Abstract
The Fe(II)- and α-ketoglutarate (αKG)-dependent dioxygenases use mononuclear nonheme iron centers to effect hydroxylation of their substrates and decarboxylation of their cosubstrate, αKG, to CO2 and succinate. Our recent dissection of the mechanism of taurine:αKG dioxygenase (TauD), a member of this enzyme family, revealed that two transient complexes accumulate during catalysis in the presence of saturating substrates. The first complex contains the long-postulated C-H-cleaving Fe(IV)-oxo intermediate, J, and the second is an enzyme·product(s) complex. Here, we demonstrate the accumulation of two transient complexes in the reaction of a prolyl-4-hydroxylase (P4H), a functional homologue of human αKG-dependent dioxygenases with essential roles in collagen biosynthesis and oxygen sensing. The kinetic and spectroscopic properties of these two P4H complexes suggest that they are homologues of the TauD intermediates. Most notably, the first exhibits optical absorption and Mössbauer spectra similar to those of J and, like J, a large substrate deuterium kinetic isotope on its decay. The close correspondence of the accumulating states in the P4H and TauD reactions supports the hypothesis of a conserved mechanism for substrate hydroxylation by enzymes in this family.
Keywords: Fe(IV)-oxo intermediates, hydroxylation, nonheme iron enzymes, oxygen activation
The Fe(II)- and α-ketoglutarate (αKG)-dependent oxygenases reductively activate molecular oxygen, cleave their cosubstrate, αKG, to CO2 and succinate, and oxidize their substrates by two electrons (1–4). In the vast majority of the reactions, the substrate is hydroxylated, but desaturation, cyclization, and halogenation reactions also are known (1–3, 5, 6). Regardless of the outcome, an unactivated C-H bond of the substrate is cleaved, and the reaction is carried out at a mononuclear nonheme Fe(II) center. In most cases, the Fe(II) is coordinated by a (His)2(Asp/Glu) “facial triad” (7), but Blasiak et al. (8) recently described a remarkable variation on this theme in which the absence of the Asp/Glu ligand creates a site for halide coordination to permit formation of an alkyl halide product.
The reactions catalyzed by these enzymes are important in the biosyntheses of collagen (9, 10), antibiotics (11), and a variety of plant and microbial secondary metabolites (2), the cellular sensing of and response to oxygen insufficiency (hypoxia) (12–19), the repair of alkylated DNA (20, 21), and the regulation of gene expression by demethylation of histones (22, 23). Hydroxylation at C4 of proline residues in proteins, the reaction catalyzed by αKG-dependent prolyl-4-hydroxylases (P4Hs), has particular importance in human health. The reaction helps to cross-link the helices of collagen, an essential component of connective tissue in higher organisms. Insufficient collagen P4H activity associated with vitamin C (ascorbic acid) deficiency is the cause of scurvy, a disease that is characterized by weak connective tissue. Cellular response to hypoxia in mammals depends on a cascade initiated by the transcriptional activator, hypoxia inducible factor (HIF). The activity of HIF is antagonized by hydroxylations of Pro-402 and Pro-564 in its α subunit, reactions catalyzed by HIFα-specific P4Hs. Insufficiency of O2 prevents the antagonizing modifications and allows HIF to promote transcription of genes involved in the global hypoxic response, which is important in a number of cardiovascular, pulmonary, and neoplastic disease states (24, 25).
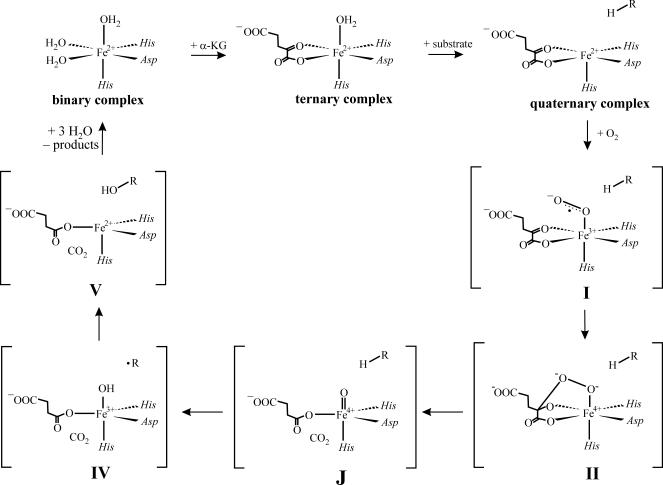
A detailed chemical mechanism for P4H first was formulated >20 years ago on the basis of theoretical considerations (4). This mechanism has served well as a general working hypothesis for the mechanisms of all of the αKG-dependent dioxygenases: extensive studies on several family members, and inorganic models thereof have failed to reveal significant inconsistencies (1–3, 26–31). However, the first direct evidence for any of the several oxidized iron intermediates invoked in this mechanism came relatively recently, with the detection by stopped-flow (SF) absorption and freeze-quench (FQ) Mössbauer spectroscopies of two transient states in the catalytic cycle of taurine:αKG dioxygenase (TauD) (see Scheme 1) (32). The first state, which was termed J, has an Fe(IV) center in the unusual high-spin (S = 2) configuration (33). The position of J in the catalytic cycle was revealed by the observation of a large deuterium kinetic isotope effect (2H-KIE, kH/kD ≈ 50; refs. 34 and 36) on its decay when the reaction was carried out with selectively deuterium-labeled substrate (34). The result suggested that J activates the substrate by H-atom abstraction, which in the mechanism is effected by the Fe(IV)–oxo (ferryl) complex. The presence of a ferryl unit in J subsequently was verified by resonance Raman (37) and x-ray absorption spectroscopies (38). The second transient state was shown to be an Fe(II)-containing product(s) complex (35).
Scheme 1.
