Abstract
Some mammalian species show an ability to discriminate between different lipopolysaccharide (LPS) partial structures (for example, lipid A and its congener LA-14-PP, which lacks secondary acyl chains), whereas others do not. Using a novel genetic complementation system involving the transduction of immortalized macrophages from genetically unresponsive C3H/HeJ mice, we now have shown that the species-dependent discrimination between intact LPS and tetra-acyl LPS partial structures is fully attributable to the species origin of Toll-like receptor 4 (Tlr4), an essential membrane-spanning component of the mammalian LPS sensor. Because Tlr4 interprets the chemical structure of an LPS molecule, we conclude that LPS must achieve close physical proximity with Tlr4 in the course of signal transduction.
Keywords: endotoxin, macrophage, C3H, HeJ, specificity
Different mammalian species show distinguishable responses to lipopolysaccharide (LPS) partial structures, in which secondary acyl chains are absent, leaving only the four primary acyl chains attached to the disaccharide backbone (tetra-acyl LPS). For example, it has been observed that tetra-acyl LPS molecules [i.e., enzymatically deacylated LPS (dLPS) or synthetic tetra-acyl lipid A molecules] (1–3) are nontoxic in the dermal Shwartzman assay in rabbits. Yet, dLPS was shown to retain immunostimulatory activity in mouse splenocyte proliferation assays (1). Further, mice (and mouse macrophages) are known to respond quite strongly to tetra-acyl molecules (4).
Human neutrophils (5), monocytes (6), and endothelial cells (7) also fail to respond to tetra-acyl LPS congeners. Indeed, dLPS acts as a competitive antagonist of intact LPS in human cellular assays (5, 7). As would be predicted, synthetic tetra-acyl lipid A structures (variously referred to as “LA-14-PP,” “lipid IVa,” or “preparation 406”) also antagonize the effects of LPS in human cellular systems (8, 9).
LPS is known to be engaged by CD14 on the surface of monocytic cells (10), and it is widely believed that this is the first step in the LPS signal transduction pathway. However, the species dependency of responsiveness to dLPS is not explained by interspecific differences in CD14 structure: both human and mouse CD14 are capable of binding dLPS (11, 12), and transfection-based analyses do not support the hypothesis that CD14 is capable of discriminating between LPS and its tetra-acyl counterpart, at least with respect to the signal that emanates from the interaction (13). Hence, an explanation for interspecific differences in dLPS activity must be sought at a more distal point in the LPS signaling pathway.
The positional cloning of the mouse Lps locus and establishment of its identity to Toll-like receptor 4 (Tlr4) (14, 15) have opened the way for inquiry into many of the biological puzzles associated with bacterial endotoxin (LPS). It now appears that Tlr4 constitutes the membrane-spanning subunit of the LPS receptor. As such, it is logical to regard Tlr4 as a potential molecular substratum for discrimination between LPS partial structures. If, indeed, interspecific differences in Tlr4 structure determine the nature of the response to different molecular forms of LPS, it would be reasonable to infer that Tlr4 and LPS are, at one point in the signaling process, in extremely close proximity with one another. However, to specifically assay transduction via Tlr4, it is necessary to devise a system in which Tlr4 of a defined structure (e.g., human Tlr4 vs. mouse Tlr4) is required to permit signal transduction to a biologically relevant endpoint.
Macrophages are of central importance to the LPS response (16, 17). They are the principal source of TNF elicited by LPS in vivo (18). TNF, in turn, contributes to the lethal effect of LPS (19). Hence, macrophages are the appropriate target for the study of LPS action and TNF production has been established clearly as a biologically relevant endpoint of the LPS response. The development of an efficient protocol for the immortalization of mouse macrophages has permitted the production of diploid macrophage lines from myeloid precursors (20–22). Hence, it was feasible to immortalize macrophages from mice of the C3H/HeJ strain, which bear a point mutation in Tlr4. The resultant amino acid substitution in the cytoplasmic domain of the protein (P712H) abolishes LPS signaling in these cells, which otherwise might be presumed competent to respond to LPS. As described below, it is possible to complement the defect in immortalized C3H/HeJ macrophages through retrovirus-mediated expression of normal mouse or human Tlr4.
