Abstract
Networks and the epidemiology of directly transmitted infectious diseases are fundamentally linked. The foundations of epidemiology and early epidemiological models were based on population wide random-mixing, but in practice each individual has a finite set of contacts to whom they can pass infection; the ensemble of all such contacts forms a ‘mixing network’. Knowledge of the structure of the network allows models to compute the epidemic dynamics at the population scale from the individual-level behaviour of infections. Therefore, characteristics of mixing networks—and how these deviate from the random-mixing norm—have become important applied concerns that may enhance the understanding and prediction of epidemic patterns and intervention measures.
Here, we review the basis of epidemiological theory (based on random-mixing models) and network theory (based on work from the social sciences and graph theory). We then describe a variety of methods that allow the mixing network, or an approximation to the network, to be ascertained. It is often the case that time and resources limit our ability to accurately find all connections within a network, and hence a generic understanding of the relationship between network structure and disease dynamics is needed. Therefore, we review some of the variety of idealized network types and approximation techniques that have been utilized to elucidate this link. Finally, we look to the future to suggest how the two fields of network theory and epidemiological modelling can deliver an improved understanding of disease dynamics and better public health through effective disease control.
Keywords: transmission, infection, contact-tracing, random network, small-world network, scale-free network
1. Standard epidemic theory
The overwhelming majority of disease models are based on a compartmentalization of individuals or hosts according to their disease status (Kermack & McKendrick 1927; Bailey 1957; Anderson & May 1992). The basic models describe the number of individuals (or proportion of the population) that are susceptible to, infected with and recovered from a particular disease. Many of the details of the progression of infection are therefore neglected, as are differences in response between individuals, but the simplification has had a long and successful history. The assumptions generate two standard sets of differential equations that provide the foundations of almost all of mathematical epidemiology: the susceptible-infectious-recovered (SIR) model
| (1.1) |
and the susceptible-infectious-susceptible (SIS) model
| (1.2) |
The SIR model is appropriate for infectious diseases that confer lifelong immunity, such as measles or whooping cough (Kermack & McKendrick 1927; Anderson & May 1992; Grenfell 1992; Rohani et al. 2000). The SIS model is predominantly used for sexually transmitted diseases (STDs), such as chlamydia or gonorrhoea, where repeat infections are common (Hethcote & Yorke 1984; Garnett & Anderson 1996). In these equations, S, I and R refer to the number of susceptible, infectious and recovered individuals, respectively, in a population of size N. The other parameters are the birth rate, b, the natural death rate, d, and the rate of recovery from infection, g. The force of infection, λ, is the rate at which susceptible individuals become infected, and is a function of the number of infectious individuals; this parameter contains information about the interactions between individuals that lead to the transmission of infection.
When the population mixes at random, so that each individual has a small and equal chance of coming into contact with any other individual, the force of infection can be calculated as follows:
| (1.3) |
This leads to a nonlinear term (βSI/N) representing the transmission of infection, generating a variety of rich dynamical behaviours (Schwartz 1985; Olsen et al. 1986; Rand & Wilson 1991; Anderson & May 1992; Earn et al. 2000; Keeling et al. 2001).
Many biologically motivated modifications have been made to this basic framework, usually involving the inclusion of more heterogeneities by further subdividing the S, I and R classification to reflect either more complex pathogen biology (Anderson 1988; Grenfell et al. 2001) or greater structure within the host population (Hethcote & Yorke 1984; Ghani et al. 1997; Keeling 1997). Often, different mixing rates are expected between the population subgroups (e.g. children mix more readily among themselves than with adults), and this can be modelled by replacing the single parameter β in equation (1.3) with a matrix of transmission parameters, β, describing the transmission of infection between different groups (Anderson & May 1992). Nevertheless, the assumption of random mixing, at least between individuals within each pair of subgroups, remains.
It is usually the case, however, that the number of contacts each individual has is considerably smaller than the population size, and in such circumstances, random mixing does not occur. Models that incorporate network structure avoid the random-mixing assumption by assigning to each individual a finite set of permanent contacts to whom they can transmit infection and from whom they can be infected. Although in network and random-mixing models, individuals may have the same number of effective contacts per unit time, within a network, this set of contacts is fixed, whereas in random-mixing models, it is continually changing. Networks thus capture the permanence of interactions.
2. Standard network theory
The historical study of networks has its grounding in two disparate fields: social sciences (Leinhardt 1977; Scott 1991; Wasserman & Faust 1994) and graph theory (Harary 1969; Bollobás 1985; West 1996). Whereas in epidemiology, we speak of ‘hosts’ and ‘contacts’, the social literature is based upon ‘actors’ and ‘relations’, while graph theory uses the terms ‘nodes’ and ‘edges’. In each case, however, it is the presence of a relationship between individuals in a population that is the issue of concern. The range of available vocabularies can hinder the transfer of ideas between these fields. We shall refer to ‘individuals’ and their ‘contacts’; the set of contacts of an individual is their ‘neighbourhood’ and the size of this neighbourhood is the individual's ‘degree’.
Research in the social sciences is often concerned with the reason behind the network connections rather than the properties of the network structure itself. However, it provides a wealth of quantitative and qualitative information about social network connections, which are related to the mixing networks for airborne diseases. Network analysis has been used as an explanatory tool to describe the evolution and spread of ideas and innovations in societies (Leinhardt 1977), and observed social dynamics can often be understood through analysis of the social networks that underlie them. Attention has been given to the nature of connections, particularly properties such as symmetry (whether a relationship between A and B implies a relationship between B and A) and transitivity (whether the friend of a friend is a friend), which together provide measures of social cohesion (Wasserman & Faust 1994; Karlberg 1997). In addition, measures of the importance of individuals have also been derived; these range from the simple (such as the number of connections) to the highly complex (number of paths between other actors in which an individual features; Scott 1991; Wasserman & Faust 1994). Because the social importance of an individual (i.e. the extent to which they dominate the network) is probably closely linked to their role in disease spread, such ideas are immediately relevant to epidemiology.
Research in graph theory has provided a wealth of quantitative tools and mechanisms for describing networks, many of which have epidemiological applications. We can use an ‘adjacency matrix’ or ‘sociomatrix’, A, to describe the connections within a population (Harary 1969; Bollobás 1979; Wasserman & Faust 1994; West 1996); Aij=1 if there is a connection such that infection could pass from individual i to individual j; otherwise, Aij=0. The matrix A summarizes all connections within the network. Most mixing networks are non-directed graphs (in the language of social sciences connections are symmetric) in which infection can pass either way across a contact and thus Aij=Aji. However, this is not necessarily the case; transmission through donated blood products is an instance when infection can only travel one way along a link. In this case, the network of relevant interactions would be a directed graph (Harary 1969; Bollobás 1979).
A number of useful network quantities can be ascertained from the adjacency matrix (Keeling 1999). For a population of size N, the average number of contacts per individual is
The matrix Am contains information about paths within the network of length m, and powers of the adjacency matrix can therefore be used to calculate measures of the amount of transitivity or clustering. One such measure is given by
This is the ratio of the number of triangles within the network to the number of connected triples; the larger this number, the greater the clustering. Similar measures can be constructed by looking at squares or larger loops.
Finally, a network (or graph) is said to be connected if any individual (or node) can be reached from any other by following network links; epidemiologically, this is equivalent to infection being able to reach the entire population from any starting point, which is the case if (or equivalently, ) has no zero terms. Zeros in either of these matrices demonstrates that the network is divided into two or more separated components, none of which has any links to any of the others.
The intuition and understanding from the social sciences coupled with the elegant formalism from graph theory provide a powerful framework within which to investigate mixing networks in epidemiology. However, the research in graph theory and social sciences generally considers an understanding of the network itself to be the ultimate goal; in contrast, epidemiological interest is focused on the spread of the disease, in which case the network forms a constraining background to the transmission dynamics.
