Abstract
Expression profiling of clinically obtainable tumor specimens has been hindered by the need for microgram quantities of RNA. In vitro transcription (IVT)-based amplifications are most commonly used to amplify small quantities of RNA for microarray analysis. However, significant drawbacks exist with IVT-based amplification, and the need for alternative amplification methods remains. Herein, we validate whole transcriptome amplification (WTA), an exponential amplification technique that produces cDNA libraries and amplified target in 3 to 4 hours from nanogram quantities of total RNA using a combination of cDNA microarrays and quantitative polymerase chain reaction (PCR). We demonstrate that WTA material can serve as a “molecular archive” because a WTA cDNA library can be faithfully amplified through multiple rounds of PCR amplification, allowing it to serve as a bankable and distributable resource. To demonstrate applicability, WTA was combined with laser capture microdissection to profile frozen prostate tissues. Unlike most IVT-based and exponential amplification techniques, WTA does not depend on the presence of a poly-A tail. Thus, we demonstrate that WTA is compatible with artificially degraded RNA and RNA isolated from formalin-fixed paraffin-embedded tissues. Taken together, WTA represents a versatile approach to profile and archive cDNA from minute tumor samples and is compatible with partially degraded RNA.
Keywords: Exponential RNA amplification, whole transcriptome amplification, prostate cancer, laser capture microdissection, FFPE tissue
Introduction
Profiling human cancer samples has become a standard technique in cancer research. With a few exceptions [1–6], most cancer profiling studies have used bulk tumors due to the requirement for at least 5 µg of total RNA. Interest in profiling clinically obtainable specimens, by needle biopsy, fine needle aspiration, or laser capture microdissection (LCM), has led to the development of techniques to amplify RNA. Currently, the most widely used and validated technique is T7 polymerase-based in vitro transcription (IVT) [7,8]. The cost, speed, labor, and requirement for multiple rounds of amplification from limiting samples have prompted the investigation of various polymerase chain reaction (PCR)-based amplification techniques and their applicability to microarray profiling [9–14]. However, these methods also have significant drawbacks, including the requirement of a poly-A tail [10,12–14], the requirement to produce double-stranded cDNA before amplification [11], extreme 3′ bias requiring custom microarrays [10], or that they are a combination of PCR and IVT-based techniques [13].
In this report, we describe the evaluation of OmniPlex whole transcriptome amplification (WTA) with respect to the amplification of RNA from specimens of interest to the cancer researcher. WTA is a simple amplification protocol that is analogous to OmniPlex whole genome amplification (WGA)—a rapid, robust, and unbiased method used to amplify genomic DNA using genome fragmentation followed by linker attachment and PCR amplification [15]. In WTA, RNA undergoes a single-step conversion into cDNA fragments flanked by universal priming sites, with subsequent PCR amplification of the resulting cDNA library using universal oligonucleotide primers. Importantly, WTA does not depend on the presence of a poly-A tail, resulting in a uniform amplification of template RNA without bias to the 3′ end.
Herein, we evaluate the reproducibility of WTA and its use for identifying differentially expressed genes using cDNA microarrays and quantitative real-time PCR (QPCR) from tissue samples and cell lines. We also demonstrate that WTA- amplified samples can serve as a reusable library or “molecular archive” because multiple rounds of WTA PCR amplification from a WTA cDNA library are reproducible and can produce enough target for thousands of microarrays. We also combine LCM with WTA to identify genes that are differentially expressed between benign epithelial cells, cancerous epithelial cells, and stroma isolated from frozen prostate tissue sections. As WTA does not depend on a poly-A tail to prime the reaction, we demonstrate that WTA can be used to profile partially degraded RNA, as well as benign and cancerous prostate sections from formalin-fixed paraffin-embedded (FFPE) tissues. Taken together, these results validate the applicability of WTA for studying gene expression from a variety of tumor specimens.
Materials and Methods
Samples
All prostate tissue specimens were obtained with written patient consent and approval of the University of Michigan Institutional Review Board. These tissues were stored in the University of Michigan Prostate SPORE Tissue Bank. For WTA reproducibility studies, we used a commercially available pool of benign prostate tissue total RNA (CPP; Clontech, Mountain View, CA). For WTA evaluation studies, both tissue samples and cell line samples were used for RNA isolation. One prostate cancer tissue specimen (PCA1) was used with the same lot of CPP as the reference for the set of experiments. Total RNA was also isolated from the prostate cancer cell line LnCAP, with cells treated with 1 nM of the synthetic androgen R1881 (LnCAP_R) or ethanol as control (LnCAP_C) for 24 hours before RNA isolation. Tissue and cell line samples were homogenized in Trizol (Invitrogen, Carlsbad, CA), and total RNA was isolated using the standard Trizol protocol. Denaturing formaldehyde agarose gel electrophoresis or Bioanalyzer 2100 (Agilent, Palo Alto, CA) was used to verify total RNA integrity. RNA was quantified using ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE) and/or Ribogreen (Molecular Probes, Eugene, OR).
