Abstract
Potassium channel activation regulates cellular excitability in cells such as neurons and heart. Ion channel activity relies on a switching mechanism between two conformations, the open and closed states, known as gating. It has been suggested that potassium channels are gated via a pivoted mechanism of the pore-lining helix. Our analysis suggests that hinging occurs at the residue immediately preceding the central glycine of the inner helix. Furthermore, we show that the highly conserved central glycine is necessary to prevent constraining interactions with critical residues in its vicinity, including those located in the selectivity filter. We show that such interactions can impair channel function, and that upon their removal channel activity can be restored.
INTRODUCTION
Potassium (K+) channel activation modulates cellular excitability in several physiological systems including that of neuronal and cardiac cells. A switching mechanism between channel open and closed states known as gating regulates activity and is triggered by a stimulus such as a change in membrane voltage or ligand binding.
Comparison of two related bacterial channels suggested that the highly conserved central glycine of the inner helix plays the role of a gating hinge (1). The MthK channel structure includes its gating machinery (the gating ring) and contains a bound ligand (a Ca2+ ion) that causes the channel to reside in an open state (PDB code 1LNQ). This structure was compared to the structure of the closed KcsA bacterial potassium channel (PDB code 1K4C) (2). A comparison of the two structures revealed large differences in the inner helices: in KcsA they are almost straight, whereas in MthK they are bent ∼30°. It was thus concluded that the bend in MthK occurs at the highly conserved central glycine, which corresponds to Gly-83 in MthK (1) and to Gly-99 in KcsA (2). This was attributed to the unique flexibility of this highly conserved glycine, which is conserved in 81% of potassium channels, following its ability to adopt a wide range of main-chain dihedral angles. The corresponding central glycine of the homomeric Kir3.4* (or GIRK4*) channel (i.e., Kir3.4(S143T) (3)), an inwardly rectifying potassium channel gated by the βγ subunits of G-proteins (4,5), was mutated to all other amino acids to investigate its role in the gating mechanism of the channel (6). These experiments showed that in most cases the mutants exhibited low-level activity with characteristic long bursts of activity and long silent periods, suggesting that the flexibility of the glycine at this position is required for ensuring the frequent gating of the helix bundle crossing (HBC) of the channel. The HBC is defined as the position in the inner helix (TM2) at which the distance between the α-carbon of the residue at this position and its counterpart on the diagonally facing subunit reaches a minimum. In Kir3.4 (GIRK4), position 187 was suggested to be the HBC and to serve as a gate (6).
Following these studies, additional studies in potassium and sodium channels have suggested that the central glycine of the inner helix is located at the position at which the inner helix undergoes a rigid body movement of a pivoted bend during gating or in its vicinity, thus playing the role of a gating hinge. These studies include structural studies that involve a comparison between KcsA and the voltage gated potassium channels, KvAP (7) (PDB code 1ORQ), functional investigations of two eukaryotic voltage dependent potassium channels, Shaker (8,9) and BK (8), and the study of the bacterial Na+ channel NaChBac (10). In the structural study of the bacterial KvAP channel, it was suggested that the pore of the channel is in an open conformation, and that the inner helix bends at the central glycine. This was attributed to crystallographic packing forces. In the functional studies that concentrated on the Shaker and the BK channels, the central glycine was mutated to Ala among other residues. The alanine mutation resulted in lack of function in the case of the Shaker and in reduced currents in the case of the BK channels, which was attributed to the loss of flexibility due to the loss of the glycine. Several observations do not fit with this interpretation. In the case of the Kir3.4* (GIRK4*) channel, mutation of the central glycine to alanine did not result in reduced currents. Among the Shaker and the BK channels themselves there is large variation in the effect of mutation of the central glycine to alanine. Also, in studies concerned with members of the KCNQ-type potassium channels (11,12), it was found that whereas KCNQ1 (Kv7.1) has an alanine at the equivalent position and is functional, mutation of the central glycine in KCNQ2 (Kv7.2) to an alanine results in a nonfunctional channel. In addition, whereas enhanced flexibility induced by glycine residues in transmembrane α-helices has been demonstrated by molecular dynamics simulations (13,14), in a simulation of the G99A mutant of KcsA, it was found that the channel opening trajectory of the mutant was very similar to that of the wild-type KcsA (15). In fact, in this study the central glycine was viewed as a part of an extended kink region that includes residues 95-105 of KcsA. Further inconsistencies are also obtained when comparing the result of the mutation of the central glycine to proline in Shaker, in Kir3.4* and in NaChBac. Whereas in Shaker and in NaChBac, this mutation resulted in enhanced currents, in Kir3.4*, the currents were significantly decreased. Furthermore, following the structural study of the bacterial KirBac1.1 potassium channel (16) (PDB code 1P7B), it was suggested that another conserved glycine located nine residues below the central glycine is the gating hinge. This glycine is conserved among ∼41% of potassium channels. Moreover, in the structural study of the mammalian Shaker family Kv1.2 channel (17) (PDB code 2A79), it was suggested that the pore of the channel is open due to bending at the PVP motif, whose second proline is located at the position that corresponds to the lower glycine. This was also suggested following the mutagenesis-based studies of this motif in the Shaker-type Kv1.5 channel (18) and of the equivalent motif, PAG, in KCNQ channels (12).
Here, we first address the question of the location of the gating hinge of potassium channels. We identify in the crystallographic structures of both the open ligand gated MthK and voltage gated KvAP channels the residue located immediately above the central glycine as the gating hinge of potassium channels. Comparison between the closed KcsA and the open MthK and KvAP channels and examination of the properties of the equivalent residue for the case of the G protein gated inwardly rectifying potassium channel, Kir3.4* (GIRK4*), further support this observation. We then address the question of the importance of the central glycine to gating through the examination of the effect of its mutation to other residues in the Kir3.4* channel. We find that mutations of the central glycine give rise to constraining interactions with critical residues in its vicinity, including those located in the signature sequence of the selectivity filter. We conclude that the small size of the glycine is key in avoiding such constraining interactions.
MATERIALS AND METHODS
Conformational memories
The starting coordinates of the 28-residue TM2 segment of Kir3.4* were modeled as α-helix following the homology model of Kir3.4* to KCSA (6). The method of conformational memories (19–20) included two phases: the exploratory phase and the biased sampling phase. The Monte Carlo (21) simulations of the TM2 were performed by varying the torsional angles. Backbone dihedral angles were restrained to ±50° from the values of dihedral angles in α-helices: (φ,ψ) = (−57°,−47°). Side-chain dihedral angles were rotated freely. In each round in the exploratory phase, repeated runs of Monte Carlo (21) simulated annealing (22) were performed from a starting temperature of T1 = 4000K, with a cooling schedule of Tn+1 = 0.9Tn, and 30,000 steps per temperature to reach 295 K. In the sampling phase, the starting temperature in each round was T1 = 897 K. Analysis of the resulting conformations was performed at Tn = 295 K. The environment was modeled by a distance-dependent dielectric. The force field for the energy calculation, was the CHARMM force field (23). To reach convergence of the resulting conformations, 200 rounds of independent random simulations were performed for each construct. 100 representative structures of each TM2 construct were selected for analysis.