Applied questions of long-term disease spread or the risk of an epidemic for a given mixing network have many similarities to results in percolation theory (Mollison 1977; Grassberger 1983; Grimmett 1989; Newman 2002). This area of mathematics examines the formation of connected structures within networks. In its most familiar form (bond percolation), a square lattice of sites is considered, in which neighbouring sites are randomly connected with some probability, p; when this probability is high enough it is possible to find a path from one side of the lattice to another. Edges within this lattice may be treated as transmission events from individual to individual, with p representing a transmission probability. Thus, the size of connected clusters that emerge in percolation models relate to the expected size of an epidemic within the network; however, percolation models do not provide a dynamic description of the epidemic process. Another model (site percolation) places individuals at lattice points with probability p—this may be considered as a representation of epidemics in a partially susceptible population. In both cases, the value of p at which large connected structures emerge is significant, for it is at this point that major epidemics can occur (Grimmett 1989). Although lattices may not be realistic representations of human mixing networks, the concepts underlying percolation theory are immediately relevant to epidemiology, and many of these ideas and the tools for understanding them have been applied in epidemiological settings (Mollison 1977; Grassberger 1983; Newman & Watts 1999; Newman 2002; Warren et al. 2002).
3. Finding ‘real’ networks
Determining a complete mixing network requires knowledge of every individual in a population and every relationship between individuals. For all but the smallest populations, this is an impractically time-consuming task. The sheer volume of data required provides the first difficulty, but even if an entire population can be sampled (Bearman et al. 2004), there are other issues that complicate network evaluation. Firstly, because individuals may have many contacts, problems of recall are probable. Secondly, evaluation of contacts requires personal information and, especially for sexual mixing networks, this may not always be readily volunteered. These two problems concern data collection, but more fundamental is the question of how a network link is defined. If networks are to be used for epidemiological purposes, then connections should only be included if they describe relationships capable of permitting the transfer of infection. However, in many cases, it is not clear how to define such a relationship; how much contact is it necessary to have with someone with influenza, say, before there is a measurable risk? The issue is likely to be most acute for infections spread by casual contact, where some degree of arbitrariness is inevitable, but even in cases where link definition should be more straightforward, such as for STDs, there are complications. Different types of relationship bestow different risks and judgments must be made about which sorts of transmission routes are likely to be significant in any given epidemic. One way around this problem is to consider valued networks in which links are not merely present or absent but are weighted according to their strength (Wasserman & Faust 1994); however, this leads to further problems with data collection and complexity of models, and is usually impractical unless considering only a small number of possible weights—for instance, monogamous and casual sexual relationships (Kretzschmar et al. 1996) or limited social settings (Eubank et al. 2004; Meyers et al. 2005).
At this point, we observe that because different infections are passed by different routes, a mixing network is necessarily disease specific. Thus, a network used in the context of HIV transmission would differ from one used to examine influenza; in such a case, we might expect the networks to be nested, with the links relevant for HIV spread to be a subset of those important for influenza. However, even for two airborne infections (such as influenza and measles), different networks may be appropriate because differing levels of interaction will be required to constitute a contact. The problems with network definition and measurement mean that any mixing networks that are obtained will depend on the assumptions and protocols of the data collection process. Nevertheless, those networks that have been explored provide important insights into interaction patterns and their implication for disease transmission.
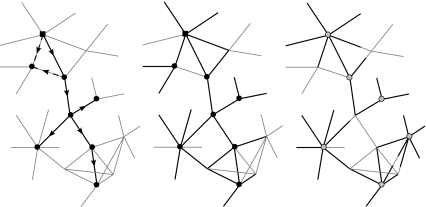
Three main techniques have been employed to gather network information: infection tracing, complete contact tracing, and diary-based studies (figure 1). These have their own advantages and disadvantages, and the method applied depends on the resources available and the purpose for which the data is being gathered.
Figure 1.
For the same simple network (thin grey lines), the type of network information that is achieved using infection tracing (left), contact tracing (middle) and diary-based studies (right). For infection and contact tracing, circles represent infected individuals, while the square shows the primary infectious case; for the diary-based study, those taking part are shown with open circles. For infection tracing, only sources of infection are traced and some individuals (e.g. top left) have multiple potential sources of infection. For contact tracing, a proportion of all contacts from infectious individuals are traced. Finally, with a diary-based study, although almost all links can be traced, the lack of a unique identifier means that often links from different individuals cannot be connected.
3.1 Infection tracing
After an epidemic, such as the recent cases of SARS in Hong Kong and Canada, field-based epidemiologists place considerable emphasis on determining the source of infection for each case (Haydon et al. 2003; Riley et al. 2003). In this way, each infected individual is linked to one other from whom they caught the infection, and additionally, to a variable number of others to whom they transmitted the disease, thus providing a ‘transmission network’ consisting of all the links through which infection spread in a single outbreak. Because all connections represent actual transmission events, this method does not suffer from problems with the definition of links; however, interactions that did not happen to lead to the transmission of infection in this particular case will be omitted from the network. The networks observed are therefore liable to be tree-like, containing no loops, and this precludes any worthwhile evaluation of the more complex network measures. However, since such tracing is often an integral component of disease control policies, these partial samples of the network can be generated a little extra cost, and provide useful information about the individuals most involved in disease transmission (Riley et al. 2003).
3.2 Contact tracing
Contact tracing aims to identify all potential transmission contacts from a source individual (known as an ‘index case’). This reveals a new set of individuals who might be infected and who can be the subject of further tracing effort (Klovdahl 1985; Kretzschmar et al. 1996; Ghani et al. 1997; Ghani & Garnett 1998; Müller et al. 2000; Wylie & Jolly 2001; Potterat et al. 2002; Eames & Keeling 2003; Fraser et al. 2004). Because it aims to identify potential transmission routes, contact tracing suffers from network definition issues; in addition, it is time consuming and relies on individuals providing complete and accurate data about personal relationships.
Contact tracing has been commonly applied not as a network evaluation device but as a control tool, most often in the case of STDs, where a contact is most easily defined (Klovdahl 1985; Rothenberg et al. 1998; Wylie & Jolly 2001; Potterat et al. 2002). In such instances, the purpose of contact tracing is to identify asymptomatic infected individuals who can then be treated or quarantined (Eichner 2003; Fraser et al. 2004). This means that the contacts of uninfected individuals are not sought, and thus only a subset of the full mixing network will be uncovered; we might expect a partial network with many dead-ends consisting of uninfected individuals (Ghani & Garnett 1998; Wylie & Jolly 2001; Potterat et al. 2002). Although a network uncovered via contact tracing is not complete and has biases, it is generally most detailed in regions of the network with the highest disease burden; thus the network data obtained is of immediate epidemiological relevance (Ghani et al. 1997; Jolly & Wylie 2002).
Several good examples of sexual mixing networks determined through contact tracing can be found in the literature (Bearman et al. 2004; De et al. 2004). Long-term and large-scale data collection has enabled large portions of sexual networks from Manitoba, Canada, and from Colorado Springs, USA, to be traced (Woodhouse et al. 1994; Rothenberg et al. 1998; Wylie & Jolly 2001; Jolly & Wylie 2002; Potterat et al. 2002). These networks highlight the heterogeneities present in sexual networks and show the importance of core groups (interconnected groups with high numbers of contacts) and ‘long-distance’ connections (linking otherwise distant parts of the network) in disease transmission.
