, and they are fragile in some species such as the major human malaria vector, A. gambiae. As a result, transposable element mobility has not been demonstrated to date in these mosquitoes. In addition, an effective marker for transformation remains to be established for these species. Because of the relatively low frequency of transformation, dominant visible markers are important. Therefore, eye color genes have been used for this purpose in other insects; but eye color mutations and the corresponding complementing genes are not well characterized in anophelines. Alternative marker candidates are genes conferring resistance to toxic chemicals and green fluorescent protein (GFP), which has been validated as transformation marker in several insects (15).
In the present study we have used Minos, the transposable element with which transformation was achieved in an insect other than Drosophila (2). We used as a marker GFP and the hygromycin B phosphotransferase gene under the control of the Drosophila Hsp70 promoter. We first demonstrated that Minos integrates in a transposase-dependent manner in the chromosomes of newly established cell lines of A. gambiae. We then confirmed that the element is also mobile in anopheline embryos, by pilot injection experiments coupled with an interplasmid transposition assay.
Materials and Methods
Plasmid Construction.
The helper plasmid pHSS6hsILMi20 has been described (16). The transformation vector pMinHyg was constructed by inserting, between the left and right arms of Minos, in the pMiNot vector (2) sequentially two fragments. A fragment, containing hygromycin B phosphotransferase gene under the control of the Hsp70 promoter from Drosophila melanogaster, and a DNA cassette, which includes a 2.6-kb fragment of the D. melanogaster act5C promoter and a 0.8-kb fragment of the Hsp70 terminator.
The plasmid pLaDHFRR used in the excision assay was constructed as follows. The left and right inverted repeats (IRs) of Minos including bases 1–350 and 1480–1777, respectively of the Minos sequences together with Drosophila hydei flanks were PCR-amplified and subcloned separately. The left IR was transferred within the PstI site of the BluescriptKSII (Stratagene). The right IR fused to sequences containing a dihydrofolate reductase (DHFR) gene linked to the proximal promoter of the Drosophila actin 5C gene (nucleotides −122 to +88) was transferred into the same plasmid as an EcoRI–SalI fragment. In this way a Minos transposon carrying internally (between the two IRs) the BluescriptKSII sequences and the actin DHFR gene was generated. Externally, a PstI–EcoRI fragment containing the tetracycline-resistance gene was introduced as spacer between the two IRs.
The plasmid pLHGR used to stably transform Sua 5.1* cells derived from the plasmid pLaDHFRR by replacing the sequences between the Minos IRs with a new BluescriptKSII plasmid carrying a rsGFP gene (Quantum Biotechnologies, Quebec, Canada) fused to the Drosophila Hsp70 promoter sequences (nucleotides −258 to +192).
Cell Culture, Transfection, and Handling.
A. gambiae cell lines Sua 4.0 and Sua 5.1* were established from neonatal larvae of the Suakoko mosquito strain, as described (17). The cell line 4a-2 was established following the same procedure from the mosquito strain 4a. All three lines were kept continuously in Schneider medium supplemented with 10% FCS.
For the excision assay cells were transfected by using the CaCl2 method essentially as described (18). The amount of helper plasmid DNA used was 1 μg and the amount of pLaDHFRR was 6 μg per transfection. PCR (1 min at 94°C, 30 sec at 55°C, 30 sec at 72°C, 35 cycles) was performed on total nucleic acids, isolated with a standard protocol (19), by using the primers (5′-AAGTGTAAGTGCTTGAAATGC-3′) and (5′-GCATCAAATTGAGTTTTGCTC-3′), which anneal to the D. hydei genomic sequences.
The Sua 5.1* cells were stably transfected by using the CaCl2 method, and the total amount of DNA used was 7 μg (1 μg of pHSS6hsILMi20, 6 μg of pLHGR plasmid). The enrichment and analysis of the Sua 5.1* cells was performed on a FACS Vantage (Becton Dickinson) cell sorter. Excitation was at 488 nm, and the emitted fluorescence was selected with a 530-nm band pass filter on a logarithmic scale. Sorting gates were set up by using forward scatter width versus side scatter height signals—to exclude dead cells, debris, and clumps—and forward scatter height versus fluorescence height signals to sort the cells that fell within a certain channel of fluorescence. Fluorescent fractions of cells were grown in Nunc 25-mm tissue culture inserts (Nalge Nunc, catalogue no. N136773) placed over a monolayer of Sua 5.1* cells, which served as feeder cells.
