Abstract
The stability of OmpA in large unilamellar vesicles of dilauroyl phosphatidylcholine was studied using different concentrations of urea. The effective energy of unfolding, as determined from refolding experiments, is greater than that for small sonicated unilamellar vesicles by an amount that is compatible with estimates of the elastic energy of highly curved vesicles. The on-rate for refolding and insertion is slower for large unilamellar vesicles than for small unilamellar vesicles, which indicates a contribution of vesicle strain also to the free energy of the transition state.
Certain β-barrel outer-membrane proteins from Gram-negative bacteria that are unfolded in urea are able, on dilution, to insert spontaneously into membranes of a variety of lipids, provided that they are in the form of small unilamellar vesicles (SUVs), which are produced by limit sonication (1–3) . However, the major outer membrane protein from Escherichia coli (OmpA), for instance, is unable to insert spontaneously into large unilamellar vesicles (LUVs), unless they are composed of short-chain phospholipids (n ≤ 12), which form highly flexible membranes (2,4).
In an elegant series of experiments, Hong and Tamm (5) established conditions for the reversible folding of OmpA in SUVs, and used this to investigate the thermodynamic stability of OmpA in a systematic range of phospholipids. Because of the dramatic differences in potential for spontaneous protein insertion between LUVs and SUVs, it is of interest to attempt studies of the stability of OmpA in LUVs. This is done here, together with consideration of the contribution of vesicle curvature elasticity to protein stability, which can at least partially explain the much higher stability of OmpA in LUVs than in SUVs.
Fig. 1 shows refolding curves for insertion of OmpA into extruded LUVs, from different concentrations of aqueous urea (solid symbols). Qualitatively similar results are obtained from LUVs of dilauroyl phosphatidylcholine (diC12PC) and of a 1:1 mol/mol mixture of diC12PC with dilauroyl phosphatidylglycerol (diC12PG). The effective free energies of folding, obtained according to Fig. 1, are 22 ± 2 and 16 ± 2 kJ.mol−1 (m′ ≈ 3.4 and 3.3 M−1) for refolding into diC12PC and into diC12PC:diC12PG, respectively, after a 1-day incubation. These values decrease to effective unfolding energies of ΔGu = 15 ± 5 and 13 ± 5 kJ.mol−1 (m′ ≈ 2.3 and 2.5 M−1), respectively, after an incubation of 12 days. On the other hand, incubation in up to 9.6 M urea for 12 days at 40°C does not induce any detectable unfolding of OmpA that is already inserted in LUVs of either lipid composition (open symbols in Fig. 1).
FIGURE 1.
Refolding (solid symbols) and denaturation (open symbols) curves of OmpA with LUVs (lipid/protein 800 mol/mol) of (A) diC12PC and (B) diC12PC/diC12PG 1:1 mol/mol. Samples were incubated at 40°C in 10 mM borax, 2 mM EDTA, pH 10.0 for 1 day (circles), or 12 days (squares). Average tryptophan fluorescence emission wavelength, 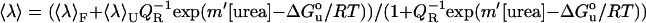 , is plotted against denaturant concentration, [urea]. For a two-state process, ΔGU is the free energy of unfolding in water alone, where 〈λ〉F and 〈λ〉U are average emission wavelengths, and QR is the ratio of the fluorescence intensities, for the folded and unfolded protein, respectively (5,6).
, is plotted against denaturant concentration, [urea]. For a two-state process, ΔGU is the free energy of unfolding in water alone, where 〈λ〉F and 〈λ〉U are average emission wavelengths, and QR is the ratio of the fluorescence intensities, for the folded and unfolded protein, respectively (5,6).
These results for LUVs differ considerably from those obtained with sonicated SUVs by Hong and Tamm (5). In the latter case, reversible folding and unfolding of OmpA was demonstrated on overnight incubation in urea at pH 10.0 and a temperature of 37.5°C. Inclusion of at least 7.5 mol % of PG in the SUVs was required to obtain these results. Also, the free energy and cooperativity of folding were found to be considerably lower than the effective values obtained here in refolding experiments with LUVs. The reversible free energy of unfolding of OmpA in SUVs at 37.5°C, extrapolated to 92.5% diC12PC plus 7.5% 1-palmitoyl-2-oleoyl PG, amounts to only 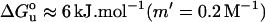 (5). Vesicle curvature therefore has a very significant effect on the stability of OmpA in SUVs of diC12PC.