Although infectious diseases motivate a significant proportion of network evaluation studies, social networks have been sought for other purposes, and these can also be useful for epidemiological modelling. Pioneering studies of social networks were carried out in Australia in the 1970s (Klovdahl et al. 1977), where study participants were questioned about their social connections and by tracing some of these contacts a picture of a city-wide network was obtained. This demonstrated the importance of setting, whether geographical, work- or leisure-related, in the formation of partnerships. Snowball sampling schemes that follow a proportion of links are a form of contact tracing that is independent of infection status (Ghani & Garnett 1998); they are therefore capable of giving information about uninfected parts of mixing networks that would not be sampled from in conventional contact tracing.
3.3 Diary-based studies
The determination of networks through tracing is highly labour intensive and relies on the subject individuals being able to recall and willing to recount their contacts. In contrast, in diary-based studies subjects record contacts as (or shortly after) they occur, shifting the work-load from the researcher to the subject and allowing a larger number of individuals to be sampled in detail (Edmunds et al. 1997a,b). The change of focus from the population approach of other tracing methods to the individual-level scale of diary-based studies has some associated problems. Firstly, the data collection is at the discretion of the subjects, thus the definition of a close contact may not be the same for all individuals. Secondly, while this method gathers detailed individual-level data, it may be difficult for the co-ordinating researcher to link this information into a comprehensive network, as the names or identifiers of contacts may not be accurately or uniquely recorded. Indeed, unless the subject individuals come from a coherent group, such as work colleagues or residents in a single small community, it is probable that the study will result in a large number of unconnected sub-networks, each one representing the personal network of a few individuals (Klovdahl 1985; Scott 1991; Wasserman & Faust 1994).
A form of diary-based study is currently occurring in the livestock industry in the UK (National Audit Office 2003). All cattle have a unique identifying ear tag, and recent legislation requires that all movements of cattle are recorded. The records derived provide a comprehensive dynamic network for diseases of cattle that are spread by animal-to-animal contact and can therefore be used to investigate patterns of livestock infections (Gilbert et al. 2005). Again, the great advantage of this network is that the responsibility for collecting the data lies with the individual rather than the researcher.
4. The use of simulated networks
While collecting network data is beset with difficulties, the simulation of disease transmission on networks is relatively straightforward (Eames & Keeling 2002; Eubank et al. 2004; Meyers et al. 2005; Read & Keeling 2003; Wallinga et al. 1999; Watts & Strogatz 1998), relying on the observation that
| (7.1) |
Many good examples exist of simulating sexually transmitted diseases on networks designed to match the available data (Garnett & Anderson 1996; Ghani et al. 1997; Morris 1997; Potterat et al. 1999; Klovdahl 2001; Rothenberg 2001; Potterat et al. 2002; Liljeros et al. 2003; Rothenberg 2003; Szendrői & Csányi 2004; Doherty et al. 2005). This work highlights the importance of network structure and, in particular, the role of core groups (interconnected individuals with a large number of contacts) in the dynamics and persistence of STDs.
Halloran et al. (2002) attempt the more difficult problem of simulating an airborne disease outbreak, which requires a network of social connections (Edmunds et al. 1997a; Wallinga et al. 1999). The networks used are generated by computer simulation to conform to several observed social characteristics. Populations of 2000 individuals are generated with a given age distribution and household size that agree with the values for the United States. Children are assigned a school, day-care centre or play group where they interact (and therefore form connections) with other children. The simulation model is used to examine the spread of smallpox, and places particular emphasis on transmission within households and family groups, which are probably the main routes of transmission for this disease. Other models (Eubank et al. 2004; Meyers et al. 2005) attempt similar tasks, using census data to determine interaction patterns; the detail of such models requires the estimation of many parameters—transmission rates in a variety of contexts, for instance—which can often only be estimated. Despite the number of approximations involved, the inherent stochasticity of such microsimulations allows a direct estimation of the variability between epidemics.
All network-based simulations are limited by fact that there is no simple way to ascertain the sensitivity of the epidemiological results to the details of the network structure. Such simulations are therefore always vulnerable to questions of ‘what if?’; for example, in the model of Halloran et al. (2002), we may ask whether the network is representative of an average American community, whether variation between communities will bias the results if large population sizes are considered and whether rare but epidemiologically important contact structures are missing from the network. It is difficult to answer such questions or gain an intuitive understanding of network structure if our experience is limited to simulations of sampled networks. Therefore, a range of idealized networks and analytical tools have been developed that can reveal the elements of network structure that are important determinants of epidemic dynamics.
5. Idealized networks
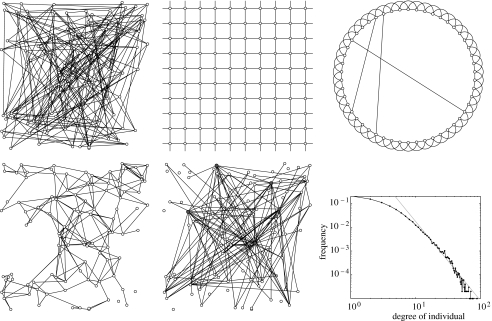
Several forms of computer-generated networks have been studied in the context of disease transmission. Each of these idealized networks can be defined in terms of how individuals are distributed in space (which may be geographical or social) and how connections are formed, thereby simplifying and making explicit the many and complex processes involved in network formation within real populations. Here, we review a range of the most popular network types and their implications for epidemic spread (figures 2 and 3).
Figure 2.
Five distinct network types containing 100 individuals. These are from left to right: random, lattice, small world (top row), spatial and scale-free (bottom row). A network generated by an exponential random graph model is not shown, as this flexible framework can encompass a huge variety of network types. For the scale-free network, the bottom right-hand graph shows the power-law distribution of individuals with a given degree from 1000 replicate networks; for this example, the power-law exponent is −3.3. The random, spatial and scale-free networks all utilize the same position of individuals, although for the random and scale-free network, the position of the individuals is irrelevant for forming connections. In all five graphs, the average number of contacts per individual is approximately four. For the scale-free network, individuals with high numbers of contacts are represented by larger dots and are shaded grey.
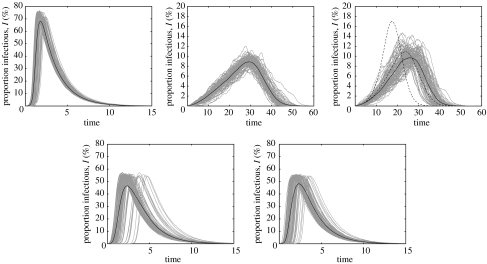
Figure 3.
Typical SIR epidemics on the five network types shown in figure 2. These are from left to right: random, lattice, small world (top row), spatial and scale-free (bottom row). Each graph shows 100 epidemic curves (grey) together with the average for all major epidemics (black) for a single example of each network type; therefore, all variability within each graph is a result of the stochastic nature of transmission and not variation in the network. All five networks contain 10000 individuals, although all individuals are not necessarily interconnected as part of a giant component. For the spatial and scale-free networks, approximately 88 and 74% are part of the giant component and can therefore potentially become infected. For these networks, the proportion of infectious individuals has been rescaled as a fraction of the giant component. In all networks, the average number of contacts per individual is approximately 4, although for the scale-free network, there is considerable heterogeneity with one individual having 85 contacts. For consistency, the small-world network is formed from a two-dimensional lattice (not a one-dimensional circle as shown in figure 2) with 10 additional random ‘long-range’ contacts. The dashed lines show the effect on the mean epidemic of increasing the number of long-range contacts to 20 and 100. (τ=1, g=0.5, b=d=0).