Construction of cumulative structures for the analysis of the differences between crystallographic structures
Before the construction of the cumulative structures, the backbone dihedral angles of inner helix residues that correspond to positions 89–110 of KcsA were determined for KcsA, MthK, and KvAP using CHARMM (23). The starting coordinates for constructing structures that represent the cumulative effect of changing the dihedral angles of a closed conformation of the channel to an open conformation was the crystallographic structure of the KcsA tetramer (2). The first structure was obtained by changing the backbone dihedral ψ angle of position 89 in the different subunits of KcsA to the value of the corresponding dihedral angle in the open conformation examined (MthK or KvAP) using the molecular graphics analysis program Quanta (Accelrys). The second structure was obtained by changing the values of the backbone dihedral φ and ψ angles of position 90 in KcsA to the value of the corresponding angles in the open conformation in addition to the change of the dihedral angle of position 89. This procedure was continued up to position 110.
Molecular modeling via restrained molecular dynamics
MD simulations were performed using CHARMM (23) version 26. The initial structure of Kir3.4* was constructed as a homology model of the KcsA crystallographic structure (6). We used structurally restrained molecular dynamics simulations that maintain the structural integrity of the helical TM domain. Specifically, we imposed NOE type restraints on α-helical backbone distances as well as harmonic restraints on the Cα atoms of residues in TM1. Following a minimization stage using the Steepest Descent and the adopted-basis Newton Raphson algorithms, a 1.2-ns MD simulation at room temperature was carried out. The environment was modeled by a distance-dependent dielectric. The force field for the energy calculation is the CHARMM force field (23). Comparison of the structure of KcsA with the structure of KirBac1.1 that was crystallized later shows that the main differences are in the TM1 helix as well as in the C-terminal part of the inner helix (Supplemental Fig. S1 A). In the simulation we have examined the region that includes the central and N-terminal parts of the inner helix as well as the N-terminal region of the signature sequence of the channel and adjacent residues in the pore helix. In this region, KcsA and KirBac1.1 almost overlap, such that the RMSD (root mean square deviation) is 0.75Å (Fig. S1 B). As a result and in view of the constraints applied in the simulations, the results obtained using a homology model based on the KcsA structure would be consistent with results that would be obtained on the basis of a homology model that is based on the more recent KirBac1.1 structure.
Expression of recombinant channels in Xenopus oocytes
Point mutations on the background of the control or GFP tagged channel Kir3.4(S143T) (Kir3.4*) (24) were generated using the Quickchange site-directed mutagenesis kit (Stratagene, La Jolla, CA). cRNAs were transcribed in vitro using the “Message Machine” kit (Ambion, Austin, TX). cRNA concentration of Kir3.4* was estimated from two successive dilutions, which were electrophoresed in parallel on formaldehyde gels and compared to known concentrations of an RNA marker (GIBCO, Gaithersburg, MD). Oocytes were isolated and microinjected as previously described (25). Expression of channel proteins in oocytes was accomplished by injection of the desired amount of cRNA into Xenopus oocytes. In all two-electrode voltage clamp experiments, oocytes were injected with cRNA, 2 ng of channel, and 2ng of each G protein subunit or βARK1-pH domain or Muscarinic type 2 (M2) receptor when required. In the single-channel experiments oocytes were injected with 0.1-1ng of channel cRNA. All oocytes were maintained at 17°C. Two-electrode voltage clamp recordings were performed 2 days following injection. Single channel recordings were performed 1–2 days after injection.
GFP surface expression
Detection of GFP-tagged channels in oocytes was performed as was previously described (26). RNA from GFP-tagged channels was injected into oocytes and, 2–3 days after injection, Xenopus oocytes were fixed in 4% paraformaldehyde overnight at room temperature. Fixed oocytes were embedded in 2.5% agarose, and 50-μm sections were cut using a Vibratome. The cut sections were mounted on cover slips and imaged using a Leica TCS confocal microscope. To compare fluorescence intensities among different oocyte sections, image acquisition parameters such as pinhole size, intensity, and offset were kept constant.
Two-electrode voltage-clamp recording and analysis
Whole-cell currents were measured by conventional two-microelectrode voltage clamp with a GeneClamp 500 amplifier (Axon Instruments, Union City, CA), as previously reported (25). A high-potassium (HK) solution was used to superfuse oocytes (in mM: 96 KCl, 1 NaCl, 1 MgCl2, 5 KOH/HEPES [pH 7.4]). Basal currents represent the difference of inward currents obtained (at −80 mV) in the presence of 3 mM BaCl2 in HK solution from those in the absence of Ba2+. To evoke receptor-dependent Kir3.4* currents (IK,ACh) (27), various concentrations of acetylcholine in high K solution were used. Kir3.4* current (IK,ACh) amplitudes from the application of five different ACh concentrations were normalized to the maximal current (Imax) elicited within each oocyte (1 μM ACh). Each experiment shown was performed on 3–5 oocytes of the same batch. A minimum of two batches of oocytes were tested for each normalized recording shown. Recordings from different batches of oocytes were normalized by the mean of whole-cell basal currents from oocytes expressing the control channel Kir3.4*. Statistics (i.e., mean and standard error of the mean) of each construct were calculated from all of the normalized data recorded from different batches of oocytes.
Single-channel recordings and analysis
The single-channel activity was recorded using an Axopatch 200A amplifier (Axon Instruments). The bath solution contained (in mM): KCl 96, EGTA 0.5, and HEPES 10 [pH 7.40]. The pipette solution contained (in mM) KCl 96, EGTA 0.5, EDTA 10 and HEPES 10 [pH 7.40]. 100 μM gadolinium was routinely included in the pipette solution to suppress native stretch channel activity in the oocyte membrane. Chemicals were purchased from Sigma (St. Louis, MO). Single-channel currents were filtered at 1–2 kHz with a six-pole low-pass Bessel filter, sampled at 5–10 kHz and stored directly into the computer's hard disk through the DIGIDATA 1200 interface (Axon Instruments). Single-channel analysis was carried out with pClamp9 (Axon Instruments).
RESULTS
Location of the gating hinge in ligand gated and voltage gated potassium channels
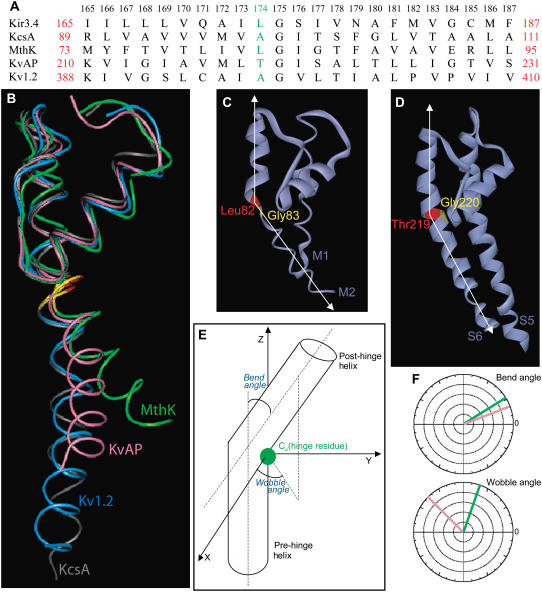
Fig. 1 A displays the alignment of residues 165–187 of the inner helix of Kir3.4 with the corresponding residues of KcsA, MthK, KvAP, and Kv1.2. We will refer to the numbers of the Kir3.4 residues in making reference to a particular position in each of the channels shown.
FIGURE 1.