Materials and Methods
Immortalization of C3H/HeJ Macrophages.
Primary cultures from thymus of C3H/HeJ newborn mice were plated at a density of 106 cells/ml. The cells were grown in RPMI 1640 medium (Sigma) supplemented with 2 mM glutamine, 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin. To immortalize macrophages, adherent cells from the primary cultures were infected with VN11 retroviruses carrying the v-myc oncogene of the avian retrovirus MH2 (23), which have been shown to immortalize mouse macrophages (20). Three to 4 weeks after infection, the growing cultures were split, established as continuous cell lines, and cloned, so that a single clonal derivative was used in these studies. The immortalized line was designated MTC3H/HeJ.
Flow Cytometric Analysis of Immortalized MTC3H/HeJ Cells.
Cells (5 × 105 per sample) were washed in PBS containing 2% FBS, 2 mM EDTA, and 0.01% sodium azide. They then were incubated for 30 min at 4°C with supernatant from the anti-FcγII/III hybridoma 2.4G2 to block nonspecific binding of immunoglobulins. They then were incubated with mAbs against the indicated surface determinants (purchased from PharMingen). Phycoerythrin-conjugated streptavidin (Sigma) and FITC-conjugated goat anti-rat Ig (Southern Biotechnology Associates) were used as secondary reagents. Analyses were performed on a FACScan machine, and analysis was performed by using cell quest software (Becton Dickinson).
PINCO Transduction System.
The PINCO vector, in which both an insertable foreign sequence and a green fluorescent protein (GFP) marker protein are expressed under the influence of separate promoters (24), was obtained from Gary Nolan (Stanford University, Stanford, CA). The 293 cell-derived amphotropic packaging cell line Phoenix Ampho was obtained, with Nolan's permission, from the American Type Culture Collection. The unmodified human and mouse (C3H/HeN) Tlr4 cDNAs were amplified from cloned cDNAs by PCR. For the human Tlr4 cDNA, the primers (5′ → 3′) ATA GGA TCC ACC ATG GTG TCT GCC TCG CGC CTG GCT G (upstream) and ATA GGA TCC TCA GAT AGA TGT TGC TTC CTG CCA ATT G (downstream) were used. For the mouse Tlr4 cDNA, the primers (5′ → 3′) ATA GGA TCC ACC ATG GTG CCT CCC TGG CTC CTG GCT A (upstream) and ATA GGA TCC TCA GGT CCA AGT TGC CGT TTC TTG (downstream) were used.
The amplification products first were cloned into the vector pT7blue-3 and then excised with BamHI and transferred to the solitary BglII site of PINCO. The orientation and integrity of each construct was verified by DNA sequencing.
The Tlr4 constructs, or empty PINCO vector, then were used to transfect the packaging line. Transfection was accomplished by using the Fugene reagent (obtained from Boehringer Mannheim). Approximately 30% of the cells were transduced, as assessed by fluorescence microscopy. The fluorescent population was enriched by two cycles of cell sorting to produce a pure producer line, secreting infectious retroviral particles.
MTC3H/HeJ macrophages (106 cells in a 6-cm plate), grown and maintained in DMEM supplemented with 10% FBS, were transduced by adding polybrene to 12-hr-conditioned medium from a confluent monolayer of the producer line at a concentration of 1 μg/ml and immediately overlaying the target cell population with this medium. After 120 min of exposure to the retrovirus at 37°C, the cells again were placed in normal medium and permitted to recover. The cells were enriched to yield a uniform population of transfectants by repeated cycles of cell sorting.
Fluorescence Microscopy.
Photographs of transfected MTC3H/HeJ cells were taken by using a confocal fluorescence microscope (MRC1024; Bio-Rad). Cells were attached to coverslips by using polylysine coating for this purpose; no fixation was used.
LPS, Lipid A, and LA-14-PP.
LPS (Escherichia coli strain 0127:B8) was obtained from Sigma. Synthetic lipid A and LA-14-PP were obtained from ICN. Each compound was maintained in solution at a concentration of 1 mg/ml in physiological saline.
Induction of Macrophages by LPS, Lipid A, and LA-14-PP.