5.1 Random networks
In random networks, the spatial position of individuals is irrelevant, and connections are formed at random (Bollobás 1985). In the most analytically tractable version of the random network, each individual has a fixed number of contacts through which infection can spread. The random network is therefore characterized by a lack of clustering and by homogeneity of individual-level network properties. The dynamics of diseases on random networks can be studied as a simple branching process (Diekmann et al. 1998), from which it is found that both the early growth rate of the disease and the final epidemic size are reduced when compared with the random-mixing model.
where τ is the transmission rate across a contact, n is the number of contacts in the network and is the effective number of contacts per unit time in a random mixing model. The reduction in the growth rate occurs for two reasons: firstly, each infectious individual has been infected by one of its contacts, reducing the number of susceptible contacts to n−1; secondly, as an infectious individual begins to infect its susceptible contacts, it depletes its local environment, even when population prevalence is low, and hence limits the rate of disease spread. These two processes are shared by all epidemics on networks (although the strength of the effects may vary). Analytical results derived from the study of simple networks can allow us to develop an intuitive understanding of the effects of more complex social structures on disease spread.
An alternative formulation of the random network is to connect any two nodes with probability p. This leads to a network with an approximately Poisson degree distribution and a mean number of contacts per node of , where N is the total number of nodes. In such a network, the growth rate is still reduced:
however, given this effective rescaling, the epidemic dynamics for this particular network are analogous to a an SIR epidemic in a randomly mixed population (Barbour & Mollison 1990).
5.2 Lattices
Lattice models are based on very different assumptions. Individuals are positioned on a regular grid of points, usually in two dimensions, and adjacent individuals are connected; therefore, contacts are localized in space. Lattices are homogeneous at the individual level and because of the localized nature of connections are highly clustered. In general, transmission through lattices is studied by computer simulation, with the contact process (Harris 1974) and the forest-fire model (Bak et al. 1990) being the two best-known examples. The contact process is an abstraction of the SIS model, with sites that can be characterized as ‘on’ or ‘off’. The forest-fire model has strong parallels with the SIR disease model: trees burn, leaving empty sites that can be recolonized, which can be interpreted as SIR infection with births.
In common with all networks, lattice models show a reduced initial growth of infection compared with random-mixing models, although this effect is much stronger than in the random networks because the spatial clustering of contacts causes a more rapid saturation of the local environment. In general, lattice models also display a wave-like spread of infection, such that, from an initial seed, infection spreads out in a roughly circular manner. Lattice-based models capture many of the aspects of the spread on infection across a spatially extended landscape where wave-like progression is common (Mollison 1977; Grenfell et al. 2001). The spatial integrity of the wavefronts relies on the highly localized nature of transmission, and the inclusion of long-range connections (described as ‘sparks’ or ‘lightening strikes’ in forest fire models) can lead to colliding waves and a much more rapid spread of infection through the system.
Another common feature of lattice models is the existence of self-organized criticality and power-law scaling, although such features can also be observed for other forms of network. The dynamics of forest-fire models are considered examples of self-organized criticality, whereby critical behaviour and power-law scalings exist for a wide range of parameter values (Bak et al. 1990). In particular, the frequency distributions of both epidemic sizes and epidemic durations obey a power-law. Rhodes and co-workers (Rhodes & Anderson 1997; Rhodes et al. 2003) have used this scaling to explain the observed behaviour of three childhood infections, measles, whooping cough and mumps in the Faroe Islands. The observed case reports were found to closely match the power-law distribution:
with α=0.265 for measles, α=0.255 for whooping cough and α=0.447 for mumps. The invariance of this scaling to precise parameter values also has practical implications for the behaviour of infection, and may explain why moderate levels of vaccination against measles (from 1970 to the late 1980s) did not significantly change the pattern of temporary extinctions (Keeling 1997).
5.3 Small-world networks
Lattices display high clustering but long path lengths, that is, it takes many steps to move between two randomly selected individuals, whereas random networks have short path lengths, since there are many long-range links, but low clustering. Small-world networks, described in the work of Watts & Strogatz (1998; see also Watts 1999), offer a means of moving between the rigid arrangement of lattices and the unstructured connections of random networks. Small worlds can be formed by adding a small number of random connections to a lattice. Rare long-range connections have a surprisingly large effect, allowing infection to reach all parts of the lattice relatively quickly—hence the term ‘small world’. Even with a few long-distance links, there are significant changes in epidemic behaviour, demonstrating that small differences in the structure of networks can dramatically alter the population-level spread of infection. Nevertheless, because these long-range links are rare, the transmission of infection remains predominantly localized, so strong saturation effects and wave-like epidemics are still observed. Both clustering of connections and long-range transmission events are likely to be significant factors in disease spread; therefore, small-world networks are an important epidemiological concept.
Small-world networks are characterized by high clustering and short path lengths and have been observed in a range of biological settings. Human social networks are thought to be small worlds; indeed, it is in this context that the expression first came to prominence (Milgram 1967; Travers & Milgram 1969). In similar settings, small worlds have been observed in the collaboration networks of scientific authors (Newman 2001) and the co-star networks of film actors (Watts & Strogatz 1998). On a much smaller scale are gene and neural networks, which display the high clustering and low path lengths associated with the small-world model (Watts & Strogatz 1998).
Disease spread through small-world networks has received considerable attention from both a theoretical and more applied context. The high level of clustering means that most infection occurs locally, but short path lengths mean that epidemic spread through the network is rapid and disease is unlikely to be contained within small regions of the population (Watts & Strogatz 1998). Percolation theory has been applied to small-world networks to calculate threshold parameter values at which epidemics can take place, demonstrating that random long-range connections within the network can dramatically increase the likelihood of an epidemic (Moore & Newman 2000). If each individual is linked to its two nearest neighbours and on average to ϕ randomly chosen other individuals, then the critical bond percolation probability is
In epidemiological terms, if the transmission probability across a contact is greater than pc, then a large-scale epidemic is possible.
Long-range contacts lead to increased synchronization within the network, with a transition from independent epidemics in distant parts of the network to synchronized patterns of infection as path lengths are reduced (Kuperman & Abramson 2001). The evolution of pathogen virulence on small-world networks has been investigated (Boots & Sasaki 1999), and the presence of long-range links can lead to the emergence of more virulent infections as the ability to transmit to distant individuals reduces the cost to a pathogen of wiping out a local host population. The fact that features of small-world networks are also present in social mixing networks means that these results may have implications for epidemics in human populations. For example, short path lengths suggest that the spatial spread of disease is likely to be rapid; effective containment is therefore liable to require drastic restrictions on population mixing behaviour.
5.4 Spatial networks
Spatial networks are one of the most flexible forms of network. Individuals are positioned within a given area (or volume) and two individuals are connected with a probability that depends on their separation defined by a connection kernel. By changing the distribution of individuals or the kernel, it is possible to generate a wide variety of networks ranging from highly clustered lattices to small-world arrangements to globally connected random networks (Eames & Keeling 2002; Read & Keeling 2003; Keeling 2005). Spatial networks generally show a reasonably high degree of heterogeneity, with the degree distribution often being approximately Poisson. In addition, when local connections are favoured, the spatial wave-like spread of infection that characterizes lattice models is seen.
5.5 Scale-free networks
One of the most standard network measures is of the degree distribution of individuals. In many observed networks, this is far from homogeneous; it is often the case that many individuals have a small number of neighbours, while a few have significantly more connections (Albert et al. 1999; Barabási & Albert 1999; Jeong et al. 2000; Liljeros et al. 2001). Small worlds, random networks and lattice models display little variation in neighbourhood sizes, while spatial networks generally have degree distributions that are approximately Poisson. However, since highly connected individuals (termed super-spreaders) are likely to be disproportionately important in disease transmission, incorporating such individuals into networks is necessary if we are to capture the complexities of disease spread (Hethcote & Yorke 1984; Anderson & May 1992). Scale-free networks provide a means of achieving such extreme levels of heterogeneity.
