Abstract
The incidence of variant Creutzfeldt–Jakob disease (vCJD) in the United Kingdom appears to be in decline, with only four deaths reported this year (to 6 September 2004). However, results of a survey of lymphoreticular tissues have suggested a substantially higher prevalence of vCJD than expected from the clinical data alone. There are two plausible explanations for this discrepancy: first, a proportion of those infected will not develop clinical disease (subclinical infection); and second, the genetic group in which no clinical cases of vCJD have yet occurred is susceptible. Using mathematical models for the primary transmission of bovine spongiform encephalopathy to humans, we explore the impact of these hypotheses on case predictions. Under the first hypothesis, the results suggest relatively few future cases will arise via primary transmission, but that these cases are a small proportion of those infected, with most having subclinical infection. Under the second hypothesis, results suggest a maximum fivefold increase in cases, but this hypothesis is unable to account for the discrepancy between clinical cases and the estimated prevalence. Predictions of future cases of vCJD therefore remain uncertain, particularly given the recent identification of additional cases infected via blood transfusion.
Keywords: bovine spongiform encephalopathy, epidemiology, mathematical model, projections, variant Creutzfeldt–Jakob disease
1. Introduction
The probable link between the outbreak of new variant Creutzfeldt–Jakob disease (vCJD) and the bovine spongiform encephalopathy (BSE) epidemic in cattle was established in March 1996 (Collinge et al. 1996; Will et al. 1996; Bruce et al. 1997). To date, there have been 147 confirmed cases of vCJD reported in the United Kingdom, only four of whom remain alive (http://www.cjd.ed.ac.uk/figures.htm—accessed 6 September 2004). Recent trends in the incidence of vCJD strongly suggest that the primary epidemic is in decline following the peak of 28 cases in 2000. This pattern has resulted in projections that suggest relatively small numbers of future cases (Cooper et al. 2000; Huillard d'Aignaux et al. 2001; Valleron et al. 2001; Ghani et al. 2003a). Projections are made by combining information on the BSE epidemic with assumptions about key parameters of vCJD, such as the incubation period distribution. However, considerable uncertainty remains regarding some of these assumptions, and hence about the projections themselves.
Some uncertainties in the understanding of vCJD have been highlighted by recent results from a study to detect the presence of the abnormal prion protein in appendix and tonsil tissues (Hilton et al. 2004a). The study protocol identified positive, or infected, samples by the pattern of immunohistochemical accumulation of infectious material in the lymphoreticular system (Hilton et al. 1998, 2002; Ironside et al. 2000). This technique is widely used in other studies of animal neurodegenerative diseases (Schreuder et al. 1998; O'Rourke et al. 2000, 2002), where, as with vCJD, there is little or no immune response and no reliable blood test available. In the study, three positive results were found among 12 674 admissible samples. Interpreted naïvely, this figure corresponds to a prevalence estimate of 235 per million (95% confidence interval (CI): 49–692 per million). If this interpretation is correct, it implies that the extent of the vCJD infection in the UK population is far greater than was expected given the current understanding of the disease and the number of clinical cases to date.
The discrepancy between prevalence estimates is important because it challenges some of the working assumptions made about key parameters of vCJD, which until now have been impossible to query. However, this advantage is offset somewhat by new uncertainties surrounding the diagnostic tests used to identify infected tissue samples. In particular, little is known about the sensitivity and specificity of the diagnostic tests over the course of the incubation period, or whether all positive tests (as defined by the study protocol) correspond to the same manifestation of vCJD as has been seen to date.
The purpose of this paper is to investigate the sensitivity of current model projections of the primary epidemic to biologically plausible alternative assumptions about the key disease parameters, and to uncertainties concerning the diagnostic tests. We begin by presenting some background information about the UK outbreak of vCJD, and describing the two data sets in more detail. We follow this in §2 by introducing in detail a model framework, which was originally developed to predict future cases, and its extension to consider the additional uncertainties highlighted by the survey of lymphoreticular tissues. In §3, we describe the projections obtained using the original models. In §4, we consider the implications of a carrier state for projections of the primary epidemic. In §5, we consider the implications of wider genetic susceptibility. We discuss our findings and draw conclusions in §6.
1.1 The UK epidemic
vCJD is a member of the transmissible spongiform encephalopathy (TSE) family of neurodegenerative diseases, affecting many species including humans (kuru, sporadic CJD, iatrogenic CJD), cattle (BSE) and sheep (scrapie). They are often called ‘prion’ diseases because infection results in conformational changes to host prion proteins, which result in abnormal protease-resistant prion proteins (PrPSc) that propagate and are markers for neuronal loss and spongiform changes (Pan et al. 1993; Jackson et al. 1999).
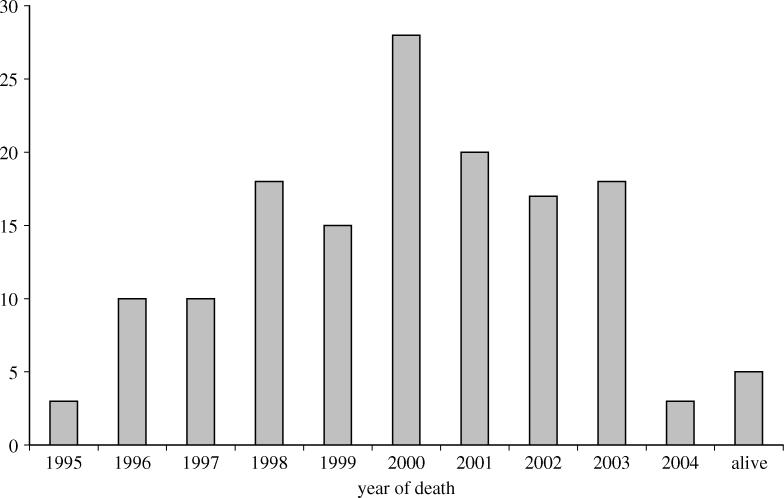
There is now substantial evidence linking vCJD to the aetiological agent causing BSE in cattle (Collinge et al. 1996; Bruce et al. 1997), with oral consumption of infected meat and meat products being hypothesized as the primary route of infection (Cooper & Bird 2002a,b, 2003). A time-series of vCJD onsets and deaths from 1995 to the current time (July 2004) is presented in figure 1. These data appear to indicate that the outbreak is in decline; the number of deaths peaked at 28 in 2000, with only four deaths so far this year (to 6 September 2004).
Figure 1.
Time series of the number of clinical cases of vCJD by date of death to the end July 2004.