For stable transfection with the pMinHyg plasmid, a subculture of Sua 4.0 cells was used. Cells grown in M199 medium supplemented with 10% FCS at 26°C were transfected by using Lipofectin reagent (GIBCO/BRL). Cells were plated at a density of 3 × 105 cells/well in 24-well/plate, 20 hr before transfection. A mixture of 4 μg of plasmid pMinHyg and 0.5 μg of plasmid pHSS6hsILMi20 was incubated with a 5% Lipofectin solution in serum-free M199 medium at room temperature for 15 min and then added to the cells. Six hours later the medium was replaced with fresh medium containing 10% FCS. Two days after transfection, the cells were subjected to selection by culturing in medium containing 250 μg/ml of hygromycin B. After 4 weeks under selection pressure, resistant cells were plated into 96-wells/plate at a density of ca. 1,000 cells/well, and 3–5 weeks later colonies were picked and expanded.
DNA Manipulations and Sequencing.
Plasmid DNA and DNA from mosquito cells and embryos were isolated according to standard protocols (19, 20). The EcoRI genomic library of clone F1 was constructed by using the Lamba Zap II predigested vector (Stratagene), according to the manufacturer's instructions, and was screened by using the probe M. Isolated clones were sequenced by using primers annealing internally to the IRs (pMR: 5′-CGAGTTAAATGCGTAATGC-3′ and pML: 5′-GCTCTTCTTGAGATTAAGG-3′). The nucleotide sequence at the junction between the Minos right end and the genomic fragments in the rescued plasmids from the Sua 5.1* experiment was determined by using the primer Mioo (5′-GTTGTTGTTGGAAATAGAGCAAAA-3′), which anneals to sequences in the right IR of Minos. In situ hybridizations of isolated genomic fragments on polytene chromosomes of ovarian nurse cell nuclei of the Suakoko strain were performed according to a previously described protocol (21).
Embryo Microinjections and Interplasmid Transposition Assay.
A. stephensi adult mosquitoes (strain sd 500) were allowed to lay eggs 48–72 hr after a blood meal. Eggs were laid in a Petri dish containing 3MM paper soaked in a p-nitrophenyl p-guanidinobenzoate (pNpGB) solution (Sigma, catalogue no. N8010) at a final concentration of 0.1 mM in isotonic buffer (150 mM NaCl/5 mM KCl/2.5 mM CaCl2/10 mM Hepes, pH 7.2). Eggs then were left on the pNpGB-soaked paper until injection. Injections were carried out between 90 and 120 min after oviposition essentially as described (1). Approximately 20 hr after injection, embryos were subjected to heat shock at 42°C for 1 hr and allowed to recover for 2 hr at room temperature. DNA was extracted and used to transform Escherichia coli supercompetent cells (XL-1 blue, Stratagene). The efficiency of recovery of the target plasmid was estimated by plating 1% of the transformation mixture onto LB plates containing chloramphenicol (30 μg/ml). The remaining 99% of the transformation mix were plated onto LB agar containing chloramphenicol (30 μg/ml) and sucrose (10%). Resistant colonies then were transferred to liquid LB media containing tetracycline (10 μg/ml). The nucleotide sequence at the junctions between the Minos element and the target DNA was determined by using one primer annealing internally to the left arm of Minos (pML: 5′-GCTCTTCTTGAGATTAAGG-3′) and one primer annealing on the 5′ end of the tetracycline gene (PTet-compl: 5′-GGATGACGATGAGCGC-3′).
Results
Minos Transposase Activity in A. gambiae Cell Lines.