Scale-free networks can be constructed dynamically by adding new individuals to a network one by one with a connection mechanism that mimics the natural formation of social contacts (Barabási & Albert 1999;Albert et al. 2000; Pastor-Satorras & Vespignani 2001). Each new individual that is added to the population connects preferentially to individuals that already have a large number of contacts, which corresponds to individuals wanting to be friends with the most popular people. This results in the number of contacts per individual taking a power-law distribution. This property was initially observed for worldwide Web connections (Albert et al. 1999), but has also been reported in power grid networks, graphs of actor collaborations (Barabási & Albert 1999) and networks of human sexual contacts (Liljeros et al. 2001).
The extreme heterogeneity in numbers of contacts displayed by scale-free networks is a feature of populations that has long been of interest to epidemiologists. Super-spreaders and core groups play a pivotal role in the spread and maintenance of infection. It is important to realize that having many contacts has two effects; it means that the individual is at greater risk of infection and, once infected, can transmit the disease to many others. Core groups of such high-risk individuals help to maintain sexually transmitted diseases in a population where the majority are in long-term monogamous relationships (Hethcote & Yorke 1984), while in the SARS epidemic, a significant proportion of all infections were caused by super-spreaders (Riley et al. 2003). These findings are in agreement with results from theoretical models of disease spread through scale-free networks where it has been shown that the infection is concentrated among individuals with highest degree (Pastor-Satorras & Vespignani 2001; Newman 2002).
In the preferential attachment model of Barabási & Albert (1999), the existence of individuals of arbitrarily large degree means that there is no level of random vaccination that is sufficient to prevent an epidemic (Albert et al. 2000; Lloyd & May 2001; Pastor-Satorras & Vespignani 2001). In contrast, when there is some upper limit imposed on the degree of individuals (Rozenfeld et al. 2002), or when a scale-free network is generated by nearest neighbour attachment within a lattice (Warren et al. 2002), it becomes possible to control infection through random vaccination. Targeted vaccination in scale-free networks is extremely efficient: the dominant role of super-spreaders means that the vaccination of only a few of these individuals can be sufficient to prevent an epidemic (Albert et al. 1999; Lloyd & May 2001; Pastor-Satorras & Vespignani 2001), reinforcing standard public-health guidelines.
5.6 Exponential random graph models
Exponential random graph models (also known as ‘p* models’; Frank & Strauss 1986) provide a method of constructing networks with a given set of properties. If we are only concerned that the mean degree is correct, then we can either add a fixed number of edges to a set of nodes or add an edge between two nodes with a fixed probability, p, independent of all other edges. For instance, if the average degree is in a population of size N, then generates a network with the desired expected number of connections (Bollobás 1985). However, such networks always display low clustering (since connected individuals are unlikely to share a neighbour), low path lengths and a binomial degree distribution.
When an alternative distribution of higher-order network structures is required, exponential random graph models allow networks with the required properties to be created. Exponential random graph models have the simple property that the probability of connection between two nodes is independent of the connection between any other pair of distinct nodes. This allows the likelihood of any nodes being connected to be calculated conditional on the graph having certain network properties. Techniques such as Markov Chain Monte Carlo can then be used to create a range of plausible networks that agree with a wide variety of information collected on network structures even if the complete network is unknown (Handcock & Jones 2004; Robins et al. 2004; Snijders 2001).
6. Pairwise approximations
The various network types mentioned above are designed as caricatures of real networks. As such, they focus on certain aspects of the population mixing behaviour (such as low path lengths, or heterogeneous degree distributions) while ignoring others. Indeed, some observed networks fall into several of the idealized categories—authorship networks having both small-world and scale-free characteristics, for instance (Newman 2001). We now turn to an alternative modelling approach—pairwise approximations—that attempts to model the spread of infection on generic networks where higher-order structure has been ignored. Rather than modelling a network of interactions in its entirety, pairwise models, as the name suggests, examine the various types of connected pairs found within a population (Keeling et al. 1997; Boots & Sasaki 1999; Keeling 1999; Ferguson & Garnett 2000).
For the motivation behind pairwise models, we return to the simple SIS model (equation (1.2)); here, the infection term is written as λS, with λ given by the transmission rate multiplied by the number of infectious contacts. The random-mixing model approximates the infection term as , but instead, this can be written exactly as τ[SI], where [SI] is the number of partnerships between susceptible and infected individuals. Within the pairwise formulation, the numbers of different pair types are included as variables rather than approximated in terms of the numbers of individuals. For example, the number of S–I pairs can change by infection from outside the pair, infection within the pair, or recovery; in an SIS model, this leads to
| (6.1) |
where [SSI] is the number of triples of individuals consisting of a central susceptible individual with both an infected and a susceptible contact. Iterating the system requires knowledge of the numbers of the various triples that appear, evaluated using a moment closure approximation:
| (6.2) |
where A, B and C represent any of the disease states and n is the degree of the central individual in the triple. This identity allows the system to be closed at the level of pairs.
Parameterization of the pairwise model requires knowledge of the distribution of pair types in the population, which is equivalent to a ‘who mixes with whom’ matrix as used in mean-field models (Anderson & May 1992; Eames & Keeling 2002). Thus, the pairwise approach makes much better use of routinely available mixing data, which can be obtained by any of the network determination methods outlined above. The advantage of this method is that the complete network is not required so long as the pairs within the network are well sampled.
By including connected individuals as its basic variables, the pairwise model can capture the correlations between neighbouring individuals that emerge in the system (Keeling et al. 1997). For example, because infection is a local process, infected individuals tend to have infected neighbours (by whom they were infected or to whom they have transmitted infection). It is this localized depletion of susceptibles that is the main difference between random-mixing and network models, and pairwise methods include this explicitly. Indeed, pairwise models have been shown to be accurate approximations of many network-based epidemics (Eames & Keeling 2002). However, although pairwise models can be adapted to allow for clustering (Keeling et al. 1997), they generally do not take into account higher order network structures such as loops, and so are generally less accurate when network connections are strongly localized.
Pairwise models have been used to examine a number of epidemiological issues: fade-out and critical community size for childhood infections (Keeling et al. 1997); evolution of pathogen virulence (Boots & Sasaki 1999); and spread and control of sexually transmitted diseases in heterogeneous populations (Ferguson & Garnett 2000; Eames & Keeling 2002). They have also been used to provide real-time predictions during the 2001 foot and mouth epidemic in the UK (Ferguson et al. 2001). While all of these problems could have been addressed through detailed simulation, the differential equation formulation of pairwise models makes them far more amenable to rapid parameterization and analytic study, allowing some rigorous results to be proved.
7. Emergent networks
All of the approaches described thus far have assumed that there is some structure behind social interactions; this structure, the mixing network, determines the relationships that are permitted and the individuals capable of transmitting infection to each other. The concept of networks is an appealing one and agrees with our ideas of how societies operate. An alternative approach, used in STD modelling, is to return to something more akin to randomly mixing models, in which any two individuals can potentially interact, but to impose predominantly monogamous partnerships (Kretzschmar et al. 1996; Dietz & Hadeler 1988; Ghani & Garnett 2000). Such partnership models have many things to recommend them as STD modelling tools but, in the context of networks, they are of interest because if all partnerships over some time period are recorded then a network of historical connections emerges.
The emergent network generated from partnership models can be used to test which network properties, such as number of partnerships, concurrent relationships or network position, are epidemiologically important. For instance, in the modelling approach of Ghani & Garnett (2000), an individual's risk of both acquiring and transmitting infection was found to depend primarily on the number of partners but was also strongly affected by partnership concurrency and distance from other individuals within the network. It was also seen that somewhat different factors determined an individual's likelihood of acquiring and of transmitting infection. Broadly speaking, local network measures matter more for acquisition of infection and global measures for transmission. Neighbourhood size, or degree, determines the likelihood of an individual acquiring infection whereas more complex network properties, such as the number of paths on which an individual, lies are significant determinants for the importance of an individual as a spreader of infection.
