Abstract
It is not clear whether APC mutations are sufficient for early colorectal adenomas to grow or whether additional mutations at other loci are required. We previously have screened 210 early colorectal adenomas from familial adenomatous polyposis patients for mutations and allelic loss at APC. Here, we determined whether allelic loss at APC had any effect on the nearby α-catenin gene. However, loss on 5q in familial adenomatous polyposis adenomas rarely extended as far as α-catenin, and no differences in α-catenin protein expression were found in tumors that showed loss encompassing both APC and α-catenin. We then screened all 210 tumors for mutations at candidate loci other than APC (K-ras, β-catenin, and allelic loss at 1p33-p35 and 1p36) and for microsatellite instability (MSI). Each of these loci has been implicated previously in early colorectal tumorigenesis. One tumor harbored a β-catenin mutation and another MSI, but none showed K-ras mutation or allelic loss at 1p33-p35 or 1p36. These data support the following hypotheses derived from sporadic colorectal tumors: β-catenin mutations are generally an alternative to mutations at APC, MSI is not usually an early phenomenon in colorectal tumorigenesis, and K-ras mutations are more typical of large- and moderate-sized adenomas. Contrary to some previous reports, chromosome 1p allelic loss is infrequent in very early adenomas. APC mutations are generally sufficient for colorectal tumors to grow to about 1-cm diameter, although chance mutations at other loci can provide these early colorectal adenomas with a selective advantage, and some colorectal tumors may develop along a pathway not involving APC.
It is generally accepted that tumors start to grow in the colorectum of patients with familial adenomatous polyposis (FAP) when an appropriate cell acquires a somatic mutation at the APC locus to accompany the preexisting germ-line mutation (1). We have shown that this somatic “second hit” is nonrandom, with germ-line APC mutations around codon 1300 associated with allelic loss in FAP polyps (and more severe disease) and germ-line mutations outside this region associated with truncating “second hits” (2). It is highly likely that different tumors in the same or different FAP patients grow at different rates, because all APC genotypes do not confer identical, selective advantages. Although the initiating role of APC mutations is part of most models of colorectal tumorigenesis (3), it is not clear whether biallelic APC mutations are sufficient for the growth of early colorectal adenomas or whether mutations at other loci are required even for early colorectal tumors to develop. Very few studies have analyzed early lesions of less than a few millimeters in diameter to address this question.
Although K-ras changes classically are the mutations subsequent to APC in the genetic pathway of colorectal tumorigenesis (3), several candidate genes have been reported to undergo mutation in early colorectal adenomas. It is not clear, however, that mutations at these loci are selected, let alone necessary, in the growth of very early lesions that already possess APC mutations. β-Catenin mutations, for example, appear to act as alternatives to APC mutations in some early colorectal tumors and to provide an additional, selective advantage to APC mutations in others (4, 5). Other loci reported to be mutated in early colorectal adenomas include the unidentified target(s) of allelic loss on the short arm of chromosome 1 (6–9) and the mismatch repair genes, resulting in microsatellite instability (MSI) (8).
We therefore have analyzed a set of 210 early colorectal adenomas from 35 patients (mean = 6 adenomas, median = 5, range = 1–22) from 26 families with known germ-line APC mutations mapping throughout the 5′ half of the APC gene (2). An advantage of analyzing these tumors is to minimize genetic heterogeneity, because it is entirely reasonable to assume that all tumors develop along a genetic pathway involving APC, although not all tumors have easily detectable somatic APC mutations. We have searched in these adenomas for β-catenin and K-ras mutations, microsatellite instability, and allelic loss at sites on chromosome 1p33-p35 and 1p36. We also have determined whether allelic loss at APC has any effect on the nearby α-catenin gene and expression of its protein. The results of these analyses suggest that APC mutations are sufficient for the growth of early colorectal adenomas.
Methods
Collection and Preparation of Samples.
Two hundred and ten fresh-frozen or paraffin-embedded colorectal adenomas were removed from colectomy specimens from FAP patients from St. Mark's Hospital. Tumors were visible by eye, but otherwise unselected. All patients had classical FAP [and one had attenuated FAP (AAPC)] on the basis of clinical features, family history, and a known germ-line APC mutation. Sections were cut from each adenoma and reviewed for histological features. The set of samples had a mean and median diameter of 3 mm (range, 0.5–10 mm). Tumor grade also was noted, although almost all lesions were mildly dysplastic tubular adenomas. Every tumor had a minimum of 50% neoplastic cells after microdissection, if necessary. DNA was isolated from frozen specimens by using the Qiagen Tissue Extraction Kit (Qiagen, Chatsworth, CA). DNA was extracted from paraffin-embedded tissue by incubation for 1–3 days in standard PCR buffer and proteinase K (250 μg/ml). For each patient, DNA was extracted from paired samples of normal tissue or blood to serve as a control.