Consensus mechanism for oxygen activation by the Fe(II)- and αKG-dependent dioxygenases. The states labeled J and V have been trapped and characterized for TauD (33–35) and P4H (this work).
The TauD work provided two reasons for optimism that the postulated ferryl intermediates in other nonheme-iron oxygenases (including the P4Hs) might be directly characterized. First, the work showed that a ferryl complex can be remarkably long-lived in the nonheme coordination environment, despite its anticipated high reactivity. Second, it showed that hydrogen abstraction by a nonheme ferryl complex can exhibit a very large 2H-KIE, providing a powerful tool to stabilize such a species and promote its accumulation even when it would otherwise be obscured by unfavorable kinetics. Here, we have applied the experimental framework developed in the TauD studies to characterize the reaction of a P4H from Paramecium bursaria Chlorella virus 1, a functional homologue of the human enzymes with roles in collagen biosynthesis and oxygen sensing. The kinetic and spectroscopic data document direct detection of a nonheme Fe(IV) (almost certainly ferryl) enzyme intermediate and confirm that the large substrate 2H-KIE associated with hydrogen abstraction by such a species can permit its accumulation, even when it is normally kinetically masked. In addition, the data reveal the accumulation of a second, Fe(II)-containing state, which is probably the homologue of the second TauD state [a product(s) complex]. The close correspondence of the accumulating states in the P4H and TauD reactions provides strong experimental confirmation that oxygen activation and substrate hydroxylation by the αKG-dependent dioxygenases follow a conserved chemical mechanism.
Results
Titration of P4H·Fe(II) with αKG [±(Pro-Ala-Pro-Lys)3 Peptide Substrate] to Form P4H·Fe(II) [αKG·(Pro-Ala-Pro-Lys)3].
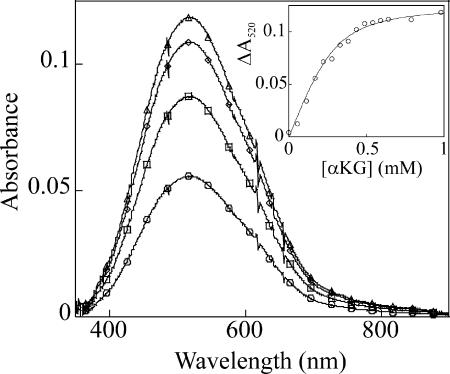
As a prelude to the mechanistic studies, the binding constant for αKG to the active site Fe(II) in P4H was determined, as described for TauD in ref. 33. Binding of αKG to the binary P4H·Fe(II) complex elicits the visible absorption feature in the ≈500-nm region (λmax = 520 nm, ε520 = 250 M−1·cm−1) that has been assigned in related systems to a metal-to-ligand charge-transfer (MLCT) transition arising from chelation of the Fe(II) by the C1-carboxylate and C2-carbonyl of αKG (Fig. 1) (39). Titrations monitoring this feature gave KD = 27 ± 6 μM (Fig. 1 Inset). The presence of (Pro-Ala-Pro-Lys)3 peptide does not affect the KD for αKG but results in a slight decrease in intensity of the MLCT band (to ε520 = 200 M−1·cm−1).
Fig. 1.
Spectrophotometrically monitored titration of αKG into a solution of the P4H·Fe(II) complex at 5°C in the absence of O2. The starting protein solution contained 650 μM P4H and 500 μM Fe(II) in P4H buffer (described in Experimental Methods). The spectra shown correspond to αKG concentrations of 0.17 mM (circles), 0.33 mM (squares), 0.54 mM (diamonds), and 0.98 mM (triangles). The points in Inset depict the change of absorbance at 520 nm. The solid line is a fit yielding a Kd of 27 ± 6 μM.
SF Absorption Evidence for Substrate Triggering and Accumulation of Two Intermediates in the P4H Reaction.
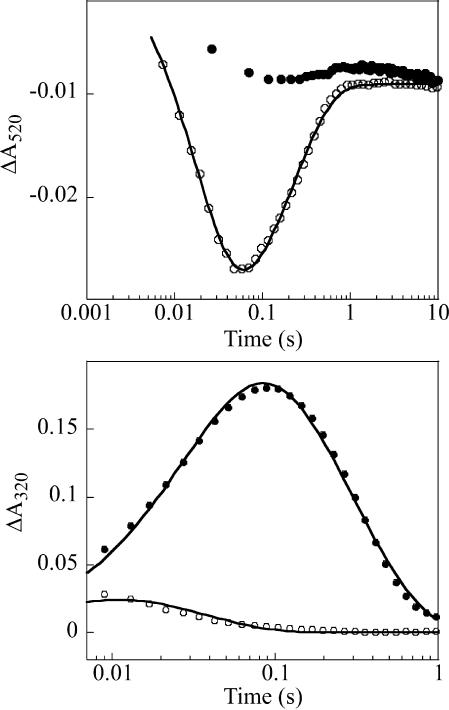
The P4H·Fe(II)·αKG ternary complex reacts very sluggishly with O2, as shown by the minor decay of the 520-nm absorption feature during the first 10 seconds after the two reactants are mixed at 5°C (Fig. 2Upper, filled circles). By contrast, the P4H·Fe(II)·αKG·(Pro-Ala-Pro-Lys)3 quaternary complex reacts rapidly with O2, resulting in loss of absorbance at 520 nm (Fig. 2 Upper, open circles). These results indicate that binding of the peptide substrate activates the Fe(II) center of P4H for reaction with O2. This “substrate triggering,” which is thought to be due (at least in part) to dissociation of a water ligand from the O2-binding site upon binding of the substrate (40), is a general feature of enzymes in this family (1). For example, binding of taurine enhances O2 reactivity by >103-fold in TauD (41). The observation of the effect in P4H provides assurance that (Pro-Ala-Pro-Lys)3 is a good mimic of the enzyme’s natural substrate(s). Two additional observations indicate that the absorbance changes reflect steps in the P4H catalytic cycle. First, A520 returns nearly to its initial value, indicating that the reactant quaternary complex is regenerated after consumption of O2, the limiting substrate in this experiment. Second, the apparent first-order rate constants of 51 s−1 and 4.3 s−1 (5°C) for the fall and rise phases (obtained as described in Experimental Methods) are both greater than the turnover number at 22°C under similar reaction conditions (3.2 s−1).