8. The future
Networks have an important role to play in shaping our understanding of epidemiological processes. The restriction of interactions to those within a network, rather than an entire population, slows and reduces the spread of infection; therefore, if we are attempting to predict population-level dynamics from individual-level observations, then it is vitally important that network structure is taken into account. In addition, many methods of control, such as contact tracing or ring vaccination, can only be accurately captured and modelled using network-based approaches. The emergence of network modelling tools allows these more sophisticated interventions to be studied and enables different strategies to be tested in an artificial environment.
The work using idealized networks and pairwise approximations has highlighted many of the differences between standard random-mixing disease models and disease spread through networks. The aim of such approaches should be to develop an intuitive understanding of network-based epidemics and the effects of network structure. Clearly, the ultimate goal is a set of robust network statistics that allow us to predict epidemic dynamics when the population structure deviates from the random-mixing ideal.
Most of the networks discussed here have been static—the connections have remained constant over time. This contrasts with our intuitive perception of human interactions breaking and forming. However, when these networks are used for epidemic modelling, this is not necessarily a problem. Provided that the turnover of connections is slow relative to the time-scale of the pathogen, the network will change little during the epidemic phase of infection. If the links represent close social or family relationships, or sexual partnerships, this might be expected to be the case. However, when long-term results are sought, care needs to be taken over changes in network structure. Furthermore, the behaviour of a population may change markedly as a consequence of an outbreak of infection, which needs to be considered when designing interventions. Although microsimulation models (Eubank et al. 2004; Meyers et al. 2005) and partnership models (Dietz & Hadeler 1988; Kretzschmar et al. 1996; Ghani & Garnett 2000; Eames & Keeling 2004) are designed to allow for changes within a network, this is not the norm and remains an important challenge for the future.
Finally, recently advances in mobile phone technology and GPS location mean that it may soon be possible to accurately track the movement of people in real time. This would allow us to build full and comprehensive networks for many airborne diseases, and also to track the changing network structure in the face of a severe epidemic, such as an influenza pandemic or bioterrorist attack, when behaviour may be radically different from the norm.
Acknowledgements
This work was funded by The Royal Society (M.J.K.), the Wellcome Trust (M.J.K.), NIH (M.J.K.), Emmanuel College, Cambridge (K.T.D.E.), and EPSRC (K.T.D.E.). The authors would like to thank three anonymous referees for their considered and constructive suggestions.
References
- Albert R, Jeong H, Barabási A.-L. Diameter of the world-wide web. Nature. 1999;401:130–131. [Google Scholar]
- Albert R, Jeong H, Barabási A.-L. Error and attack tolerance of complex networks. Nature. 2000;406:378–381. doi: 10.1038/35019019. [DOI] [PubMed] [Google Scholar]
- Anderson R.M. The epidemiology of HIV infection: variable incubation plus infectious periods and heterogeneity in sexual activity. J.R. Stat. Soc. A. 1988;151:66–98. [Google Scholar]
- Anderson R.M, May R.M. Oxford University Press; Oxford: 1992. Infectious diseases of humans. [Google Scholar]
- Bailey N.T.J. Griffin; London: 1957. The mathematical theory of epidemics. [Google Scholar]
- Bak P, Chen K, Tang C. A forest-fire model and some thoughts on turbulence. Phys. Lett. A. 1990;147:297–300. [Google Scholar]
- Barabási A.L, Albert R. Emergence of scaling in random networks. Science. 1999;286:509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- Barbour A, Mollison D. Epidemics and random graphs. In: Gabriel J.-P, Lefèvre C, Picard P, editors. Stochastic processes in epidemic theory. Springer; New York: 1990. pp. 86–89. [Google Scholar]
- Bearman P.S, Moody J, Stovel K. Chains of affection: the structure of adolescent romantic and sexual networks. Am. J. Sociol. 2004;110:44–91. [Google Scholar]
- Bollobás B. Springer; New York: 1979. Graph theory. [Google Scholar]
- Bollobás B. Academic Press; London: 1985. Random graphs. [Google Scholar]
- Boots M, Sasaki A. ‘Small worlds’ and the evolution of virulence: infection occurs locally and at a distance. Proc. R. Soc. B. 1999;266:1933–1938. doi: 10.1098/rspb.1999.0869. doi:10.1098/rspb.1999.0869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De P, Singh A.E, Wong T, Yacoub W, Jolly A.M. Sexual network analysis of a gonorrhoea outbreak. Sex. Transm. Infect. 2004;80:280–285. doi: 10.1136/sti.2003.007187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekmann O, Heesterbeek J.A.P, Metz J.A.J. A deterministic epidemic model taking account of repeated contacts between the same individuals. J. Appl. Prob. 1998;35:462–468. [Google Scholar]
- Dietz K, Hadeler K.P. Epidemiological models for sexually transmitted diseases. J. Math. Biol. 1988;26:1–25. doi: 10.1007/BF00280169. [DOI] [PubMed] [Google Scholar]
- Doherty I.A, Padian N.S, Marlow C, Aral S.O. Determinants and consequences of sexual networks as they affect the spread of sexually transmitted infections. J. Infect Dis. 2005;191:S42–S54. doi: 10.1086/425277. [DOI] [PubMed] [Google Scholar]
- Eames K.T.D, Keeling M.J. Modeling dynamic and network heterogeneities in the spread of sexually transmitted diseases. Proc. Natl Acad. Sci. USA. 2002;99:13330–13335. doi: 10.1073/pnas.202244299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eames K.T.D, Keeling M.J. Contact tracing and disease control. Proc. R. Soc. B. 2003;270:2565–2571. doi: 10.1098/rspb.2003.2554. doi:10.1098/rspb.2003.2554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eames K.T.D, Keeling M.J. Monogamous networks and the spread of sexually transmitted diseases. Math. Biosci. 2004;189:115–130. doi: 10.1016/j.mbs.2004.02.003. [DOI] [PubMed] [Google Scholar]
- Earn D.J.D, Rohani P, Bolker B.M, Grenfell B.T. A simple model for complex dynamical transitions in epidemics. Science. 2000;287:667–670. doi: 10.1126/science.287.5453.667. [DOI] [PubMed] [Google Scholar]
- Edmunds W.J, Medley G.F, O'Callaghan C.J. Social ties and susceptibility to the common cold. J. Am. Med. Assoc. 1997;278:1231. doi: 10.1001/jama.278.15.1231b. [DOI] [PubMed] [Google Scholar]
- Edmunds W.J, O'Callaghan C.J, Nokes D.J. Who mixes with whom? A method to determine the contact patterns of adults that may lead to the spread of airborne infections. Proc. R. Soc. B. 1997;264:949–957. doi: 10.1098/rspb.1997.0131. doi:10.1098/rspb.1997.0131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichner M. Case isolation and contact tracing can prevent the spread of smallpox. Am. J. Epidemiol. 2003;158:118–128. doi: 10.1093/aje/kwg104. [DOI] [PubMed] [Google Scholar]