The age distribution of vCJD cases has remained constant over time, suggesting that the effect of age on exposure and susceptibility for vCJD is far stronger than its effect on the incubation period (Ghani et al. 2000, 2003b). Genetic factors also play a strong part, with polymorphisms at codon 129 of the prion protein (PrP) known to influence susceptibility to infection and the length of the incubation period in other prion diseases (Alperovitch et al. 1999; Brown et al. 2000; Lee et al. 2001). To date, the 113 clinical cases that have been genotyped have been methionine (MM) homozygous at codon 129 of the PrP gene. The Caucasian population is comprised of approximately 40% MM homozygotes, 10% valine homozygotes (VV), and 50% heterozygotes (MV) (Owen et al. 1990; Collinge et al. 1991).
1.2 Results of appendix survey
Tests to detect vCJD infection prior to the onset of clinical symptoms have been recently developed, based on the pattern of PrPSc accumulation in lymphoreticular tissue (Hilton et al. 1998; Maissen et al. 2001). Two large-scale studies have been undertaken in the UK using such tests to investigate the prevalence of vCJD infection, while a further survey is underway in Switzerland. The only results currently available are from a large retrospective survey of stored tonsil and appendix tissues removed from operations in the UK between 1996 and 2000 (Hilton et al. 2004a). Under the study protocol, the survey detected three positive appendix tissues in a sample of 12 674 tissues, the majority of which were appendix tissues derived from the 10–30 age group. This translates to a detectable prevalence of 237 per million (95% CI: 49–692 per million) in this age group if we assume that the tests are 100% sensitive and specific throughout the course of the incubation period. The tests are known to be highly sensitive for individuals who have developed clinical disease: Hilton et al. (2004a) found 19 of 20 tissues from clinical cases tested positive. However, it is likely that the test sensitivity is much lower early in the incubation period, either because only small quantities of PrPSc would have accumulated in the tonsil and appendix tissues, or because PrPSc appears in tonsil and appendix tissues only towards the end of the incubation period.
2. Model framework
In this section, the models used to make projections based on the clinical cases and the results from the survey of lymphoreticular tissues are presented. The discrepancy between the two data sets, which motivates the extensions of the model presented here, is discussed in §3.
2.1 Original model
The probability that a susceptible, MM genotype individual dies from clinical disease at time u and age a is given by
| (2.1) |
and the infection prevalence among susceptible individuals at (u, a) by
| (2.2) |
where S(u, a) is the all-cause survival probability estimated from UK census data, f(u) is the incubation period distribution, β is the transmission coefficient, and
| (2.3) |
The age- and time-dependent hazard I(t, a) in equation (2.3) is given by
| (2.4) |
where v(t) is the effectiveness of control measures limiting the bovine tissues allowed into the human food supply, g(a) an age-dependent susceptibility-exposure function, Ω(z) the relative infectiousness of a bovine with BSE slaughtered at time z into its incubation period, and w(t, z) the proportion of cattle slaughtered at time t and time z from disease onset.
Further details on the assumed parametric form of the incubation period, exposure-susceptibility function, ban effectiveness, relative bovine infectiousness and slaughtered cattle have previously been published (Ghani et al. 1998, 2003a). In summary, f(u) follows a four-parameter generalized lambda distribution, g(a) is a piecewise uniform distribution with gamma-distributed tails, v(t) is a binary step-function, Ω(z) is a distribution which increases exponentially in the upper tail, and w(t, z) is obtained from recent estimates of the number of BSE-infected animals that entered the food supply over time (Ferguson et al. 2002).
To facilitate estimation, it is necessary to derive expressions for the expected values of the clinical cases (incidence) and survey data (prevalence). Under model I, the original model, the expected number of cases in the entire population aged a at time u can be written
| (2.5) |
where B(u−a) is the number of individuals born at time u−a (also obtained from UK census data), and π=0.4 is the probability of an individual having the MM genotype. Before considering the infection prevalence, it is necessary to extend equation (2.2) to incorporate a plausible scenario for the diagnostic test sensitivity. In the absence of any concrete evidence about the true test sensitivity, a simple time-dependent scenario is chosen here, following a step function where the sensitivity is 0% until the last 100τ% of the incubation period, after which it is 100σ% sensitive. For this scenario, the detectable infection prevalence can be written
| (2.6) |
where and δ(z)=1 if z is true and 0 otherwise. For example, the choice τ=σ=1 corresponds to a test with 100% sensitivity. It follows that
| (2.7) |
are the expected infection prevalence and detectable infection prevalence frequencies, respectively, of all individuals aged a at time u.
The model, and its extensions to be defined in §§2.2 and 2.3, is fitted to the clinical cases and survey data using maximum likelihood estimation, and approximate 95% confidence intervals for the parameters obtained using the profile likelihood method. See Appendix A for further details on maximum likelihood estimation and profile likelihood confidence intervals.
2.2 Inclusion of a carrier/subclinical state
To relax the assumption that all individuals will go on to develop clinical disease, an extra parameter ω is included in the model defined above, to represent the probability that an individual is infected but does not go on to develop disease (i.e. subclinical infection). Denote this by model II, the carrier model. The expected case and prevalence frequencies under model II are
| (2.8) |
and
| (2.9) |
where f(u) is now the incubation period distribution among those who do not develop subclinical infection.
The expression for the detectable infection prevalence under the carrier model requires a scenario for the test sensitivity to be specified for subclinical infections. For preclinical infections, the time-dependent sensitivity scenario in equation (2.6) follows a simple step-function allowing the diagnostic test to become more sensitive towards the end of the incubation period, which can be specified as before with the test 100σ2% sensitive in the last 100τ% of the incubation period. However, a time-dependent sensitivity function for the subclinical infections cannot be specified in this way because there is no incubation period. Instead, the test is fixed to be 100σ1% sensitive from the time of infection, where σ1 corresponds to the average sensitivity over the course of the infection period. Thus, the expected detectable infection prevalence frequency in the population is
| (2.10) |
2.3 Wider genetic susceptibility
To extend the model to allow all genotypes to be susceptible to infection, we begin by splitting the population into two groups based on whether the host is MM or non-MM homozygous () at codon 129 of the prion protein gene. It is assumed that no other genetic traits are involved in determining individual susceptibility, and that the effects on susceptibility of the MV and VV genotypes within the group are equal. It is possible to specify a more complex model of genetic heterogeneity but, without information on these genetic characteristics in the vCJD patients to date, it is not possible to constrain any generated scenarios.
We can now extend the original model to allow for differential susceptibility to infection and disease pathology by genotype. First, assume that the consumption and exposure to infected food products is the same for the as for the MM groups, implying that v(t), Ω(z), w(t, z) do not vary by genotype. The age-dependent exposure-susceptibility function from equation (2.4) can be written ; namely, the product of the susceptibility, the additional risk of consuming infectious products and mean food consumption frequency for individuals aged a (Ghani et al. 1998). The assumption that consumption of, and exposure to, infected food products is genotype-independent implies that gr(a) and gc(a) are also equal for the MM and genotypes.