Allelic Loss and MSI Analysis.
Microsatellite markers were chosen from the Location Database (http://cedar.genetics.soton.ac.uk/) to map to sites of frequent allelic loss on 1p33-p35 (D1S513) and 1p36 (D1S243). Loss at α-catenin was assessed by using microsatellite D5S2117 (also chosen from the Location Database). These markers and three previously analyzed microsatellites near APC (D5S656, D5S489, and D5S82) were used to assess MSI. For each tumor and paired normal DNA, PCR was performed by using fluorescently labeled oligonucleotides in a 25-μl reaction volume in standard PCR buffer (Perkin–Elmer). Cycling conditions were 95°C for 6 min, followed by 35 cycles of 95°C/55°C/72°C for 1 min each, and a final 10-min incubation at 72°C. Products were detected by using the Applied Biosystems 377 sequencer and results were analyzed by using genescan and genotyper software. In informative cases, allelic loss was scored if the area under either allele peak was reduced in a tumor to <50% of its normal value, relative to the other allele. MSI was scored if one or more markers showed “laddered” novel alleles typical of replication slippage in the tumor DNA.
Mutation Screening.
Exon 1 (codons 12 and 13) contains the great majority of K-ras mutations found in colorectal tumors. This exon was amplified in the PCR from all tumor DNAs (10) and was screened for K-ras mutations by direct sequencing of PCR products in forward and reverse orientations by using the Applied Biosystems 377 sequencer. All samples with possible mutations were sequenced again from a new PCR product to confirm the change. For β-catenin, mutations have been reported to cluster in exons 3 and 5, although the former probably are associated with tumors lacking APC mutations. Exons 3 and 5 of β-catenin were amplified from tumor DNA in the PCR by using the following pairs of oligonucleotides (BCATX5.F, 5′-GGTGGTTAATAAGGCTGCAGTT-3′, and BCATX5.R, 5′-ATTTTCACCAGGGCAGGAAT-3′; BCATX3.F, 5′-ATTTGATGGAGTTGGACATGGC-3′, and BCATX3.R 5′-CCAGCTACTTGTTCTTGAGTGAAGG-3′) and standard PCR conditions (Ta = 55°C in both cases). PCR products were screened for mutations by using SSCP analysis, using midi-gels and silver staining as described (11). For all samples with bandshifts, a new PCR product from that exon was sequenced in forward and reverse orientations by using the Applied Biosystems 377 sequencer.
Immunohistochemistry.
α-Catenin immunohistochemistry was performed on paraffin-embedded sections from tumors showing allelic loss at α-catenin, on five tumors not showing α-catenin loss, on five sections of “normal” FAP colon, and on five sections of colon from individuals without colon tumors by using the standard methods described by Hao et al. (12).
Results and Discussion
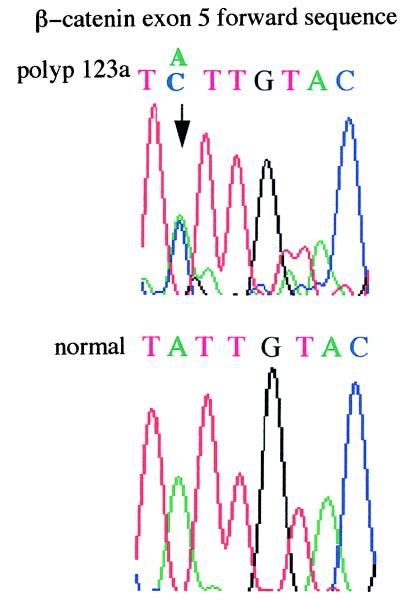
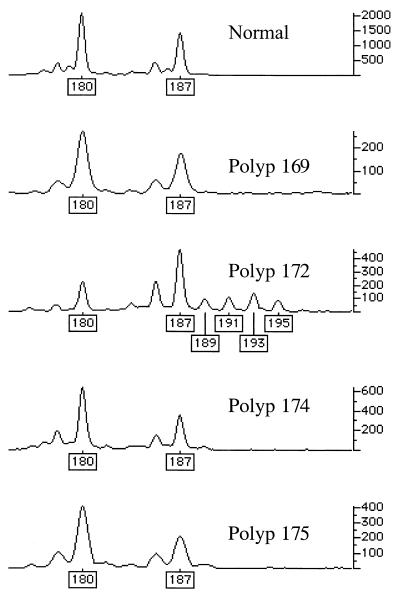
Genetic changes additional to those at APC were found infrequently in the colorectal adenomas studied (Figs. 1 and 2). One tumor (no. 123a, 6 mm, mildly dysplastic tubular adenoma), from patient 956 with an R554X germ-line APC mutation and no identified somatic mutation at APC, had an exon 5 β-catenin mutation (I198L). No tumor had an exon 3 β-catenin mutation. One tumor (no. 172, 3 mm, mildly dysplastic tubular adenoma), from a patient with a codon 1309 frameshift (del5bp) germ-line mutation and allelic loss at APC, showed microsatellite instability at D1S513. No tumor harbored a K-ras mutation or showed allelic loss at 1p33-p35 or at 1p36.
