Abstract
Cajal bodies are small nuclear organelles with a number of nuclear functions. Here we show that FLICE-associated huge protein (FLASH), originally described as a component of the apoptosis signaling pathway, is mainly localized in Cajal bodies and is essential for their structure. Reduction in FLASH expression by short hairpin RNA results in disruption of the normal architecture of the Cajal body and relocalization of its components. Because the function of FLASH in the apoptosis receptor signaling pathway has been strongly questioned, we have now identified a clear function for this protein.
Keywords: coiled bodies, nuclear organelles
Cajal bodies (CBs) are small nuclear organelles described in vertebrate cells a century ago by Ramon y Cajal and which have since been observed in a variety of animal and plant nuclei. Many components of CBs are shared with the nucleolus, and CBs frequently localize to the nucleolar periphery or within the nucleoli (1–3). CBs disappear from prophase nuclei and reappear in late G1 after resumption of transcription in the daughter nuclei (for review see refs. 1, 4, and 5). Although their function is still in part elusive, recent work suggests that they are involved in several nuclear functions, including modification of small nuclear RNAs and small nuclear ribonucleoproteins, important for spliceosome formation, and assembly of the three eukaryotic RNA polymerases (pol I, pol II, and pol III) with their respective transcription and processing factors that are then transported as multiprotein complexes to the sites of transcription (1). More recently CBs have been implicated in replication-dependent histone gene transcription and mRNA maturation (1, 6–10), and a subset of CBs is physically associated with histone gene clusters on chromosomes 1 and 6 (11). Here we identify FLASH (FLICE-associated huge protein) (12) as a new component of CBs and show that it is essential for their structure.
FLASH was initially identified as a component of the apoptosis signaling complex known as the death-inducing signaling complex (12, 13), which is associated with caspase 8 in the death-inducing signaling complex and thus essential for caspase 8 activation. However, this role of FLASH has been questioned (14). More recently it has been shown that, in response to TNFα, FLASH translocates to the nucleus and binds the glucocorticoid receptor-interacting protein (GRIP-1), inhibiting both its interaction with, and the transcriptional activity of, the glucocorticoid receptor (15, 16).
Results
FLASH Has a Nuclear Localization.
Despite the presence of three nuclear localization signals FLASH was originally described as a cytoplasmic protein (12, 17). Staining endogenous FLASH with four different anti-FLASH antibodies, however, showed that FLASH only localized to the nucleus with a clear punctate appearance (Fig. 5a, which is published as supporting information on the PNAS web site). The specificity of the antibodies used was confirmed by the disappearance of the staining after short hairpin RNA for FLASH (Fig. 4a). Western blot after nuclear cytoplasmic fractionation confirmed that FLASH was expressed only in the nuclear fraction (Fig. 1d). To further confirm the nuclear localization, we overexpressed GFP-tagged FLASH in different cell lines (HeLa, SAOS-2, MCF-7, and Hek293). Again, overexpressed GFP-tagged FLASH had a clear punctate nuclear localization (Fig. 5b and data not shown). Similar results were obtained by using FLAG-tagged FLASH (data not shown). Because both GFP and FLAG are N-terminal tags, we could not exclude the possibility that processing of FLASH released a C-terminal fragment that translocated into the cytoplasm. To address this question we expressed C-terminal V5-tagged FLASH. Similar to the previous constructs, V5 FLASH was also exclusively expressed in the nucleus (data not shown). Massive overexpression using a Tet-on inducible cell line resulted in a more diffuse nuclear localization and in some cells a lighter cytoplasmic staining (Fig. 5c).
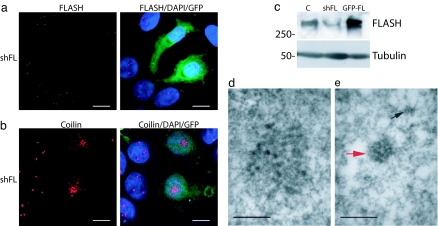
Fig. 4.