Second, note that the average probability of an individual aged a being infected is βgs(a) (under a linear dose–response model and other relatively minor assumptions). We shall further assume, for the sake of parsimony in the forthcoming analysis, that the relative susceptibility gs(a) is genotype-independent, but that the transmission coefficient is not, and denote the coefficient for the group by β*. In other words, the ratio between any two age-specific infection probabilities is genotype-independent, but the absolute risk of infection is genotype-dependent.
Finally, we allow the incubation period distribution to vary by genotype. Denote the probability density function for group by f*(u). Under model III for wider genetic susceptibility, the expected cases frequency can be written
| (2.11) |
where p(u, a) is defined by equation (2.1), and
is its analogue for the group. The expected infection prevalence frequency is
| (2.12) |
where . Using the same scenario and parameters for the test sensitivity as used in equation (2.6), the expected detectable infection frequency can be written
| (2.13) |
under the minor assumption that average sensitivity does not vary by genotype.
To date, no vCJD clinical cases in the primary outbreak have been found in the group, and none of the appendix and tonsil samples have been genotyped. Thus, neither the incidence nor the survey data contain information about β* and f *(u). Any exploration into the impact of genetic heterogeneity therefore requires a sensitivity analysis across a range of plausible choices of β* and f *(u). The approach to assessing sensitivity is to fix
| (2.14a) |
and
| (2.14b) |
where Θ and Ψ are fixed constants and T is a random variable for incubation period that follows a four-parameter generalized lambda distribution with expected value
for individuals with genotype g=MM, ; are the parameters of the incubation period distribution, which has inverse cumulative distribution function (Ramberg et al. 1979; Ghani et al. 1998). In other words, through (2.14b) we constrain the transmission probability to be Θ times larger or smaller than the MM tranmission probability; and through (2.14b) we constrain the mean incubation period to be Ψ times larger or smaller than the mean MM incubation period distribution.
As no genotype clinical vCJD cases have yet been identified, the biologically plausible values of Θ and Ψ are 0≤Θ≤1 (i.e. the transmission probability for the group is smaller than for MM group) and Ψ≥1 (i.e. the mean incubation period is longer for genotype individuals). In practice, constraint (2.14a) is easily imposed by replacing β* by Θβ in the likelihood function. Imposing constraint (2.14b) involves fixing to depend on the remaining incubation period distribution parameters by setting
in the likelihood function. The parameters of interest, namely the future cases and future prevalence defined below, were found to be estimable under the constraints defined by equation (2.14a,b); we give a justification of this in Appendix B.
2.4 Future case-number projections and infection prevalence frequency
In the following analysis, we wish to compare estimates of two population parameters under the three different models. The most important of these parameters is the number of future cases between 2004 and 2080, which under model m is written
| (2.15) |
where m=I, II, III indicates the original, the carrier and the wider genetic susceptibility models, respectively. We shall also consider estimates of the number of infected individuals in 2004
| (2.16) |
under model m=I, II, III. Note that dm(u, a) does not appear in equation (2.16) because we are interested in the underlying and not the detectable infection prevalence frequency.
3. Discrepancy between clinical case numbers and PrPSc prevalence estimates
The original model was fitted to the clinical case data alone assuming that the diagnostic test is 100% sensitive in the last 100τ% of the incubation period, where τ is a free parameter to be estimated from the data (Ghani et al. 2003a,b). The likelihood profile for the future number of deaths from 2004 to 2080 is shown in figure 2a. All the projections and estimates under the three models defined above are presented in table 1. From table 1, it can be seen that the maximum likelihood estimate under the original model fitted to the clinical cases data is 70 future cases (95% CI: 10–190). This estimate is clearly inconsistent with that from the survey data, where a crude estimate of 3800 can be obtained by applying the survey results to the population to the 10–30 age group alone, or 1850 if applied to the 20–30 age group, again assuming 100% test sensitivity (Hilton et al. 2004a).
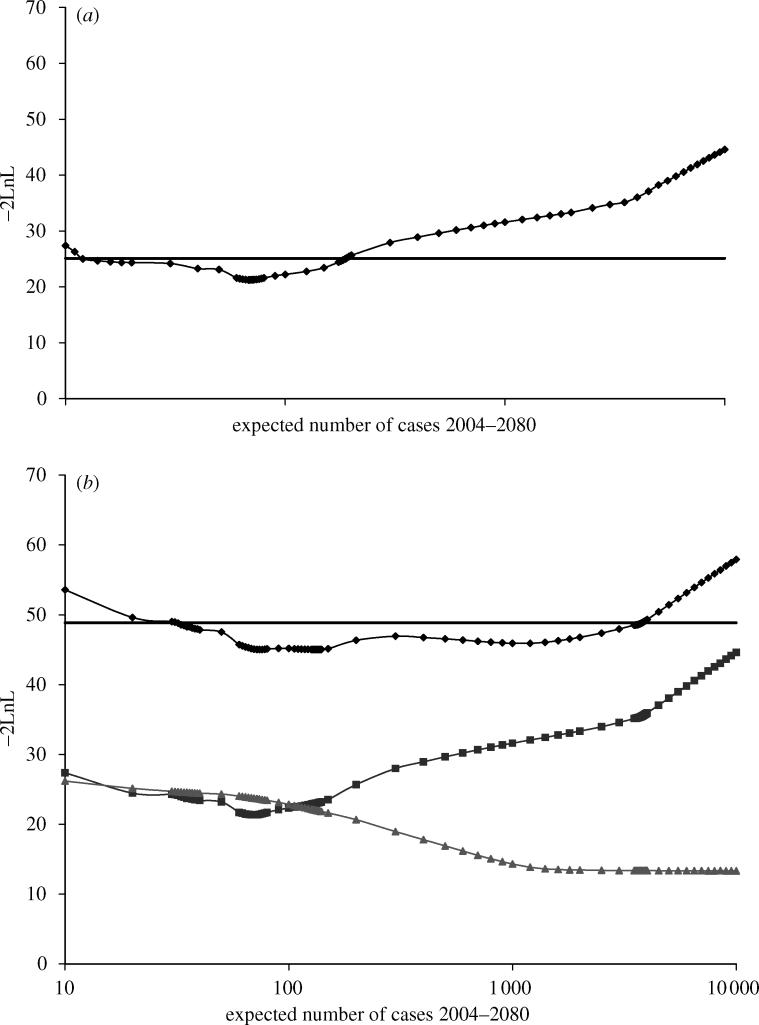
Figure 2.