Figure 1.
I198L β-catenin mutation in tumor 123a. The A→C change is indicated by an arrow.
Figure 2.
MSI in tumor 172 at D1S513. genotyper outputs for normal tissue and four polyps from the same patient are shown. Only polyp 172 shows evidence of MSI (new alleles at sizes 189, 191, 193, and 195, and change in relative sizes of the two wild-type allele peaks).
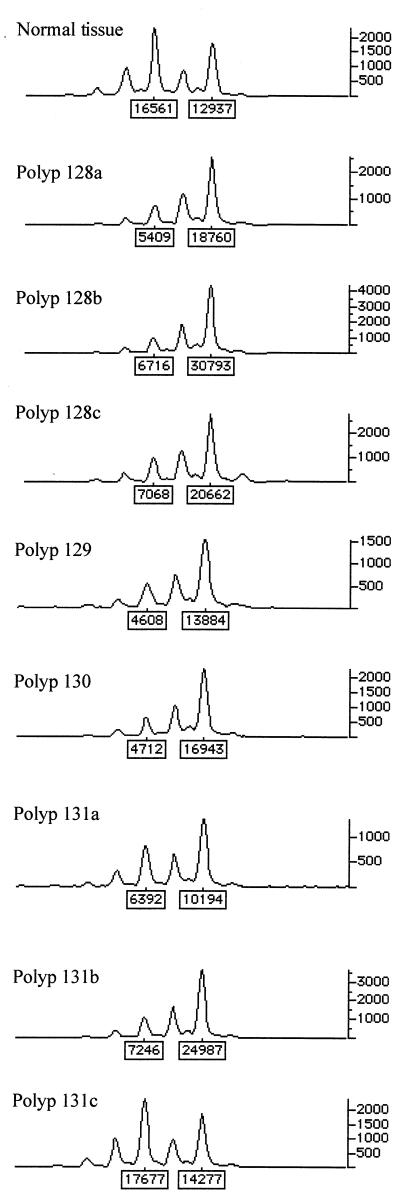
Forty-two (20%) adenomas showed allelic loss at APC. In 26 (62%) of these tumors, the region of loss was large enough to extend along chromosome 5q to involve the marker next to α-catenin (Fig. 3). Previous data have indicated that allele loss at APC in FAP polyps usually results from deletion. In no case, however, was abnormal α-catenin protein expression detected by immunohistochemistry in these tumors. Thus, despite the fact that α-catenin has a candidate role as an adhesion molecule in colorectal tumorigenesis and abnormal α-catenin expression has been found in colorectal adenomas (12), in early adenomas there appears to be neither a dosage effect caused by loss of one copy of the α-catenin gene nor mutation of the remaining wild-type allele resulting in loss of expression. These data agree with our failure to find any α-catenin mutations in a set of 38 colorectal cancer cell lines (details not shown).
Figure 3.
Allelic loss at α-catenin (D5S2117) in seven tumors from one FAP patient. genotyper outputs for normal tissue and polyps from the same patient are shown. All polyps except 131c show loss.