Down-regulation of FLASH results in CB disruption. (a) Immunofluorescence showing that MCF-7 cells cotransfected with GFP-spectrin and pSUPER-FLASH-1 (green) (shFL) and analyzed after 72 h show complete disappearance of endogenous FLASH (red) staining. (Scale bars: 10 μm.) (b) Immunofluorescence using antibodies against coilin (red) showing that cells in which FLASH has been down-regulated (green) show an altered coilin distribution. (Scale bars: 10 μm.) (c) Western blot showing that transfection of MCF-7 cells with pSUPER-FLASH-1 (shFL) but not with a scrambled vector (C) results in reduction of FLASH protein levels. Overexpressed GFP-FLASH was loaded as a positive control (GFP-FL). (d and e) Ultrastructural morphology of HeLa cells transfected with GFP-spectrin and pSUPER-scrambled (d) or pSUPER FLASH-1 (e). Cells transfected with pSUPER-FLASH-1 and sorted for the expression of GFP contain small (<200 nm) structures that exhibit the morphological characteristics of CB. Many of these structures are rounded in profile (red arrow) whereas others (black arrow) are much smaller and irregular and resemble fragments of CB. (Scale bars: 250 nm.)
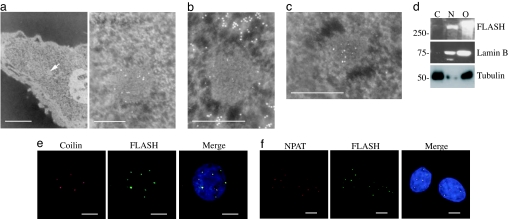
Fig. 1.
FLASH is a component of CBs. (a) Ultrastructural ImmunoGold cytochemistry of MCF-7 cells showing FLASH in bodies (denoted by an arrow in Left and enlarged in Right) with the structural characteristics of coiled bodies. (Scale bars: 1 μm in Left and 250 nm in Right.) (b) Ultrastructural ImmunoGold cytochemistry showing that anti-FLASH label (5 nm of gold) is concentrated over a body whereas DNA (10 nm of gold) is detected only in the surrounding euchromatin. (Scale bar: 250 nm.) DNA exclusion confirms identity of these bodies as CBs. (c) Ultrastructural ImmunoGold labeling of MCF-7 cells with anti-FLASH (5 nm of gold) and anti-coilin (10 nm of gold) antibodies confirms colocalization. (Scale bar: 250 nm.) (d) Western blot of endogenous FLASH after separation of nuclear (N) and cytoplasmic (C) fractions shows that FLASH is only in the nuclear fraction. Cells overexpressing GFP-tagged FLASH were used as a positive control (O). Lamin B was used as a control of nuclear fraction purity, and β-tubulin was used as a control of the cytoplasmic fraction purity. (e) Coimmunostaining using anti-FLASH (green) and anti-coilin (red) antibodies in MCF-7 cells shows that the endogenous proteins colocalize. (Scale bars: 5 μm.) (f) Coimmunostaining using anti-FLASH (green) and anti-NPAT (red) antibodies in MCF-7 cells showing that the endogenous proteins colocalize. (Scale bars: 5 μm.)
FLASH Localizes to CBs.
Ultrastructural ImmunoGold cytochemistry only detected endogenous FLASH in the nucleus, where it was sometimes associated with the nucleolus but, much more strikingly, in bodies 200–800 nm in diameter (Fig. 1a), with the structure and staining characteristics of “coiled bodies” (18, 19). Like coiled bodies, the FLASH-labeled structures were also devoid of detectable levels of DNA (Fig. 1b). Coiled bodies are synonymous with CBs, are associated with the nucleolus, and are characterized by the presence of the protein p80-coilin (1, 4). Therefore, to confirm that FLASH localizes into CBs we performed costaining using anti-FLASH together with either anti-coilin or anti-p220/NPAT (known component of CBs). FLASH colocalized with coilin and p220/NPAT in MCF-7 and SAOS-2 cells (Figs. 1 e and f and 5d) and in all other cell lines tested (Hek293, HeLa, IMR90, and primary human fibroblasts) (ref. 20 and data not shown). These results were further confirmed by ImmunoGold labeling showing colocalization of coilin and FLASH (Fig. 1c). Staining with antibodies against BRCA1, MRE11, and TRF2 showed that FLASH did not colocalize with these proteins in the corresponding speckled structures (Fig. 6, which is published as supporting information on the PNAS web site). Similarly, staining with antibodies against promyelocytic leukemia (PML) showed that FLASH does not localize in PML bodies (Fig. 7, which is published as supporting information on the PNAS web site); however, as previously reported for CBs (21, 22), we observed a low-frequency association between PML bodies and FLASH-positive bodies. This association was most evident in HeLa cells, where ≈75% of cells contained at least one CB associated with PML bodies. In some cases the two bodies overlapped, but in most instances they appeared next to each other (Fig. 7). Whereas FLASH and NPAT showed 100% colocalization in all cell lines studied (including HeLa), this was not always true for FLASH and coilin (Table 1). Most of the primary and cancer cell lines studied showed a high percentage of FLASH/coilin colocalization ranging from 56% to 89%. However, in all cases, although a certain percentage of FLASH positive/coilin-negative bodies was observed, very few coilin-positive/FLASH-negative bodies were seen (Table 1), possibly suggesting a role for FLASH in the assembly of the bodies. HeLa cells were the only exception to this staining pattern, where only 33% FLASH/coilin colocalization was observed and 48% of bodies were coilin-positive and FLASH-negative (Table 1). Fig. 2a shows the distribution of FLASH/coilin double-positive bodies in the different cell lines studied. Interestingly, all cells analyzed (including HeLa) showed an average number of double-positive bodies per cell below four with a range between one and four bodies per cell.