Fig. 2.
SF absorption kinetic traces from reactions of P4H·Fe(II)·αKG [±(Pro-Ala-Pro-Lys)3] containing either unlabeled (Pro-Ala-Pro-Lys)3 or deuteriated substrate (d7-Pro-Ala-d7-Pro-Lys)3 with O2. Final reaction conditions were 0.75 mM P4H, 0.50 mM Fe(II), 5 mM αKG, 5 mM ascorbate, 5 mM substrate, and 0.2 mM O2. (Upper) Absorbance at 520 nm for the reactions lacking any peptide substrate (squares) and with (Pro-Ala-Pro-Lys)3 (open circles). The solid line is a fit of the equation given in Experimental Methods to the data. (Lower) Absorbance at 320 nm in the reactions of (Pro-Ala-Pro-Lys)3 (open circles) and (d7-Pro-Ala-d7-Pro-Lys)3 (closed circles). The solid lines are fits, as described in Experimental Methods.
The transient decrease of A520 is evidence for accumulation of a state that absorbs less than the reactant complex at this wavelength, as was also observed for the TauD reaction. Decay of A520 exhibits no obvious lag phase in the P4H reaction. By contrast, the pronounced lag phase observed for the TauD reaction is due to accumulation of J (which itself absorbs in the 500-nm region to compensate partially for loss of the contribution of the reactant complex) before the transparent product(s) complex (33). Thus, the absence of such a lag in the P4H trace suggests that the intermediate corresponding to J accumulates minimally, if at all. Formation and decay of J in TauD was detected directly at 320 nm, because of its increased absorption (Δε320 ≈ 1,500 M−1·cm−1); under conditions similar to those used in the experiment of Fig. 2, A320 rises to a maximum of ≈0.17 at ≈20 ms before decaying to its original value in the TauD reaction. The P4H reaction shows a much smaller transient increase (Fig. 2 Lower, open circles) and reaches its maximum value of A320 at a much earlier reaction time. The data suggest that, in the P4H reaction, the presumptive Fe(IV) intermediate accumulates minimally, perhaps as a result of its rapid decay to an optically transparent product(s) complex.
The large substrate deuterium kinetic isotope effect (2H-KIE) on decay of J in the TauD reaction suggests a test of the above interpretation of the P4H kinetic data; use of deuterium-substituted (Pro-Ala-Pro-Lys)3 {(Pro-Ala-Pro-Lys)3 peptide containing six 2,3,3,4,4,5,5-[2H]7-proline residues (d7-Pro-Ala-d7-Pro-Lys)3} would be expected to slow decay of the presumptive Fe(IV) intermediate and permit its accumulation to higher levels (34, 36). The amplitude of the A320 transient is indeed markedly increased and its tmax shifted to longer reaction time (tmax = 87 ms) in the reaction with (d7-Pro-Ala-d7-Pro-Lys)3 (Fig. 2 Lower, filled circles), demonstrating accumulation of an intermediate that absorbs in the UV region. The A320 kinetic traces for the reactions of the protium- and deuterium-containing substrates were analyzed together as described in Experimental Methods. It was assumed that only k2, the rate constant for decay of the intermediate, is sensitive to the presence of deuterium in the substrate, as was found to be true for J in the TauD work (34). The fits suggest that the 2H-KIE on decay of the intermediate is ≈60 (kH/kD = ≈210/3.4 ≈ 60). However, this value is only an estimate. More accurate determination will require modification of the reaction conditions to permit greater accumulation of the intermediate in the reaction with the protium-containing substrate so that the decay rate constant can be more accurately determined. Nevertheless, the SF data provide support for the hypothesis that the C-H/D cleaving intermediate accumulates in the P4H reaction only with the deuterium-labeled substrate.
FQ Mössbauer-Spectroscopic Evidence for Accumulation of a Product(s) Complex in the P4H Reaction with Unlabeled (Pro-Ala-Pro-Lys)3 and a High-Spin Fe(IV) Intermediate in the Reaction with (d7-Pro-Ala-d7-Pro-Lys)3.