(A) Sequence alignment between residues 165–187 of the inner helix of Kir3.4 with the corresponding residues of KcsA, MthK, KvAP, and Kv1.2. Leu-174 of Kir3.4, Ala-98 of KcsA, Leu-82 of MthK, Thr-219 of KvAP, and Ala-397 of Kv1.2 are highlighted. The numbers above the alignment refer to Kir3.4 numbers. (B) Relative orientation of the inner helix of KcsA (gray), MthK (green), KvAP (pink), and Kv1.2 (blue). (C) The TM pore domain of the open conformation of MthK (PDB code 1LNQ), showing the bending of the inner helix at Leu-82 (red), the position that precedes the central Gly-83 (yellow). (D) The TM pore domain of the open conformation of KvAP (PDB code 1ORQ), showing the bending of the inner S6 helix at Thr-219 (red), the position that precedes the central Gly-220 (yellow). (E) Diagram displaying the bend and wobble angles in a hinged helix. (F) Bend and Wobble angles of the inner helices of MthK (green) and KvAP (pink) as obtained using the program Simulaid (29).
Compared with the closed structure of KcsA (PDB code 1K4C), MthK (PDB code 1LNQ), KvAP (PDB code 1ORQ), and Kv1.2 (PDB code 2A79) all show some degree of opening. Fig. 1 B displays the relative conformations of the four channels. To compare the extent of opening in these structures, we examined the distance between the α-carbons of opposite subunits at the helix bundle crossing (HBC), i.e., at the equivalent position to position 187 in Kir3.4 (6). The crystallographic structure of MthK portays an open conformation in which the distance at the HBC between the α-carbons of opposite subunits is ∼40 Å. In the crystallographic structure of the voltage-gated KvAP the corresponding distance is ∼27 Å. As the open conformation obtained for KvAP is not achieved through its native mechanism but rather most probably due to crystal packing, this structure may represent an intermediate state between the closed and open state of KvAP. In the recent crystallographic structure of the voltage gated Kv1.2 channel, the distance between the α-carbons of opposite subunits is ∼17 Å, which is significantly less than the opening in KvAP, and which is closer to the HBC distance of ∼9 Å in KcsA, as can be seen in Fig. 1 B. This result casts doubt that the crystallographic structure of Kv1.2 displays an actual open conformation of the inner helix. Furthermore, as can be seen in Fig. 1, C and D, MthK and KvAP show a clear bend in the inner helix, at the vicinity of the central glycine of the inner helix. In fact, within the resolution limitations of crystallographic structures, both structures suggest that the bend occurs at the residue that precedes the central glycine, position 174 in Kir3.4. Kv1.2, on the other hand, shows no bend in the vicinity of the central glycine whereas some bending is observed at the PVP motif, which is located at the corresponding positions to 182–184 of Kir3.4.
As both the MthK and KvAP crystallographic structures suggest that there is a bend at the residue that precedes the central glycine, it is possible that the residue above the glycine and not the glycine itself serves as the hinge in the transition from the closed to the open channel conformations. In both Kir3.4 and MthK there is a leucine in position 174 that precedes the central glycine. In KcsA and Kv1.2 there is an alanine and in KvAP a threonine in the equivalent position.
The structural features of a hinge can be characterized by two parameters, the bend and wobble angles, which can be defined in terms of a prehinge helix and a posthinge helix similarly to the characterization of proline kinks (28), see Fig. 1 E. These refer to the part of the helix from the N-terminus to the hinge residue and to the part from the hinge residue to the C-terminus, respectively. The bend angle is the angle between the two parts of the helix when it is bent along its axis. It ranges from 0° to 180°, such that the closer its value to 0°, the smaller the bend in the helix is. The wobble angle is the angle that defines the orientation of the post-hinge helix in 3D, with respect to the pre-hinge helix. It ranges from 0° to 360°. The wobble angle is close to 0° (or 360°) when the axis of the post-hinge helix is bent toward the α-carbon of the hinge residue, and it is close to 180° when it is moved away from the hinge. Fig. 1 F depicts the bend and wobble angles for MthK and KvAP (29). MthK displays a larger bend angle of ∼32° compared to KvAP in which the bend angle is 21°. In addition, the orientation of the post-hinge helix differs in the two structures. The wobble angle in the KvAP structure is ∼134°, whereas in MthK it is ∼71°.
To quantify the details of the bending motion of the inner helix in potassium channels, we examined the differences in the dihedral angles of the inner helix residues in the crystallographic structures of the bacterial KcsA channel (2), representing the closed state and the bacterial MthK structure (1), representing the open state. It has been suggested that MthK represents the open conformation of potassium channels. Previous EPR studies of the conformational changes of the KcsA transmembrane domain that occur when the channel opens (30–31) are consistent with the crystallographic structure of MthK. Similarly to the MthK structure, the EPR studies showed that TM2 undergoes tilting away from the permeation pathway and twisting around its helical axis with a pivot point near the middle of TM2. We have chosen to compare MthK to KcsA and not to the more recently determined KirBac1.1 potassium channel (16) as MthK is more closely related to KcsA than KirBac1.1, and currently the open state conformation structure from KirBac channels is not available. Furthermore, KcsA and KirBac1.1 are superimposable in the key regions discussed below, including the relevant part of the inner helix, in which the bending motion occurs (Supplemental Fig. S1). Moreover, modeling of the open state of KirBac1.1 on the basis of electron microscopy data on two-dimensional crystals of the inwardly rectifying potassium channel KirBac3.1 are also in agreement with the open configuration of MthK (32). Fig. 2 A displays the values of the backbone dihedral angles of the inner helix residues of KcsA and MthK. The first largest change in dihedral angles between KcsA and MthK occurs at position 174 that precedes the central glycine in agreement with the bend seen in the crystallographic structure of MthK (Fig. 1 C).
FIGURE 2.
(A) Backbone dihedral angles of inner helix residues of KcsA (residues 89–110) and MthK (residues 73–95) that correspond to positions 165–186 of Kir3.4. (B) Effect of the cumulative changes of the backbone dihedral angles of the inner helix of KcsA to those in MthK on the distances between the α-carbons of opposite subunits at the HBC and (C) on the distances between the positions of the α-carbon of the HBC and its position in KcsA and MthK. (D) Ribbon representation of the backbones of two diagonally facing subunits of the homology model of Kir3.4 based on the KcsA structure, and the effect of changing the backbone dihedral angles of positions 165–186 of the inner helices to their value in the MthK channel. The connected lines portray the accumulating effect of the changes in the backbone dihedral angles of the inner helix residues from the KcsA conformation to the MthK conformation on the position of the α-carbon of the HBC, for the two subunits. (E) Enlargement from Fig. 1 D for subunit A. The numbers correspond to Kir3.4 numbering.
To examine the effect of the differences in the dihedral angles we examined the accumulating effect of changing the backbone dihedral angles of the inner helix residues from the KcsA conformation to the MthK conformation on the following measures: a), the distances between the α-carbons of opposite subunits at the equivalent position to 187 in Kir3.4, the helix bundle crossing (HBC), which is a measure of the extent of the opening of the channel and b), the distances between the position of the α-carbon of the HBC in KcsA and its position in MthK, which enables determination of the position of the transition from a closed conformation to an open conformation. Fig. 2 B depicts the accumulating changes in the distance between the α-carbons of the HBC, as the dihedral angles of KcsA are changed to those of MthK (Fig. 2 A). The accumulated opening distance obtained following the change in the dihedral angles of the residue located immediately above the central glycine is 38.9 Å. When changing the dihedral angles of this position alone, the distance obtained at the HBC is 40.7 Å. In contrast, the distance at the HBC obtained following a change in the dihedral angles of the central glycine alone leads to an opening of 12.9 Å only. This is comparable to the distance of ∼8.9 Å at the HBC for the closed KcsA channel.