- Eubank S, Guclu H, Kumar V.S.A, Marathe M.V, Srinivasan A, Toroczkai Z, Wang N. Modelling disease outbreaks in realistic urban social networks. Nature. 2004;429:180–184. doi: 10.1038/nature02541. [DOI] [PubMed] [Google Scholar]
- Ferguson N.M, Garnett G.P. More realistic models of sexually transmitted disease transmission dynamics: sexual partnership networks, pair models, and moment closure. Sex. Transm. Dis. 2000;27:600–609. doi: 10.1097/00007435-200011000-00008. [DOI] [PubMed] [Google Scholar]
- Ferguson N.M, Donnelly C.A, Anderson R.M. The foot-and-mouth epidemic in Great Britain: pattern of spread and impact of interventions. Science. 2001;292:1155–1160. doi: 10.1126/science.1061020. [DOI] [PubMed] [Google Scholar]
- Frank O, Strauss D. Markov Graphs. J. Am. Stat. Soc. 1986;81:832–842. [Google Scholar]
- Fraser C, Riley S, Anderson R.M, Ferguson N.M. Factors that make an infectious disease outbreak controllable. Proc. Natl Acad. Sci. USA. 2004;101:6146–6151. doi: 10.1073/pnas.0307506101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett G.P, Anderson R.M. Sexually transmitted diseases and sexual behavior: insights from mathematical models. J. Infect. Dis. 1996;174:S150–S161. doi: 10.1093/infdis/174.supplement_2.s150. [DOI] [PubMed] [Google Scholar]
- Ghani A.C, Garnett G.P. Measuring sexual partner networks for transmission of sexually transmitted diseases. J. R. Stat. Soc. A. 1998;161:227–238. [Google Scholar]
- Ghani A.C, Garnett G.P. Risks of acquiring and transmitting sexually transmitted diseases in sexual partner networks. Sex. Transm. Dis. 2000;27:579–587. doi: 10.1097/00007435-200011000-00006. [DOI] [PubMed] [Google Scholar]
- Ghani A.C, Swinton J, Garnett G.P. The role of sexual partnership networks in the epidemiology of gonorrhea. Sex. Transm. Dis. 1997;24:45–56. doi: 10.1097/00007435-199701000-00009. [DOI] [PubMed] [Google Scholar]
- Gilbert M, Mitchell A, Bourn D, Mawdsley J, Clifton-Hadley R, Wint W. Cattle movements and bovine tuberculosis in Great Britain. Nature. 2005;435:491–496. doi: 10.1038/nature03548. [DOI] [PubMed] [Google Scholar]
- Grassberger P. On the critical behaviour of the general epidemic process and dynamical percolation. Math. Biosci. 1983;63:157–172. [Google Scholar]
- Grenfell B.T. Chance and chaos in measles dynamics. J. R. Stat. Soc. B. 1992;54:383–398. [Google Scholar]
- Grenfell B.T, Bjornstad O.N, Kappey J. Travelling waves and spatial hierarchies in measles epidemics. Nature. 2001;414:716–723. doi: 10.1038/414716a. [DOI] [PubMed] [Google Scholar]
- Grimmett G. Springer; Berlin: 1989. Percolation. [Google Scholar]
- Halloran M.E, Longini I.M, Jr., Nizam A, Yang Y. Containing bioterrorist smallpox. Science. 2002;298:1428–1432. doi: 10.1126/science.1074674. [DOI] [PubMed] [Google Scholar]
- Handcock M.S, Jones J.H. Likelihood-based inference for stochastic models of sexual network formation. Theor. Popul. Biol. 2004;65:413–422. doi: 10.1016/j.tpb.2003.09.006. [DOI] [PubMed] [Google Scholar]
- Harary F. Addison-Wesley; Reading, MA: 1969. Graph theory. [Google Scholar]
- Harris T.E. Contact interactions on a lattice. Ann. Probab. 1974;2:969–988. [Google Scholar]
- Haydon D.T, Chase-Topping M, Shaw D.J, Matthews L, Friar J.K, Wilesmith J, Woolhouse M.E.J. The construction and analysis of epidemic trees with reference to the 2001 UK foot-and-mouth outbreak. Proc. R. Soc. B. 2003;270:121–127. doi: 10.1098/rspb.2002.2191. doi:10.1098/rspb.2002.2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hethcote H.W, Yorke J.A. Springer Lecture Notes in Biomathematics. Springer; Berlin: 1984. Gonorrhea transmission dynamics and control. [Google Scholar]
- Jeong H, Tombar B, Albert R, Oltvai Z.N, Barabási A.-L. The large-scale organization of metabolic networks. Nature. 2000;407:651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- Jolly A.M, Wylie J.L. Gonorrhoea and chlamydia core groups and sexual networks in Manitoba. Sex. Transm. Infect. 2002;78:i45–i51. doi: 10.1136/sti.78.suppl_1.i145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlberg M. Testing transitivity in graphs. Soc. Networks. 1997;19:325–343. [Google Scholar]
- Keeling M.J. Modelling the persistence of measles. Trends Microbiol. 1997;5:513–518. doi: 10.1016/S0966-842X(97)01147-5. [DOI] [PubMed] [Google Scholar]
- Keeling M.J. The effects of local spatial structure on epidemiological invasions. Proc. R. Soc. B. 1999;266:859–867. doi: 10.1098/rspb.1999.0716. doi:10.1098/rspb.1999.0716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling M.J. Implications of network structure for epidemic dynamics. Theor. Popul. Biol. 2005;67:1–8. doi: 10.1016/j.tpb.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Keeling M.J, Rand D.A, Morris A.J. Correlation models for childhood epidemics. Proc. R. Soc. B. 1997;264:1149–1156. doi: 10.1098/rspb.1997.0159. doi:10.1098/rspb.1997.0159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling M.J, Rohani P, Grenfell B.T. Seasonally-forced disease dynamics explored as switching between attractors. Physica D. 2001;148:317–335. [Google Scholar]
- Kermack W.O, McKendrick A.G. A contribution to the mathematical theory of epidemics. Proc. R. Soc. A. 1927;115:700–721. [Google Scholar]
- Klovdahl A.S. Social networks and the spread of infectious diseases: the AIDS example. Soc. Sci. Med. 1985;21:1203–1216. doi: 10.1016/0277-9536(85)90269-2. [DOI] [PubMed] [Google Scholar]
- Klovdahl A.S. Networks and pathogens. Sex. Transm. Dis. 2001;28:25–28. doi: 10.1097/00007435-200101000-00006. [DOI] [PubMed] [Google Scholar]
- Klovdahl A.S, Dhofier Z, Oddy G, O'Hara J, Stoutjesdijk S, Whish A. Social networks in an urban area: first Canberra study Aust. N. Z. J. Sociol. 1977;13:169–172. [Google Scholar]
- Kretzschmar M, van Duynhoven Y.T.H.P, Severijnen A.J. Modeling prevention strategies for gonorrhea and chlamydia using stochastic network simulations. Am. J. Epidem. 1996;144:306–317. doi: 10.1093/oxfordjournals.aje.a008926. [DOI] [PubMed] [Google Scholar]
- Kuperman M, Abramson G. Small world effects in an epidemiological model. Phys. Rev. Lett. 2001;86:2909–2912. doi: 10.1103/PhysRevLett.86.2909. [DOI] [PubMed] [Google Scholar]
- Leinhardt S, editor. Social networks: a developing paradigm. Academic Press; New York: 1977. [Google Scholar]
- Liljeros F, Edling C.R, Amaral L.A.N, Stanley H.E, Aberg Y. The web of human sexual contacts. Nature. 2001;411:907–908. doi: 10.1038/35082140. [DOI] [PubMed] [Google Scholar]
- Liljeros F, Edling C.R, Amaral L.A.N. Sexual networks: implications for the transmission of sexually transmitted infections. Microbes Infect. 2003;5:189–196. doi: 10.1016/s1286-4579(02)00058-8. [DOI] [PubMed] [Google Scholar]
- Lloyd A.L, May R.M. How viruses spread among computers and people. Science. 2001;292:1316–1317. doi: 10.1126/science.1061076. [DOI] [PubMed] [Google Scholar]