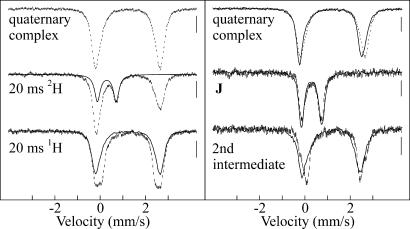
Comparison of the 4.2 K/40 mT Mössbauer spectra of samples frozen (freeze-quenched) at 20 ms in the reactions with the deuterium- and protium-containing substrates provides strong support for the above interpretation of the SF data (Fig. 3). The spectrum of the sample from the reaction with unlabeled (Pro-Ala-Pro-Lys)3 (Fig. 3 Left Bottom) exhibits only peaks attributable to Fe(II) ions, but distinct differences relative to the spectrum of the reactant complex (Fig. 3 Left Top) confirm that a reaction has occurred. The spectrum can be analyzed as a superposition of the spectrum of the reactant complex (62% of total intensity, solid line) and a new quadrupole doublet with δ = 1.26 mm/s, |ΔEQ| = 2.39 mm/s, and line width = 0.35 mm/s. These parameters are typical of high-spin Fe(II) and similar to those associated with the product(s) complex in TauD, suggesting that a cognate of this state accumulates after 20 ms in the P4H reaction. No features attributable to an Fe(IV)-containing intermediate are detectable. By contrast, a new peak at ≈0.7 mm/s, near the position of the high-energy line of J in TauD, is readily seen in the spectrum of the 20-ms sample from the reaction with the deuterium-labeled (d7-Pro-Ala-d7-Pro-Lys)3 substrate (Fig. 3 Left Middle). The spectrum can be analyzed as a superposition of the spectrum of the quaternary complex (62%) and a new quadrupole doublet (38%) with parameters (δ = 0.30 mm/s, |ΔEQ| = 0.82 mm/s, line width = 0.30 mm/s) that are typical of a high-spin Fe(IV) complex (42). Moreover, these parameters are nearly identical to those of J in TauD (33). The similarity of the spectra of the quaternary complexes (Fig. 3 Right Top) and the two states that accumulate during the reaction [the Fe(IV) intermediate (Fig. 3 Right Middle), and the product(s) complex (Fig. 3 Right Bottom)] in P4H (vertical lines) and TauD (solid lines) is striking. The Mössbauer data thus provide strong independent support for the hypothesis that the P4H intermediates are cognates of those previously detected in TauD.
Fig. 3.
4.2-K/40-mT Mössbauer spectra of selected samples of P4H. (Left) O2-free P4H·Fe(II)·αKG·(Pro-Ala-Pro-Lys)3 complex (Top) and 20-ms freeze-quenched samples from the reaction of this complex with O2 at 5°C [Middle, deuteriated (d7-Pro-Ala-d7-Pro-Lys)3; Bottom, unlabeled (Pro-Ala-Pro-Lys)3]. The solid line in the middle spectrum is the contribution of the Fe(IV) intermediate (38%) simulated according to the parameters quoted in the text. The solid line in Bottom is the contribution of the P4H·Fe(II)·αKG·(Pro-Ala-Pro-Lys)3 complex (62%). (Right) Comparison of derived spectra of the three accumulating states from the P4H (vertical lines) and TauD (solid lines) catalytic cycles: quaternary complex (Top), Fe(IV) intermediate (Middle), and product(s) complex (Bottom). For the 20-ms samples, the concentrations after mixing were 1.4 mM P4H, 1.25 mM Fe(II), 5 mM αKG, 5 mM ascorbate, 3.3 mM (Pro-Ala-Pro-Lys)3 (unlabeled or deuteriated), and 1.2 mM O2.
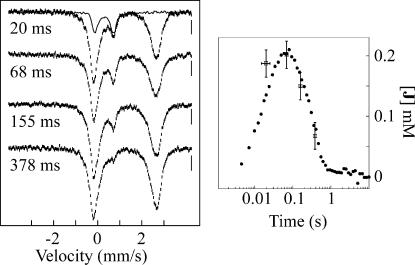
SF and FQ experiments with identical reaction conditions were carried out on the (d7-Pro-Ala-d7-Pro-Lys)3 reaction to verify that the transient 320-nm absorption and δ = 0.30 mm/s Mössbauer quadrupole doublet are associated with the same intermediate (Fig. 4Left). The kinetics of the intermediate obtained by the two methods agree well. Notably, the plot assumes a molar absorptivity of ε320 = 1,500 M−1·cm−1 for the Fe(IV) intermediate, a value identical to that determined for J in TauD (33). The combined absorption and Mössbauer data thus confirm that reaction of O2 with the P4H reactant complex formed with unlabeled (Pro-Ala-Pro-Lys)3 leads to rapid accumulation of a product(s) complex, whereas a large 2H-KIE on conversion of the substrate-hydroxylating Fe(IV) intermediate to the product(s) complex allows the former to accumulate to much higher levels in the reaction with deuterium-labeled substrate.
Fig. 4.
Kinetics of the P4H reaction by SF absorption and FQ Mössbauer. Left, 4.2-K/40-mT Mössbauer spectra of samples quenched at 20 ms, 68 ms, 160 ms, and 370 ms. The solid line in the top spectrum is the derived spectrum of J. Right, Comparison of the deduced concentration of J with the A320 kinetic trace from the SF experiment with identical reaction conditions. Error bars on the concentrations of J deduced from Mössbauer spectra reflect the uncertainty of ±3% of the absorption area of the spectra (ordinate) and the uncertainty of the reaction time of ±5 ms (abscissa). Optimal agreement between concentration and absorbance required Δε320 = 1,500 M−1·cm−1 for the Fe(IV) intermediate. Concentrations were 1 mM P4H, 0.75 mM Fe(II), 5 mM αKG, 5 mM ascorbate, 5 mM (d7-Pro-Ala-d7-Pro-Lys)3, and 0.65 mM O2.