WTA Amplification
The OmniPlex WTA described in this report was performed using a TransPlex WTA kit (Rubicon Genomics, Inc., Ann Arbor, MI). The indicated amounts of total RNA in a volume of 25 µl were converted into OmniPlex WTA cDNA libraries and amplified by WTA PCR using reagents and protocols supplied with or recommended by beta-commercial TransPlex WTA kits (www.rubicongenomics.com). Briefly, RNAse-free water was added with the indicated amount of total RNA to a volume of 16.5 µl. A 5x library synthesis buffer (1 x final concentration) and a 10x library stabilization buffer were added; the mixture was heated at 70°C for 5 minutes and immediately cooled. One microliter of library synthesis enzyme was added, and WTA cDNA libraries were synthesized using the following thermocycler program: 24°C for 15 minutes, 42°C for 2 hours, and 95°C for 5 minutes. Aliquots were WTA PCR-amplified using Titanium Taq polymerase (Clontech) in the presence of amino-allyl deoxyuridine triphosphate (dUTP) for postamplification labeling. For hybridizations using reamplified WTA products, multiple 1- to 5-ng aliquots of the initial WTA PCR reaction were subjected to an additional WTA PCR amplification in the presence of amino-allyl dUTP, and products were pooled before hybridization. Yields after all WTA PCR amplifications were between 2 and 5 µg per reaction, and reaction progress was monitored in real time using SYBR Green I (Molecular Probes) on iCycler IQ (BioRad, Hercules, CA) or ABI 7300 Real-Time PCR System (Applied Biosystems, Foster City, CA). Real-time PCR amplifications were terminated at or before plateau phase, as measured by fluorescence incorporation, to preserve maximum representation. A range of 10 to 30 µg of Cy3- and Cy5-labeled product was evaluated for hybridization, as described in Table W1.
cDNA Microarray Procedures
The 20K-element spotted cDNA microarrays used were constructed essentially as described [16–18]. The complete list of sequence-verified clones is presented in Table W2, and all clones are available from Research Genetics (Huntsville, AL) (www.resgen.com).
The complete details of the printing, postprocessing, labeling, and hybridization of the arrays are available in the MIAME checklist (supplementary materials). For microarray experiments using unamplified total RNA, 30 µg of total RNA from PCA1, CPP, LnCAP_C, and LnCAP_R was used for target synthesis in the presence of oligo-dT and random primers. For WTA samples, after incorporation of dUTP in the final WTA PCR reaction and purification, amplified target was labeled and hybridized as for the unamplified samples.
Data Analysis
Arrays were scanned with an Axon 4000B scanner, and primary analysis was performed using Genepix 4.0 software package (Axon Instruments, Sunnyvale, CA). For all arrays described in this study, features flagged by GenePix as not found during grid alignment and areas of obvious defects were excluded from further analysis. For reproducibility studies using self-self hybridizations, features were ranked based on the sum of the medians (Cy3 + Cy5 intensity), and the top 50% was used. All included features were normalized to set the aggregate median of ratios (Cy5/Cy3) to one. Background-corrected mean Cy3 and Cy5 intensities for all included features (Table W3) were plotted using Microsoft Excel.
For all other arrays, features on each array were filtered based on the rank of the sum of the medians (tissue samples and cell line samples, top 50% used; LCM and FFPE samples, top 25% used), and the median of ratios (log2 of Cy5/Cy3) was normalized using locally weighted regression (LOWESS) with a window of 0.6 using custom software written in Perl and R. Finally, to remove unreliable data, features showing an average normalized median of ratios of >1.5 or <0.75 across a series of 11 self-self hybridizations (including dye swaps and spanning print runs used in this study) were removed from all arrays. Cy5/Cy3 (normalized log2) ratios for these hybridizations can be found in Table W2.
To create the composite array for the tissue samples from the five unamplified hybridizations, features were flagged as described above and only features present on at least four of five arrays were included. The normalized median of ratios for each feature was then averaged to create the composite array. The intensity-dependent Z score analysis used for the cell line hybridizations was performed essentially as described [19], using a sliding window of 50 features to calculate the local mean, standard deviation, and Z score for each feature on the array using custom software written in Perl. For all analyses, features were clustered using Cluster or were visualized directly with Tree View, as indicated in the text [20].
QPCR
We performed QPCR using SYBR Green, as previously described [21,22]. Briefly, WTA and (directly) reverse-transcribed cDNA, as indicated, were analyzed. The amount of each target gene relative to the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) for each sample was determined using the comparative threshold cycle (Ct) method (Applied Biosystems user bulletin 2; http://docs.appliedbiosystems.com/pebiodocs/04303859.pdf), with the unamplified CPP cDNA sample for each gene serving as calibrator. All primers were synthesized by Integrated DNA Technologies, and sequences are available on request. AMACR[21], EZH2 [22], TPD52 [23], HMBS, and GAPDH [24] primers were as described. Approximately equal efficiencies of the primers were confirmed previously using serial dilutions of prostate cDNA to use the comparative Ct method.
LCM
LCM was used to isolate approximately 5000 to 10,000 cells from frozen prostate tissue specimens from separate patients. One to three serial sections were used per sample. Epithelia from cancerous glands were isolated from four samples (LCM_PCA1-4), epithelia from benign glands were isolated from five samples (LCM_NOR1-5), and stroma adjacent to cancerous glands (LCM_STROMA1) or stroma from a nodule of benign prostatic hyperplasia (LCM_STROMA2) were captured. To prepare slides for LCM, a 6-mm slice was cut with a cryostat from the prostate cancer tissue block embedded in OCT (Sakura, Tokyo, Japan) and placed on a specially manufactured membrane slide (MMI, Knoxville, TN). The slide was placed in distilled water and stained with Harris hematoxylin for 50 seconds, followed by two short water washes in distilled water. Eosin staining was performed for 30 seconds. Excessive eosin was rinsed off with distilled water, and slides were air-dried. Fixation or dehydration with ethanol was avoided. The SL Microtest device (MMI) using the µCUT software was used for LCM. Areas of interest were circled and cut by UV laser. Isolated cells were picked up using the adhesive surface of the lid of specially manufactured tubes (MMI). Total RNA was isolated from each sample using the RNAqueous Micro Kit, treated with DNase I (Ambion Diagnostics, Austin, TX) according to the manufacturer's instructions, quantified with Ribogreen (Molecular Probes), subjected to WTA cDNA synthesis and two rounds of WTA PCR amplification in parallel with an equal amount of CPP, and hybridized (LCM sample versus CPP) as described above.