(5). Vesicle curvature therefore has a very significant effect on the stability of OmpA in SUVs of diC12PC.
The free energy of bending that is associated with each monolayer of a curved vesicle is given by (7)
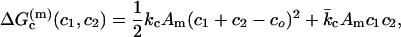 |
(1) |
where c1 and c2 are the principal curvatures, kc and 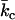 are the elastic moduli of mean and Gaussian curvature of the monolayer, respectively, co is the spontaneous curvature of the constituent lipid monolayers, and Am is the area of the monolayer surface at the neutral plane. For a spherical vesicle, the areas at the neutral surfaces of the outer and inner monolayers are given, respectively, by:
are the elastic moduli of mean and Gaussian curvature of the monolayer, respectively, co is the spontaneous curvature of the constituent lipid monolayers, and Am is the area of the monolayer surface at the neutral plane. For a spherical vesicle, the areas at the neutral surfaces of the outer and inner monolayers are given, respectively, by:
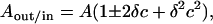 |
(2) |
where A and c (= 1/Rves) are the area and curvature, respectively, at the bilayer midplane, and ±δ are the distances of the neutral surfaces from the midplane (see Fig. 2). Correspondingly, the curvatures at the neutral planes of the outer and inner monolayers of a spherical vesicle are given, respectively, by:
 |
(3) |
where an outward curvature is defined as positive, and an inward curvature (as in lipid HII phases) is negative.
FIGURE 2.
Geometry of outer (out) and inner (in) monolayers in a spherical vesicle. Curvatures of the two neutral surfaces are given by cout = 1/(Rves + δ) and cin = −1/(Rves − δ). Areas at the two neutral surfaces are Aout = A(1 + δc)2 and Ain = A(1 − δc)2, where c = 1/Rves is the curvature of the vesicle. The asymmetric environment in which a transmembrane protein, P, finds itself in a SUV is indicated schematically by the different degree of projection from the vesicle surface in the outer and inner monolayers.
Summing free energy contributions from the outer and inner monolayers of the bilayer membrane (taking due account of both sign and magnitude of the different curvatures, and of the different numbers of lipids in the two monolayers) yields the following result for the bending free energy of a spherical vesicle:
 |
(4) |
This value is expressed relative to a planar bilayer of the same area, for which c = 0 and the elastic curvature energy is simply 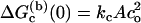 . For SUVs with diameters of 30 nm, this elastic energy corresponds to ∼950 kJ/mol, assuming values typical for dioleoyl phosphatidylcholine (diC18:1PC) of
. For SUVs with diameters of 30 nm, this elastic energy corresponds to ∼950 kJ/mol, assuming values typical for dioleoyl phosphatidylcholine (diC18:1PC) of  ,
, 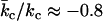 , co ≈ −0.1 nm−1, and δ ≈ 1.5 nm for a monolayer (8–11). This is a very conservative estimate because sonicated vesicles are highly strained and the effective bending rigidity, under these conditions, is possibly much higher.
, co ≈ −0.1 nm−1, and δ ≈ 1.5 nm for a monolayer (8–11). This is a very conservative estimate because sonicated vesicles are highly strained and the effective bending rigidity, under these conditions, is possibly much higher.
In a fluid bilayer of diC18:1PC, the area per lipid molecule is AL ≈ 0.72 nm2 (12). The curvature free energy of diC18:1PC SUVs thus amounts to minimally ∼120 J.mol−1 per lipid molecule. The cross-sectional area of OmpA corresponds to ∼15 lipid molecules (13), and the number of first-shell lipids that can be accommodated around the intramembranous perimeter is nb ≈ 20 (14). Perturbations of lipid curvature by incorporation and folding of OmpA therefore can influence the protein stability in SUVs appreciably. Alleviation of curvature stress by protein incorporation in SUVs of diC18:1PC would hence tend to stabilize the inserted protein. This is in accordance with the observation that OmpA inserts spontaneously into SUVs of diC18:1 (or dimyristoyl) PC, but not into LUVs composed of either of these lipids (2,4).