Our findings show that a small proportion of early colorectal adenomas from FAP patients harbors mutations that are additional to the germ-line “first hit” and somatic “second hit” at APC, which are presumed to initiate the growth of these tumors. These additional genetic changes include β-catenin mutations and MSI in addition to “third hits,” which have been described previously at APC (2, 13, 14). The mutation frequencies that we have detected probably underestimate the true frequencies for four reasons: (i) the polyclonal origin of many adenomas in FAP, (ii) contaminating normal tissue present in some tumors, (iii) the fact that our tumors were almost all very small lesions and technically difficult to study, and (iv) the existence of other uncharacterized genes selected in colorectal tumorigenesis, although the loci we have studied are the best candidates currently available. It appears likely, therefore, that mutations at other loci can be selected in early colorectal adenomas with biallelic APC mutations, although most small colorectal adenomas possess only APC mutations. Thus, even in tumors that have APC genotypes providing suboptimal selective advantages (2), the advantage conferred by their APC mutations is sufficient for that tumor to grow to a size of at least a few millimeters. (Presumably, the tumors with “fitter” genotypes have grown faster, but we cannot observe this.) As far as can be determined from this study, additional “hits” are not necessary for these tumors to initiate neoplastic growth and to undergo (at least) tens of abnormal cell divisions. We also have preliminary evidence to support this contention from comparative genomic hybridization (details not shown), which failed to detect any deletion or amplification of genetic material (apart from loss on chromosome 5q at APC) in one polyp from a patient with a codon 1309 (del5bp) germ-line APC mutation. It is not clear to what size colorectal adenomas can grow without mutations additional to “two hits” at APC.
Previous studies have found that K-ras mutations can occur in histologically normal mucosa (15) and aberrant crypt foci (16), as well as in colorectal tumors. K-ras mutations generally are detected infrequently in small adenomas and in adenomas from young patients, as we have found, but are detected in up to 50% of late adenomas. By contrast, β-catenin mutations in exon 3 have been reported to occur more frequently in small adenomas than in larger lesions, contrary to our findings. It has been hypothesized that exon 3 β-catenin mutations are alternatives to APC mutations, perhaps initiating tumorigenesis. Samowitz et al. (17) suggested that fewer adenomas with β-catenin than APC mutations progress to later-stage lesions, presumably because additional mutations are required for growth to persist. It also has been shown that β-catenin mutant tumors are also more likely to be MSI-positive. Our data showing no β-catenin exon 3 mutations in FAP adenomas mutations are in agreement with these hypotheses, because all adenomas in FAP are almost certain to possess two APC mutations and nearly all are nearly all are MSI-negative (see below). It has been proposed that exon 5 mutations in β-catenin are not initiating events in tumorigenesis (5) and, for reasons that are unclear, can be selected in addition to APC mutations in MSI-negative tumors. Again, our finding of an exon 5 β-catenin mutation supports this proposition. We have argued previously that MSI is unlikely to be an early event in colorectal tumorigenesis outside hereditary nonpolyposis colorectal cancer, given that “two hits” at APC are likely to initiate the growth of most sporadic colorectal tumors (18, 19). Because it is most probable that FAP adenomas start to grow as a result of a “second hit” at APC, our finding of low-level MSI in even one early adenoma perhaps is unexpected. MSI detection is much more sensitive—down to a level of less than 20% of the cells in a tumor sample—than most mutation screenings, so we are confident that little if any undetected MSI exists in our tumor set. The stage of colorectal tumorigenesis at which allelic loss on chromosome 1p becomes frequent is unclear, but it has been reported in early lesions of less than 1 cm (8) and it has been proposed as a very early event in colorectal tumorigenesis. 1p loss in colorectal carcinomas has been associated with higher Dukes' stage and with poorer prognosis (20). Our results show that loss at either of two reported sites on 1p loss is very rare in colorectal adenomas, at least in tumors developing along a genetic pathway initiated by APC mutations.
The growth of most sporadic colorectal cancers, and probably most colorectal adenomas, is initiated as a result of “two hits” at APC, and FAP, therefore, is a good model for the pathogenesis of most sporadic colorectal tumors. Early adenomas (mildly dysplastic lesions up to 1 cm) in the FAP colon appear to derive all their necessary selective advantage from two APC mutations, even though some APC genotypes probably provide less of an advantage than others. It is even possible that a single APC mutation can, under favorable circumstances, initiate tumor growth (21). With the exception of “third hits” at APC (2, 12, 13), subsequent events in the genetic pathway—including mutations at K-ras and in exon 5 of β-catenin, loss of chromosome 1p, and MSI—occur very infrequently in these early tumors. Selection in the FAP colon is likely to be more stringent than in the normal bowel, because competition between adenomas will be much more severe in the former case. It is likely, therefore, that most early, sporadic colorectal tumors also possess just two mutations at the APC locus, but it must be borne in mind that a minority of sporadic tumors develops along a pathway that does not involve APC and may include alternatives such as mutations in β-catenin.
Acknowledgments
We are grateful to the Equipment Park, Imperial Cancer Research Fund, for invaluable help.