RNA Hydrolysis
Artificial degradation of total RNA was performed essentially as described [25]. Total RNA (2.5 µg in 2.5 µl) isolated from LnCAP_C and LnCAP_R cells, as described above, was added to 2.5 µl of 5x first-strand cDNA synthesis buffer (Invitrogen). RNA was then heated to 80°C for 5 or 15 minutes. After heating, 500 ng of each sample was analyzed on an RNA Nano LabChip using Bioanalyzer 2100 (Agilent), and RNA was quantified with an ND-1000 spectrophotometer. Twenty-five nanograms of each artificially degraded sample was used for WTA cDNA synthesis and two rounds of WTA PCR amplification, and hybridized (LnCAP_R versus LnCAP_C for each time point) as described above.
FFPE Tissue RNA Isolation
Total RNA was extracted from FFPE tissue samples using an Optimum FFPE RNA Isolation Kit (Ambion Diagnostics) according to the manufacturer's instructions. Total RNA was isolated from three cases, with two containing clinically localized prostate adenocarcinoma (FFPE_PCA1-2) and one containing only benign prostate tissue (FFPE_NOR1). FFPE_PCA1 and FFPE_PCA2 were embedded in 2003, and FFPE_NOR1 was embedded in 1995. Briefly, two 10-µm sections were cut from the paraffin block, immediately placed in a microcentrifuge tube, and stored at -80°C until RNA isolation. Sections were twice incubated in xylene for 10 minutes, followed by an ethanol gradient wash. After overnight incubation with proteinase K, RNA was isolated, treated with DNase I, and quantified using an ND spectrophotometer. RNA integrity was determined using denaturing formaldehyde agarose gel electrophoresis. Twenty-five nanograms of each sample was used for WTA cDNA synthesis and two rounds of WTA PCR amplification, and hybridized (FFPE_PCA1-2 or FFPE_NOR1 vs CPP) as described above.
Results
Reproducibility of WTA for Gene Expression Profiling and Construction of Molecular Archives
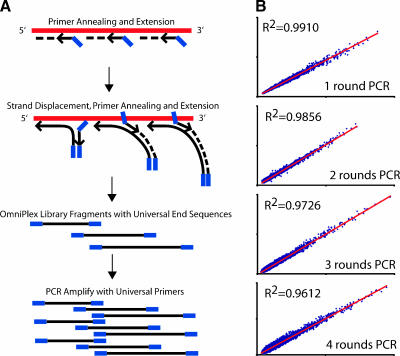
In this report, we describe the evaluation of WTA, a novel non-T7-based exponential RNA amplification method (Figure 1A). Initially, the transcriptome is converted to a cDNA library of controlled fragment length with identical linkers on the 5′ and 3′ ends. To synthesize the library, sample RNA is incubated with a reverse transcriptase in the presence of non-self-complementary primers comprised of a quasi-random 3′ end and a universal 5′ end. When annealed primers are extended by a polymerase, displaced single strands are generated, which become new templates for primer annealing and extension. This process creates a cDNA library comprised of random, overlapping 100- to 1000-base fragments flanked by a universal end sequence. Universal primer PCR is then used to amplify the cDNA library and to produce WTA products.
Figure 1.
Non-T7-polymerase based WTA strategy and reproducibility. (A) Initially, the RNA transcriptome (red) is converted to a WTA cDNA library using a reverse transcriptase in the presence of non-self-complementary primers consisting of a quasi-random 3′ end (black arrow) and a universal 5′ end (blue). Extension of annealed primers by a polymerase generates single strands, which become new templates for primer annealing and extension. This process creates a cDNA library of random, overlapping 100- to 1000-base fragments with universal end sequences. These fragments are then amplified by one or more rounds of WTA PCR using primers specific for the universal ends. Amino-allyl dUTP is incorporated in the WTA PCR amplification for subsequent labeling, and WTA PCR amplification can be monitored in real time using SYBR Green to ensure maximum representation and to avoid saturation. The entire library creation and one round of WTA PCR amplification take 3 to 4 hours using the manufacturer's protocol. (B) Scatter plots of self-self hybridizations demonstrate the reproducibility of WTA. Two identical 25-ng aliquots of a benign prostate tissue pool (CPP) were converted into WTA cDNA libraries and subjected to one, two, three, or four rounds of WTA PCR amplification, as indicated before labeling with Cy3 or Cy5 and hybridization together on 20K-element spotted cDNA microarrays. Background-corrected mean Cy3 intensity vs. background-corrected mean Cy5 intensity was plotted, and Pearson correlation coefficients (R2) are indicated.
To validate WTA for expression profiling, we began by evaluating the reproducibility of WTA using a series of self-self hybridizations on 20K-element cDNA microarrays. Two identical 25-ng aliquots of total RNA from a commercially available pool of benign prostate tissue (CPP) were converted into WTA cDNA libraries and subjected to one round of WTA PCR amplification. Fifteen micrograms of amplified cDNA from each aliquot was labeled with Cy3 or Cy5, and hybridized competitively on the same array. This experiment was also repeated with separate RNA aliquots subjected to WTA cDNA synthesis and two, three, or four rounds of WTA PCR amplification before labeling and hybridization. Background-corrected mean Cy5 vs Cy3 intensities were plotted and displayed as scatter plots (Figure 1B). Pearson correlation coefficients (R2; one-round WTA PCR = 0.9910; two-round WTA PCR = 0.9856; three-round WTA PCR = 0.9726; four-round WTA PCR = 0.9612) reveal excellent reproducibility for WTA cDNA library synthesis and one, two, three, or four rounds of WTA PCR amplification. When amplified products produced from different rounds of WTA product were competitively hybridized (Cy3, four rounds; Cy5, two rounds), they showed reduced correlation (R2 = 0.8361), demonstrating that samples, to be hybridized, should be subjected to the same number of WTA PCR rounds.