Minos transposase activity can be detected through a fast and accurate excision assay. Minos transposon excisions from a donor plasmid, which is transiently transfected or injected into cells or embryos, result in circularization of “empty” plasmid, from which a diagnostic excision band can then be generated by PCR (16). This excision assay was applied to three different A. gambiae cell lines. Lines Sua 4.0 and Sua 5.1* are derived from the same Suakoko mosquito strain whereas 4a-2 originates from the mosquito strain 4a. The cells were transfected with a mixture of donor and helper (transposase-producing) plasmids. The donor plasmid pLaDHFRR (see Materials and Methods) carried a Minos transposon flanked by D. hydei genomic sequences. The helper plasmid pHSS6hsMi20 (16) contained a Minos transposase gene, uninterrupted by the normal 60-bp intron, and placed under the control of the Hsp70 promoter. Transfected cells were harvested and total nucleic acids were extracted for use as template in PCRs. In these reactions, transposase-induced excisions are detectable as a 167-bp long excision band, derived from the empty donor plasmids; the band can be amplified by PCR using primers annealing to the D. hydei genomic sequences (Fig. 1, black arrows). Such a band was detected from the mosquito cells transfected with the mixture of helper/donor plasmid and subjected to a heat shock at 42°C for 1 hr to express transposase (Fig. 1, lanes +). The same band was not detected from cells transfected with the donor plasmid alone (Fig. 1, lanes −) or when heat shock was omitted (data not shown). Evidently, the overproduction of Minos transposase massively mobilizes Minos transposons from donor plasmids in A. gambiae cells.
Figure 1.
Diagram of the excision assay and results from three A. gambiae cell lines (4a-2, Sua 4.0, and Sua 5.1*) as well as the mbn2 hemocyte cell line of D. melanogaster (25), transformed with donor plasmid with (+) or without (−) a transposase-producing plasmid. Excision results in a DNA circle from sequences outside the Minos ends. Primers annealing to sequences within the circle (black arrows) can drive a PCR, resulting in a 167-bp excision band in + but not in − lanes. Lane C shows a control PCR without template. The doublet of bands at ca. 400 bp is derived from ectopic priming from donor plasmid sequences and is competed by the specific priming (compare lanes + and −).
Mobility of Minos Element in A. gambiae Sua 4.0 Cells.
In an initial attempt to introduce Minos element insertions in the mosquito cell genome, the Sua 4.0 cell line was transfected with plasmid pMinHyg in the presence of the helper plasmid pHSS6ILM20. The plasmid pMinHyg (Fig. 2A) was derived from the previously described Minos vector pMiNot (2) and was marked with an Hsp70-hygromycin B phosphotransferase fusion gene. Stably transformed cells were selected by using hygromycin, and genomic DNA was isolated from eight different clones. Southern analysis of the DNA, digested with EcoRI and NotI and hybridized with the probe INT (data not shown), indicated the presence of the MinHyg transposon sequences in the genome of all the cell clones except for B12. This clone probably developed spontaneous resistance to the antibiotic and was included as negative control in the further analysis. The transposon sequences in the other clones could have been integrated in the genome of the cells either by a transposase-mediated mechanism or by random integration of the plasmid, which is not uncommon in cultured cells.
Figure 2.
(A) Schematic representation of plasmid pMinHyg used for genetic transformation of Sua 4.0 cells. The hatched boxes represent Minos sequences located internally to the Minos ends (boxed arrowheads). Minos sequences were interrupted by the insertion of an Hsp70-hygromycin B phosphotransferase fusion gene (Hsp70-Hyg), and a stuffer DNA sequence that includes a 2.6-kb fragment of the D. melanogaster act5C promoter and a 0.8-kb fragment of the Hsp70 terminator (marked as A–G). The ampicillin resistance gene derived from plasmid pTZ18R also is indicated. The black bars delineate the probes used for Southern analysis. N, NotI; E, EcoRI; H, HincII. (B) Southern analysis of HincII-digested genomic DNA of the Sua 4.0 clones hybridized with probe M. B1, C1, C2, B11, B12, C12, F1, and D2 represent eight different hygromycin-resistant clones of Sua 4.0 cells, transfected with a mixture of the pMiHyg plasmid and the helper plasmid pHSS6hsMi20. The probe verifies the presence of Minos transposon sequences in the genomic DNA of these stable tranfected cells. Clone B12 is a spontaneously resistant clone that lacks insertions and is included as a negative control. The arrowheads indicate bands of size and intensity consistent with expectation for illegitimate integrations of pMiHyg plasmid concatamers in the chromosomes of the cells. * indicates a band expected for concatamers of the helper plasmid. (C) Southern analysis of the same clones digested with HincII restriction enzyme and probed with probe Amp. A 2.9-kb band indicates the presence of sequences derived from the plasmid backbone in all of the clones except F1.