As seen from Eq. 4, the elastic free energy of SUVs depends directly on the spontaneous curvature, co, of the constituent lipids. For lipids with  (∼+0.4 nm−1), the sign of the overall bending energy changes and becomes negative. The reason for this is that the outer monolayer of an SUV, which contains 50% more lipids than the inner monolayer, has a positive curvature. There are no experimental measurements of the spontaneous curvature for diC12PC. However, considerations of lipid molecular shape suggest that the spontaneous curvature for diC12PC is positive and greater in absolute magnitude than that for diC18:1PC (9,15). It is therefore perfectly conceivable that incorporation of OmpA in SUVs of diC12PC could be unfavorable as regards bending free energy of the vesicle and hence destabilize the protein relative to the state in LUVs.
(∼+0.4 nm−1), the sign of the overall bending energy changes and becomes negative. The reason for this is that the outer monolayer of an SUV, which contains 50% more lipids than the inner monolayer, has a positive curvature. There are no experimental measurements of the spontaneous curvature for diC12PC. However, considerations of lipid molecular shape suggest that the spontaneous curvature for diC12PC is positive and greater in absolute magnitude than that for diC18:1PC (9,15). It is therefore perfectly conceivable that incorporation of OmpA in SUVs of diC12PC could be unfavorable as regards bending free energy of the vesicle and hence destabilize the protein relative to the state in LUVs.
There is another aspect of the transbilayer asymmetry in SUVs that also could affect the stability of inserted proteins. An approximately cylindrical β-barrel protein is not readily compatible with the transmembrane lipid packing in SUVs (see Fig. 2), particularly at the inner monolayer, which is less extensive (Ain < Aout) and more highly curved (|cin| > |cout|) than is the outer monolayer. Almost certainly, protein incorporation will require transfer of some lipid molecules between the two monolayers. The change in curvature free energy on transfer of a lipid molecule from the inner to the outer monolayer is −4 kcALcco per lipid, according to Eqs. 1 and 3. This amounts to ∼−5.0 kJ.mol−1.nm × co for a 30-nm vesicle and is determined directly by the magnitude and sign of the intrinsic curvature, co, of the lipid molecule. This contribution from vesicle rearrangement may therefore have either a stabilizing or a destabilizing effect depending on the particular lipid and on the direction of lipid transfer.
It is thus quite possible that the above effects of curvature elasticity could together account for the apparent destabilization (i.e., reduction in unfolding energy) of OmpA in SUVs, relative to LUVs, which is ΔΔGu ≈ −(7–9 ± 5) kJ.mol−1. In this connection, it should be taken into account that the effective bending rigidity for highly curved, sonicated SUVs may be considerably larger than assumed here (see above) and that equilibrium refolding might not have been achieved in LUVs, even after such a long incubation time.
The unfolding energy ΔGu contributes also to the activation free energy for unfolding: 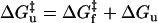 , where
, where 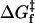 is the activation free energy for folding. A change of the size measured here, i.e., ΔΔGu = 7–9 kJ.mol−1 relative to SUVs, would decrease the unfolding rate by a factor of ≤30-fold. This alone would be insufficient to account for the very slow (i.e., undetectable) unfolding rates in LUVs, relative to SUVs. Most probably also the free energy of the transition state
is the activation free energy for folding. A change of the size measured here, i.e., ΔΔGu = 7–9 kJ.mol−1 relative to SUVs, would decrease the unfolding rate by a factor of ≤30-fold. This alone would be insufficient to account for the very slow (i.e., undetectable) unfolding rates in LUVs, relative to SUVs. Most probably also the free energy of the transition state 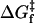 is reduced in SUVs, relative to LUVs, which should then be reflected in the on-rates. Fig. 3 shows results on folding kinetics for insertion of OmpA into diC12PC vesicles of different diameters. The forward rate constant is 0.067 min−1 for 30-nm sonicated SUVs, and has a mean value of 0.013 ± 0.004 min−1 for extruded LUVs of diameter 100 nm and greater. Taken together with the experimental difference in unfolding energy, this factor of 5× in on-rate would predict a difference in off-rates (i.e., in rates of unfolding) of maximally 150-fold. This could be sufficient to explain the difference between overnight incubation of SUVs and a 12-day incubation of LUVs, i.e., the apparent irreversibility of folding in LUVs may simply be a kinetic limitation.