As each WTA PCR amplification produces microgram quantities of cDNA products and only nanograms are required for subsequent PCR amplifications, WTA cDNA products can serve as “molecular archives,” which can be faithfully reamplified. To demonstrate the stability of these “molecular archives,” a separate 25-ng aliquot of CPP total RNA was converted into a WTA cDNA library and subjected to one round of WTA PCR amplification. One aliquot of this WTA-amplified cDNA was subjected to a second round of WTA PCR amplification immediately, whereas another aliquot was taken 6 months later and subjected to a second round of WTA PCR. The products were labeled with Cy3 or Cy5, and hybridized together as above, with R2 = 0.9923.
Fidelity of WTA for Detecting Differentially Expressed Genes
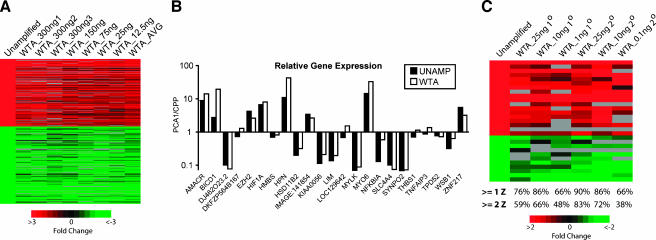
As one of the main uses for microarrays is the detection of genes differentially expressed between benign and cancerous tissues, we sought to validate WTA for this application. We began by profiling tissue samples representing benign and cancerous prostate tissues, which we expected to have a large number of differentially expressed genes based on our previous profiling studies [16,17]. Total RNA from the same clinically localized prostate cancer specimen (PCA1) and CPP was used directly or WTA-amplified before hybridization. Five replicate hybridizations from unamplified total RNA were performed with PCA1 labeled with Cy5, and with CPP labeled with Cy3, to create a composite unamplified array, as described in the Materials and Methods section. We then performed seven hybridizations after WTA cDNA library synthesis and a single round of WTA PCR amplification on a range of input total RNA (300 ng-12.5 ng) from the same samples, PCA1 (labeled with Cy5) and CPP (labeled with Cy3). After filtering, as described in the Materials and Methods section, features from each WTA hybridization were compared to the composite array. We identified 345 of 7564 features on the composite unamplified array that showed a >3-fold differential expression, and their expression ratios were visualized along with their corresponding values from WTA hybridizations (Figure 2A). The correlation coefficient (R) between the expression ratios of all 7564 features on the composite unamplified array and the average expression ratio of the seven WTA hybridizations is 0.84. The correlation coefficient for the 345 features on the composite array showing a >3-fold differential expression is 0.99. From these initial experiments, we determined that hybridization of 10 µg of amplified products (both control and experimental) is sufficient starting with >100 ng of input RNA, whereas 15 to 20 µg is optimal for hybridizations starting with less RNA.
Figure 2.
Concordance of differentially expressed genes between unamplified and WTA-amplified total RNA. (A) Unamplified total RNA from a prostate cancer tissue specimen (PCA1, Cy5) was hybridized against unamplified total RNA from benign prostate tissues (CPP, Cy3), and five replicate hybridizations were averaged to create a composite array. The indicated amounts of the same total RNA were then converted into WTA cDNA libraries, amplified by one round of WTA PCR and hybridized for comparison. The 345 features identified as being differentially expressed by >3-fold on the composite array (Unamplified) and their corresponding expression values from the indicated WTA hybridizations and the average of the seven WTA arrays (WTA_AVG) were visualized using TreeView. Columns represent individual hybridizations using the indicated amount of input total RNA for WTA (as described in Table W1), and rows represent individual features. Red and green cells represent upregulation or downregulation, respectively (see scale at the bottom), in PCA1 compared to CPP. Black cells indicate probes with roughly equivalent expression, and grey cells indicate features not passing filtering. (B) Validation of WTA fidelity using QPCR. Directly reverse-transcribed cDNA (UNAMP) or WTA cDNA (WTA; from 25 ng of input total RNA, WTA cDNA library synthesis, and one round of WTA PCR amplification) from the same samples used for microarray analysis in (A) PCA 1 and CPP was assessed using QPCR. The amount of each target gene in PCA 1 and CPP was normalized to the amount of GAPDH for the sample. Data are presented as the ratio of PCA 1 to CPP for each target gene on a log scale. (C) Unamplified total RNA from the prostate cancer cell line LnCAP treated with the synthetic androgen R1881 (LnCAP_R, Cy5) was hybridized against unamplified total RNA from LnCAP cells treated with vehicle (LnCAP_C). The indicated amounts of the same total RNA were used for WTA cDNA library synthesis followed by one (1°) or two (2°) rounds of WTA PCR amplification and hybridized for comparison. The top 29 differentially expressed features from the unamplified array (≥4 Z score units, minimum of 1.97-fold upregulated, or 1.78-fold downregulated LnCAP_R/LnCAP_C) and their corresponding expression values from the WTA hybridizations were visualized using TreeView as above. The number below each column indicates the percentage of these top 29 differentially expressed features identified in the unamplified array that had Z scores ≥1 or ≥2 on the indicated WTA hybridization. The complete gene lists from all heat maps are available in Table W4.
To further evaluate WTA technology, QPCR was used to determine the expression of 23 genes. These genes were selected to represent both unchanged and differentially expressed genes that spanned the intensity spectrum of the composite unamplified array to assess the fidelity of WTA for both high- and low-copy-number transcripts. Relative quantities of these genes, compared to the housekeeping gene GAPDH, were determined in unamplified reverse-transcribed cDNA and WTA cDNA (Figure 2B). Twenty five nanograms of input total RNA was used for WTA cDNA synthesis and one round of WTA PCR amplification.