To define the molecular structure of the pMinHyg insertions, genomic DNA from the transformed cells was digested with HincII and hybridized with probe M, which encompasses the left (ML) and right (MR) Minos arms. HincII sites are present several times inside the transposon and in the backbone of plasmid pMinHyg, just outside both the left and right IRs of Minos (Fig. 2A). This Southern analysis revealed the presence of multiple bands in all clones examined except for the clone B12 (Fig. 2B). Two intense bands of approximately 0.8 and 1.3 kb (Fig. 2B, marked with arrowheads) were consistent with the expected size of bands derived from concatameric insertions of the plasmid, including vector sequences flanking the Minos ends. These bands were detected in all positive clones except C1 and F1, indicating that in most of the cell clones random integrations coexist with multiple potential transposase-induced insertions. Within the limits of Southern blots the similarities in the hybridization patterns of B1, C2, and C12 indicated a common origin of these cell clones. In contrast the hybridization patterns of C1 and F1 were quite different and clone specific. It is also noteworthy that an approximately 4-kb intense band (Fig. 2B, marked by *) migrating at the predictable size for a HincII digest of the helper plasmid was detected.
The presence of sequences derived from the plasmid backbone was revealed with probe AMP: a hybridization signal of variable intensity was detected in six of the clones, confirming the presence of plasmid integrations (Fig. 2C). The variable weak band in C1 indicated a lesser extent of random recombinations in this clone relative to others. Only clone F1 did not hybridize with probe AMP, strongly suggesting that the integration events were mainly transposase mediated. The nature of these integration events was confirmed subsequently (see below).
Mobility of Minos Element in Sua 5.1* Cells.
A different strategy was devised to introduce insertions in the Anopheles Sua 5.1* cells. In this case a Minos “plasmid rescue” transposon (pLHGR), which allows the direct cloning of the insertions, was generated (Fig. 3A). Between the two ends of the Minos element an Hsp70-rsGFP fusion gene and a whole plasmid (BluescriptKSII; Stratagene) was introduced. Cells were transfected with this plasmid, with or without helper plasmid pHSS6hsMi20, and 36 h after transfection the cells were harvested and sorted in a fluorescence activated cell sorter. Before sorting the cells were subjected to a heat shock for 1 hr at 42°C to produce sufficient transposase and GFP. A similar fraction of cells was found to be fluorescent in both cell populations (transfected with or without helper plasmid), because of transient expression of GFP (data not shown). Fluorescent fractions from both populations were further expanded by culturing for 40 days. The expanded cell populations were then heat-shocked and resorted. A cell fraction with significant levels of fluorescence was present only in the sample derived from cells transfected in the presence of helper plasmid (Fig. 3B, +H), but not in the absence of helper (Fig. 3B, −H). This finding suggested that transposase-dependent stable integrations of the transposon had been introduced in the genome of these cells.
Figure 3.
(Aa) Schematic representation of the plasmid pLHGR used to transfect the Sua 5.1* cells. Between the Minos ends (boxed arrowheads), an Hsp70-GFP fusion gene and a whole Bluescript plasmid (PS) were incorporated. Outside the Minos ends, D. hydei genomic sequences and a DNA fragment corresponding to the tetracycline resistance gene generate a stuffer between the two ends. (Ab) Predicted legitimate insertion of the above transposon in genomic DNA (crosshatched lines). P, PstI; S, SacII restriction sites. Letters in parenthesis indicate the same restriction sites that may be present in the A. gambiae flanking genomic DNA. (Ac) Predicted structure of rescued plasmids corresponding to canonical Minos insertions in the genome of Sua 5.1* cells. Such plasmids should contain a common Bluescript-bearing DNA fragment (PS) linked to a variable size fragment extending into A. gambiae flanking genomic DNA. (B) Comparison of histograms of cell populations derived from Sua 5.1* cells initially transfected with plasmid pLHGR in the presence (+H) or the absence (−H) of helper plasmid. The transfected cells were expanded and resorted after 40 days posttransfection. A fluorescent cell fraction could be sorted from the +H cells. The horizontal axis corresponds to increasing fluorescence intensity expressed on a logarithmic scale. The vertical axis corresponds to cell counts per fluorescence channel. (Inset) A pair of fluorescent cells. (C) Restriction patterns of rescued plasmids representing 10 different Minos-mediated insertion events. The plasmids were digested with a PstI/SacII enzyme combination. A common Bluescript restriction band, 2.9 kb long (PS), is present in all of the plasmids, together with bands of variable size deriving from the flanking genomic DNA. A second variable band in the same lane indicates the presence of a SacII site in the flanking DNA (see Ac). In the case of E6, the second band was caused by a PstI fragment derived from the X chromosome that was trapped in the plasmid during the rescue procedure.