Examination of the accumulating effect of changing the backbone dihedral angles of the inner helix residues from the KcsA conformation to the MthK conformation on the position of the α-carbon of the HBC (see Fig. 2, C–E), shows that before the change in the dihedral angles of the position that precedes the central glycine, the position of the α-carbon at the HBC remains close to its position in the KcsA structure. In fact, after incorporating the changes in the dihedral angles of two helical turns that correspond to positions 165–172, the HBC returns to a distance of 0.75 Å from its position before these changes. Following changes in the backbone dihedral angles of position 173, the α-carbon at the HBC moves toward the open conformation of the channel, 5.0 Å from its position before these dihedral angle changes, and ∼19 Å from to its position after incorporating the dihedral angle changes of positions 165–186. When the dihedral angles at 174, the position that precedes the central glycine, are changed the distance from the position of the α-carbon of the HBC in KcsA increases to ∼24 Å, whereas the distance from its position in the MthK structure decreases to ∼2 Å. After changes in the backbone dihedral angles of three helical turns that correspond to positions 175–185, the HBC returns to approximately the same distances from its position in the KcsA structure (24–25 Å), and from its position in the open MthK structure (1–2 Å). Thus, until the incorporation of the change in the backbone dihedral angles of the residue located above the central glycine, the position of the HBC fluctuates around its location in the closed KcsA conformation. After incorporating the changes in the backbone dihedral angles of position 174, however, the position of the α-carbon at the HBC moves closer to its position in the MthK conformation and fluctuates around this position. This observation reinforces the role of the residue that precedes the central glycine in the inner helix as the hinge of the channel.
To further explore the location of the gating hinge we have proceeded to compare the structure of the voltage-gated potassium channel, KvAP (7), with the KcsA (2) structure.
KvAP is opened wider than the closed KcsA structure possibly due to crystal packing interactions. Comparison between the dihedral angles of KcsA and KvAP (see Fig. 3 A) shows that the differences between them are smaller than the differences between KcsA and MthK. As noted above, the crystallographic structure, however, shows a clear bend in the S6 inner helix, at the residue that precedes the central glycine (Fig. 1 D). Fig. 3 B depicts the accumulating changes in the distance between the α-carbons of the equivalent position to 187 in Kir3.4, the HBC, between two opposite subunits as the dihedral angles of KcsA are changed to those of KvAP. Comparison of this figure with Fig. 2 A, shows that apart from the fact that the opening in KvAP (27.4 Å after changing the dihedral angles of positions 165–186) is significantly smaller compared to the opening in MthK (40.5 Å), the figures portray a similar trend. Similarly to the analysis of the open conformation of MthK, we have also examined the accumulating effect of changing the backbone dihedral angles of the inner helix residues from the KcsA conformation to the KvAP conformation on the position of the α-carbon of the HBC. As can be seen in Fig. 3 C, similarly to the result obtained in the case of MthK (Fig. 2 C), before the change in the dihedral angles of the position that precedes the central glycine, the position of the α-carbon at the HBC remains close to its position in the KcsA structure.
FIGURE 3.
(A) Backbone dihedral angles of residues located in the inner helix of KcsA (residues 89–110) and KvAP (residues 210–231) that correspond to positions 165–186 of Kir3.4. (B) Effect of the cumulative changes of the backbone dihedral angles of the inner helix of KcsA to those in KvAP on the distances between the α-carbons of opposite subunits at the HBC and (C) on the distances between the positions of the α-carbon of the HBC and its position in KcsA and KvAP.
Stimulus application leads to full activation regardless of the hinge size
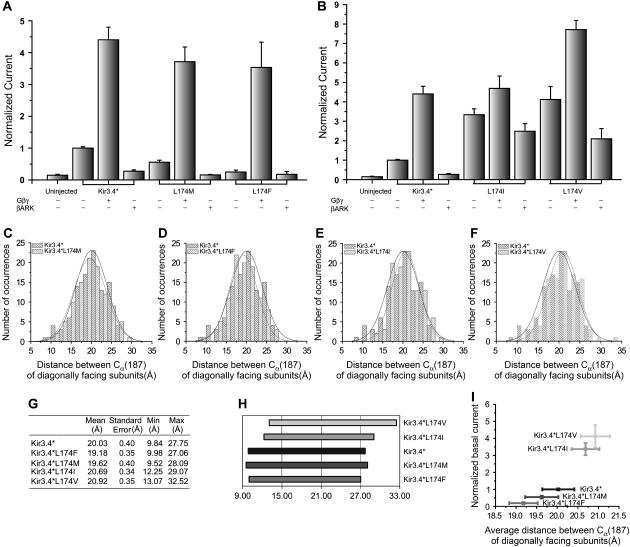
To experimentally examine the characteristics of this position, we mutated (see Methods) Leu-174 in the Kir3.4* channel to methionine and phenylalanine. The results show that when the size of the residue at position 174 increases, the basal current decreases (p < 0.05 for each pair, see Fig. 4 A). Furthermore, coexpression of the channel with Gβγ (see Methods) leads to similar current activity for the control and the mutants (p > 0.1 for the mutants coexpressed with Gβγ). These results suggest that when the size of the residue at position 174 increases above the size of the leucine, the barrier for the transition from the closed conformation to the open conformation of the channel increases, making it more difficult to open the channel. Following coexpression of the channel with Gβγ this difference is overcome, resulting in similar activity levels for the control Kir3.4* and the two mutants.
FIGURE 4.
(A) Summary of normalized whole-cell, currents from control Kir3.4*, Kir3.4*L174M, and Kir3.4*L174F expressed alone or with Gβγ or βARK-PH recorded in Xenopus oocytes at −80 mV (* p < 0.05, Unpaired t-test). (B) Summary of whole-cell currents from control Kir3.4* and Kir3.4*L174I, and Kir3.4*L174V expressed alone or with Gβγ or βARK-PH recorded in Xenopus oocytes at −80 mV. (C–F) Histograms of the distances between the Cα's of position 187, the HBC, between two opposite subunits for Kir3.4*, Kir3.4*L174M, Kir3.4*L174F, Kir3.4*L174I, and Kir3.4*L174V as obtained from alignment of the results of the conformational memories simulations with two opposite subunits of the KcsA based homology model of Kir3.4*. (G) Statistics of the data presented in (C–F) for Kir3.4* and the mutants Kir3.4*174F, Kir3.4*L174M, Kir3.4*174I, and Kir3.4*174V regarding the distribution of the distances between the α carbons of the helix bundle crossing residues (F187) of two diagonally facing subunits in Kir3.4*. (H) Graphical presentation of the range of the distances summarized in (G). (I) Normalized basal Kir3.4* currents as a function of the average distance between the α carbons at the HBC of two diagonally facing subunits.