We then sought to determine the fidelity of WTA for samples with limited numbers of differentially expressed genes. We used total RNA from the prostate cancer cell line LnCAP treated with the synthetic androgen R1881 (LnCAP_R) or control (LnCAP_C). Unamplified total RNA was used to produce hybridization with LnCAP_R labeled with Cy5, and with LnCAP_C labeled with Cy3. Six hybridizations were then performed after WTA cDNA library synthesis and one or two rounds of WTA PCR amplification on a range of input total RNA (25 ng–100 pg) from the same samples, LnCAP_R (labeled with Cy5) and LnCAP_C (labeled with Cy3). As expected, a much smaller set of features was differentially expressed (eight features ≥3-fold differential expression) compared to the tissue sample experiments described above. Thus, we used intensity-dependent Z scores, a more sensitive measure than standard fold change [19], to identify the top 29 differentially expressed features (≥4 Z score units, a minimum of 1.97-fold upregulated or 1.78-fold downregulated LnCAP_R/LnCAP_C) on the unamplified array. The expression values for these 29 features from the unamplified array and the corresponding values from the WTA hybridizations were visualized (Figure 2C). To quantify the ability of WTA to maintain differential expression, we determined the percentage of these top 29 differentially expressed features identified in the unamplified array that had Z scores ≥1 or ≥2 (in the same direction) on each WTA hybridization, as shown below each column in Figure 2C. These experiments demonstrate that WTA maintains expression patterns of differentially expressed genes with as little as 100 pg to 1 ng of input total RNA, although hybridizations from <10 ng of input total RNA had decreased correlation to the unamplified composite array. Hybridizations after WTA cDNA synthesis and two rounds of WTA PCR also improved the correlation with unamplified total RNA. Taken together, these experiments demonstrate that WTA faithfully maintains differential expression over a wide range of input RNA and is applicable to profiling cancerous tissue or cell line samples.
LCM and WTA for Profiling Benign Prostate Epithelium, Cancerous Epithelium, and Stroma
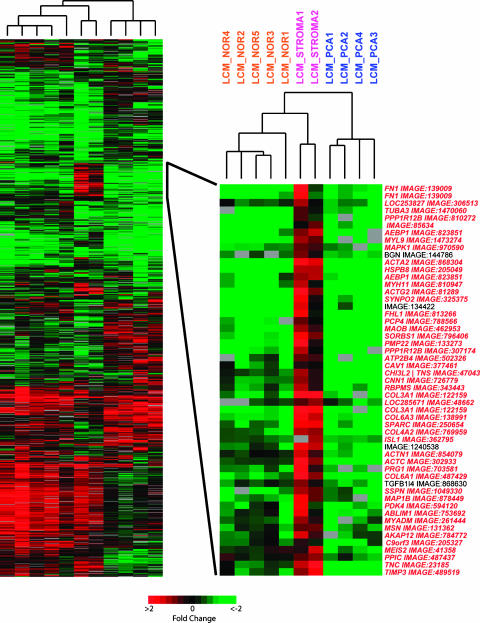
After validating WTA with minimal amounts of cell line and tissue samples, we investigated the ability of WTA to detect differentially expressed genes from material obtained by LCM. Total RNA was isolated from 5000 to 10,000 cells obtained by LCM from 11 samples. Separate cases were used for each sample, with five samples of epithelium from benign glands (LCM_NOR1-5) and four samples of cancerous epithelium (LCM_PCA1-4). Two stromal samples were also captured: one isolated from stroma adjacent to cancerous glands (LCM_STROMA1) and a second sample from a nodule of benign prostatic hyperplasia (BPH) (LCM_STROMA2). Ten to 45 ng of total RNA was isolated per sample. WTA cDNA library synthesis and two rounds of WTA PCR amplification were used to produce labeled targets for hybridization from 10 ng of isolated total RNA. Fifteen micrograms of the amplified target was labeled and hybridized with 15 µg of the amplified target produced from an equivalent amount of CPP amplified in parallel. After filtering the data as described in the Materials and Methods section to include only high-quality features, 1002 features that passed filtering in at least 9 of 11 arrays and showed a standard deviation of values ≥0.3 were used for unsupervised average-linkage hierarchical clustering with Cluster and visualized with TreeView (Figure 3). The four cancerous epithelial samples clustered on a separate branch from the benign and stromal samples, with the stromal samples on a distinct terminal branch from the benign samples. Although a complete examination of differentially expressed genes is outside the scope of this study, a region of interest that demonstrates the usefulness of LCM and WTA for profiling studies was highlighted. Gene names denoted in red in the inset region have been identified in at least one profiling study of grossly dissected samples as being significantly downregulated (P < .05) in prostate cancer compared to benign samples (including normal adjacent tissue and BPH), according to Oncomine 2.0 (www.oncomine.org), a compendium of microarray profiling studies [26]. In our study, rather than being downregulated in cancerous epithelium, these transcripts were highly expressed in stroma compared to both cancerous and normal epithelia.
Figure 3.
WTA amplification from RNA isolated from LCM prostate tissue reveals unique signatures. Approximately 5000 to 10,000 cells from different cases containing cancerous epithelia (LCM_PCA1-4), benign epithelia (LCM_NOR1-5), stroma adjacent to cancerous glands (LCM_STROMA1), and stroma from a nodule of benign prostatic hypertrophy (LCM_STROMA2) from frozen prostate specimens were captured by LCM. Isolated total RNA was used for WTA cDNA library synthesis and two rounds of WTA PCR amplification in parallel with an equal amount of CPP and hybridized. For each experiment, the LCM sample was labeled with Cy5 (red), and CPP was labeled with Cy3 (green). The 1002 features that passed filtering on at least 9 of 11 arrays and showed a standard deviation of ≥0.3 were clustered using unsupervised average-linkage hierarchical clustering with Cluster and visualized with TreeView (see Figure 2 for the matrix color scheme). Names denoted in red in the inset region represent genes identified as being significantly (P < .05) underexpressed in tumor samples compared to benign prostate in at least one previous profiling study using grossly dissected tissues according to Oncomine 2.0 (www.oncomine.org)—a compendium of cancer microarray profiling studies.