Likelihood profile (diamonds) for future cases from fitting the model to (a) clinical case data alone and (b) simultaneously to the clinical case data and the results from the survey of lymphoreticular tissues. The bold line shows the cut-off corresponding to the 95% confidence limit (based on 1 degree of freedom). In (b), the lines with squares and triangles show the contribution to the likelihood from the vCJD case data and survey data respectively.
Table 1.
Summary of maximum likelihood estimates and 95% confidence intervals (in parentheses) under the original, carrier, and wider genetic susceptibility models. (Original model diagnostic test sensitivity scenario: 100% sensitive for last 100τ% of incubation period. n/a: prevalence estimates unreliable without survey data. Carrier and wider genetic susceptibility models fitted to both clinical cases and survey data. Scenario I for diagnostic test sensitivity: preclinical infections—100% sensitive for last 100τ% of incubation period; subclinical infections—100% sensitive. Scenario II for diagnostic test sensitivity: preclinical infections—90% sensitive for last 50% of incubation period; subclinical infections—50% sensitive. Optimistic scenario for group: incubation period distribution—3.2 times that for MM group; susceptibility—0.05 times that for MM group. Pessimistic scenario for group: incubation period distribution—1.9 times that for MM group; susceptibility—equal to that for MM group.)
| case numbers, 2004–2080 | number infected, 2004 | probability of subclinical infection | −2×log-likelihood | |
|---|---|---|---|---|
| original model | ||||
| clinical cases only | 70 (10–190) | n/a | n/a | 21.3 |
| clinical cases and survey data | 133 (32–3780) | 140 (36–10 050) | n/a | 45.0 |
| carrier model | ||||
| scenario I | 69 (10–190) | 3000 (520–6810) | 0.93 (0.7–0.97) | 34.7 |
| scenario II | 69 (10–190) | 5413 (1130–13 440) | 0.96 (0.88–0.99) | 34.8 |
| wider genetic susceptibility | ||||
| optimistic | 54 | 47.5 | ||
| pessimistic | 363 | 40.3 |
The discrepancy between the results from the clinical cases (incidence) and the survey (prevalence) data is further highlighted by the results from fitting the original model simultaneously to both data sets. Figure 2b shows the likelihood profile for the expected number of cases; diamonds denote the profile likelihood based on both data sets, with the squares and triangles denoting the contribution to the overall profile likelihood from the clinical cases data and from the survey data, respectively. The future case numbers estimate is 133 (95% CI: 32–3780). However, the clinical case data clearly point towards a relatively small future epidemic, whereas the survey data suggest a larger potential epidemic. The overall estimate is a trade-off between the two.
There are three plausible explanations for the discrepancy between these two data sets. The first explanation concerns the observation that two of the three positive samples had different patterns of lymphoreticular accumulation (Hilton et al. 2004a). It may be that the two differentially patterned positives are false positives not indicating vCJD infection. This possibility remains, although lymphoreticular accumulation has not been found in any disease other than vCJD, and the specificity of lymphoreticular accumulation in diagnosing vCJD has been found to be very high (Hilton et al. 2004b).
The second explanation is that the high prevalence estimate from the survey data, as compared with the number of clinical cases, indicates the presence of asymptomatic infection. There are two possibilities: subclinical and preclinical infections. The former refers to a subgroup of the population who are infected but will not develop clinical disease; whereas the latter refers to a subgroup of individuals who are infected and will die but have yet to develop the clinical symptoms of vCJD. To date, the evidence favours the subclinical explanation: subclinical forms of prion disease have been identified in animal experiments (Hill & Collinge 2003), and experiments, in which MM transgenic mice have been inoculated with the BSE agent, have found a high incidence of subclinical infection (Asante et al. 2002).
The third explanation is that the different patterns of lymphoreticular accumulation are the result of genetic heterogeneity. While all the clinical cases to date have been identified as MM, it is possible that other genotypes will develop disease with longer incubation periods and/or lower susceptibility to infection. Support for this hypothesis comes from experimental work, which has found a relationship between genotype and lymphoreticular patterning (Parchi et al. 1999) and from the recent identification of infection in the spleen of an individual with MV genotype (Peden et al. 2004). In other words, an excess of preclinical infections owing to the effect of different genotypes may be the cause of the discrepancy.
In the following sections, we explore the impact of the final two explanations on projections of the future size of the vCJD epidemic.
4. Inclusion of a carrier/subclinical state
Figure 3a shows the likelihood profile for the number of future vCJD cases obtained by fitting the carrier state model to the clinical cases and survey data. The scenario used for the diagnostic test sensitivity is referred to as ‘scenario I’, a straightforward extension of that used for the original model, with the test 100% sensitive for subclinical infections as well as 100% sensitive over the last 100τ% of the incubation period for preclinical infections. The fit of this model is an improvement on the original model (change in −2×log-likelihood=45.0−34.7=11.3 on 1 degree of freedom gives a p-value <0.001 using the likelihood ratio test; see Appendix A.2 for further details about model testing). The maximum likelihood estimate for future case numbers under this scenario is 69 (95% CI: 10–190), which is almost equal to the projection by the original model fitted to the clinical cases alone. The likelihood profile demonstrates the dependence of this estimate on the clinical cases, with the contribution to the overall fit from the survey data almost non-informative (i.e. flat) about the future case numbers.
Figure 3.
Likelihood profiles obtained by fitting the carrier model to the clinical case data and the results from the survey of lymphoreticular tissues. (a) Likelihood profile for future cases—the lines with squares and triangles show the contribution to the likelihood from the vCJD case data and survey data respectively and the line with diamonds the overall fit. (b) Likelihood profile for the proportion of those infected that develop clinical disease under the two scenarios considered for the sensitivity of the tests to detect PrPSc in appendix tissues. (c) Likelihood profile for prevalence of infection in 2004 under the two scenarios for the sensitivity of the tests to detect PrPSc in appendix tissues. In all three plots, the bold line shows the cut-off corresponding to the 95% confidence limit (based on 1 degree of freedom).
Figure 3b shows the likelihood profile for the proportion of individuals with subclinical infection for this model. The first is scenario I defined above, from which the probability of subclinical infection is estimated as 0.93 (95% CI: 0.70–0.97), which is very high. To explore whether this unrealistic estimate was a result of an unrealistic test sensitivity profile, a more realistic ‘scenario II’ was considered, in which the test is 50% sensitive for subclinical infections, and 0% sensitive for preclinical infections, until the final 50% of the incubation period, after which it is 90% sensitive. The second profile in figure 3b is for scenario II, under which the subclinical probability estimate is 0.96 (95% CI: 0.84–0.99), which is also very high. We can conclude from this, therefore, that the high subclinical infection probability estimate is not the result of over-optimistic assumptions about the test sensitivity.