Genomic DNA was isolated from the resorted fluorescent fraction of Sua 5.1* cells, PstI-digested, religated under high dilution conditions, and used to transform E. coli cells. PstI cuts once inside the Hsp70-GFP gene but leaves the plasmid sequences intact. In this way the three ends of insertions including plasmid sequences together with flanking genomic sequences could be rescued (Fig. 3Ab).
Plasmid DNA from independent colonies was subjected to restriction analysis using the restriction enzymes PstI and SacII (Fig. 3C). SacII cuts once in the pLHGR sequence adjacent to MR sequence (Fig. 3A). Therefore, rescued plasmids representing Minos-mediated insertions should show a common Bluescript restriction band, 2.9 kb long, as well as a band of variable size extending into flanking genomic DNA. More than one variable band is expected if extra SacII site(s) are present in the flanking genomic sequences. This pattern was indeed observed (Fig. 3C). In contrast, rescued plasmids representing random integration of the pLHGR plasmid in the genome would include pLHGR sequences outside the Minos ends. Because two PstI sites flank the transposon ends in pLHGR (Fig. 3Aa), two bands, 2.9 kb and 370 bp long, are expected. None of 48 analyzed plasmids showed this restriction pattern (Fig. 3C, data not shown).
Insertions in Both Sua 5.1* Cells and Sua 4.0 Clone F1 Are Canonical, Minos-Dependent Integrations in Mosquito Chromosomes.
Cloning, sequence analysis, and in situ hybridization of rescued flanking genomic fragments to polytene chromosomes gave definitive evidence that the persistent transposons are indeed integrated in the mosquito genome by a legitimate transposase-dependent mechanism. Ten rescued plasmids from transfected, cultured, and resorted Sua 5.1* cells were selected for this analysis, based on their unique restriction patterns, that implied different integration events. When sequenced, using a primer annealing to the MR end, all 10 plasmids were found to represent characteristic transposase-induced insertions. The element's precise end is bordered by the TA dinucleotide where Minos is known to integrate and which it duplicates. The known D. hydei sequences immediately flanking the Minos end in pLHGR are absent, as expected for Minos-mediated insertions. Instead the Minos end is attached to variable, presumably chromosomal, sequences (Fig. 4A). The latter point was confirmed, because the rescued plasmids all hybridized to individual and different sites of A. gambiae chromosomes (Fig. 4 A and B).
Figure 4.
(A) Sequences of the Minos insertion sites in the genome of Sua 5.1* and Sua 4.0 cells. Chromosomal flanking sequences are represented with capital letters in italics. Small lettering represents the sequences of the Minos end. The expected TA dinucleotide of the insertion site is shown in bold. The chromosomal divisions and subdivisions from which the flanking sequences were derived are indicated with the chromosomal arm listed in parenthesis. (B) Typical results of determining the location of origin of the rescued genomic fragments by in situ localization to polytene chromosomes of the Suakoko mosquito strain.
In parallel, insertions in the Sua 4.0 cell clone F1 were recovered from a genomic library, constructed from EcoRI-digested F1 chromosomal DNA and screened with the M probe. As EcoRI cuts twice within the pMiHyg transposon (Fig. 2A), the two arms of Minos, ML and MR, only could be cloned separately. Sequencing of four isolated clones by using primers annealing to the Minos IRs documented transposase-mediated events (Fig. 4A). Again, Minos ends were adjacent to the diagnostic TA dinucleotide, which was followed by variable sequences. Hybridization to polytene chromosomes showed that flanking sequences were chromosomal and corresponded to widely scattered genomic locations (Fig. 4A).