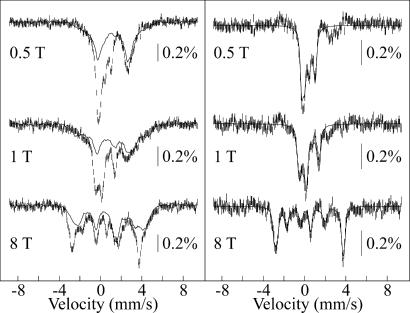
To probe the spin-state and electronic structure of the Fe(IV) intermediate in P4H in greater depth, Mössbauer spectra of the 20-ms sample from the (d7-Pro-Ala-d7-Pro-Lys)3 reaction were acquired in applied magnetic fields of 0.5 T, 1 T, and 8 T (Fig. 5Left, vertical lines). Removal of the contribution of the reactant complex (solid line in each spectrum) yields spectra for the Fe(IV) intermediate in P4H (Fig. 5 Right, vertical lines). The overall splitting of the peaks increases as the magnetic field increases, which is typical of a paramagnetic complex with a (nearly) axial integer-spin electronic ground state with a positive zero-field splitting parameter, D. The large separation of the peaks in the 8-T spectrum indicates that the ground state of the Fe site is in the high-spin configuration (S = 2). Analysis of the spectra allows for determination of D and the x and y components of A/gNβN, as described in ref. 43. Theoretical simulations according to the spin-Hamiltonian formalism in the slow relaxation limit and the parameters given in Table 1 are shown as solid lines. For the analysis, parameters determined from the 40-mT spectrum (δ and ΔEQ) were kept constant, and it was assumed that the A-tensor is axial. Similar spectra were observed for J in TauD (43) and a high-spin Fe(IV)–oxo model complex (31), but the magnetic splitting in the spectra of the P4H complex is smaller than that for J in TauD. The diminished splitting reflects smaller internal magnetic fields and is a consequence of the larger zero-field splitting parameter, D. Neese and colleagues (42, 44) showed that the magnitude of D is primarily governed by the energy separation between the S = 2 ground state and the low-lying S = 1 excited state. It is not yet known what structural changes are reflected by the difference in the D values of the two Fe(IV) complexes.
Fig. 5.
4.2-K/variable-field Mössbauer spectra of the 20-ms sample from the (d7-Pro-Ala-d7-Pro-Lys)3 reaction shown in Fig. 3. (Left) Experimental spectrum (vertical lines) and spectrum of O2-free control, scaled to 62% of the total intensity (solid line). (Right) Derived spectra of the Fe(IV) intermediate from P4H (vertical lines) and spin Hamiltonian simulations (solid lines) according to the parameters given in Table 1.
Table 1.
Comparison of the Mössbauer parameters of high-spin Fe(IV)–oxo complexes
| Fe(IV)–oxo complex | D, cm−1 | (E/D) | δ, mm/s | ΔEQ, mm/s | η | A/gNβN, T | Ref. |
|---|---|---|---|---|---|---|---|
| J of P4H | 15.5 | 0.02 | 0.30 | −0.82 | −0.2 | (−18.0, −18.0, n.d.) | This work |
| J of TauD | 10.5 | 0.03 | 0.31 | −0.88 | 0 | (−18.0, −18.0, n.d.) | 43 |
| [Fe(O)(H2O)5]2+ | 9.7 | 0 | 0.38 | 0.15 | 0 | (−20.3, −20.3, n.d.) | 31 |
The coordinate systems of the zero-field splitting tensor and the electric field gradient are assumed to be collinear.
Discussion
To assess the generality of the insight obtained in the TauD studies and to provide a foundation for characterizing the medically important collagen-cross-linking and hypoxia-sensing P4Hs, we sought a readily available, stable, monomeric P4H with a simple, synthetically accessible substrate. The single-subunit, 27-kDa P4H from Paramecium bursaria Chlorella virus 1 (PBCV-1), which had been produced in Escherichia coli in active form and shown to hydroxylate short peptide substrates, met these criteria. Although neither the natural substrate of PBCV-1 P4H nor the functional significance of Pro hydroxylation for the virus has been determined, several likely substrates, hypothetical proteins containing proline-rich motifs, were recognized from the sequence of the viral genome (45). Eriksson et al. (46) showed that peptides having multiple repeats of these proline-rich motifs [(Pro-Ala-Pro-Lys)n, (Ser-Pro-Lys-Pro-Pro)n, (Pro-Glu-Pro-Pro-Ala)n, and (Lys-Pro-Ala)n] are substrates for PBCV-1 P4H. For example, both Pro residues in the repeating unit of (Pro-Ala-Pro-Lys)3 are hydroxylated, with those preceding Ala being hydroxylated more readily (with the exception for the N-terminal Pro, which is not hydroxylated) (46). On the basis of the steady-state kinetic parameters for (Pro-Ala-Pro-Lys)1–5 peptides reported in this previous study, we selected the 12-mer as the shortest substrate likely to properly trigger the enzyme and permit accumulation of reaction intermediates. The results of Fig. 2 confirm that the 12-mer indeed is sufficient to trigger rapid O2 activation by PBCV-1 P4H.
The combined SF absorption and FQ Mössbauer data for the reaction of the P4H·Fe(II)·αKG·(Pro-Ala-Pro-Lys)3 complex with O2 demonstrate the accumulation of two kinetically competent intermediates. The first intermediate is a high-spin Fe(IV) complex, which exhibits a large 2H-KIE on its decay, suggesting that it is the species that activates the substrate by H-atom abstraction. Because of the spectroscopic and kinetic analogy of this species to J in the TauD reaction, we assign it as a high-spin Fe(IV)–oxo (ferryl) complex, although we as yet have no direct spectroscopic evidence for the presence of the oxo group. The second accumulating state contains a high-spin Fe(II) center and is presumably an Fe(II)·product(s) complex. The correspondence of the two accumulating complexes in the P4H and TauD reactions suggests that these distantly related αKG-dependent dioxygenases employ the same chemical mechanism, consistent with the widely held notion of a conserved mechanism for the hydroxylases.
The high-spin ground-state configuration of the nonheme Fe(IV)-oxo intermediates in TauD and P4H contrasts with the low-spin (S = 1) configuration of the many ferryl heme complexes that have been studied. The difference in spin state can be rationalized by the difference in the strength of the equatorial ligands (42, 47). In the heme complexes, strong donation by the N ligands in the x–y plane destabilizes the dx2-y2 orbital (z axis is along the Fe-oxo bond). Computational studies by Neese (42) and Decker et al. (48) suggest that the Fe(IV)–oxo bond is similarly covalent for high-spin and low-spin Fe(IV)–oxo complexes.