WTA Is Compatible with Partially Degraded RNA
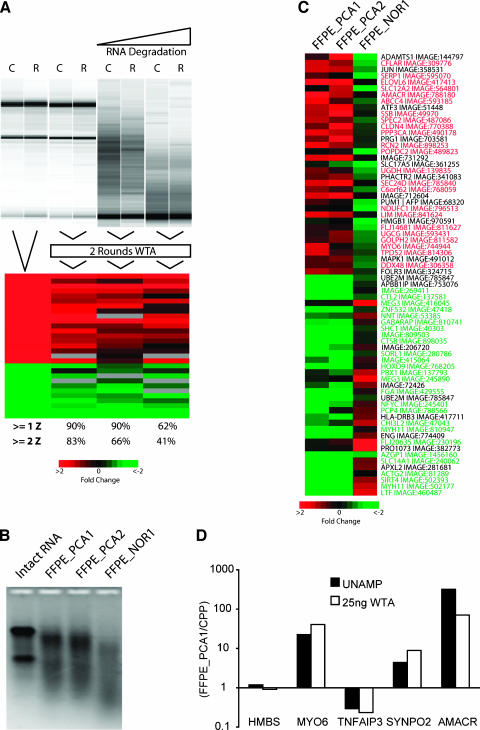
As WTA does not depend on the presence of a poly-A tail on the RNA transcript, we sought to determine whether WTA is compatible with degraded RNA, including RNA isolated from FFPE samples. To assess the compatibility of WTA with artificially degraded RNA, we used the same total RNA from LnCAP cells treated with the androgen analogue R1881 or control (LnCAP_R or LnCAP_C, respectively), as in our initial validation experiments (Figure 2C). Total RNA from LnCAP_R and LnCAP_C was base-hydrolyzed for 5 or 15 minutes, and 25 ng was used for WTA cDNA library synthesis and two rounds of WTA PCR amplification, labeling (LnCAP_R, Cy5; LnCAP_C, Cy3), and hybridization. Results of hybridization from 25 ng of the intact samples subjected to WTA cDNA library synthesis and two rounds of WTA PCR amplification, and these two degraded samples were compared to the top 29 differentially expressed features identified from intact unamplified samples (Figure 4A). The numbers below each column again indicate the percentage of the top 29 differentially expressed features identified in the unamplified array that had Z scores ≥1 or ≥2 on each WTA hybridization. After 5 minutes of degradation resulting in the absence of 28S and 18S ribosomal bands, 90% and 66% of the most differentially expressed genes identified on the unamplified hybridization still had Z scores >1 or >2, respectively. With further degradation, WTA-amplified samples still showed similar expression patterns to unamplified intact RNA; however, there was decreased correlation to the unamplified sample.
Figure 4.
WTA gives high-fidelity results with partially degraded RNA and RNA isolated from FFPE samples. (A) Total RNA from the prostate cancer cell line LnCAP treated with R1881 (R) or control (C), as described in Figure 2C, was hydrolyzed, and RNA integrity was determined. Twenty-five nanograms of hydrolyzed RNA was used for WTA cDNA library synthesis and two rounds of WTA PCR before hybridization, as described in Figure 2C. Results were visualized with TreeView along with corresponding hybridizations from intact unamplified total RNA and 25 ng of intact RNA subjected to two rounds of WTA (see Figure 2 for the matrix color scheme). The number below each column indicates the percentage of these top 29 differentially expressed features identified in the unamplified array that had Z scores ≥1 or ≥2 on the indicated WTA hybridization. (B) Total RNA was isolated from two sections from FFPE specimens containing prostate cancer (FFPE_PCA1-2) or benign prostate (FFPE_NOR1). RNA integrity was determined using denaturing formaldehyde agarose gel electrophoresis and compared to intact total RNA as indicated. (C) Twenty-five nanograms of total RNA from each FFPE sample was used for WTA cDNA library synthesis and two rounds of WTA PCR in parallel with an equal amount of CPP, and hybridized. For each experiment, the FFPE sample was labeled with Cy5 (red), and CPP was labeled with Cy3 (green). The 70 genes showing the greatest differential expression between the cancer and normal samples were visualized using TreeView. Genes denoted in red or green represent genes identified as being significantly (P < .05) dysregulated in the same direction in bulk tumor samples compared to normal prostate tissues (upregulated or downregulated, respectively) in at least one profiling study using Oncomine 2.0. (D) Validation of differentially expressed genes using QPCR. Directly reverse-transcribed cDNA (UNAMP) or WTA cDNA, as indicated from FFPE_PCA1 and CPP, was assessed using QPCR. The amount of each target gene for FFPE_PCA1 and CPP was normalized to the corresponding amount of GAPDH for the sample. Data are presented as the ratio of FFPE_PCA1 to CPP for each target gene on a log scale.