Constitutive activity via strongly similar mutations of the gating hinge
Two interesting mutations at position 174 of Kir3.4*, L174V and L174I, result in constitutively active channels (Fig. 4 B). In contrast to the control Kir3.4* and to the Kir3.4*L174M and Kir3.4*L174F mutants, whose basal current is completely inhibited by the Gβγ scavenger PH domain of the β-adrenergic receptor kinase (βARK-PH) (33), both Kir3.4*L174V and Kir 3.4*L174I showed reduced sensitivity to βARK-PH. To examine the effect of these mutations on the structure of TM2 we used the method of conformational memories (see Methods), and calculated the distance between the α-carbons of the HBC. The results presented in Figs. 4, C–F, show that unlike in the cases of Kir3.4*L174M and Kir3.4*L174F, in both Kir3.4*L174V and Kir 3.4*L174I the distribution of the opening distance is shifted to larger values (see Figs. 4, G–H). Whereas for Kir3.4*L174M and Kir3.4*L174F, the average of the distribution shifts to a lower value, but the range of the distribution (9.5 Å–28.1 Å for Kir3.4*L174M and 10.0–27.1 Å for Kir3.4*L174F) resembles that of the control Kir3.4* (9.8–27.8 Å), the range of the distribution shifts to 12.3–29.1 Å for Kir3.4*L174I and to 13.1–32.5 Å for Kir3.4*L174V. Since the open probability of Kir3.4* coexpressed with exogenous Gβγ is ∼2.5% (34), a subtle shift in the distribution of the distance at the HBC between two opposite subunits may be sufficient to produce the level of constitutive activity observed. Fig. 4 I depicts the normalized basal currents of Kir3.4* and the L174 mutants discussed above as a function of the average opening distance at the HBC. These results suggest that the basal currents increase with the opening distance at the HBC implying that the opening of the channel can be controlled from position 174. This conclusion is consistent with the observation obtained from comparing the crystallographic structures of KcsA and MthK and KvAP, namely that the position above the central glycine can play the role of the gating hinge.
The highly conserved central glycine of TM2 is needed to avoid constraining interactions with the selectivity filter gate
When the highly conserved central glycine of the inner helix in Kir3.4*, Gly-175, is mutated to other amino acids, in most cases the resulting mutants show decreased basal currents. To understand the molecular details of the effect of mutation of Gly-175, we have carried out molecular dynamics simulations of Kir3.4* for the different mutants using a KcsA (2) based homology model of Kir3.4 (6). As the structure in the region examined in the simulation is superimposable to the KirBac1.1 structure (16), the results of the simulations are likely to be applicable also to a KirBac1.1 based homology model of Kir3.4 (see Methods and Supplemental Fig. S1). We used structurally restrained molecular dynamics simulations that maintain the structural integrity of the TM domain (see Methods). For several mutants in which Gly-175 was replaced by charged or polar residues such as aspartate, glutamate, lysine, and glutamine, the side chains of these residues interacted with residues in their vicinity. Depending on the mutant, the interactions observed included the side chain of Asn-179 as well as of Thr-149, which is a part of the selectivity filter, see for example Fig. 5, A–B.
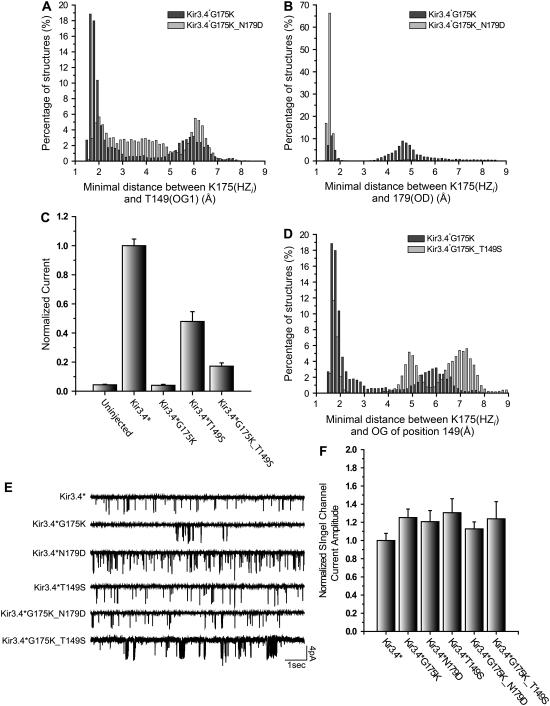
FIGURE 5.
(A) Representative conformations of the TM domain of one subunit of the Kir3.4*G175K mutant showing the interactions observed between K175 and the side chain of N179 and (B) the side chain of T149, which is a part of the selectivity filter. The inserts display the details of the nomenclature of the atoms that participate in the interaction. (C) Whole-cell basal currents from Kir3.4*, Kir3.4*N179D, Kir3.4*G175D, Kir3.4*G175E, Kir3.4*G175K, Kir3.4*G175Q and the double mutants Kir3.4*G175D_N179D, Kir3.4*G175E_N179D, Kir3.4*G175K_N179D, and Kir3.4*G175Q_N179D recorded in Xenopus oocytes at −80 mV. (D) Confocal images from oocyte sections expressing GFP-tagged Kir3.4* subunits or Kir3.4*G175K. An image from an uninjected oocyte section is shown for comparison. Black and white images of the oocyte sections are also shown. (E) Examples of single channel bursts obtained from Kir3.4*, Kir3.4*N179D, Kir3.4*G175D, Kir3.4*G175E, Kir3.4*G175K, and Kir3.4*G175Q recorded at −80 mV in a cell-attached mode.
To test the importance of these interactions to the effect of the Gly-175 mutants on channel activity, we mutated Asn-179 to aspartate. Kir3.4*N179D shows similar level of activity compared to Kir3.4* (Fig. 5 C), and similar single channel current amplitude (Figs. 5 E and 6 E–F). In addition, both asparagine and aspartate appear in this position in different inward rectifiers: asparagine appears in Kir1.1, Kir3.2–Kir3.4, Kir5.1, and Kir6.1–Kir6.2, and aspartate appears in Kir2.1–Kir2.4 and Kir3.1. Whole-cell recordings of the double mutants of N179D with different mutants of Gly-175 show significant increase in the activity level of the doubly mutated channels compared with the activity of the 175 mutants; see Fig. 5 C.
FIGURE 6.
(A) H-bonding between HZi (i = 1,2,3) of the lysine with the side chain OG1 of T149 in Kir3.4*G175K and Kir3.4*G175K_N179D. (B) H-bonding between HZi (i = 1,2,3) of the lysine with the side chain of position 179 in Kir3.4*G175K and Kir3.4*G175K_N179D. (C) Whole-cell basal currents from Kir3.4*, Kir3.4*G175K, Kir3.4*T149S, and Kir3.4*G175K_T149S recorded in Xenopus oocytes at −80 mV. (D) H-bonding between HZi (i = 1,2,3) of the lysine with the side chain OG of position 149 in Kir3.4*G175K and Kir3.4*G175K_T149S. (E) Representative traces of single channel recordings obtained from Kir3.4*, Kir3.4*G175K, Kir3.4*N179D, Kir3.4*T149S, Kir3.4*G175K_N179D, and Kir3.4*G175K_T149S recorded at −80 mV in a cell-attached mode. (F) Normalized single channel current amplitudes obtained by fitting the amplitude histograms of the data recorded from the patches whose representative traces are depicted in Fig. 6 E.
The most significant enhancement of channel activity following the addition of the second mutation N179D was obtained for the Kir3.4*G175K mutant, whose whole-cell currents have suggested that it is not functional (6). The double mutant, Kir3.4*G175K_N179D, showed significant increase in the activity level compared with Kir3.4*G175K (Fig. 5 C). In fact, the double mutant displayed whole-cell currents of ∼50% of the control Kir3.4* channel. Since GFP tagging (see Methods) showed that lack of activity in the case of the G175K mutation was not due to lack of expression (see Fig. 5 D), and since this mutant did not reduce single channel conductance (see Figs. 5 E and 6 E–F), we have proceeded to analyze the details of the interactions of the lysine with its surroundings. Molecular Dynamics simulations showed that whereas in the single mutant Kir3.4*G175K, HZi (i = 1,2,3) of the lysine hydrogen bonded with the side chain OG1 of Thr-149 in ∼60% of the structures, the double mutant displayed this interaction in only ∼25% of the structures (Fig. 6 A). On the other hand, the double mutant interacts with the side chain of position 179 in ∼96% of the structures, whereas the single mutant displays interaction with the side chain of position 179 only in ∼25% of the structures (Fig. 6 B). This analysis shows that following the N179D mutation, significant changes in the balance in the interaction pattern between position 175 and positions 149 and 179 occur such that the interactions between Lys-175 and Thr-149 are reduced and Lys-175 shows higher probability of interacting with the side chain of Asp-179 instead. These results led us to hypothesize that the reason Kir3.4*G175K displays very low basal currents is due to its interaction with Thr-149, which is a part of the signature sequence of the selectivity filter.