Table 1.
Distribution of nuclear foci containing coilin and FLASH
| Cell line | No. of cells | Total no. of bodies | Average no. of bodies per cell | No. of F+/C− bodies (%) | No. of F−/C+ bodies (%) | No. of F+/C+ bodies (%) | Average no. of F+/C+ bodies per cell |
|---|---|---|---|---|---|---|---|
| Fibroblasts | 100 | 229 | 2.3 | 96 (42) | 4 (2) | 129 (56) | 1.3 |
| IMR90 | 100 | 162 | 1.6 | 61 (38) | 2 (1) | 99 (61) | 1 |
| MCF7 | 100 | 556 | 5.6 | 180 (32) | 13 (3) | 363 (65) | 3.6 |
| SAOS2 | 100 | 171 | 1.7 | 15 (9) | 3 (2) | 153 (89) | 1.5 |
| HeLa | 100 | 756 | 7.6 | 142 (19) | 356 (48) | 250 (33) | 2.5 |
Total number of bodies counted in 100 cells positive for FLASH (F+), coilin (C+), or both (total bodies) are reported as well as the number of bodies positive only for FLASH (F+/C−), positive only for coilin (F−/C+), or positive for both (F+/C+).
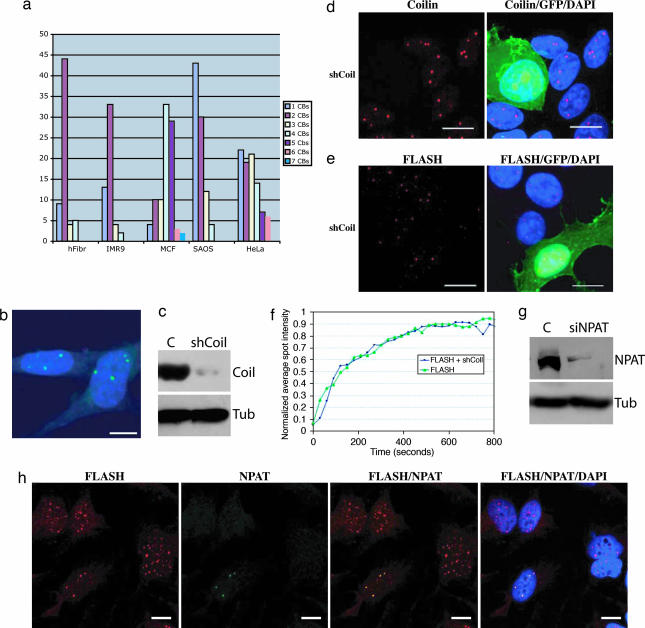
Fig. 2.