MTC3H/HeJ macrophages transduced with each construct were plated at a density of 106 cells per well in 24-well plates. After allowing cells to adhere to the plastic, stimulation was carried out by adding each compound over the specified range of concentrations (1 ng/ml to 1 μg/ml). The cells were allowed to incubate in LPS for 15 hr. Medium then was harvested and tumor necrosis factor (TNF) accumulation was measured by biological assay.
TNF Bioassay.
L-929 cells (obtained from the American Type Culture Collection) were plated at a density of 80,000 cells per well in 96-well plates. Assay of TNF activity was carried out precisely as described elsewhere (25, 26), and results were calculated with reference to a standard curve that was generated by using recombinant mouse TNF. TNF content of macrophage medium was determined by means of nonlinear interpolation, using the program prism 3.0.
Northern Blot Analysis.
Cells containing each of the two Tlr4 constructs and cells transduced with the empty PINCO vector were grown to confluence in 10-cm plates and harvested by trypsinization. Total cellular RNA was prepared by using a variant of the standard Nonidet P-40 lysis method (27), in which heparin was employed to block endogenous RNase activity, and nuclear sedimentation was used to remove genomic DNA. Cytoplasmic RNA was subjected to electrophoresis in a 1.2% agarose gel in the presence of formaldehyde and transferred to nylon for hybridization. As a probe, a random-primed mouse Tlr4 sequence, representing the bulk of the coding region in the third exon of the gene, was used. Hybridization was performed by using ExpressHyb solution (CLONTECH), with 30 min of prehybridization and 1 hr of hybridization at 65°C. The membrane was washed once at room temperature in 1× SSC/0.1% SDS and once at 70°C in 0.1× SSC/0.1% SDS. It then was exposed to film with intensifying screens at −70°C.
Miscellaneous Reagents.
Polybreen was obtained from Sigma, crystal violet was obtained from Fluka, and all tissue culture reagents were obtained from GIBCO.
Results
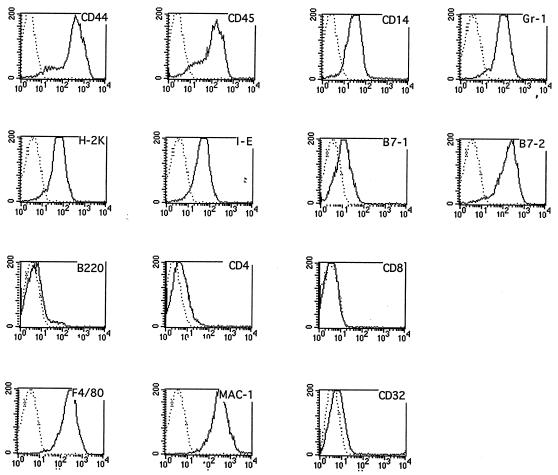
MTC3H/HeJ cells were analyzed by flow cytometry to assess their expression of characteristic macrophage markers (Fig. 1). The cells essentially were negative for B220 and CD8, excluding a B lymphoid or cytotoxic T lymphoid phenotype. Class II (I-E) MHC antigen, F4/80, B7–1, B7–2, MAC-1, Gr-1, and CD14 all were detected on these cells, consistent with a macrophage phenotype. Moreover, the cells were adherent to plastic and were morphologically similar to macrophages. The cells were karyotyped and found to be diploid (not shown). Hence, they had characteristics suggestive of primary macrophages, but were immortal, with a doubling time of approximately 1 day.
Figure 1.
Phenotypic characterization of the myc-immortalized MTC3H/HeJ macrophage line by flow cytometric analysis. Fourteen antigenic determinants were surveyed by using a panel of monoclonal reagents that distinguish T, B, and macrophage cell lineages. Open histograms show binding of specific antibodies, whereas isotype-matched control antibodies or secondary reagents are represented by dotted histograms. Abscissa, for all graphs: fluorescence intensity (logarithmic scale). Ordinate, for all graphs: cell number.