Figure 3c contains the likelihood profiles for the infection prevalence in 2004 under the two scenarios for the diagnostic test sensitivity discussed above. Under scenario I, the maximum likelihood estimate for the prevalence frequency is 3000 (95% CI: 520–6810). The relatively low value obtained here (compared with the estimate of 3800 obtained by applying the survey results to the 10–30 age group) is owing to the best-fitting age-dependent susceptibility function, which peaks strongly in the 10–20 age group and suggests that most infected individuals in 2000 (10 years on from the peak risk of becoming infected) are in the 20–30 age group. Relaxing the assumptions regarding sensitivity increases the estimates, with scenario II giving the estimate 5413 (95% CI: 1130–13 440).
5. Wider genetic susceptibility
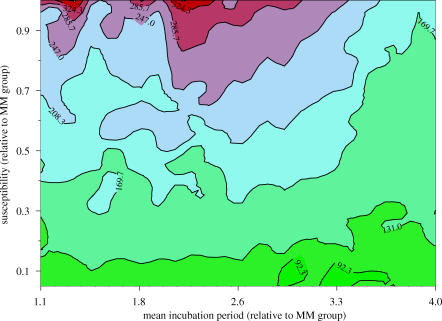
The results of a sensitivity analysis into the impact of wider genetic susceptibility using diagnostic test sensitivity scenario I are shown in the contour plot in figure 4. The plot shows how the best estimate of epidemic size (represented by colour, with larger epidemics shown in red colours) varies according to two parameters: the first, Ψ, is the scaling of the transmission coefficient (1 indicates that the group is equally susceptible, and values less than 1 that the group has reduced susceptibility compared with the MM group); the second, Θ, is the scaling of the mean incubation period in the group compared with the MM group (1 again indicates the same mean incubation period in the two groups, and values greater than 1 fix longer incubation periods in the group). Owing to the lack of data in the group, it is not possible to estimate (Ψ, Θ) because the data are unable to discriminate between different values. However, all points on the plot are plausible scenarios with which we can perform a sensitivity analysis.
Figure 4.
Contour plot for sensitivity analysis to explore the impact of wider genetic susceptibility on predictions. The x-axis shows the scaling of the mean incubation period in the non-MM genotypes compared with the MM genotypes (e.g. 2 indicates a mean incubation period double that in the MM genotypes). The y-axis shows the relative susceptibility to infection of the non-MM genotypes compared with the MM genotypes, where a value of 1 indicates identical susceptibility and a value less than 1 indicates lower susceptibility. The colours and contour lines represent the total number of future cases of vCJD.
The estimates of the total number of future vCJD cases ranges from 54 at (Ψ, Θ)=(0.05, 3.2) to 363 at (Ψ, Θ)=(1.0, 1.9), compared with the original model predictions of 70 future cases. Thus inclusion of wider genetic susceptibility in the model results in a maximum fivefold increase in projections. It is not possible to ascertain which of the scenarios presented in figure 4 is most likely given the data, because the data contain no information about (Ψ, Θ). However, we can compare values of −2×log-likelihood, or −2LnL as described in Appendix A.1. For example, the original model corresponds to any special case of the wider genetic susceptibility model with Θ=0, which we know from previously gives −2LnL=45.0. The most pessimistic scenario regarding future case numbers is 363 cases when (Ψ, Θ)=(1.0, 1.9), with −2LnL=40.3. Although we cannot use the deviance difference to test whether this difference is significant (since the likelihood is not adjusted for the relative likelihood of the (Ψ, Θ) values), it is interesting to note that neither conditional fit is as good as for the carrier model (−2LnL=34.7).
6. Discussion
Clinical cases of vCJD in the UK have continued to decline since their peak in 2000, with only four deaths reported to date in 2004 (6 September 2004). This decline is in line with projections made over the past two years based on epidemiological models linking the pattern of clinical cases in humans to past exposure to BSE-infected animals (Huillard d'Aignaux et al. 2001; Valleron et al. 2001; Ghani et al. 2003a,b). The results from fitting the original epidemiological model to deaths from vCJD to the end of 2003 are similar to those obtained a year ago (Ghani et al. 2003a), with a best estimate of 70 future deaths (95% CI: 10–190).
Considerable debate regarding the validity of these projections has arisen following the publication of results of a large-scale survey of lymphoreticular tissues, from which a much higher estimate of the prevalence of asymptomatic infection is obtained than suggested by the pattern of clinical cases or the epidemiological models. However, the survey data introduce further uncertainty to the analysis. First, only one of the three positive tissues showed a pattern of staining similar to that observed in tissues taken from those with clinical disease, with interpretation of the remaining two positive tissues less certain. Second, relatively little is known about the sensitivity or specificity of the tests used to detect the prion protein in these tissues. In particular, uncertainty in the sensitivity of the tests relates not only to the ability of the test to detect prion protein when it is present in the tissue (classical sensitivity), but also to the distribution of prion protein throughout the lymphoreticular system at different stages in the incubation period. It is worth noting that less than perfect sensitivity, which is highly likely, only further widens the discrepancy between the clinical cases and the survey results.
If the survey results do indeed represent a higher prevalence of infection than expected, this does not necessarily invalidate the projections made for future clinical cases. Instead, it questions some of the underlying assumptions that are made regarding the link between prevalence of asymptomatic infection and clinical cases. One of the most plausible explanations for the discrepancy between clinical case numbers and this estimated prevalence is the possibility that a proportion of infected individuals do not go on to develop clinical disease within their normal lifespan. Distinction is often made between preclinical infections (those animals or humans in whom neuropathological and biochemical changes and accumulation of prion protein in the brain can be observed, but who do not yet have overt symptoms of disease) and subclinical infections (in which infectivity and accumulation of prion protein are observed, but who do not go on to develop clinical disease within normal lifespan). Given our still limited understanding of disease pathogenesis for TSEs, it remains difficult to distinguish between these two states (Hill & Collinge 2003). One hypothesis for the differential pattern of staining observed in two of the three positive appendices could be that these individuals were subclinically infected. Inclusion of a subclinical state in our model significantly improves the fit of the model to both data sets, with projections of future clinical cases similar to those obtained by fitting the model to the clinical case data alone. However, even assuming that the tests in tonsil and appendix tissues are 100% sensitive throughout the course of the incubation period, the estimate of the proportion of individuals that do not go on to develop clinical disease is 93% (95% CI: 60–97%). For more realistic values for the sensitivity of tests through the course of the incubation period (90% in the last 50% of the incubation period, 0% prior to this) and for subclinical infections (50%), this estimate is much higher (estimate 96, 95% CI: 84–99%). While this model best fits the data, it remains debatable as to whether such a high proportion of infections not resulting in clinical disease is biologically reasonable.