Coilin down-regulation does not affect FLASH localization to CBs. (a) Distribution of FLASH-positive/coilin-positive bodies in the different cell lines tested. (b) staining of coilin −/− MEFs with anti-FLASH antibody shows a normal punctate staining. (Scale bar: 10 μm.) (c) Western blot of MCF-7 cells transfected with pSUPER coilin (shCoil) or with a control vector (C) and collected 48 h after transfection by using an anti-coilin antibody. A strong reduction of coilin protein levels was observed. MCF-7 cells were transfected with pSUPER-Coilin (shCoil) together with GFP-spectrin (5:1 ratio) and stained for coilin (d) or FLASH (e) in red 48 h after transfection. Cells in which coilin has been down-regulated (green, GFP-positive cells) show a normal staining pattern for FLASH. (Scale bars: 10 μm.) (f) Fluorescence recovery after photobleaching analysis of SAOS-2 cells transfected for 36 h with GFP-FLASH alone (green line) or together with pSUPER coilin (blue line). Recovery after bleaching was followed for 800 s. (g) Western blot of HeLa cells transfected with Hs_NPAT_2Hp siRNA oligos (siNPAT) or with a control oligo (C) and collected 15 h after transfection by using an anti-NPAT antibody. (h) HeLa cells were transfected with Hs_NPAT_2Hp siRNA oligos and stained after 15 h by using antibodies against FLASH (red) and NPAT (green). Cells in which p220/NPAT is down-regulated show nuclear relocalization of FLASH, and cells in which p220/NPAT is still expressed have a normal distribution of FLASH in CBs. (Scale bars: 10 μm.)
Localization of FLASH to CBs, however, was not affected by coilin, because FLASH also showed a punctate staining in coilin knockout mouse embryonic fibroblasts (MEFs) (Fig. 2b) and after coilin knock-down (Fig. 2 c–e). To compare dynamic behavior of FLASH within CBs we performed fluorescence recovery after photobleaching experiments in cells transiently expressing GFP-FLASH before and after coilin short hairpin RNA. As shown in Fig. 2f, GFP-FLASH signal showed a 50% recovery within 90 s and reached a 84% recovery plateau after 450 s. This recovery time is comparable with that of the CB components with the highest residence time and thus compatible with FLASH being a structural component of CBs (2). Down-regulation of coilin did not affect GFP-FLASH recovery, confirming that FLASH localization to CBs is coilin-independent. On the contrary, down-regulation of NPAT by two different siRNA resulted in FLASH delocalization, suggesting that the two proteins are tightly connected (Fig. 2 g and h and data not shown).
FLASH Is a Short-Lived Protein.
Transcriptional inhibition as well as a block of protein synthesis resulted in disruption of the normal CB structure and relocalization of coilin (19, 23). Treatment of SAOS-2 cells (and MCF-7; data not shown) with actinomycin D (data not shown) or with cycloheximide resulted in the expected coilin relocalization (Fig. 3a), and within 5 h of treatment 50% of coilin-positive bodies disappeared (Fig. 3b). This finding was paralleled by a disappearance of FLASH- and NPAT-stained bodies. However, although coilin protein levels did not change after 5 h of treatment with either drug (Fig. 3c and data not shown), under the same conditions there was loss of FLASH staining (Fig. 3a) consistent with the rapid down-regulation of FLASH observed by Western blot (Fig. 3c).
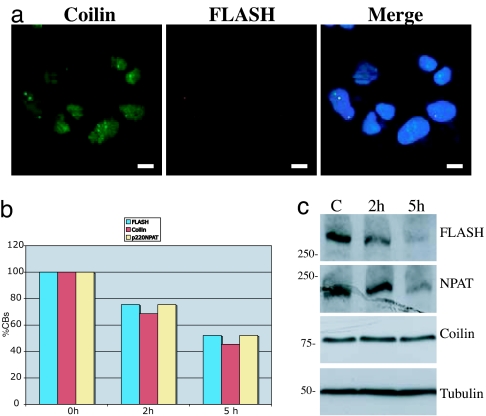
Fig. 3.
Protein synthesis block results in rapid FLASH down-regulation. (a) Treatment of SAOS-2 cells with 30 μg/ml cycloheximide for 5 h results in relocalization of coilin to the nucleus (green) and in a disappearance of FLASH staining (red). (Scale bars: 10 μm.) (b) Number of FLASH (blue), coilin (red), and p220/NPAT (yellow) bodies after treatment with 30 μg/ml cycloheximide for 0, 2, and 5 h. (c) Western blot of SAOS-2 cells untreated (C) or treated with 30 μg/ml cycloheximide for 2 or 5 h showing down-regulation of FLASH and NPAT but not coilin.
Down-Regulation of FLASH Results in CB Disruption.