To test this hypothesis, we mutated Thr-149 to several other residues, which have been shown not to affect the selectivity properties of the channel in the case of Shaker voltage-gated potassium channel, and include alanine, asparagine, glycine, and serine (35). All four Thr-149 mutants in Kir3.4* showed that the selectivity of the channel was not affected by this mutation (See Fig. 7 A for Kir3.4*T149S). These mutations, however, affect the activity level of the channel, possibly due to an effect on the gating of the channel. It has been recently shown that a point mutation on the Shaker voltage-gated potassium channel at the corresponding position of 149 of Kir3.4* displayed substantial energetic coupling with mutations that affect the activation gate located at the HBC (36). Among the four residues mentioned above, serine is the closest residue to threonine. Thus, to avoid introduction of additional factors, we have chosen the T149S mutation to further investigate the effect of the G175K mutation. In addition, as ACh signaling through Muscarinic type 2 (M2) receptors activates Kir3 channels specifically via the βγ subunits of G-proteins, we examined the effect of this mutation on the ACh dose-response curve of Kir3.4* (see Methods). As can be seen in Fig. 7 B, the Kir3.4*T149S mutation did not have a significant effect on the ACh dose-response curve. In addition, as can be seen in Fig. 6, E–F, this mutant did not have a significant effect on single channel conductance as well.
FIGURE 7.
(A) Selectivity characteristics of Kir3.4*T149S. HK is a high-potassium solution (in mM: 96 KCl, 1 NaCl, 1 MgCl2, 5 KOH/HEPES [pH 7.4]), and LK is a low-potassium solution (in mM: 2KCl, 91 NaCl, 1 MgCl2, 5 NaOH/HEPES [pH 7.4]). (B) ACh dose-response curve of Kir3.4* and Kir3.4*T149S.
Kir3.4*T149S displayed basal currents of ∼50% compared to Kir3.4* (Fig. 6 C). Mutation of G175K in addition to T149S resulted in a decrease in basal current to a level of ∼40% compared to the Kir3.4*T149S mutation (see Fig. 6 C). This ratio between the whole-cell currents of Kir3.4*T149S_G175K and Kir3.4*T149S is comparable to the ratio of ∼50% obtained for Kir3.4*N179D_G175K and Kir3.4*N179D. Simulation of the Kir3.4*T149S_G175K double mutant shows that the double mutant displays H-bonding between HZi (i = 1,2,3) of the lysine side chain and the side chain OG of the serine at position 149 in only 24% of the structures compared to the single mutant Kir3.4*G175K, that displays H-bonding between HZi (i = 1,2,3) of the lysine and the side chain OG1 of Thr-149 in over 60% of the structures, see Fig. 6 D. This is comparable to the percentage of H-bonding of 25% between HZi (i = 1,2,3) of Lys-175 and the side chain OG1 of Thr-149 in the Kir3.4*N179D_G175K double mutant (Fig. 6 A). These results support the observation obtained from the simulation, namely that there is interaction between Lys-175 and Thr-149. Furthermore, they also support the hypothesis that this interaction plays a significant role in reducing the basal current of Kir3.4* upon mutation of Gly-175 to a lysine.
Basal currents in the absence of the highly conserved glycine
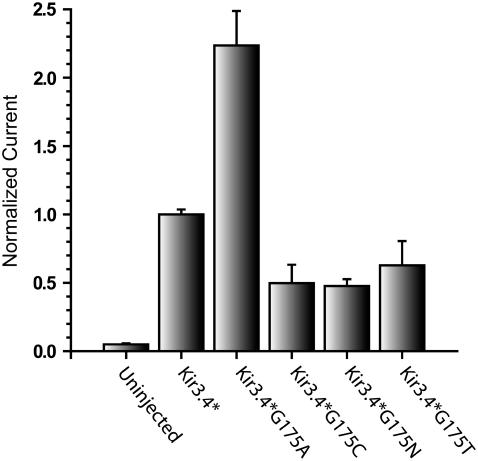
The results presented above also point to the fact that from the point of view of the ability to activate the channel, glycine can be replaced by other residues as long as they do not interact with residues that play a crucial role in channel activation. Examination of four Gly-175 mutants that do not display significant interactions with position 149, shows that they display significant basal currents. These include alanine, cysteine, asparagine, and threonine, as can be seen in Fig. 8. Specifically, the alanine mutant, displayed basal currents, which were twice as large as Kir3.4*, and the cysteine, asparagine, and threonine mutants exhibited reduced basal currents which were 50% or higher compared to the control current. This result further reinforces the conclusion that glycine can be replaced by other residues. In fact, in 7.5% of the 478 potassium channels that were examined (6) there was an asparagine instead of the glycine, in 4% there was an alanine, and in 1% of the channels there was a threonine including, for example, the inward rectifiers Kir4.1 and Kir4.2.
FIGURE 8.
Whole-cell basal currents of Kir3.4*, Kir3.4*G175A, Kir3.4*G175C, Kir3.4*G175N, and Kir3.4*G175T recorded in Xenopus oocytes at −80 mV.
DISCUSSION
Comparison of the crystallographic structures of the closed conformation of KcsA with those of the open conformations of MthK and KvAP suggests that within the resolution of crystallographic structures, the residue that precedes the highly conserved central glycine in the inner helix plays the role of a gating hinge. As a result of a bend at the position that precedes the central glycine, MthK and KvAP exhibit an opening of 40.5 Å and 27.4 Å, respectively, at the HBC compared to the distance of 8.9 Å in KcsA. As the open conformation obtained for the voltage-gated KvAP is not achieved through its native mechanism but rather most probably due to crystal packing, it is possible that this structure represents some intermediate state between the closed and open state of KvAP, and that therefore the open structure is not completely developed. In spite of that, the structure already exhibits similar characteristics to those observed in the case of the Ca2+ activated MthK channel, namely that the bending of the inner helix occurs immediately above the central glycine.