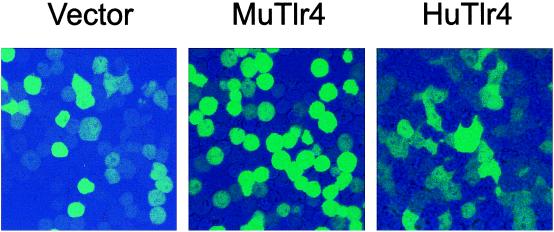
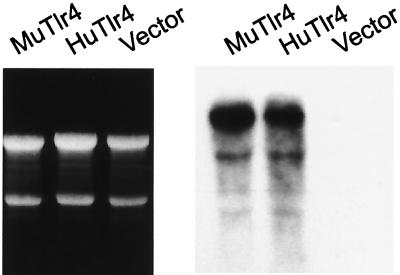
Approximately 5% of each macrophage population was transduced, as assessed by fluorescence microscopy 2 days after retroviral infection. After two cycles of cell sorting, nearly 100% of each population was found to be transduced (Fig. 2). The morphology of the cells transduced to express mouse Tlr4 was readily distinguishable from that of cells transduced to express human Tlr4. Whereas cells expressing the mouse Tlr4 protein were rounded (and essentially identical to the nontransfected culture), cells expressing the human Tlr4 protein were flattened, scalloped, and moderately vacuolated. The transduction process yielded high expression of both human and mouse Tlr4 mRNA, readily visualized with a 5-hr exposure to film (Fig. 3). The probe derived from the murine sequence was suitable for detecting the mRNA of both species (as was a probe based on the human sequence; not shown). Notably, the endogenous Tlr4 mRNA was not visualized on brief exposure, indicating that it was expressed at far lower levels than the recombinant mRNA.
Figure 2.
Pure populations of retrovirally transduced MTC3H/HeJ cells expressing human or mouse Tlr4. Cells were enriched by two cycles of cell sorting, based on GFP expression originating from the bicistronic PINCO vector. More than 99% of the population carries the transducing retroviral vector as assessed by fluorescence microscopy. (×400.) Fluorescence intensity was adjusted so that only the most intensely fluorescent cells are visible. Note the difference in morphology between cells transduced to express the mouse protein (Center) as compared with the human protein (Right).
Figure 3.
Expression of human and mouse Tlr4 mRNAs in MTC3H/HeJ cells transduced with each cDNA. Northern blot was probed with the mouse Tlr4 cDNA. RNA was obtained from cells transfected with the mouse Tlr4 cDNA, the human Tlr4 cDNA, or the PINCO vector as indicated and stained with ethidium bromide (Left). The blot was exposed to film for 5 hr to produce the autoradiogram (Right). Under these conditions, only the recombinant mRNA (which is present in great excess over the native Tlr4 mRNA) is visualized.
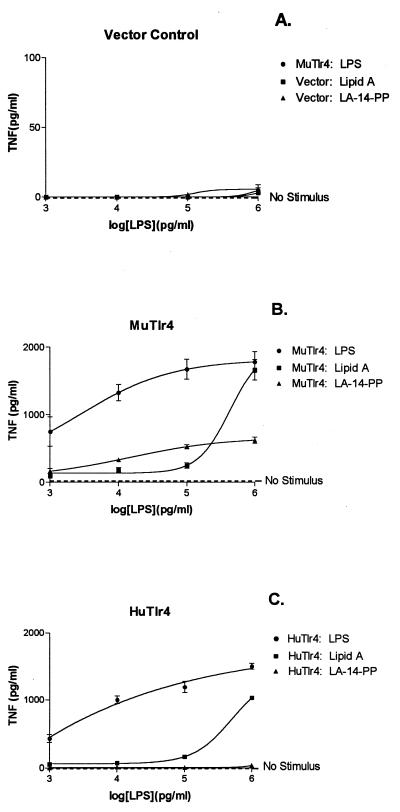
Tests of LPS responsiveness were carried out by using cells transduced with the empty PINCO vector (control) and cells transduced with the human and mouse Tlr4 constructs. LPS response was assayed over a 1,000-fold range of concentrations. Normal (acylated) E. coli LPS, synthetic lipid A, and LA-14-PP were used as stimuli (Fig. 4).
Figure 4.