An alternative hypothesis for the differential pattern of staining observed in two of the three positive appendix tissues is that these could represent infection in non-MM homozygous individuals. The genotype of these tissues is not currently known. While to date no clinical cases of vCJD have been observed in either valine homozygous individuals or heterozygous individuals, recent identification of subclinical infection in a heterozygous individual infected via blood transfusion suggests that future cases are possible in these genotypes (Peden et al. 2004). In particular, it is well documented that the incubation period in homozygous individuals is generally shorter than that in heterozygous individuals for other human TSEs such as kuru and sporadic CJD (Cervenakova et al. 1998; Huillard d'Aignaux et al. 2002). In addition, it is possible that VV and MV individuals could be less susceptible to infection (Cervenakova et al. 1998; Brown et al. 2000; Lee et al. 2001). Our sensitivity analyses suggest that, even in the worst-case scenario, when non-MM homozygous individuals are equally susceptible but have longer mean incubation periods than MM-homozygous individuals, the best estimate of the potential scale of the epidemic is unlikely to exceed 400 future cases. Furthermore, inclusion of wider genetic susceptibility in the model is unable to explain the large discrepancy between the numbers of clinical cases and the results from the survey of lymphoreticular tissues.
The analyses presented here do not consider the potential role of secondary transmission via blood or surgical instruments. Recent reports of the probable transmission of vCJD via blood transfusion have highlighted the potential for a secondary epidemic of vCJD (Llewelyn et al. 2004; Peden et al. 2004). Public health measures, including leucodepletion of blood introduced in 1999 and the ban on blood donations from those who have received blood transfusions initiated in March 2004 and further increased in July 2004, are now in place to minimize the current and future risk of transmission via this route (Department of Health 2004). However, the high estimate of prevalence of infection, whether preclinical or subclinical, from the survey of lymphoreticular tissues could have important implications for the potential for future cases of vCJD arising via past exposure to infected blood. Because of the many uncertainties in the transmissibility and extent of exposure via this route, the magnitude of any future epidemic arising via secondary transmission remains highly uncertain.
Acknowledgments
We are grateful to Bob Will, James Ironside and David Hilton for providing data on the vCJD cases and the survey of lymphoreticular tissues. We also thank Neil Ferguson and Christl Donnelly for helpful comments. This work was supported by the Department of Health. The views expressed in this publication are those of the authors and not necessarily those of the Department of Health. A.C.G. acknowledges fellowship support from the Royal Society.
Appendix A. Statistical methods
A.1 Maximum likelihood estimation
Data are available from two independent sources from which the likelihood function can be developed. First, there is time- and age-stratified data on the number of vCJD deaths to the end of December 2003. The frequency of cases at each time and age are assumed to follow a Poisson distribution, from which the log-likelihood function (LnL1) can be derived. Maximum likelihood estimation proceeds by maximizing LnL1, or equivalently minimizing
| (A 1) |
for model m=I, II, III, where x(u, a) is the frequency of observed cases at time u and age a, and cm(u, a) is defined in §2. The approximations on which statistical inference from equation (A 1) are based require that all x(u, a)≥5, which is ensured by classifying cases by age group.
The second data source is from the tissue survey. The reported results arose from 13 independent batches, with three positive results in two of the batches and no positives in the other 11. Switching to discrete time, within each batch the available information is: the sample size nb, the number of positives xb, the time interval during which the samples were taken Ub=[a1b, a2b], and the age range of individuals Ab=[a1b, a2b]. If we assume that the time and age of each sampled individual were known, then the ‘complete data’ likelihood for the batch would be
under a multinomial sampling model, where xu,a is the unknown frequency of positive samples and yu,a the unknown frequency of negative samples in time-age group (u, a), and αb(u, a) is the probability of selecting a batch b individual from this time-age group. From Dempster et al. (1977), the ‘observed data’ likelihood is calculated by summing the above expression over all xu,a that sum to xb and all yu,a that sum to nb−xb. Evaluating this expression and applying the multinomial rule gives
for batch b's observed data likelihood. By noting that
and substituting this into the above expression for the likelihood, it can be shown that the overall contribution of the survey data is
| (A 2) |
where
for model m=I, II, III and batch b=1, 2,…,13.
As both data sets are independent, the model can be fitted to the combined function
or fitted to either component individually. Model fitting is performed via optimization techniques for non-linear objective functions using custom-written code (Press et al. 1992). Fitting involves an intensive search of plausible parameter space regions from multiple starting points to ensure that the global maximum is found.
A.2 Model selection using deviance change
The value of −2LnL is also used to compare the relative fit of two models, where one model is an elaboration of the other. Denote these models by M1 and M2, where the simplest model M2 is nested within M1 in the sense that its parameter space is a subspace of the parameter space of M1. To test whether the extra parameters added to M2 fit the data better, compare the deviance change
with a chi-square distribution on Δ degrees of freedom, where Δ= difference between number of parameters in M1 and M2, and −2LnL(M) is the minimum of −2LnL for model M=M1, M2.
A.3 Profile likelihood confidence intervals
Let θ denote the free model parameters. The maximum likelihood estimate for arbitrary population parameter K=K(θ) can be written , where . In this paper, K=Tm(2004, 2080) and K=Pm(2004), as defined by equations (2.15) and (2.16), respectively. The maximum likelihood at is called the profile likelihood at κ and written
An approximate 95% CI for K can be obtained by finding (κL, κU) such that κL<κU and , where χ2(0.95) is the 95-percentile of a χ2 random variable with 1 degree of freedom. Note that this method assumes both −2LnL1 and −2LnL2 are well approximated by concave functions.
Appendix B. Parameter identifiability
B.1 Original model
Given data on clinical cases and choice of probability distributions for the exposure-susceptibility and incubation period distributions, all future case number projections are identified (Isham 1989). However, it is impossible to distinguish between different model assumptions that fit the clinical cases data equally well but give different projections—hence the use of experimental results on other prion diseases to determine the choice of parametric distribution.
The expected prevalence is also identified because, under the model assumption that all infected hosts go on to develop the disease, it can be written as a weighted sum of future case numbers among those infected prior to the time at which the diagnostic tests were performed. However, any prevalence estimate based only on the clinical cases is strongly model-assumption dependent, and cannot be tested. In other words, it is impossible to distinguish between two different assumptions which fit the clinical cases equally well but give very different prevalence estimates. Introducing the survey data permits discrimination because model fit can be assessed with respect to the joint fit between the clinical cases and survey data.