In Kir3.4* as well as in MthK, the position that precedes the central glycine is occupied by a leucine. In KcsA it is occupied by an alanine and in KvAP by a threonine. Examination of 478 different potassium channels shows that in 76.4% of the channels the equivalent position is occupied by a hydrophobic residue: alanine (11.3%), isoleucine (14.2%), leucine (15.3%), methionine (2.7%), phenylalanine (17.8%), and valine (15.1%). In 21.1% of the channels this position is occupied by a polar residue: asparagine (0.6%), cysteine (3.8%), serine (11.7%), threonine (3.8%), tryptophan (0.4%), and tyrosine (0.8%). In 2.5% of the channels a glycine occupies this position. We have examined the effect of the mutation of Leu-174 in Kir3.4* to other hydrophobic residues. In all cases, the channel could be activated by Gβγ (Fig. 4). This result implies that an appropriate stimulus could induce the activation of potassium channels, where a large residue plays the role of a gating hinge. In two cases Kir3.4*L174I and Kir3.4*L174V we have shown that the channel becomes constitutively active (see Fig. 4 B). This further supports the notion that activation of potassium channels could be controlled through this position. Thus, activation of Kir3.4* can be induced through a leucine located at position 174, which corresponds to Ala-98 in KcsA, to Leu-82 in MthK, and to Thr-219 in KvAP. This position precedes the central glycine of the inner transmembrane (TM) helices of potassium channels, and following the comparison between the crystallographic structure of KcsA with that of MthK and of KvAP, it is likely to constitute the gating hinge of potassium channels. In view of this observation and of the fact that 19% of potassium channels do not possess glycine at the center of the inner helix, our finding indicates that channels that do not possess glycines in their pore-lining helix could also be gated by a pivoted bending mechanism of this helix.
Yet, glycine is known to possess unique characteristics due to its lack of a side chain. As a result it is regarded as a residue that can confer flexibility to α-helices, as has been shown in molecular dynamics simulations (e.g., 13,14). When examined in the context of the KcsA channel (15), the major difference between the channel opening trajectory of the G99A mutant and that of the wild-type was that the number of hydrogen bonds decreased sharply in the mutant as the channel opened, whereas in the wild-type the change was smoother. Apart from this, however, it was found that the channel opening trajectory of the mutant was very similar to that of the wild-type KcsA, which led to the suggestion that although Gly-99 weakens the central region of the inner helix in KcsA, it is not a critical determinant of the activation mechanism of the channel (15). In this context it is important to note that although in general glycine is able to acquire unique dihedral angles, these are not manifested upon activation in the case of the highly conserved glycine located in the center of the inner TM helices of potassium channels. Comparison of the Ramachandran plots (37) that depict the distribution of the backbone dihedral angles of the different amino acids shows that glycine can display several combinations of dihedral angles. These include distributions centered around (ϕ,ψ) of approximately: a), (−70°,−50°); b), (−90°,180°); and c), (75°,0°). Among these, the distributions around b and c are significantly smaller in other amino acids, and in some cases the distribution around c is completely absent. These combinations of the dihedral angles correspond approximately to conformational angles found in α-helices (a), β-strands (b), and left-handed α-helices (c). As can be seen in Fig. 1 B, the backbone dihedral angles of the central glycine in the KcsA structure are (−76°,−51°), and in MthK they are (−78°,−23°). These angles are in the vicinity of distribution center (a) that corresponds to a α-helical structure, which implies that the ability of glycine to acquire unique dihedral angles in other structures is not utilized for channel gating. Furthermore, although it has been shown (38) that whereas alanine is one of the best helix forming residues, and glycine is one of the most helix destabilizing residues, especially when located in the center of an α-helix, helix propensities of amino acids are widely distributed. It was thus suggested that the results of mutagenesis experiments would depend on the glycine's neighboring residues (38). This may be the reason that in certain cases mutation of the highly conserved central glycine to alanine results in nonfunctional channels (8,9,11,12), whereas others can function in the absence of the central glycine (6,12). For example, this may be the reason for the fact that although the wild-type KCNQ1 is functional with an alanine in the center of the inner helix at the corresponding position of the highly conserved central glycine, KCNQ2 becomes nonfunctional when its central glycine is mutated to alanine, but a chimera of KCNQ2 that includes the inner helix of KCNQ1, is functional (12).
In this study, we examined the effect of the mutation of the highly conserved central glycine of the inner helix. Our results indicate that interactions introduced by mutations at position 175 can have a substantial effect on channel activity. In particular, interactions with residues in the signature sequence of the selectivity filter may be especially constraining, and understanding the exact consequence of these interactions will require further studies. Upon reduction in the occurrence of such interactions through the mutation of Asn-179 to aspartate, the performance of a number of Gly-175 mutated channels increased substantially (see Fig. 5 C). Reduction in the occurrence of interactions with position 149 could also be obtained by mutating the threonine to a serine, as was demonstrated for the case of Kir3.4*G175K (see Fig. 6 C). Thus, our results suggest that from the point of view of the ability to activate the channel, glycine can be replaced by other residues as long as they do not affect crucial residues. In particular mutation of this glycine to alanine resulted in basal currents, which were twice as large as the control Kir3.4* (Fig. 8). Our results suggest that a major reason that glycine is highly conserved among potassium channels is that it prevents interactions with residues in the signature sequence of the selectivity filter and its vicinity.
SUPPLEMENTARY MATERIAL
An online supplement to this article can be found by visiting BJ Online at http://www.biophysj.org.
Acknowledgments
We thank members of the Logothetis Lab for assistance throughout the work and Dr. T. Jin for critical feedback on the manuscript.
Confocal laser scanning microscopy was performed at the MSSM-Microscopy Shared Resource Facility, supported with funding from National Institutes of Health-National Cancer Institute shared resources grant (R24 CA095823) and National Science Foundation Major Research Instrumentation grant (DBI-9724504). This work was supported by a NIH grant (HL-54185) to D.E.L. D.E.L is an Established Investigator of the American Heart Association.
References
- 1.Jiang, Y., A. Lee, J. Chen, M. Cadene, B. T. Chait, and R. MacKinnon. 2002. The open pore conformation of potassium channels. Nature. 417:523–526. [DOI] [PubMed] [Google Scholar]
- 2.Doyle, D. A., J. Morais Cabral, R. A. Pfuetzner, A. Kuo, J. M. Gulbis, S. L. Cohen, B. T. Chait, and R. MacKinnon. 1998. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 280:69–77. [DOI] [PubMed] [Google Scholar]