To investigate the functional role of FLASH, we down-regulated its expression. Transfection of MCF-7 cells with a vector containing the sequence for a short hairpin RNA against FLASH (pSUPER-FLASH-1) resulted in down-regulation of FLASH protein, evident by both immunofluorescence (Fig. 4a) and Western blot (Fig. 4c). Down-regulation of FLASH resulted in the disappearance of the large CBs and in the appearance of a diffuse micropunctate nuclear and perinucleolar localization of coilin (Fig. 4b). These results were confirmed by using another short hairpin RNA (pSUPER-FLASH-2) targeting a different sequence of the FLASH gene (Fig. 8 a and b, which is published as supporting information on the PNAS web site). To investigate whether down-regulation of FLASH also results in a morphological change of CB structure, we transfected HeLa cells with GFP-spectrin together with pSUPER-FLASH-1 or a scrambled control vector and sorted GFP-positive cells by flow cytometry 48 h after transfection before subjecting them to ultrastructural analysis. Fig. 4e shows that cells transfected with pSUPER-FLASH contained structures that exhibit the morphological characteristics of CBs but are much smaller (<200 nm) compared with the normal-size CBs observed in cells transfected with the control vector (Fig. 4d). Many of these structures were rounded in profile (red arrow), whereas others (black arrow) were significantly smaller and irregular while still resembling fragments of CBs. These findings are consistent with the staining pattern observed by confocal analysis. We examined 100 nuclear profiles from each group and found no fragmented CBs in the controls, whereas fragmented CBs were found in 68 of the profiles from pSUPER-FLASH-1 transfected cells, and only two of these profiles contained normal-size CBs. Consistent with a disruption of the normal CB structure, other proteins known to localize to CBs (SMN and p220/NPAT) also showed an altered staining pattern after FLASH interference (Fig. 8c and data not shown). Altogether these results demonstrate that FLASH is an important component of CBs and that its depletion results in altered localization of other CB components.
Discussion
FLASH was originally identified as a component of the apoptotic signaling complex known as the death-inducing signaling complex, which is assembled in response to Fas ligand binding (12, 13). Formation of the death-inducing signaling complex results in activation of a protease, caspase 8, and consequent activation of the proteolytic caspase cascade that leads to apoptosis. The precise mechanism through which FLASH acts to trigger apoptosis is not well understood, and a direct role for FLASH in this process has been questioned (14).
Although FLASH has been reported to localize primarily to the cytoplasm by Western blotting using polyclonal antibodies (12), immunofluorescence analyses indicated that a fraction of the protein could also be detected in the nucleus (15). Furthermore, upon treatment with TNFα or UV irradiation, the distribution of FLASH was shifted primarily to the nucleoplasm (15). Curiously, ectopic expression of GFP-FLASH revealed that the protein was primarily nuclear and sometimes displayed focal accumulations (15). Another group generated anti-FLASH antibodies but was unable to detect the endogenous protein altogether (17). In summary, the subcellular distribution of endogenous FLASH was not well understood up to now, and the mechanism by which it activates apoptosis is similarly unclear. To elucidate these and other questions, we used four different polyclonal anti-FLASH antibodies.
Our data clearly reveal that FLASH is a nuclear protein, making it unlikely that it participates directly in death receptor signaling. Endogenous FLASH always shows a clear punctate nuclear staining, and overexpressed FLASH has a very similar distribution despite the different tags used to detect it. The use of both amino- and carboxyl-terminal tags also excludes that truncated forms of FLASH translocate to the cytoplasm. FLASH appears in the cytoplasm only when highly overexpressed, suggesting that in this case we are observing an overexpression artifact. More importantly, we show that FLASH is clearly localized in CBs and not in other nuclear organelles and is essential for maintenance of their structure. In fact, down-regulation of FLASH results in disruption of the normal structure of CBs. Very few intact CBs can be detected by EM analysis in cells where FLASH is down-regulated, but only what appear like small fragments of CBs are found. The exact nature of these fragments is still obscure, and further work will be required to establish whether they are incompletely assembled CBs or fragmented bodies and whether they exist thanks to residual undetectable FLASH molecules. Possibly only a complete knockout study will provide answers to these questions.
Although further work will be required to show whether the functions of CBs, such as maturation of small nuclear ribonucleoproteins and assembly of transcription machinery components, are also FLASH-dependent, we have clearly established that FLASH is essential for the correct assembly of CBs in a manner analogous to the essential requirement for PML in the structure of PML bodies (24). This provides a plausible context for interpretation of recently published data linking FLASH to transcriptional activity of the glucocorticoid and mineralocorticoid receptors (15, 16).
Materials and Methods
Cell Cultures and Transfections.