B.2 Carrier model
The first model assumption to be relaxed, using the carrier model, is that all infected individuals will eventually develop the disease. The model parameter ω indexing the proportion of subclinical infections can only be identified when the model is fitted to both data sets.
B.3 Sensitivity analysis of wider genetic susceptibility
As discussed in §2, it is necessary to perform a sensitivity analysis for the possible impact of wider genetic susceptibility because the parameters of the incubation period distribution are inestimable, owing to the lack of data on incidence or prevalence in the group. However, for the results of the sensitivity analysis to make sense, it is necessary to establish that the constraints defined in equation (2.14b) are sufficient to ensure the remaining parameters of the group, , are identified.
It is difficult to establish analytically that the parameters of the incubation period are identified. This difficulty arises because the clinical cases data contain information about the parameters of the incubation period distribution for the MM group, and the survey data contain information about both the MM and groups, and it is not trivial to construct a contrast between the data sets which could be used to identify the group parameters. A simpler approach is to construct likelihood profiles for each of the three parameters using the method described in Appendix A.3. A flat likelihood profile plot indicates non-informativity and inestimable parameters.
We found that , indexing the spread of the incubation period distribution, has a curved profile plot and is estimable. Conversely, , indexing the negative and positive skew of the distribution, both have flat profile plots. However, the 2004–2080 projection was found to be insensitive to values of the two parameters: the projection estimates vary by less than 10 for any value in that range. This finding was the same for other, randomly chosen, values of (Θ, Ψ).
References
- Alperovitch A, Zerr I, Pocchiari M, Mitrova E, de Pedro Cuesta J, Hegyi I, Collins S, Kretzschmar H, van Dujin C, Will R.G. Codon 129 prion protein genotype and sporadic Creutzfeldt–Jakob disease. Lancet. 1999;353:1673–1674. doi: 10.1016/S0140-6736(99)01342-2. [DOI] [PubMed] [Google Scholar]
- Asante E.A, et al. BSE prions propagate as either variant CJD-like or sporadic CJD-like prion strains in transgenic mice expressing human prion protein. EMBO J. 2002;21:6358–6366. doi: 10.1093/emboj/cdf653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown P, et al. Iatrogenic Creutzfeldt–Jakob disease at the millennium. Neurology. 2000;55:1075–1081. doi: 10.1212/wnl.55.8.1075. [DOI] [PubMed] [Google Scholar]
- Bruce M.E, et al. Transmissions to mice indicate that ‘new variant’ CJD is caused by the BSE agent. Nature. 1997;389:498–501. doi: 10.1038/39057. [DOI] [PubMed] [Google Scholar]
- Cervenakova L, Goldfarb L.G, Garruto R, Lee H.S, Gajdusek D.C, Brown P. Phenotype–genotype studies in kuru: implications for new variant Creutzfeldt–Jakob disease. Proc. Natl. Acad. Sci. USA. 1998;95:13 239–13 241. doi: 10.1073/pnas.95.22.13239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinge J, Palmer M.S, Dryden A.J. Genetic predisposition to iatrogenic Creutzfeldt–Jakob disease. Lancet. 1991;337:1441–1442. doi: 10.1016/0140-6736(91)93128-V. [DOI] [PubMed] [Google Scholar]
- Collinge J, Sidle K.C.L, Meads J, Ironside J, Hill A.F. Molecular analysis of prion strain variation and the aetiology of ‘new variant’ CJD. Nature. 1996;383:685–690. doi: 10.1038/383685a0. [DOI] [PubMed] [Google Scholar]
- Cooper J.D, Bird S.M. UK bovine carcass meat consumed as burgers, sausages and other meat products: by birth cohort and gender. J. Cancer Epidemiol. Prev. 2002a;7:49–57. doi: 10.1080/147666502321082719. [DOI] [PubMed] [Google Scholar]
- Cooper J.D, Bird S.M. UK dietary exposure to BSE in head meat: by birth cohort and gender. J. Cancer Epidemiol. Prev. 2002b;7:71–83. doi: 10.1080/147666502321082737. [DOI] [PubMed] [Google Scholar]
- Cooper J.D, Bird S.M. Predicting incidence of vCJD from UK dietary exposure to BSE for the 1940 to 1969 and post-1969 birth cohorts. Int. J. Epidemiol. 2003;32:784–791. doi: 10.1093/ije/dyg248. [DOI] [PubMed] [Google Scholar]
- Cooper J.D, Bird S.M, de Angelis D. Prevalence of detectable abnormal prion protein in persons incubating vCJD: plausible incubation periods and cautious inference. J. Epidemiol. Biostat. 2000;5:209–219. [PubMed] [Google Scholar]
- Dempster A.P, Laird N.M, Rubin D.B. Maximum likelihood estimation from incomplete data via the EM algorithm (with discussion) J. R. Stat. Soc. B. 1977;39:1–38. [Google Scholar]
- Department of Health. 2004 Update on precautions to protect blood supply. Reference: 2004/0270. London: Department of Health.