In all, we documented 14 different canonical Minos insertions in four of five polytene chromosomal arms of the mosquito genome. We were not able to detect by blast analysis any homology of the flanking sequences to sequences deposited in the databanks except for the E9 insertion. In this case a significant similarity of the host-derived sequence to the sequence of the nonmuscle myosin II heavy chain was detected.
Minos Activity in A. stephensi Embryos.
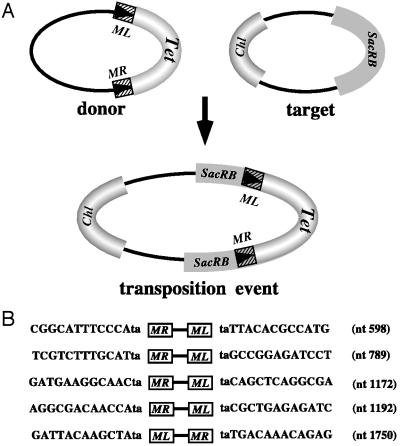
The ability of Minos to transpose in A. stephensi embryos also was assessed, by applying an interplasmid transposition assay essentially as described (16). This species was chosen because it grows more robustly and the embryos are more resistant to manipulations than the A. gambiae embryos. Freshly laid embryos were injected with a DNA mixture containing the helper pHSS6hsILMi20, the donor pMiLRTetR(L), and the target pBCSacRB plasmids at concentrations of 100/200/400 μg/ml, respectively. In total 216 embryos were injected, and the injection procedure was much facilitated by pretreating the embryos with p-nitrophenyl p-guanidinobenzoate, which delayed the hardening of the vitelline membrane and chorion. The target plasmid contains the Bacillus subtilis SacRB gene and the donor plasmid contains a Minos transposon marked with the tetracycline-resistance gene (Fig. 5). Transpositions of the marked transposon to the target plasmid, which disrupt the function of the SacRB gene, can be selected, by using a triple selection scheme (chloramphenicol, sucrose, and tetracycline).
Figure 5.
(A) Schematic representation of interplasmid transposition events. Transposition of a Minos transposon from the donor plasmid into the sucrase gene carried by the target plasmid results in a combined plasmid that can be selected by using a triple selection scheme (Chl, chloramphenicol; Tet, tetracycline; SacRB, sucrase). (B) Sequences flanking the ends of Minos insertions in the sucrase gene, in events that occurred in A. stephensi embryos. The nucleotide positions in the sucrase gene (Genbank accession nos. XO2730 and KO1987), at which the insertion took place are indicated. The orientation of the insertion relative to the sucrase sequence also is indicated (ML-MR or MR-ML).
Five such independent interplasmid transposition events were isolated among 36,000 donor plasmids recovered after transformation of low molecular weight DNA derived from injected embryos. Thus the frequency of transposition was estimated as 1.3 × 10−4. When the same assay was applied in Drosophila embryos a transposition frequency of 2.3 × 10−3 was observed, and in Aedes aegypti Mos20 cells the recorded frequency was 8.7 × 10−5 (16).
Transposition events were not recorded in the absence of helper plasmid, indicating that active transposase enzyme is required. Sequencing of all five interplasmid transposition events again revealed the characteristic features of transposase-mediated integrations: the transposed sequences were delimited by the Minos IRs, and the insertions occurred at TA dinucleotides that were duplicated on insertion. The sequences of Minos insertion sites in the sucrase gene are indicated in Fig. 5B. Two of the TA dinucleotide sites were hit by the element also in previous experiments and one of them (nucleotide position 1172) has been described as a hot spot (16). The other three insertions (nucleotide positions 598, 1192, and 1750) were first recovered in the present experiment.
Discussion
The present study has established that Minos transposase is active in Anopheles cells and embryos, where it can mediate canonical DNA excisions and integrations. As such, it represents an important step toward the establishment of Minos-based transformation methods, for cell lines as well as the germ line of the malaria vector mosquitoes.