Responses of MTC3H/HeJ cells to LPS and LPS partial structures: effect of Tlr4 species origin. (A) MTC3H/HeJ cells transfected with vector control. (B) MTC3H/HeJ cells transfected to express mouse Tlr4. (C) MTC3H/HeJ cells transfected to express human Tlr4. Each point represents the mean of four TNF assays, and error bars reflect SD. Curves were drawn to fit a one-site hyperbola model by using prism 3.0. Note that the y axis in A is drawn to a different scale than in B and C to highlight the complete nonresponsiveness of the control-transfected cells. The dashed line parallel to the abscissa indicates the level of TNF produced without the addition of any ligand (No Stimulus).
MTC3H/HeJ macrophages transfected with the empty PINCO vector did not respond to any agonist preparation at any of the concentrations used (Fig. 4A). Both mouse Tlr4 and human Tlr4 expression conferred strong responsiveness to LPS and to synthetic lipid A (Fig. 4 B and C). Indeed, the level of TNF production witnessed in these cells, previously entirely refractory to LPS, was quite comparable to that observed in cultures of LPS-induced RAW 264.7 macrophages (X. Du, A.P., M. Silva, and B.B., unpublished results) (26). Although no significant TNF production occurred in the absence of an inducing stimulus (Fig. 4, dashed line), very low concentrations of LPS were sufficient to yield an impressive response. Hence, in cells transfected with the mouse Tlr4 isoform, TNF accumulated to a mean concentration of 700 pg/ml after induction by LPS at a concentration of 1 ng/ml (Fig. 4B), and about 400 pg/ml was achieved in cultures transduced with the human Tlr4 isoform (Fig. 4C).
Whether human or murine Tlr4 protein was expressed, the response to lipid A was less intense than the response to intact LPS at low concentrations of agonist, but at 1 μg/ml it approached parity with LPS, much as has been reported previously and ascribed to the physicochemical state of the agonists (28). The shape of the response curve was similar regardless of Tlr4 species origin.
Strikingly, only mouse Tlr4, and not human Tlr4, conferred responsiveness to LA-14-PP. Hence, LA-14-PP evoked impressive TNF accumulation at all concentrations used in cells transfected with mouse Tlr4. TNF was induced to a concentration of 200 pg/ml in cultures exposed to LA-14-PP at a concentration of 1 ng/ml, and a maximum response (600 pg/ml TNF) was achieved after exposure to LA-14-PP at a concentration of 1 μg/ml. However, no response could be detected in cells expressing human Tlr4 at any concentration of LA-14-PP (Fig. 4C).
Discussion
Using an immortalized macrophage line derived from C3H/HeJ mice, we have devised a system for the assay of LPS-transducing activity via the Tlr4 protein. The cells ordinarily are unable to signal the presence of LPS because they were derived from mice homozygous for the Tlr4Lps-d allele, which are highly refractory to LPS. Both human and mouse Tlr4 sequences confer strong LPS responsiveness in these cells, suggesting that both upstream (e.g., CD14) and downstream (e.g., MyD88) components of the mouse LPS-signaling pathway interact with Tlr4 to transduce a signal, irrespective of Tlr4 species origin. Hence, these cells may be used to examine the properties of human Tlr4 mutations that we have identified recently (I. Smirnova, A.P., E. K. L. Chan, E. Alejos, C. McBride, and B.B., unpublished results) and other mutations yet to be reported (I. Smirnova, A.P., E. K. L. Chan, C. McBride, and B.B., unpublished data). Facile expression of mutant Tlr4 proteins may be achieved by using the PINCO retroviral expression vector, which permits immediate enrichment of clones that express the exogenous protein.
Interestingly, the morphology of MTC3H/HeJ cells transfected with the human isoform of Tlr4 is readily distinguishable from the morphology of the same cells transfected with the mouse isoform of Tlr4. This observation might be taken to indicate that constitutive signal transduction of some type occurs when the human protein is expressed in a murine cellular context. However, no TNF production can be detected in cells expressing the human Tlr4 protein without the addition of LPS or lipid A. Hence, if signaling occurs, it is likely to involve pathways collateral to those that lead to TNF biosynthesis or, at least, not all of the pathways that are required to elicit TNF biosynthesis.