Abbreviations
- FAP
familial adenomatous polyposis
- MSI
microsatellite instability
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.040564697.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.040564697
References
- 1.Miyaki M, Tanaka K, Kikuchi Yanoshita R, Muraoka M, Konishi M. Crit Rev Oncol Hematol. 1995;19:1–31. doi: 10.1016/1040-8428(94)00129-h. [DOI] [PubMed] [Google Scholar]
- 2.Lamlum H, Ilyas M, Rowan A, Clark S, Johnson V, Bell J, Frayling I, Efstathiou J, Pack K, Payne S, et al. Nat Med. 1999;5:1071–1075. doi: 10.1038/12511. [DOI] [PubMed] [Google Scholar]
- 3.Kinzler K, Vogelstein B. Cell. 1997;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 4.Morin P J, Sparks A B, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler K W. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 5.Ilyas M, Tomlinson I P M, Rowan A, Pignatelli M, Bodmer W F. Proc Natl Acad Sci USA. 1997;94:10330–10334. doi: 10.1073/pnas.94.19.10330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bardi G, Johansson B, Pandis N, Bakjensen E, Orndal C, Heim S, Mandahl N, Andren-Sandberg A, Mitelman F. Cancer. 1993;71:306–314. doi: 10.1002/1097-0142(19930115)71:2<306::aid-cncr2820710207>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka K, Yanoshita R, Konishi M, Oshimura M, Maeda Y, Mori T, Miyaki M. Oncogene. 1993;8:2253–2258. [PubMed] [Google Scholar]
- 8.Lothe R A, Andersen S N, Hofstad B, Meling G I, Peltomaki P, Heim S, Brogger A, Vatn M, Rognum T O, Borresen A L. Genes Chromosomes Cancer. 1995;14:182–188. doi: 10.1002/gcc.2870140305. [DOI] [PubMed] [Google Scholar]
- 9.Praml C, Finke L H, Herfarth C, Schlag P, Schwab M, Amler L. Oncogene. 1995;11:1357–1362. [PubMed] [Google Scholar]
- 10.Span M, Moerkerk P, De Goeij A, Arends J. Int J Cancer. 1996;69:241–245. doi: 10.1002/(SICI)1097-0215(19960621)69:3<241::AID-IJC15>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 11.Beck N E, Tomlinson I P M, Homfray T F R, Frayling I M, Hodgson S V, Bodmer W F. Gut. 1997;41:335–338. doi: 10.1136/gut.41.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hao X, Palazzo J P, Ilyas M, Tomlinson I, Talbot I C. Anticancer Res. 1997;17:2241–2247. [PubMed] [Google Scholar]
- 13.Miyaki M, Konishsi M, Kikuchi-Yanoshita R, Enomoto M, Igari T, Tanaka K, Muraoka M, Takahashi H, Amada Y, Fukayama M, et al. Cancer Res. 1994;54:3011–3020. [PubMed] [Google Scholar]
- 14.Spirio L, Samowitz W, Robertson J, Robertson M, Burt R, Leppert M, White R. Nat Genet. 1998;20:385–388. doi: 10.1038/3865. [DOI] [PubMed] [Google Scholar]
- 15.Minamoto T, Yamashita N, Ochiai A, Mai M, Sugimura T, Ronai Z, Esumi H. Cancer. 1995;75, Suppl. 6:1520–1526. doi: 10.1002/1097-0142(19950315)75:6+<1520::aid-cncr2820751523>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 16.Smith A J, Stern H S, Penner M, Hay K, Mitri A, Bapat B V, Gallinger S. Cancer Res. 1994;54:5527–5530. [PubMed] [Google Scholar]
- 17.Samowitz W, Powers M, Spirio L, Nollet F, van Roy F, Slattery M. Cancer Res. 1999;59:1442–1444. [PubMed] [Google Scholar]
- 18.Tomlinson I, Novelli M, Bodmer W. Proc Natl Acad Sci USA. 1996;93:14800–14803. doi: 10.1073/pnas.93.25.14800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Homfray T F R, Cottrell S E, Ilyas M, Rowan A, Talbot I C, Bodmer W F, Tomlinson I P M. Hum Mutat. 1998;11:114–120. doi: 10.1002/(SICI)1098-1004(1998)11:2<114::AID-HUMU3>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 20.Ogunbiyi O, Goodfellow P, Gagliardi G, Swanson P, Birnbaum E, Fleshman J, Kodner I J, Moley J F. Gastroenterology. 1997;113:761–766. doi: 10.1016/s0016-5085(97)70169-0. [DOI] [PubMed] [Google Scholar]
- 21.Bodmer W. Cytogenet Cell Genet. 1999;86:99–104. doi: 10.1159/000015360. [DOI] [PubMed] [Google Scholar]