Our initial experiments in A. stephensi embryos demonstrated that the injected plasmids are stable in this environment, that transposase can be produced from the helper plasmid, and that it can mediate transposition of Minos-based transposons at frequencies comparable to those reported previously with this assay in other species. Because of the difficulty of injecting anopheline eggs, it would be helpful to increase further the Minos transposition frequency as monitored by the interplasmid transposition assay. Potential improvements include the use of alternative promoters, as well as in vitro-transcribed transposase RNA or transposase protein, (thus circumventing the question of optimizing promoters). The heat shock promoter is evidently workable, but heat shock may decrease the viability of the injected embryos. Leaky Hsp70 promoter activity has been used for Minos transformation of C. capitata (2) as well as in the Sua 4.0 cells (this report).
The present study also has validated two markers for use in cell transformation: hygromycin and GFP. Their usefulness for anopheline embryos remains to be tested, but the success of GFP as a marker for other insect species (15) is encouraging.
If we assume that cell lines Sua 4.0 and 5.1* are comparable in susceptibility to transformation, the results with transposon pLHGR are particularly satisfactory. We were able to recover multiple canonical insertions without any indication of random integrations, unlike the case of transposon pMinHyg where numerous putative concatamer integrations were observed. We suspect that selection of random integrations is partly caused by the antibiotic pressure, as has been observed in other stable transformed cell lines (22, 23). It is also possible that increasing the levels of transposase by heat shock (as was done in the pLHGR but not pMinHyg experiment) may shift the ratio of random recombinations vs. transposase-mediated events in favor of the latter. In the case of P element, control of transposase-mediated insertions was documented previously in Drosophila Kc cells (24). We expect that use of RNA as a transposase source (which is possible in these mosquito cells; T.G.L., unpublished observations) will permit finer control of the rate of integration. Control of the number of Minos insertions per cell genome should lead to a series of applications. For example, a promoter strength in the genomic environment would be quickly testable. In addition, expression cDNA libraries directly constructed in Minos cell transformation vectors could be functionally screened by expression in the mosquito cell lines. Furthermore, the use of plasmid rescue Minos transposons as mutagenesis agents in mosquito cells would permit the generation of enhancer/gene trap and transposon-tagged lines. Such lines could permit identification of genes that respond to the presence of Plasmodium parasites or chemical and environmental stimuli. Efficient application of the above methodologies also will require the employment of techniques for efficient cloning of individual transformed cell clones and for massive screens that are routinely applied to other cells, including insect cells. Such applications will greatly enhance the value of the available mosquito cell lines (ref. 17 and this report) for molecular genetic studies, offering a reliable and widely useable experimental system as an important complementary tool even when germ-line transformation is achieved.
Acknowledgments
We thank Dr C. Savakis for sharing results before publication. T.G.L. thanks B. Miñana for helping with the sequencing and A. Atzberger (Biochemical Instrumentation Program at the European Molecular Biology Laboratory) for introducing him to FACS analysis and for help with cell sorting. This research was supported by a Network Grant from the Training and Mobility of Researchers Program of the European Community, the John D. and Catherine T. MacArthur Foundation, and Grant SFB 544 from the Deutsche Forschungsgemeinschaft. T.G.L. was supported by an individual fellowship (BIO4-CT97–5065) from the Biotechnology Research and Technological Development Programme of the European Union.