Despite the difference in spin state, the nonheme ferryl complexes may exhibit the same chemical versatility often attributed to their heme counterparts. Whereas the two nonheme ferryl enzyme intermediates characterized to date mediate alkyl hydroxylations for which hydrogen abstraction followed by hydroxyl radical rebound is the most likely mechanism, similar complexes have been postulated in distinct enzymatic transformations that necessarily proceed by different mechanisms. For example, in the pterin-dependent hydroxylases, the hypothetical Fe(IV)-oxo intermediate is believed to perform an electrophilic attack on the aromatic ring of the substrate (phenylalanine, tyrosine, or tryptophan) rather than hydrogen abstraction (49, 50). A single ferryl species even may be capable of several different reactivities. The enzyme thymine hydroxylase, named for its physiological methyl hydroxylation reaction, also catalyzes further oxidation of the hydroxymethyluracil initial product to the corresponding aldehyde and acid upon subsequent O2 activation events. In addition, Thornburg et al. (51, 52) demonstrated that the enzyme can catalyze epoxidation of an olefin, conversion of an alkyne to a ketene, and successive S oxidations of a thioether to sulfoxide and then sulfone. These latter transformations are not hydroxylations and do not involve hydrogen atom abstraction. It appears that alternative transformations are possible even subsequent to the “standard” hydrogen atom abstraction. The postulated Fe(IV)-oxo centers in isopenicillin-N-synthase and the αKG-dependent halogenases are expected on the basis of x-ray crystal structures of stable enzyme forms to be coordinated by a thiolate derived from the substrate and a halide ion, respectively (8, 53). In both cases, it is thought that the Fe(IV)-oxo group abstracts a hydrogen atom from the substrate. Combination of the resulting substrate radical with either a sulfur radical or a halogen radical leads to a group transfer reaction (thiolation or chlorination) distinct from hydroxylation. Because it has not yet been shown which of these various alternative transformations involves a ferryl intermediate and which may proceed by a fundamentally different mechanism [e.g., interception of an earlier intermediate, such as an Fe(III)–superoxide complex like I in Scheme 1], dissection of their mechanisms by direct detection of reactive intermediates should provide significant additional fundamental insight.
Experimental Methods
Materials.
PBCV-1 genomic DNA was obtained from James L. van Etten (University of Nebraska, Lincoln, NE). The pET15b plasmid was obtained from Novagen (Darmstadt, Germany). DNA primers were purchased from Integrated DNA Technologies (Coralville, IA). (Pro-Ala-Pro-Lys)3 peptides were synthesized by Anne Stanley in the Macromolecular Core Facility of Penn State Milton S. Hershey Medical Center (Hershey, PA). 2,3,3,4,4,5,5-[2H]7-l-proline was purchased from Cambridge Isotopes Laboratories (Andover, MA), and 9-fluorenylmethoxycarbonyl protecting group was appended by AnaSpec (San Jose, CA) for peptide synthesis. All other reagents were obtained from Sigma (St. Louis, MO).
Preparation of apo P4H.
His6-tagged PBCV-1 P4H was overexpressed in E. coli BL21(DE3) from the pET15b overexpression vector. The gene was amplified from genomic DNA by PCR according to procedures in ref. 46. P4H was purified at 5°C by affinity chromatography on Ni(II)-nitrilotriacetate agarose resin, as described in ref. 46. The purified protein was exhaustively dialyzed against 50 mM Tris·Cl, pH 7.6, at 5°C/500 mM NaCl/10% (vol/vol) glycerol. All reactions were carried out in this buffer.
SF Absorption and FQ Mössbauer Experiments.
An O2-free solution of P4H·Fe(II)·αKG·(Pro-Ala-Pro-Lys)3 was mixed at 5°C with a buffered solution containing O2. Reactant concentrations after mixing are given in the figure legends. Sample preparation and spectroscopic measurements were carried out as described in ref. 33.
Analysis of Kinetic Data.
SF absorption kinetic traces were analyzed with the program Kaleidagraph (Synergy Software, Reading, PA) according to the equation, At = A0 + [R]0·(ΔεI)·{k1/(k2 − k1)}·{exp(−k1·t) − exp(−k2·t)}. This equation gives absorbance as a function of time (At) for a sequence of two irreversible reactions (R → I → P) with rate constants k1 and k2, isosbestic reactant (R) and product (P), and an intermediate (I) that absorbs more or less intensely by ΔεI. For the A320-versus-time traces (Fig. 2 Bottom), the (d7-Pro-Ala-d7-Pro-Lys)3 trace was analyzed first, and the amplitude {[R]0·(ΔεI)} and formation rate-constant (k1) from this fit were assumed in fitting the (Pro-Ala-Pro-Lys)3 trace to obtain an estimate for the decay rate constant (k2) necessary to account for the lesser accumulation and faster decay in the (Pro-Ala-Pro-Lys)3 reaction.
Simulation of Mössbauer Spectra.
Simulations were carried out with the program WMOSS (Web Research, Edina, MN). Some of the simulations are based on the spin Hamiltonian,
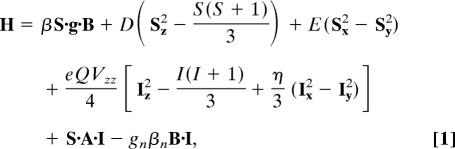 |
in which the first three terms describe the electron Zeeman effect and zero-field splitting of the S = 2 ground state, the fourth term represents the interaction between the electric field gradient and the nuclear quadrupole moment, the fifth term describes the magnetic hyperfine interactions of the electronic spin with the57Fe nucleus, and the last term represents the nuclear Zeeman interaction.