- Ferguson N.M, Ghani A.C, Donnelly C.A, Hagenaars T.J, Anderson R.M. Estimating the risk to human health posed by possible entry of BSE infection into the GB sheep flock. Nature. 2002;415:420–425. doi: 10.1038/nature709. [DOI] [PubMed] [Google Scholar]
- Ghani A.C, Ferguson N.M, Donnelly C.A, Hagenaars T.J, Anderson R.M. Epidemiological determinants of the pattern and magnitude of the vCJD epidemic in Great Britain. Proc. R. Soc. Lond. B. 1998;265:2443–2452. doi: 10.1098/rspb.1998.0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghani A.C, Ferguson N.M, Donnelly C.A, Anderson R.M. Predicted vCJD mortality in Great Britain. Nature. 2000;406:583–584. doi: 10.1038/35020688. [DOI] [PubMed] [Google Scholar]
- Ghani A.C, Donnelly C.A, Ferguson N.M, Anderson R.M. Updated projections of future vCJD deaths in the UK. BMC Infect. Dis. 2003a;3:4. doi: 10.1186/1471-2334-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghani A.C, Ferguson N.M, Donnelly C.A, Anderson R.M. Factors determining the pattern of the variant Creutzfeldt–Jakob disease (vCJD) epidemic in Great Britain. Proc. R. Soc. B. 2003b;270:689–698. doi: 10.1098/rspb.2002.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill A.F, Collinge J. Subclinical prion infection in humans and animals. Br. Med. Bull. 2003;66:161–170. doi: 10.1093/bmb/dg66.161. [DOI] [PubMed] [Google Scholar]
- Hilton A.F, Fathers E, Edwards P, Ironside J.W, Zajick J. Prion immunoreactivity in appendix before clinical onset of variant Creutzfeldt–Jakob disease. Lancet. 1998;352:703–704. doi: 10.1016/S0140-6736(98)24035-9. [DOI] [PubMed] [Google Scholar]
- Hilton D.A, Ghani A.C, Conyers L, Edwards P, McCardle L, Penney M, Ritchie D, Ironside J.W. Accumulation of prion protein in tonsil and appendix: review of tissue samples. Br. Med. J. 2002;325:633–634. doi: 10.1136/bmj.325.7365.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton D.A, Ghani A.C, Conyers L, Edwards P, McCardle L, Ritchie D, Penney M, Hegazy D, Ironside J.W. Prevalence of lymphoreticular prion protein accumulation in UK tissue samples. J. Pathol. 2004a;203:733–739. doi: 10.1002/path.1580. [DOI] [PubMed] [Google Scholar]
- Hilton D.A, Sutak J, et al. Specificity of lymphoreticular accumulation of prion protein for variant Creutzfeldt–Jakob disease. J. Clin. Pathol. 2004b;57:300–302. doi: 10.1136/jcp.2003.012278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huillard d'Aignaux J.N, Cousens S.N, Smith P.G. Predictability of the UK variant Creutzfeldt–Jakob disease epidemic. Science. 2001;294:1729–1731. doi: 10.1126/science.1064748. [DOI] [PubMed] [Google Scholar]
- Huillard d'Aignaux J.N, Cousens S.N, Maccario J, Costagliola D, Alpers M.P, Smith P.G, Alpérovitch A. An analysis of the incubation period of kuru by sex and age at infection. Epidemiology. 2002;13:402–408. doi: 10.1097/00001648-200207000-00007. [DOI] [PubMed] [Google Scholar]
- Ironside J.W, Hilton D.A, Ghani A, Johnston N.J, Conyers L, McCardle L.M, Best D. Retrospective study of prion-protein accumulation in tonsil and appendix tissues. Lancet. 2000;355:1693–1694. doi: 10.1016/S0140-6736(00)02243-1. [DOI] [PubMed] [Google Scholar]
- Isham V. Estimation of the incidence of HIV infection. Phil. Trans. R. Soc. B. 1989;325:113–121. doi: 10.1098/rstb.1989.0076. [DOI] [PubMed] [Google Scholar]
- Jackson G.S, Hosszu L.L.P, Power A, Hill A.F.K.J, Saibil H, Craven C.J, Waltho J.P, Clarke A.R, Collinge J. Reversible conversion of monomeric human prion protein between native and fibrilogenic conformations. Science. 1999;283:1935–1937. doi: 10.1126/science.283.5409.1935. [DOI] [PubMed] [Google Scholar]
- Lee H.S, Brown P, Cervenakova L, Garruto R.M, Alpers M.P, Gajdusek D.C, Goldfarb L.G. Increased susceptibility to Kuru of carriers of the PRNP 129 methionine/methionine genotype. J. Infect. Dis. 2001;183:192–196. doi: 10.1086/317935. [DOI] [PubMed] [Google Scholar]
- Llewelyn C.A, Hewitt P.E, Knight R.S, Amar K, Cousens S, Mackenzie J, Will R.G. Possible transmission of variant Creutzfeldt–Jakob disease by blood transfusion. Lancet. 2004;363:417–421. doi: 10.1016/S0140-6736(04)15486-X. [DOI] [PubMed] [Google Scholar]
- Maissen M, Roeckl C, Glatzel M, Goldmann W, Aguzzi A. Plasminogen binds to disease-associated prion protein of multiple species. Lancet. 2001;357:2026–2028. doi: 10.1016/S0140-6736(00)05110-2. [DOI] [PubMed] [Google Scholar]
- O'Rourke K.I, et al. Preclinical diagnosis of scrapie by immunohistochemistry of third eyelid lymphoid tissue. J. Clin. Microbiol. 2000;38:3254–3259. doi: 10.1128/jcm.38.9.3254-3259.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke K.I. Active surveillance for scrapie by third eyelid biopsy and genetic susceptibility testing of flocks of sheep in Wyoming. Clin. Diagn. Lab. Immunol. 2002;9:966–971. doi: 10.1128/CDLI.9.5.966-971.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen F, Poulter M, Collinge J, Crow T.J. Codon 129 changes in the prion protein gene in Caucasians. Am. J. Hum. Genet. 1990;46:1215–1216. [PMC free article] [PubMed] [Google Scholar]
- Pan K.M, et al. Conversion of alpha-helices into beta-sheets features in the formation of the scrapie prion proteins. Proc. Natl Acad. Sci. USA. 1993;90:10 962–10 966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parchi P, et al. Classification of sporadic Creutzfeldt–Jakob disease based on molecular and phenotypic analysis of 300 subjects. Ann. Neurol. 1999;46:224–233. doi: 10.1002/1531-8249(199908)46:2:224:AID-ANA12>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Peden A, Head M.W, Ritchie D.L, Bell J.E, Ironside J.W. Preclinical vCJD after blood transfusion in a PRNP codon 129 heterozygous patient. Lancet. 2004;364:527–528. doi: 10.1016/S0140-6736(04)16811-6. [DOI] [PubMed] [Google Scholar]
- Press W.H, Teukolsky S.A, Vetterling W.T, Flannery B.P. Cambridge University Press; Cambridge: 1992. Numerical recipes in C. [Google Scholar]
- Ramberg J.S, Tadikamalia P.R, Dudewicz E.J, Mykytka E.F. A probability distribution and its uses in fitting data. Technometrics. 1979;21:201–209. [Google Scholar]
- Schreuder B.E.C, Van Keulen L.J.M, Vromans M.E.W, Langeveld J.P.M, Smits M.A. Tonsillar biopsy and PrPSc detection in the preclinical diagnosis of scrapie. Vet. Rec. 1998;142:564–568. doi: 10.1136/vr.142.21.564. [DOI] [PubMed] [Google Scholar]
- Valleron A.J, Boelle P.Y, Will R, Cesbron J.Y. Estimation of epidemic size and incubation time based on age characteristics of vCJD in the United Kingdom. Science. 2001;294:1726–1728. doi: 10.1126/science.1066838. [DOI] [PubMed] [Google Scholar]
- Will R.G, et al. A new variant of Creutzfeldt–Jakob disease in the UK. Lancet. 1996;347:921–925. doi: 10.1016/S0140-6736(96)91412-9. [DOI] [PubMed] [Google Scholar]