- 3.Vivaudou, M., K. W. Chan, J. L. Sui, L. Y. Jan, E. Reuveny, and D. E. Logothetis. 1997. Probing the G-protein regulation of GIRK1 and GIRK4, the two subunits of the KACh channel, using functional homomeric mutants. J. Biol. Chem. 272:31553–31560. [DOI] [PubMed] [Google Scholar]
- 4.Logothetis, D. E., Y. Kurachi, J. Galper, E. J. Neer, and D. E. Clapham. 1987. The beta gamma subunits of GTP-binding proteins activate the muscarinic K+ channel in heart. Nature. 325:321–326. [DOI] [PubMed] [Google Scholar]
- 5.Reuveny, E., P. A. Slesinger, J. Inglese, J. M. Morales, J. A. Iniguez-Lluhi, R. J. Lefkowitz, H. R. Bourne, Y. N. Jan, and L. Y. Jan. 1994. Activation of the cloned muscarinic potassium channel by G protein beta gamma subunits. Nature. 370:143–146. [DOI] [PubMed] [Google Scholar]
- 6.Jin, T., L. Peng, T. Mirshahi, T. Rohacs, K. W. Chan, R. Sanchez, and D. E. Logothetis. 2002. The βγ subunits of G proteins gate a K+ channel by pivoted bending of a transmembrane segment. Mol. Cell. 10:469–481. [DOI] [PubMed] [Google Scholar]
- 7.Jiang, Y., A. Lee, J. Chen, V. Ruta, M. Cadene, B. T. Chait, and R. MacKinnon. 2003. X-ray structure of a voltage-K1 channel. Nature. 423:33–41. [DOI] [PubMed] [Google Scholar]
- 8.Magidovich, E., and O. Yifrach. 2004. Conserved gating hinge in ligand- and voltage-dependent K+ channels. Biochemistry. 43:13242–13247. [DOI] [PubMed] [Google Scholar]
- 9.Ding, S., L. Ingleby, C. Ahern, and R. Horn. 2005. Investigating the putative glycine hinge in Shaker potassium channel. J. Gen. Physiol. 126:213–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao, Y., V. Yarov-Yarovoy, T. Scheuer, and W. A. Catterall. 2004. A gating hinge in Na+ channels; a molecular switch for electrical signaling. Neuron. 41:859–865. [DOI] [PubMed] [Google Scholar]
- 11.Wuttke, T. V., G. Seebohm, S. Bail, S. Maljevic, and H. Lerche. 2005. The new anticonvulsant Retigabine favors voltage-dependent opening of the Kv7.2 (KCNQ2) channel by binding to its activation gate. Mol. Pharmacol. 67:1009–1017. [DOI] [PubMed] [Google Scholar]
- 12.Seebohm, G., N. Strutz-Seebohm, O. N. Ureche, R. Baltaev, A. Lampert, G. Kornichuk, K. Kamiya, T. V. Wuttke, H. Lerche, M. C. Sanguinetti, and F. Lang. 2006. Differential roles of S6 domain hinges in the gating of KCNQ potassium channels. Biophys. J. 90:2235–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bright, J. N., I. H. Shrivastava, F. S. Cordes, and M. S. Sansom. 2002. Conformational dynamics of helix S6 from Shaker potassium channel: simulation studies. Biopolymers. 64:303–313. [DOI] [PubMed] [Google Scholar]
- 14.Bright, J. N., and M. S. Sansom. 2003. The flexing/twirling helix: exploring the flexibility about molecular hinges formed by proline and glycine motifs in transmembrane helices. J. Phys. Chem. B. 107:627–636. [Google Scholar]
- 15.Tikhonov, D. B., and B. S. Zhorov. 2004. In silico activation of KcsA K+ channel by lateral forces applied to the C-termini of inner helices. Biophys. J. 87:1526–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuo, A., J. M. Gulbis, J. F. Antcliff, T. Rahman, E. D. Lowe, J. Zimmer, J. Cuthbertson, F. M. Ashcroft, T. Ezaki, and D. A. Doyle. 2003. Crystal structure of the potassium channel KirBac1.1 in the closed state. Science. 300:1922–1926. [DOI] [PubMed] [Google Scholar]
- 17.Long, S. B., E. B. Campbell, and R. Mackinnon. 2005. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 309:897–903. [DOI] [PubMed] [Google Scholar]
- 18.Labrom, A. J., A. L. Raes, I. Bellens, N. Ottschytsch, and D. J. Snyders. 2003. Gating of shaker-type channels requires the flexibility of S6 caused by prolines. J. Biol. Chem. 278:50724–50731. [DOI] [PubMed] [Google Scholar]
- 19.Guarnieri, F., and S. R. Wilson. 1995. Conformational memories and a simulated annealing program that learns. Application to LTB4. J. Comput. Chem. 16:648–653. [Google Scholar]
- 20.Guarnieri, F., and H. Weinstein. 1996. Conformational memories and the exploration of biologically relevant peptide conformations: an Illustration for the Gonadotropin releasing hormone. J. Am. Chem. Soc. 118:5580–5589. [Google Scholar]
- 21.Metropolis, N., A. W. Rosenbluth, M. N. Rosenbluth, A. H. Teller, and E. Teller. 1953. Equation of state calculation by fast computing machines. J. Chem. Phys. 21:1087–1092. [Google Scholar]
- 22.Kirkpatrick, S., C. D. Gelatt, Jr., and M. P. Vecchi. 1983. Optimization by simulated annealing. Science. 220:671–680. [DOI] [PubMed] [Google Scholar]
- 23.Brooks, B. R., R. E. Bruccoleri, B. D. Olafson, D. J. States, S. Swaminathan, and M. Karplus. 1983. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 4:187–217. [Google Scholar]
- 24.Chan, K. W., J. L. Sui, M. Vivaudou, and D. E. Logothetis. 1997. Specific regions of heteromeric subunits involved in enhancement of G protein-gated K+ channel activity. J. Biol. Chem. 272:6548–6555. [DOI] [PubMed] [Google Scholar]
- 25.He, C., X. Yan, H. Zhang, T. Mirshahi, T. Jin, A. Huang, and D. E. Logothetis. 2002. Identification of critical residues controlling G protein-gated inwardly rectifying K+ channel activity through interactions with the beta gamma subunits of G proteins. J. Biol. Chem. 277:6088–6096. [DOI] [PubMed] [Google Scholar]
- 26.Mirshahi, T., D. E. Logothetis, and M. Sassaroli. 2001. Localization and quantification of GFP-tagged ion channels expressed in Xenopus oocytes. In Ion Channel Localization. A. N. Lopatin and C. G. Nichols, editors. Humana Press, Totowa, NJ. 215–231.
- 27.Zhang, Q., A. Dickson, and C. A. Doupnik. 2004. Gβγ-activated inwardly rectifying K+ (GIRK) channel activation kinetics via Gαi and Gαo-coupled receptors are determined by Gα-specific interdomain interactions that affect GDP release rates. J. Biol. Chem. 279:29787–29796. [DOI] [PubMed] [Google Scholar]
- 28.Visiers, I., B. B. Braunheim, and H. Weinstein. 2000. Prokink: a protocol for numerical evaluation of helix distortions by proline. Protein Eng. 13:603–606. [DOI] [PubMed] [Google Scholar]
- 29.Mezei, M. Simulaid. http://fulcrum.physbio.mssm.edu/∼mezei/simulaid/simulaid.html. [Online].
- 30.Perozo, E., D. M. Cortes, and L. G. Cuello. 1999. Structural rearrangements underlying K+-channel activation gating. Science. 285:73–78. [DOI] [PubMed] [Google Scholar]
- 31.Liu, Y. S., P. Sompornpisut, and E. Perozo. 2001. Structure of the KcsA channel intracellular gate in the open state. Nat. Struct. Biol. 8:883–887. [DOI] [PubMed] [Google Scholar]
- 32.Domene, C., D. A. Doyle, C. Venien-Bryan. 2005. Modeling of an ion channel in its open conformation. Biophys. J. 89:L01–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He, C., H. Zhang, T. Mirshahi, and D. E. Logothetis. 1999. Identification of a potassium channel site that interacts with G Protein betagamma subunits to mediate agonist-induced signaling. J. Biol. Chem. 274:12517–12524. [DOI] [PubMed] [Google Scholar]
- 34.Yakubovich, D., V. Pastushenko, A. Bitler, C. W. Dessauer, and N. Dascal. 2000. Slow modal gating of single G protein-activated K+ channels expressed in Xenopus oocytes. J. Physiol. 524:737–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heginbotham, L., Z. Lu, T. Abramso, and R. MacKinnon. 1994. Mutations in the K+ channel signature sequence. Biophys. J. 66:1061–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yifrach, O., and R. MacKinnon. 2002. Energetics of pore opening in a voltage-gated K+ channel. Cell. 111:231–239. [DOI] [PubMed] [Google Scholar]
- 37.Kleywegt, G. J., and T. A. Jones. 1996. Phi/psi-chology: Ramachandran revisited. (Supplementary material). Structure. 4:1395–1400. [DOI] [PubMed] [Google Scholar]
- 38.Chakrabartty, A., J. A. Schellman, and R. L. Baldwin. 1991. Large differences in the helix propensities of alanine and glycine. Nature. 351:586–588. [DOI] [PubMed] [Google Scholar]