Acknowledgments
We thank Ms. Elizabeth A. Forbes, Ms. Khyati A. Valand, and Ms. Anne Stanley for technical assistance and Prof. James L. van Etten (University of Nebraska) for providing PBCV-1 genomic DNA. This work was supported by National Institutes of Health Grant GM-69657 (to J.M.B. and C.K.), the donors of American Chemical Society Petroleum Research Fund Grant ACS-PRF 41170-G3 (to C.K.), the Arnold and Mabel Beckman Foundation Young Investigator Award (to C.K.), and the Pennsylvania Department of Health Tobacco Settlement Funds (to J.M.B. and C.K.).
Abbreviations
- αKG
α-ketoglutarate
- (d7-Pro-Ala-d7-Pro-Lys)3
(Pro-Ala-Pro-Lys)3 peptide containing six 2,3,3,4,4,5,5-[2H]7-proline residues
- FQ
freeze-quench
- 2H-KIE
deuterium kinetic isotope effect
- PBCV-1
Paramecium bursaria Chlorella virus 1
- P4H
prolyl-4-hydroxylase
- SF
stopped-flow
- TauD
taurine:α-ketoglutarate dioxygenase.
Footnotes
The authors declare no conflict of interest.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Solomon EI, Brunold TC, Davis MI, Kemsley JN, Lee S-K, Lehnert N, Neese F, Skulan AJ, Yang Y-S, Zhou J. Chem Rev. 2000;100:235–349. doi: 10.1021/cr9900275. [DOI] [PubMed] [Google Scholar]
- 2.Hausinger RP. Crit Rev Biochem Mol Biol. 2004;39:21–68. doi: 10.1080/10409230490440541. [DOI] [PubMed] [Google Scholar]
- 3.Costas M, Mehn MP, Jensen MP, Que L., Jr Chem Rev. 2004;104:939–986. doi: 10.1021/cr020628n. [DOI] [PubMed] [Google Scholar]
- 4.Hanauske-Abel HM, Günzler V. J Theor Biol. 1982;94:421–455. doi: 10.1016/0022-5193(82)90320-4. [DOI] [PubMed] [Google Scholar]
- 5.Vaillancourt FH, Yin J, Walsh CT. Proc Natl Acad Sci USA. 2005;102:10111–10116. doi: 10.1073/pnas.0504412102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vaillancourt FH, Yeh E, Vosburg DA, O’Connor SE, Walsh CT. Nature. 2005;436:1191–1194. doi: 10.1038/nature03797. [DOI] [PubMed] [Google Scholar]
- 7.Que L., Jr Nat Struct Biol. 2000;7:182–184. doi: 10.1038/73270. [DOI] [PubMed] [Google Scholar]
- 8.Blasiak LC, Vaillancourt FH, Walsh CT, Drennan CL. Nature. 2006;440:368–371. doi: 10.1038/nature04544. [DOI] [PubMed] [Google Scholar]
- 9.Hutton JJ, Jr, Tappel AL, Udenfriend S. Biochem Biophys Res Comm. 1966;24:179–184. doi: 10.1016/0006-291x(66)90716-9. [DOI] [PubMed] [Google Scholar]
- 10.Kivirikko KI, Myllyharju J. Matrix Biol. 1998;16:357–368. doi: 10.1016/s0945-053x(98)90009-9. [DOI] [PubMed] [Google Scholar]
- 11.Kershaw NJ, Caines MEC, Sleeman MC, Schofield CJ. Chem Comm. 2005;34:4251–4263. doi: 10.1039/b505964j. [DOI] [PubMed] [Google Scholar]
- 12.Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG., Jr Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 13.Jaakkola P, Mole DR, Tian Y-M, Wilson MI, Gielbert J, Gaskell SJ, von Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, et al. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 14.Epstein ACR, Gleadle JM, McNeill LA, Hewitson KS, O’Rourke J, Mole DR, Mukherji M, Metzen E, Wilson MI, Dhanda A, et al. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 15.Bruick RK, McKnight SL. Science. 2001;294:1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- 16.Yu F, White SB, Zhao Q, Lee FS. Proc Natl Acad Sci USA. 2001;98:9630–9635. doi: 10.1073/pnas.181341498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masson N, Willam C, Maxwell PH, Pugh CW, Ratcliffe PJ. EMBO J. 2001;20:5197–5206. doi: 10.1093/emboj/20.18.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lando D, Peet DJ, Gorman JJ, Whelan DA, Whitelaw ML, Bruick RK. Genes Dev. 2002;16:1466–1471. doi: 10.1101/gad.991402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hewitson KS, McNeill LA, Riordan MV, Tian Y-M, Bullock AN, Welford RW, Elkins JM, Oldham NJ, Bhattacharya S, Gleadle JM, et al. J Biol Chem. 2002;277:26351–26355. doi: 10.1074/jbc.C200273200. [DOI] [PubMed] [Google Scholar]
- 20.Trewick SC, Henshaw TF, Hausinger RP, Lindahl T, Sedgwick B. Nature. 2002;419:174–178. doi: 10.1038/nature00908. [DOI] [PubMed] [Google Scholar]
- 21.Falnes PØ, Johansen RF, Seeberg E. Nature. 2002;419:178–182. doi: 10.1038/nature01048. [DOI] [PubMed] [Google Scholar]