Abbreviations
- GFP
green fluorescent protein
- IR
inverted repeat
- DHFR
dihydrofolate reductase
- ML
Minos left
- MR
Minos right
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.040568397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.040568397
References
- 1.Miller L H, Sakai R K, Romans P, Gwadz R W, Kantoff P, Coon H G. Science. 1987;237:779–781. doi: 10.1126/science.3039658. [DOI] [PubMed] [Google Scholar]
- 2.Loukeris T G, Livadaras I, Arca B, Zabalou S, Savakis C. Science. 1995;270:2002–2005. doi: 10.1126/science.270.5244.2002. [DOI] [PubMed] [Google Scholar]
- 3.Coates C J, Jasinskiene N, Miyashiro L, James A A. Proc Natl Acad Sci USA. 1998;95:3748–3751. doi: 10.1073/pnas.95.7.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jasinskiene N, Coates C J, Benedict M Q, Cornel A J, Rafferty C S, James A A, Collins F H. Proc Natl Acad Sci USA. 1998;95:3743–3747. doi: 10.1073/pnas.95.7.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lidholm D A, Lohe A R, Hartl D L. Genetics. 1993;134:859–868. doi: 10.1093/genetics/134.3.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O'Brochta D A, Warren W D, Saville K J, Atkinson P W. Genetics. 1996;142:907–914. doi: 10.1093/genetics/142.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lohe A R, Hartl D L. Genetics. 1996;143:365–374. doi: 10.1093/genetics/143.1.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lozovskaya E R, Nurminsky D I, Hartl D L, Sullivan D T. Genetics. 1996;142:173–177. doi: 10.1093/genetics/142.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Handler A M, McCombs S D, Fraser M J, Saul S H. Proc Natl Acad Sci USA. 1998;95:7520–7525. doi: 10.1073/pnas.95.13.7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Brochta D A, Warren W D, Saville K J, Attkinson P W. Mol Gen Genet. 1994;244:9–14. doi: 10.1007/BF00280181. [DOI] [PubMed] [Google Scholar]
- 11.Sarkar A, Coates C J, Whyard S, Willhoeft U, Atkinson P W, O'Brochta D A. Genetica. 1997;99:15–29. doi: 10.1007/BF02259495. [DOI] [PubMed] [Google Scholar]
- 12.Sarkar A, Yardley K, Atkinson P W, James A A, O'Brochta D A. Insect Biochem Mol Biol. 1997;27:359–363. doi: 10.1016/s0965-1748(97)00018-0. [DOI] [PubMed] [Google Scholar]
- 13.Coates C J, Turney C L, Frommer M, O'Brochta D A, Atkinson P W. Mol Gen Genet. 1997;253:728–733. doi: 10.1007/s004380050377. [DOI] [PubMed] [Google Scholar]
- 14.Lobo N, Li X, Fraser M J., Jr Mol Gen Genet. 1999;261:803–810. doi: 10.1007/s004380050024. [DOI] [PubMed] [Google Scholar]
- 15.Berghammer A J, Klingler M, Wimmer E A. Nature (London) 1999;402:370–371. doi: 10.1038/46463. [DOI] [PubMed] [Google Scholar]
- 16.Klinakis, A. G., Loukeris, T. G., Pavlopoulos, A. & Savakis, C. (2000) Insect Mol. Biol., in press. [DOI] [PubMed]
- 17.Muller H M, Dimopoulos G, Blass C, Kafatos F C. J Biol Chem. 1999;274:11727–11735. doi: 10.1074/jbc.274.17.11727. [DOI] [PubMed] [Google Scholar]
- 18.Bieber A J. Methods Cell Biol. 1994;44:683–696. doi: 10.1016/s0091-679x(08)60938-3. [DOI] [PubMed] [Google Scholar]
- 19.Holmes D S, Bonner J. Biochemistry. 1973;12:2330–2338. doi: 10.1021/bi00736a023. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 21.Kumar V, Collins F H. Insect Mol Biol. 1994;3:41–47. doi: 10.1111/j.1365-2583.1994.tb00149.x. [DOI] [PubMed] [Google Scholar]
- 22.Monroe T J, Muhlmann-Diaz M C, Kovach M J, Carlson J O, Bedford J S, Beaty B J. Proc Natl Acad Sci USA. 1992;89:5725–5729. doi: 10.1073/pnas.89.13.5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lycett G J, Crampton J M. Gene. 1993;136:129–136. doi: 10.1016/0378-1119(93)90456-d. [DOI] [PubMed] [Google Scholar]
- 24.Segal D, Cherbas L, Cherbas P. Somat Cell Mol Genet. 1996;22:159–165. doi: 10.1007/BF02369906. [DOI] [PubMed] [Google Scholar]
- 25.Gateff E, Gissmann L, Shrestha R, Plus N, Pfister H, Schroder J, Zur Hausen H. In: Invertebrate Systems In Vitro. Kurstak E, Maramorosch K, Dubendorfer A, editors. Amsterdam: Elsevier; 1980. pp. 517–533. [Google Scholar]