We also directly tested the usefulness of WTA with FFPE samples, which are known to yield degraded RNA, due to cross-linking between nucleic acids and proteins. Total RNA was isolated from two 10-µm sections from grossly dissected tumor samples (FFPE_PCA1 and FFPE_PCA2) and one grossly dissected sample containing only benign tissue (FFPE_NOR1) that had been fixed in formalin and embedded in paraffin. Denaturing formaldehyde agarose gel electrophoresis revealed degradation in each FFPE RNA sample compared to intact RNA (Figure 4B), as expected. Twenty-five nanograms of isolated RNA was used to produce labeled target for hybridization after WTA cDNA library synthesis and two rounds of WTA PCR amplification. These samples were hybridized against an equivalent amount of CPP subjected to WTA cDNA library synthesis and two rounds of WTA PCR amplification in parallel. Seventy features showing the greatest differential expression between the average of the two cancer samples and the normal sample were visualized with TreeView (Figure 4C). Genes denoted in green or red have been identified in profiling studies as being significantly dysregulated (P < .05), in the same direction, in at least one profiling study comparing grossly dissected cancer and benign samples (downregulated or upregulated, respectively) according to Oncomine 2.0. A subset of differentially expressed genes was validated using QPCR on directly reverse-transcribed cDNA or WTA-amplified cDNA from FFPE_PCA1 (Figure 4D). Taken together, these results demonstrate the ability of WTA to faithfully amplify partially degraded RNA, including materials obtained from FFPE tissues.
Discussion
In this report, we describe the evaluation and validation of OmniPlex WTA, a novel, exponential RNA amplification technique. Using cDNA microarrays, we demonstrate that WTA cDNA synthesis and multiple rounds of exponential amplification are highly reproducible. The reproducibility of additional rounds of WTA PCR amplification allows WTA to be used to archive valuable samples and serve as a distributable molecular library for reamplification. We also show that WTA can be used to reliably detect differential expression of low- and high-copy transcripts in profiling studies using a combination of cDNA microarrays and real-time PCR. Although detailed comparison studies to standard IVT techniques are still needed, we demonstrate that WTA provides an attractive alternative for many cancer biology applications where RNA amplification is desired. Furthermore, we have recently reported the discovery of recurrent gene fusions of TMPRSS2 and ETS family members in prostate cancer, based in part on a large-scale profiling study using samples subjected to WTA [27].
To demonstrate the applicability of WTA in this study, we combined WTA with LCM to profile normal epithelia, cancerous epithelia, and stroma from frozen prostate tissues. Comparing our results to previous profiling studies using grossly dissected tissues, we identified a cluster of genes that had been identified in profiling studies using grossly dissected samples as being downregulated in prostate cancer compared to benign tissue. However, our results show that these transcripts are expressed at lower levels in both cancerous and normal epithelia, with much stronger expression in the stromal compartment. Many of these features, including smooth muscle actin (ACTA2, ACTC, ACTN1, and ACTG2), smooth muscle myosin (MYL9 and MYH11), and smooth muscle calponin (CNN1), represent transcripts expressed predominantly in smooth muscle cells, which are known to be a major component of prostate stroma [28]. A recent study using in silico analysis also determined that the majority of transcripts identified in profiling studies as being “downregulated” from normal prostate to prostate cancer in fact represents stromal transcripts [29]. Taken together, these results show that WTA, in conjunction with LCM, can be used for profiling studies where increased specificity over grossly dissected tissue is desired. This specificity may be crucial for studies profiling tumors such as prostate cancer, where both benign and cancerous cells and multiple tissue types may be present.
WTA does not require the presence of a poly-A tail, as the cDNA synthesis step is not restricted to oligo-dT priming, as is the case with most IVT-based and exponential amplification techniques. We confirmed that WTA amplifies sequences distal to the 3′ terminus by selecting QPCR amplicons for TNFAIP3, AMACR, ZNF217, THBS1, WSB1, HIF1A, and MYO6 located 1.7 to 2.5 kb from any known poly-A sites, all of which were robustly amplified from WTA cDNA. Preliminary experiments using Affymetrix GeneChips also demonstrated no 3′ bias (data not shown). In this report, we show that WTA can be used to reliably amplify artificially degraded RNA, including samples with no detectable 18S and 28S ribosomal bands. As a final demonstration of applicability, we used WTA to profile cancerous and benign FFPE prostate tissue samples and confirmed the fidelity of WTA using QPCR. These results suggest that WTA may be used to profile cohorts of FFPE tissue samples with longterm clinical follow-up, which are often unavailable in frozen tissue banks. Studies are underway to evaluate the effects of the age of samples and the extent of RNA degradation in FFPE samples on the reproducibility and fidelity of WTA. Taken together, these experiments demonstrate that WTA and expression profiling could be ideal for many diagnostic and prognostic applications with limiting sample size, such as needle biopsies, LCM, and FFPE. As the WTA methodology leads to the generation of a reamplifiable cDNA library, it is ideally suited for the development of a molecular archive that can serve as a distributable resource.
Acknowledgements
The authors would like to thank Lei Wang, Terrence Barrette, Anjana Menon, Xuhong Cao, Saravana Dhanasekaran, and Ira Maine for technical assistance. The raw microarray data are available at GEO (GSE2618).
Footnotes
This work was supported, in part, by the American Cancer Society (RSG-02-179-MGO to A.M.C. and M.A.R.), the National Institutes of Health (Prostate SPORE P50CA69568 to A.M.C, M.A.R., and R.B.S.), R01AG21404 (to M.A.R. and A.M.C.), Early Detection Research Network (UO1 CA111275-01 to A.M.C. and M.A.R.), the Department of Defense (PC040517 to R.M.), and the V Foundation (to A.M.C.). A.M.C. is a Pew Biomedical Scholar, whereas S.A.T. and D.R.R. are Fellows of the Medical Scientist Training Program.
This article refers to supplementary material, which is designated by “W” (i.e., Table W1, and Figure W1) and is available online at www.bcdecker.com.