SAOS-2 and IMR90 cells were grown in RPMI medium 1640 (Invitrogen, Carlsbad, CA), HeLa, and MCF-7 cell lines, and MEFs were grown in D-MEM (Invitrogen) at 37°C in a humidified atmosphere of 5% (vol/vol) CO2 in air. All of the media were supplemented with 10% (vol/vol) FBS (Invitrogen). Coilin −/− MEFs (25) were a kind gift of A. G. Matera (Case Western Reserve University, Cleveland, OH). Transient transfections were performed with the Calcium Phosphate Transfection Kit (Invitrogen) according to the manufacturer's protocol.
The inducible cell line was generated by transfection of GFP-FLASH-pTRE2Hyg plasmid in HeLa TET-ON cells (BD Clontech, Mountain View, CA) according to the BD TET-ON Gene Expression Systems user manual.
Plasmids and siRNA Oligos.
FLASH c-DNA was amplified by RT-PCR. Forward primer (5′-atggcagcagatgat-3′) and reverse primer (5′-cagttttacgtctatt-3′) (GenBank accession no. 16306505) from normal epidermal human keratinocytes (American Type Culture Collection, Manassas, VA) RNA were cloned in-frame with the HA tag into pcDNA by using the NheI and XhoI unique restriction sites. The insert was then subcloned in-frame with amino-terminal FLAG tag and carboxyl-terminal V5 tag in pcDNA3 (Invitrogen) and in frame with an amino-terminal GFP tag in pEGFP-C3 vector (BD Clontech) and in pTRE2Hyg (BD Clontech).
The pSUPER-FLASH-1, pSUPER-FLASH-2, pSUPER-Coilin, and pSUPER-scrambled vectors were generated by insertion in pSUPER vector (OligoEngine, Seattle, WA) of oligos targeting the following sequences: FLASH-1, 5′-gattgtctgagtttccaca-3′ (this sequence is 100% identical both in human and mouse FLASH); FLASH-2, 5′-aagggagaagtccttgataat-3′; coilin, 5′- agttgctgagaattctg-3′; scrambled, 5′-aattctccgaacgtgtcacgt-3′. pGFP-spectrin was kindly provided by R. F. Kalejta (University of Wisconsin, Madison, WI) (26). NPAT was down-regulated by using siRNA oligos (Qiagen, Valencia, CA) targeting the following sequences: Hs_NPAT_2_Hp siRNA (catalog no. SI00660814), 5′-aagcaacatcttcaaacaata-3′; Hs_NPAT_3_Hp siRNA (catalog no. SI00660821), 5′-caagataattcttgtcttcaa-3′. As a negative control we used the negative control siRNA from Qiagen (catalog no. 1022563) targeting the sequence 5′-aattctccgaacgtgtcacgt-3′.
EM and ImmunoGold Cytochemistry.
Cells were processed and labeled as previously described (27, 28) by using anti-FLASH ab8420 (Abcam, Cambridge, U.K.) and anti-coilin ab11822 (Abcam). DNA was localized by a modified in situ hybridization technique using ApoTag reagents S7100 (Chemicon, Temecula, CA) (28).
Immunofluorescence.
Immunofluorescence was performed as described previously (29). The following antibodies were used: anti-coilin antibody ab11822 (Abcam); anti-NPAT antibody 611344 (BD Transduction Laboratories, San Jose, CA); anti-BRCA1 (D-9) sc-6954 (Santa Cruz Biotechnology, Santa Cruz, CA); anti-TRF2 IMG-124 (Imgenex, San Diego, CA); anti-MRE11 MS-MRE11-PX1 (Genetex, San Antonio, TX); anti-PML (PG-M3) SC-966 (Santa Cruz Biotechnology).
For FLASH we used anti-FLASH antibody M300, sc-9088 (lot no. B040; Santa Cruz Biotechnology) for all of the immunofluorescence shown. In addition, similar staining (data not shown) was obtained by using the rabbit anti-FLASH antibody-purified clone 522 generated by Thomas Hofmann (German Cancer Research Center, Heidelberg, Germany) and the rabbit anti-FLASH antibodies SL1133 and SL1134 generated in our laboratory.
Fluorescence Recovery After Photobleaching.