- Meyers L.A, Pourbohloul B, Newman M.E.J, Skowronski D.M, Brunham R.C. Network theory and SARS: predicting outbreak diversity. J. Theor. Biol. 2005;232:71–81. doi: 10.1016/j.jtbi.2004.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milgram S. The small world problem. Psychol. Today. 1967;1:61–67. [Google Scholar]
- Mollison D. Spatial contact models for ecological and epidemic spread. J. R. Stat. Soc. 1977;39:283–326. [Google Scholar]
- Moore C, Newman M. Epidemics and percolation in small-world networks. Phys. Rev. E. 2000;61:5678–5682. doi: 10.1103/physreve.61.5678. [DOI] [PubMed] [Google Scholar]
- Morris M. Sexual networks and HIV. AIDS. 1997;11:S209–S216. [PubMed] [Google Scholar]
- Müller J, Kretzschmar M, Dietz K. Contact tracing in stochastic and deterministic epidemic models. Math. Biosci. 2000;164:39–64. doi: 10.1016/s0025-5564(99)00061-9. [DOI] [PubMed] [Google Scholar]
- National Audit Office . The Stationary Office; London: 2003. Identifying and tracking livestock in England. [Google Scholar]
- Newman M.E.J. The structure of scientific collaboration networks. Proc. Natl Acad. Sci. USA. 2001;98:404–409. doi: 10.1073/pnas.021544898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman M.E.J. Spread of epidemic disease on networks. Phys Rev. E. 2002;66:016128. doi: 10.1103/PhysRevE.66.016128. [DOI] [PubMed] [Google Scholar]
- Newman M.E.J, Watts D.J. Scaling and percolation in the small-world network model. Phys Rev. E. 1999;60:7332–7342. doi: 10.1103/physreve.60.7332. [DOI] [PubMed] [Google Scholar]
- Olsen L.F, Truty G.L, Schaffer W.M. Oscillations and chaos in epidemics: a nonlinear dynamic study of six childhood diseases in Copenhagen, Denmark. Theor. Pop. Biol. 1986;33:344–370. doi: 10.1016/0040-5809(88)90019-6. [DOI] [PubMed] [Google Scholar]
- Pastor-Satorras R, Vespignani A. Epidemic spreading in scale-free networks. Phys. Rev. Lett. 2001;86:3200–3203. doi: 10.1103/PhysRevLett.86.3200. [DOI] [PubMed] [Google Scholar]
- Potterat J.J, Rothenberg R.B, Muth S.Q. Network structural dynamics and infectious disease propagation. Int. J. STD AIDS. 1999;10:182–185. doi: 10.1258/0956462991913853. [DOI] [PubMed] [Google Scholar]
- Potterat J.J, Philips-Plummer L, Muth S.Q, Rothenberg R.B, Woodhouse D.E, Maldonado-Long T.S, Zimmerman H.P, Muth J.B. Risk network structure in the early epidemic phase of HIV transmission in Colorado Springs. Sex. Transm. Infect. 2002;78:i159–i163. doi: 10.1136/sti.78.suppl_1.i159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand D.A, Wilson H.B. Chaotic stochasticity—a ubiquitous source of unpredictability in epidemics. Proc. R. Soc. B. 1991;246:179–184. doi: 10.1098/rspb.1991.0142. [DOI] [PubMed] [Google Scholar]
- Read J.M, Keeling M.J. Disease evolution on networks: the role of contact structure. Proc. R. Soc. B. 2003;270:699–708. doi: 10.1098/rspb.2002.2305. doi:10.1098/rspb.2002.2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes C.J, Anderson R.M. Epidemic thresholds and vaccination in a lattice model of disease spread. Theor. Popul. Biol. 1997;52:101–118. doi: 10.1006/tpbi.1997.1323. [DOI] [PubMed] [Google Scholar]
- Rhodes C.J, Jensen H.J, Anderson R.M. On the critical behaviour of simple epidemics. Proc. R. Soc. B. 1997;264:1639–1646. doi: 10.1098/rspb.1997.0228. doi:10.1098/rspb.1997.0228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley S, et al. Transmission dynamics of the etiological agent of SARS in Hong Kong: impact of public health interventions. Science. 2003;300:1961–1966. doi: 10.1126/science.1086478. [DOI] [PubMed] [Google Scholar]
- Robins G, Pattison P, Woolcock J. Missing data in networks: exponential random graph (p*) models for networks with non-respondents. Soc. Networks. 2004;26:257–283. [Google Scholar]
- Rohani P, Earn D.J.D, Grenfell B.T. The impact of immunisation on pertussis transmission in England and Wales. Lancet. 2000;355:285–286. doi: 10.1016/S0140-6736(99)04482-7. [DOI] [PubMed] [Google Scholar]
- Rothenberg R. Commentary—how a net works—implications of network structure for the persistence and control of sexually transmitted diseases and HIV. Sex. Transm. Dis. 2001;28:63–68. doi: 10.1097/00007435-200102000-00001. [DOI] [PubMed] [Google Scholar]
- Rothenberg R. STD transmission dynamics: some current complexities—2002 Thomas Parran Award Lecture. Sex. Transm. Dis. 2003;30:478–482. doi: 10.1097/00007435-200306000-00002. [DOI] [PubMed] [Google Scholar]
- Rothenberg R.B, Potterat J.J, Woodhouse D.E, Muth S.Q, Darrow W.W, Klovdahl A.S. Social network dynamics and HIV transmission. AIDS. 1998;12:1529–1536. doi: 10.1097/00002030-199812000-00016. [DOI] [PubMed] [Google Scholar]
- Rozenfeld A.F, Cohen R, ben-Avraham D, Havlin S. Scale-free networks on lattices. Phys. Rev. Lett. 2002;89:218701. doi: 10.1103/PhysRevLett.89.218701. [DOI] [PubMed] [Google Scholar]
- Schwartz I.B. Multiple recurrent outbreaks and predictability in seasonally forced nonlinear epidemic models. J. Math. Biol. 1985;18:233–253. doi: 10.1007/BF00276232. [DOI] [PubMed] [Google Scholar]
- Scott J. SAGE Publications; London: 1991. Social network analysis: a handbook. [Google Scholar]
- Snijders T.A.B. The statistical evaluation of social network dynamics. Sociol. Methodol. 2001;31:361–395. [Google Scholar]
- Szendrői B, Csányi G. Polynomial epidemics and clustering in contact networks. Proc. R. Soc. B. 2004;271:S364–S366. doi: 10.1098/rsbl.2004.0188. doi:10.1098/rsbl.2004.0188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers J, Milgram S. An experimental study of the small world problem. Sociometry. 1969;32:425–443. [Google Scholar]
- Wallinga J, Edmunds W.J, Kretzschmar M. Perspective: human contact patterns and the spread of airborne infectious diseases. Trends Microbiol. 1999;7:372–377. doi: 10.1016/s0966-842x(99)01546-2. [DOI] [PubMed] [Google Scholar]
- Warren C.P, Sander L.M, Sokolov I.M. Geography in a scale-free network model. Phys. Rev. E. 2002;66:056105. doi: 10.1103/PhysRevE.66.056105. [DOI] [PubMed] [Google Scholar]
- Wasserman S, Faust K. Cambridge University Press; Cambridge: 1994. Social network analysis. [Google Scholar]
- Watts D.J. Princeton University Press; Princeton: 1999. Small worlds: the dynamics of networks between order and randomness. [Google Scholar]
- Watts D.J, Strogatz S.H. Collective dynamics of ‘small-world’ networks. Nature. 1998;393:440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- West D.B. Prentice Hall; Upper Saddle River, NJ: 1996. Introduction to graph theory. [Google Scholar]
- Woodhouse D.E, et al. Mapping a social network of heterosexuals at high risk for HIV infection. AIDS. 1994;8:1331–1336. doi: 10.1097/00002030-199409000-00018. [DOI] [PubMed] [Google Scholar]
- Wylie J.L, Jolly A. Patterns of chlamydia and gonorrhea infection in sexual networks in Manitoba, Canada. Sex. Transm. Dis. 2001;28:14–24. doi: 10.1097/00007435-200101000-00005. [DOI] [PubMed] [Google Scholar]