Of central importance to the present study, the Tlr4 activity assay that we have described permits an informed assessment of the physical relationship between LPS and Tlr4. A tetra-acyl congener of lipid A (LA-14-PP) does not yield productive signaling via human Tlr4, whereas lipid A does. By contrast, mouse Tlr4 transduces a signal initiated by both intact lipid A and tetra-acyl lipid A. It follows, then, that human Tlr4 (but not mouse Tlr4) is capable of discriminating between lipid A and tetra-acyl lipid A. In effect, human Tlr4 “reads” the agonist structure and assesses whether secondary acyl chains are present.
Several hypotheses might be offered to explain this phenomenon. One might envision a situation in which entirely separate signaling cascades are initiated by lipid A and its tetra-acyl counterpart, each of which would necessarily begin at the level of CD14 and ultimately converge upon Tlr4 (because, in the absence of Tlr4, no LPS-like agonist is capable of evoking a signal). However, it must be granted that the likelihood of multiple upstream pathways, all necessarily emanating from CD14 and then triggering Tlr4, is remote. More plausibly, one might postulate that physical interaction with LPS evokes a conformational change in a protein (for example, CD14) or in the membrane lipid, which, in turn, contacts Tlr4. In this scheme, it would follow that distinguishable conformational changes are evoked by lipid A and tetra-acyl lipid A and that human Tlr4 (but not mouse Tlr4) would be capable of discerning the difference between these conformers, though it would have no direct contact with the agonist itself.
With respect to the size of the molecular groups involved (the one to three secondary acyl chains present on each molecule), the “reading” of LPS structure would require exceedingly fine resolution. The exclusion of secondary acyl chains yields a minimal change in the overall dimensions of the lipid A monomer (29). Although human Tlr4 is capable of making a distinction between lipid A and tetra-acyl lipid A, mouse Tlr4—which displays approximately 70% identity to human Tlr4 at the amino acid level—is less proficient at doing so. Indeed, mouse Tlr4 confers a stronger response to LA-14-PP than to lipid A at low agonist concentrations. For these reasons, it seems most probable that LPS enters into direct physical contact with Tlr4. In this model—the simplest of the three—LPS would either be transferred to Tlr4 or would be recognized by Tlr4 as part of a ternary complex (i.e., CD14/LPS/Tlr4).
At the amino acid level, the structural difference between human and mouse Tlr4 proteins that imparts the difference in ligand specificity is not known, but it could be traced potentially through mutagenesis or gene-shuffling experiments. The likelihood that Tlr4 directly engages LPS is concordant with the observation that the Tlr4 ectodomain has been driven to high levels of polymorphism in both humans and in mice (I. Smirnova, A.P., E. K. L. Chan, E. Alejos, C. McBride, and B.B., unpublished results) and with the observation that the ectodomain of Tlr4 is less conserved among species than the cytoplasmic domain. If LPS indeed has direct contact with Tlr4, it is reasonable to believe that the structure of the protein is subject to selective pressures exerted by Gram-negative organisms bearing structurally distinct LPS molecules. Hence, recognition of a given LPS may best be served by a particular allelic form of Tlr4, whereas recognition of another LPS optimally may depend on a different Tlr4 allele. LPS recognition is known to be beneficial, insofar as mice bearing destructive mutations of Tlr4 are abnormally susceptible to Gram-negative infection (30, 31).
The plasma protein LBP appears to catalyze the transfer of LPS from solution or from micelles to CD14 (32–35), which was identified some years ago as a high-affinity ligand for LPS on the surface of mononuclear cells (10). Photoaffinity crosslinking studies (36) succeeded in identifying CD14 as an LPS ligand on the surface of mononuclear cells but have not, as yet, similarly pinpointed Tlr4. However, successful identification of Tlr4 as an LPS-binding protein is dependent on the affinity of the LPS interaction (which remains unknown), on the stability of the complex once formed, and on the number of Tlr4 molecules per cell [also unknown, but believed to be very low (Du et al., unpublished results); ref. 26]. More detailed information concerning the relationship between CD14, Tlr4, and LPS might emerge from studies of mutated forms of Tlr4, coupled with direct structural analysis of the three molecules. Insofar as physical interaction between LPS and Tlr4 seems to be critically important for LPS signal transduction to occur, Tlr4 may be regarded as an essential molecular target in strategies aimed at interdiction of endotoxic responses that occur during Gram-negative infection.