is reduced in SUVs, relative to LUVs, which should then be reflected in the on-rates. Fig. 3 shows results on folding kinetics for insertion of OmpA into diC12PC vesicles of different diameters. The forward rate constant is 0.067 min−1 for 30-nm sonicated SUVs, and has a mean value of 0.013 ± 0.004 min−1 for extruded LUVs of diameter 100 nm and greater. Taken together with the experimental difference in unfolding energy, this factor of 5× in on-rate would predict a difference in off-rates (i.e., in rates of unfolding) of maximally 150-fold. This could be sufficient to explain the difference between overnight incubation of SUVs and a 12-day incubation of LUVs, i.e., the apparent irreversibility of folding in LUVs may simply be a kinetic limitation.
FIGURE 3.
Folding kinetics of OmpA into diC12PC vesicles of different diameters at 20°C. (A) SDS-PAGE of OmpA (17 μM) at different times of incubation after dilution from urea into 7 mM lipid with vesicle diameters indicated. Folding is accompanied by a downward gel shift to lower effective molecular mass. (b) Fraction of folded OmpA with time determined from gel densitometry. For further details of methods, see Kleinschmidt and Tamm (2).
References
- 1.Surrey, T., and F. Jähnig. 1992. Refolding and oriented insertion of a membrane protein into a lipid bilayer. Proc. Natl. Acad. Sci. USA. 89:7457–7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kleinschmidt, J. H., and L. K. Tamm. 2002. Secondary and tertiary structure formation of the β-barrel membrane protein OmpA is synchronized and depends on membrane thickness. J. Mol. Biol. 324:319–330. [DOI] [PubMed] [Google Scholar]
- 3.Pocanschi, C. L., H. J. Apell, P. Puntervoll, B. Hogh, H. B. Jensen, W. Welte, and J. H. Kleinschmidt. 2006. The major outer membrane protein of Fusobacterium nucleatum (FomA) folds and inserts into lipid bilayers via parallel folding pathways. J. Mol. Biol. 355:548–561. [DOI] [PubMed] [Google Scholar]
- 4.Marsh, D., B. Shanmugavadivu, and J. H. Kleinschmidt. 2006. Membrane elastic fluctuations and the insertion and tilt of β-barrel proteins. Biophys. J. 91:227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hong, H., and L. K. Tamm. 2004. Elastic coupling of integral membrane protein stability to lipid bilayer forces. Proc. Natl. Acad. Sci. USA. 101:4065–4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Royer, C. A., C. J. Mann, and C. R. Matthews. 1993. Resolution of the fluorescence equilibrium unfolding profile of Trp aporepressor using single tryptophan mutants. Protein Sci. 2:1844–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Helfrich, W. 1973. Elastic properties of lipid bilayers: theory and possible experiments. Z. Naturforsch. [C]28:693–703. [DOI] [PubMed] [Google Scholar]
- 8.Chen, Z., and R. P. Rand. 1997. The influence of cholesterol on phospholipid membrane curvature and bending elasticity. Biophys. J. 73:267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marsh, D. 1996. Intrinsic curvature in normal and inverted lipid structures and in membranes. Biophys. J. 70:2248–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Templer, R. H., B. J. Khoo, and J. M. Seddon. 1998. Gaussian curvature modulus of an amphiphilic monolayer. Langmuir. 14:7427–7434. [Google Scholar]
- 11.Siegel, D. P., and M. M. Kozlov. 2004. The Gaussian curvature elastic modulus of N-monomethylated dioleoylphosphatidylethanolamine: relevance to membrane fusion and lipid phase behavior. Biophys. J. 87:366–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagle, J. F., and S. Tristram-Nagle. 2000. Structure of lipid bilayers. Biochim. Biophys. Acta. 1469:159–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pautsch, A., and G. E. Schulz. 1998. Structure of the outer membrane protein A transmembrane domain. Nat. Struct. Biol. 5:1013–1017. [DOI] [PubMed] [Google Scholar]
- 14.Páli, T., D. Bashtovyy, and D. Marsh. 2006. Stoichiometry of lipid interaction with transmembrane proteins, deduced from the 3-D structures. Protein Sci. 15:1153–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marsh, D. 1997. Nonlamellar packing parameters for diacylglycerols. Biophys. J. 72:2834–2836. [DOI] [PMC free article] [PubMed] [Google Scholar]