Fluorescence recovery after photobleaching experiments were performed on a Nikon C1 laser confocal microscope (Nikon, Tokyo, Japan) equipped with a 405-nm diode laser, with a 488-nm argon ion laser and with a 543-nm He-Ne laser. All images were acquired through a Plan Apochromat ×60.0/1.40/0.21 oil objective. Cells were maintained throughout the whole experiment in a CO2-independent medium (GIBCO-BRL, Carlsbad, CA) on a 37°C disk incubator. Regions of interest were bleached by using five complete scansions in the selected area (9 μm2) with the 488-nm laser at full power (25 mW). Immediately after bleaching recovery was monitored by acquiring one frame every 30 s. To minimize “out-of-focus” problems, every image analyzed was obtained from the 3D projection of three different focal levels 1 μm from each other. To minimize bleaching, images were acquired at 0.25-mW laser power with a pixel dwell time of 1.7 μs per pixel. Quantification was performed by using EZ-C1 software (Nikon). For each time point the normalized spot intensity was calculated relative to a nonbleached area and considering as 100% of the normalized spot intensity the initial intensity of the same area before bleaching. Background fluorescence was subtracted and measured in a random field outside the cell. Data shown are representative of three independent experiments.
Western Blotting.
Western blots were performed as previously described (29) by using the following antibodies: anti-FLASH, M300, catalog no. sc-9088 (lot no. B040; Santa Cruz Biotechnology); rabbit anti-FLASH antibodies SL1133 and SL1134 (generated in our laboratory); anti-coilin antibody ab11822 (Abcam); and anti-NPAT antibody 611344 (BD Transduction Laboratories).
Supplementary Material
Acknowledgments
We thank Dr. A. G. Matera for helpful discussion and for providing coilin −/− MEFs. We thank Roger Snowden for help with FACS analysis and cell sorting. The work was supported by a grant from the Associazione Italiana Ricerca sul Cancro (to V.D.L.); Associazione Italiana Ricerca sul Cancro; European Union Grant QLK-CT-2002-01956; a European Union Epistem grant; an Active p53 grant; Progetto Genomica Funzionale COMETA; Investment Fund for Basic Research Grant FIRB-2001; Ministry of Education, University, and Research Grant MIUR-2002; a grant from the Ministero della Sanità; a Telethon grant (to G.M.); and a grant from the Medical Research Council (to G.M.).
Abbreviations
- CB
Cajal body
- MEF
mouse embryonic fibroblast
- PML
promyelocytic leukemia.
Footnotes
The authors declare no conflict of interest.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Gall JG. Annu Rev Cell Dev Biol. 2000;16:273–300. doi: 10.1146/annurev.cellbio.16.1.273. [DOI] [PubMed] [Google Scholar]
- 2.Dundr M, Hebert MD, Karpova TS, Stanek D, Xu H, Shpargel KB, Meier UT, Neugebauer KM, Matera AG, Misteli T. J Cell Biol. 2004;164:831–842. doi: 10.1083/jcb.200311121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ochs RL, Stein TW, Jr, Tan EM. J Cell Sci. 1994;107:385–399. doi: 10.1242/jcs.107.2.385. [DOI] [PubMed] [Google Scholar]
- 4.Gall JG. Nat Rev Mol Cell Biol. 2003;4:975–980. doi: 10.1038/nrm1262. [DOI] [PubMed] [Google Scholar]
- 5.Ogg SC, Lamond AI. J Cell Biol. 2002;159:17–21. doi: 10.1083/jcb.200206111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma T, Van Tine BA, Wei Y, Garrett MD, Nelson D, Adams PD, Wang J, Qin J, Chow LT, Harper JW. Genes Dev. 2000;14:2298–2313. doi: 10.1101/gad.829500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang A, Ikura T, Eto K, Ota MS. Biochem Biophys Res Commun. 2004;325:1509–1516. doi: 10.1016/j.bbrc.2004.10.198. [DOI] [PubMed] [Google Scholar]