Acknowledgments
We are grateful to Dr. Robert S. Munford for his critical review of this work and for his gift of lipid A and LA-14-PP. We are also grateful to Dr. Weiping Han for his assistance with fluorescence microscopy.
Abbreviations
- LPS
lipopolysaccharide
- dLPS
deacylated LPS
- LA-14-PP
tetra-acyl lipid A
- Tlr4
toll-like receptor 4
- TNF
tumor necrosis factor
- GFP
green fluorescent protein
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.040565397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.040565397
References
- 1.Munford R S, Hall C L. Science. 1986;234:203–205. doi: 10.1126/science.3529396. [DOI] [PubMed] [Google Scholar]
- 2.Kanegasaki S, Kojima Y, Matsuura M, Homma J Y, Yamamoto A, Kumazawa Y, Tanamoto K, Yasuda T, Tsumita T, Imoto M. Eur J Biochem. 1984;143:237–242. doi: 10.1111/j.1432-1033.1984.tb08364.x. [DOI] [PubMed] [Google Scholar]
- 3.Galanos C, Lehmann V, Luderitz O, Rietschel E T, Westphal O, Brade H, Brade L, Freudenberg M A, Hansen-Hagge T, Luderitz T. Eur J Biochem. 1984;140:221–227. doi: 10.1111/j.1432-1033.1984.tb08090.x. [DOI] [PubMed] [Google Scholar]
- 4.Birkland T P, Cornwell R D, Golenbock D T, Proctor R A. Adv Exp Med Biol. 1990;256:399–402. doi: 10.1007/978-1-4757-5140-6_35. [DOI] [PubMed] [Google Scholar]
- 5.Dal Nogare A R, Yarbrough W C., Jr J Immunol. 1990;144:1404–1410. [PubMed] [Google Scholar]
- 6.Golenbock D T, Hampton R Y, Qureshi N, Takayama K, Raetz C R. J Biol Chem. 1991;266:19490–19498. [PubMed] [Google Scholar]
- 7.Pohlman T H, Munford R S, Harlan J M. J Exp Med. 1987;165:1393–1402. doi: 10.1084/jem.165.5.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loppnow H, Brade H, Durrbaum I, Dinarello C A, Kusumoto S, Rietschel E T, Flad H D. J Immunol. 1989;142:3229–3238. [PubMed] [Google Scholar]
- 9.Wang M H, Flad H D, Feist W, Brade H, Kusumoto S, Rietschel E T, Ulmer A J. Infect Immunol. 1991;59:4655–4664. doi: 10.1128/iai.59.12.4655-4664.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wright S D, Ramos R A, Tobias P S, Ulevitch R J, Mathison J C. Science. 1990;249:1431–1433. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- 11.Kitchens R L, Ulevitch R J, Munford R S. J Exp Med. 1992;176:485–494. doi: 10.1084/jem.176.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kitchens R L, Munford R S. J Biol Chem. 1995;270:9904–9910. doi: 10.1074/jbc.270.17.9904. [DOI] [PubMed] [Google Scholar]
- 13.Delude R L, Savedra R, Zhao H L, Thieringer R, Yamamoto S, Fenton M J, Golenbock D T. Proc Natl Acad Sci USA. 1995;92:9288–9292. doi: 10.1073/pnas.92.20.9288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poltorak A, Smirnova I, He X L, Liu M Y, Van Huffel C, McNally O, Birdwell D, Alejos E, Silva M, Du X, et al. Blood Cells Mol Dis. 1998;24:340–355. doi: 10.1006/bcmd.1998.0201. [DOI] [PubMed] [Google Scholar]
- 15.Poltorak A, He X, Smirnova I, Liu M-Y, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, et al. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 16.Michalek S M, Moore R N, McGhee J R, Rosenstreich D L, Mergenhagen S E. J Infect Dis. 1980;141:55–63. doi: 10.1093/infdis/141.1.55. [DOI] [PubMed] [Google Scholar]