- 22.Tsukada Y-i, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 23.Schneider J, Shilatifard A. Am Chem Soc Chem Biol. 2006;1:75–81. doi: 10.1021/cb600030b. [DOI] [PubMed] [Google Scholar]
- 24.Schofield CJ, Ratcliffe PJ. Nat Rev Mol Cell Biol. 2004;5:343–354. doi: 10.1038/nrm1366. [DOI] [PubMed] [Google Scholar]
- 25.Semenza GL. Nat Rev Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 26.Wu M, Moon H-S, Begley TP, Myllyharju J, Kivirikko KI. J Am Chem Soc. 1999;121:587–588. [Google Scholar]
- 27.Chiou Y-M, Que L., Jr J Am Chem Soc. 1995;117:3999–4013. [Google Scholar]
- 28.Mehn MP, Fujisawa K, Hegg EL, Que L., Jr J Am Chem Soc. 2003;125:7828–7842. doi: 10.1021/ja028867f. [DOI] [PubMed] [Google Scholar]
- 29.Rohde J-U, In J-H, Lim MH, Brennessel WW, Bukowski MR, Stubna A, Münck E, Nam W, Que L., Jr Science. 2003;299:1037–1039. doi: 10.1126/science.299.5609.1037. [DOI] [PubMed] [Google Scholar]
- 30.Decker A, Rohde J-U, Que L, Jr, Solomon EI. J Am Chem Soc. 2004;126:5378–5379. doi: 10.1021/ja0498033. [DOI] [PubMed] [Google Scholar]
- 31.Pestovsky O, Stoian S, Bominaar EL, Shan X, Münck E, Que L, Jr, Bakac A. Angew Chem Int Ed. 2005;44:6871–6874. doi: 10.1002/anie.200502686. [DOI] [PubMed] [Google Scholar]
- 32.Bollinger JM, Jr, Price JC, Hoffart LM, Barr EW, Krebs C. Eur J Inorg Chem. 2005;2005:4245–4254. [Google Scholar]
- 33.Price JC, Barr EW, Tirupati B, Bollinger JM, Jr, Krebs C. Biochemistry. 2003;42:7497–7508. doi: 10.1021/bi030011f. [DOI] [PubMed] [Google Scholar]
- 34.Price JC, Barr EW, Glass TE, Krebs C, Bollinger JM., Jr J Am Chem Soc. 2003;125:13008–13009. doi: 10.1021/ja037400h. [DOI] [PubMed] [Google Scholar]
- 35.Price JC, Barr EW, Hoffart LM, Krebs C, Bollinger JM., Jr Biochemistry. 2005;44:8138–8147. doi: 10.1021/bi050227c. [DOI] [PubMed] [Google Scholar]
- 36.Bollinger JM, Jr, Krebs C. J Inorg Biochem. 2006;100:586–605. doi: 10.1016/j.jinorgbio.2006.01.022. [DOI] [PubMed] [Google Scholar]
- 37.Proshlyakov DA, Henshaw TF, Monterosso GR, Ryle MJ, Hausinger RP. J Am Chem Soc. 2004;126:1022–1023. doi: 10.1021/ja039113j. [DOI] [PubMed] [Google Scholar]
- 38.Riggs-Gelasco PJ, Price JC, Guyer RB, Brehm JH, Barr EW, Bollinger JM, Jr, Krebs C. J Am Chem Soc. 2004;126:8108–8109. doi: 10.1021/ja048255q. [DOI] [PubMed] [Google Scholar]
- 39.Pavel EG, Zhou J, Busby RW, Gunsior M, Townsend CA, Solomon EI. J Am Chem Soc. 1998;120:743–753. [Google Scholar]
- 40.Zhou J, Kelly WL, Bachmann BO, Gunsior M, Townsend CA, Solomon EI. J Am Chem Soc. 2001;123:7388–7398. doi: 10.1021/ja004025+. [DOI] [PubMed] [Google Scholar]
- 41.Price JC. PhD thesis. University Park, PA: Pennsylvania State University; 2005. [Google Scholar]
- 42.Neese F. J Inorg Biochem. 2006;100:716–726. doi: 10.1016/j.jinorgbio.2006.01.020. [DOI] [PubMed] [Google Scholar]
- 43.Krebs C, Price JC, Baldwin J, Saleh L, Green MT, Bollinger JM., Jr Inorg Chem. 2005;44:742–757. doi: 10.1021/ic048523l. [DOI] [PubMed] [Google Scholar]
- 44.Schöneboom JC, Neese F, Thiel W. J Am Chem Soc. 2005;127:5840–5853. doi: 10.1021/ja0424732. [DOI] [PubMed] [Google Scholar]
- 45.van Etten JL, Meints RH. Ann Rev Microbiol. 1999;53:447–494. doi: 10.1146/annurev.micro.53.1.447. [DOI] [PubMed] [Google Scholar]
- 46.Eriksson M, Myllyharju J, Tu H, Hellman M, Kivirikko KI. J Biol Chem. 1999;274:22131–22134. doi: 10.1074/jbc.274.32.22131. [DOI] [PubMed] [Google Scholar]
- 47.Decker A, Solomon EI. Angew Chem Int Ed. 2005;44:2252–2255. doi: 10.1002/anie.200462182. [DOI] [PubMed] [Google Scholar]
- 48.Decker A, Clay MD, Solomon EI. J Inorg Biochem. 2006;100:697–706. doi: 10.1016/j.jinorgbio.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 49.Kappock TJ, Caradonna JP. Chem Rev. 1996;96:2659–2756. doi: 10.1021/cr9402034. [DOI] [PubMed] [Google Scholar]
- 50.Fitzpatrick PF. Biochemistry. 2003;42:14083–14091. doi: 10.1021/bi035656u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thornburg LD, Lai MT, Wishnok JS, Stubbe J. Biochemistry. 1993;32:14023–14033. doi: 10.1021/bi00213a036. [DOI] [PubMed] [Google Scholar]
- 52.Thornburg LD, Stubbe J. Biochemistry. 1993;32:14034–14042. doi: 10.1021/bi00213a037. [DOI] [PubMed] [Google Scholar]
- 53.Roach PL, Clifton IJ, Hensgens CMH, Shibta N, Schofield CJ, Hajdu J, Baldwin JE. Nature. 1997;387:827–830. doi: 10.1038/42990. [DOI] [PubMed] [Google Scholar]