- 8.Wei Y, Jin J, Harper JW. Mol Cell Biol. 2003;23:3669–3680. doi: 10.1128/MCB.23.10.3669-3680.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ye X, Wei Y, Nalepa G, Harper JW. Mol Cell Biol. 2003;23:8586–8600. doi: 10.1128/MCB.23.23.8586-8600.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao J, Kennedy BK, Lawrence BD, Barbie DA, Matera AG, Fletcher JA, Harlow E. Genes Dev. 2000;14:2283–2297. [PMC free article] [PubMed] [Google Scholar]
- 11.Frey MR, Matera AG. Proc Natl Acad Sci USA. 1995;92:5915–5919. doi: 10.1073/pnas.92.13.5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imai Y, Kimura T, Murakami A, Yajima N, Sakamaki K, Yonehara S. Nature. 1999;398:777–785. doi: 10.1038/19709. [DOI] [PubMed] [Google Scholar]
- 13.Medema JP. Nature. 1999;398:756–757. doi: 10.1038/19638. [DOI] [PubMed] [Google Scholar]
- 14.Koonin EV, Aravind L, Hofmann K, Tschopp J, Dixit VM. Nature. 1999;401:662–663. doi: 10.1038/44317. [DOI] [PubMed] [Google Scholar]
- 15.Kino T, Chrousos GP. J Biol Chem. 2003;278:3023–3029. doi: 10.1074/jbc.M209234200. [DOI] [PubMed] [Google Scholar]
- 16.Obradovic D, Tirard M, Nemethy Z, Hirsch O, Gronemeyer H, Almeida OF. Mol Pharmacol. 2004;65:761–769. doi: 10.1124/mol.65.3.761. [DOI] [PubMed] [Google Scholar]
- 17.Choi YH, Kim KB, Kim HH, Hong GS, Kwon YK, Chung CW, Park YM, Shen ZJ, Kim BJ, Lee SY, Jung YK. J Biol Chem. 2001;276:25073–25077. doi: 10.1074/jbc.M102941200. [DOI] [PubMed] [Google Scholar]
- 18.Monneron A, Bernhard W. J Ultrastruct Res. 1969;27:266–288. doi: 10.1016/s0022-5320(69)80017-1. [DOI] [PubMed] [Google Scholar]
- 19.Raska I, Ochs RL, Andrade LE, Chan EK, Burlingame R, Peebles C, Gruol D, Tan EM. J Struct Biol. 1990;104:120–127. doi: 10.1016/1047-8477(90)90066-l. [DOI] [PubMed] [Google Scholar]
- 20.Barcaroli D, Bongiorno-Borbone L, Terrinoni A, Hofmann TG, Rossi M, Knight RA, Matera AG, Melino G, De Laurenzi V. Proc Natl Acad Sci USA. 2006;103:14808–14812. doi: 10.1073/pnas.0604227103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun J, Xu H, Subramony SH, Hebert MD. J Cell Sci. 2005;118:4995–5003. doi: 10.1242/jcs.02613. [DOI] [PubMed] [Google Scholar]
- 22.Grande MA, van der Kraan I, van Steensel B, Schul W, de The H, van der Voort HT, de Jong L, van Driel R. J Cell Biochem. 1996;63:280–291. doi: 10.1002/(sici)1097-4644(19961201)63:3<280::aid-jcb3>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 23.Rebelo L, Almeida F, Ramos C, Bohmann K, Lamond AI, Carmo-Fonseca M. Mol Biol Cell. 1996;7:1137–1151. doi: 10.1091/mbc.7.7.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salomoni P, Pandolfi PP. Cell. 2002;108:165–170. doi: 10.1016/s0092-8674(02)00626-8. [DOI] [PubMed] [Google Scholar]
- 25.Tucker KE, Berciano MT, Jacobs EY, LePage DF, Shpargel KB, Rossire JJ, Chan EK, Lafarga M, Conlon RA, Matera AG. J Cell Biol. 2001;154:293–307. doi: 10.1083/jcb.200104083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalejta RF, Shenk T, Beavis AJ. Cytometry. 1997;29:286–291. doi: 10.1002/(sici)1097-0320(19971201)29:4<286::aid-cyto4>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 27.Dwyer-Nield LD, Dinsdale D, Cohen JJ, Squier MK, Malkinson AM. Methods Mol Med. 2003;74:283–297. doi: 10.1385/1-59259-323-2:283. [DOI] [PubMed] [Google Scholar]
- 28.Dinsdale D, Lee JC, Dewson G, Cohen GM, Peter ME. Am J Pathol. 2004;164:395–407. doi: 10.1016/S0002-9440(10)63130-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Munarriz E, Barcaroli D, Stephanou A, Townsend PA, Maisse C, Terrinoni A, Neale MH, Martin SJ, Latchman DS, Knight RA, et al. Mol Cell Biol. 2004;24:10593–10610. doi: 10.1128/MCB.24.24.10593-10610.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.