- 17.Freudenberg M A, Keppler D, Galanos C. Infect Immunol. 1986;51:891–895. doi: 10.1128/iai.51.3.891-895.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mannel D N, Moore R N, Mergenhagen S E. Infect Immunol. 1980;30:523–530. doi: 10.1128/iai.30.2.523-530.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beutler B, Milsark I W, Cerami A. Science. 1985;229:869–871. [PubMed] [Google Scholar]
- 20.Lutz M B, Granucci F, Winzler C, Marconi G, Paglia P, Foti M, Assmann C U, Cairns L, Rescigno M, Ricciardi-Castagnoli P. J Immunol Methods. 1994;174:269–279. doi: 10.1016/0022-1759(94)90031-0. [DOI] [PubMed] [Google Scholar]
- 21.Pirami L, Stockinger B, Corradin S B, Sironi M, Sassano M, Valsasnini P, Righi M, Ricciardi-Castagnoli P. Proc Natl Acad Sci USA. 1991;88:7543–7547. doi: 10.1073/pnas.88.17.7543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Righi M, Mori L, DeLibero G, Sironi M, Biondi A, Mantovani A, Donini S D, Ricciardi-Castagnoli P. Eur J Immunol. 1989;19:1443–1448. doi: 10.1002/eji.1830190815. [DOI] [PubMed] [Google Scholar]
- 23.Righi M, Pierani A, Boglia A, De L G, Mori L, Marini V, Ricciardi-Castagnoli P. Oncogene. 1989;4:223–230. [PubMed] [Google Scholar]
- 24.Grignani F, Kinsella T, Mencarelli A, Valtieri M, Riganelli D, Lanfrancone L, Peschle C, Nolan G P, Pelicci P G. Cancer Res. 1998;58:14–19. [PubMed] [Google Scholar]
- 25.Cseh K, Beutler B. J Biol Chem. 1989;264:16256–16260. [PubMed] [Google Scholar]
- 26.Du X, Poltorak A, Silva M, Beutler B. Blood Cells Mol Dis. 1999;25:328–338. doi: 10.1006/bcmd.1999.0262. [DOI] [PubMed] [Google Scholar]
- 27.Favaloro J, Treisman R, Kamen R. Methods Enzymol. 1980;65:718. doi: 10.1016/s0076-6879(80)65070-8. [DOI] [PubMed] [Google Scholar]
- 28.Luderitz T, Brandenburg K, Seydel U, Roth A, Galanos C, Rietschel E T. Eur J Biochem. 1989;179:11–16. doi: 10.1111/j.1432-1033.1989.tb14514.x. [DOI] [PubMed] [Google Scholar]
- 29.Mukerjee P, Kastowsky M, Obst S, Takayama K. In: Lipopolysaccharide Preparations in Aqueous Media: Implications for Solution versus Suspension. Brade H, Opal S M, Vogel S N, Morrison D C, editors. Vol. 12. New York: Dekker; 1999. pp. 221–228. [Google Scholar]
- 30.O'Brien A D, Rosenstreich D L, Scher I, Campbell G H, MacDermott R P, Formal S B. J Immunol. 1980;124:20–24. [PubMed] [Google Scholar]
- 31.Rosenstreich D L, Weinblatt A C, O'Brien A D. CRC Crit Rev Immunol. 1982;3:263–330. [PubMed] [Google Scholar]
- 32.Yu B, Hailman E, Wright SD. J Clin Invest. 1997;99:315–324. doi: 10.1172/JCI119160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schromm A B, Brandenburg K, Rietschel E T, Flad H D, Carroll S F, Seydel U. FEBS Lett. 1996;399:267–271. doi: 10.1016/s0014-5793(96)01338-5. [DOI] [PubMed] [Google Scholar]
- 34.Yu B, Wright S D. J Biol Chem. 1996;271:4100–4105. doi: 10.1074/jbc.271.8.4100. [DOI] [PubMed] [Google Scholar]
- 35.Hailman E, Lichenstein H S, Wurfel M M, Miller D S, Johnson D A, Kelley M, Busse L A, Zukowski M M, Wright S D. J Exp Med. 1994;179:269–277. doi: 10.1084/jem.179.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kirkland T N, Virca G D, Kuus-Reichel T, Multer F K, Kim S Y, Ulevitch R J, Tobias P S. J Biol Chem. 1990;265:9520–9525. [PubMed] [Google Scholar]