References
- 1.Su H, Hu N, Shih J, Hu Y, Wang QH, Chuang EY, Roth MJ, Wang C, Goldstein AM, Ding T, et al. Gene expression analysis of esophageal squamous cell carcinoma reveals consistent molecular profiles related to a family history of upper gastrointestinal cancer. Cancer Res. 2003;63:3872–3876. [PubMed] [Google Scholar]
- 2.Sanchez-Carbayo M, Socci ND, Lozano JJ, Li W, Charytonowicz E, Belbin TJ, Prystowsky MB, Ortiz AR, Childs G, Cordon-Cardo C. Gene discovery in bladder cancer progression using cDNA microarrays. Am J Pathol. 2003;163:505–516. doi: 10.1016/S0002-9440(10)63679-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vasselli JR, Shih JH, Iyengar SR, Maranchie J, Riss J, Worrell R, Torres-Cabala C, Tabios R, Mariotti A, Stearman R, et al. Predicting survival in patients with metastatic kidney cancer by gene-expression profiling in the primary tumor. Proc Natl Acad Sci USA. 2003;100:6958–6963. doi: 10.1073/pnas.1131754100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tay ST, Leong SH, Yu K, Aggarwal A, Tan SY, Lee CH, Wong K, Visvanathan J, Lim D, Wong WK, et al. A combined comparative genomic hybridization and expression microarray analysis of gastric cancer reveals novel molecular subtypes. Cancer Res. 2003;63:3309–3316. [PubMed] [Google Scholar]
- 5.Ma XJ, Salunga R, Tuggle JT, Gaudet J, Enright E, McQuary P, Payette T, Pistone M, Stecker K, Zhang BM, et al. Gene expression profiles of human breast cancer progression. Proc Natl Acad Sci USA. 2003;100:5974–5979. doi: 10.1073/pnas.0931261100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ashida S, Nakagawa H, Katagiri T, Furihata M, Iiizumi M, Anazawa Y, Tsunoda T, Takata R, Kasahara K, Miki T, et al. Molecular features of the transition from prostatic intraepithelial neoplasia (PIN) to prostate cancer: genome-wide gene-expression profiles of prostate cancers and PINs. Cancer Res. 2004;64:5963–5972. doi: 10.1158/0008-5472.CAN-04-0020. [DOI] [PubMed] [Google Scholar]
- 7.Van Gelder RN, von Zastrow ME, Yool A, Dement WC, Barchas JD, Eberwine JH. Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc Natl Acad Sci USA. 1990;87:1663–1667. doi: 10.1073/pnas.87.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang E, Miller LD, Ohnmacht GA, Liu ET, Marincola FM. High-fidelity mRNA amplification for gene profiling. Nat Biotechnol. 2000;18:457–459. doi: 10.1038/74546. [DOI] [PubMed] [Google Scholar]
- 9.Livesey FJ, Furukawa T, Steffen MA, Church GM, Cepko CL. Microarray analysis of the transcriptional network controlled by the photoreceptor homeobox gene Crx. Curr Biol. 2000;10:301–310. doi: 10.1016/s0960-9822(00)00379-1. [DOI] [PubMed] [Google Scholar]
- 10.Iscove NN, Barbara M, Gu M, Gibson M, Modi C, Winegarden N. Representation is faithfully preserved in global cDNA amplified exponentially from sub-picogram quantities of mRNA. Nat Biotechnol. 2002;20:940–943. doi: 10.1038/nbt729. [DOI] [PubMed] [Google Scholar]
- 11.Makrigiorgos GM, Chakrabarti S, Zhang Y, Kaur M, Price BD. A PCR-based amplification method retaining the quantitative difference between two complex genomes. Nat Biotechnol. 2002;20:936–939. doi: 10.1038/nbt724. [DOI] [PubMed] [Google Scholar]
- 12.Puskas LG, Zvara A, Hackler L, Jr, Van Hummelen P. RNA amplification results in reproducible microarray data with slight ratio bias. Biotechniques. 2002;32:1330–1334. doi: 10.2144/02326mt04. (1336, 1338, 1340) [DOI] [PubMed] [Google Scholar]
- 13.Aoyagi K, Tatsuta T, Nishigaki M, Akimoto S, Tanabe C, Omoto Y, Hayashi S, Sakamoto H, Sakamoto M, Yoshida T, et al. A faithful method for PCR-mediated global mRNA amplification and its integration into microarray analysis on laser-captured cells. Biochem Biophys Res Commun. 2003;300:915–920. doi: 10.1016/s0006-291x(02)02967-4. [DOI] [PubMed] [Google Scholar]
- 14.Petalidis L, Bhattacharyya S, Morris GA, Collins VP, Freeman TC, Lyons PA. Global amplification of mRNA by template-switching PCR: linearity and application to microarray analysis. Nucleic Acids Res. 2003;31:e142. doi: 10.1093/nar/gng142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langmore JP. Rubicon Genomics, Inc. Pharmacogenomics. 2002;3:557–560. doi: 10.1517/14622416.3.4.557. [DOI] [PubMed] [Google Scholar]
- 16.Dhanasekaran SM, Barrette TR, Ghosh D, Shah R, Varambally S, Kurachi K, Pienta KJ, Rubin MA, Chinnaiyan AM. Delineation of prognostic biomarkers in prostate cancer. Nature. 2001;412:822–826. doi: 10.1038/35090585. [DOI] [PubMed] [Google Scholar]
- 17.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 18.Dhanasekaran SM, Dash A, Yu J, Maine IP, Laxman B, Tomlins SA, Creighton CJ, Menon A, Rubin MA, Chinnaiyan AM. Molecular profiling of human prostate tissues: insights into gene expression patterns of prostate development during puberty. FASEB J. 2005;19:243–245. doi: 10.1096/fj.04-2415fje. [DOI] [PubMed] [Google Scholar]
- 19.Yang IV, Chen E, Hasseman JP, Liang W, Frank BC, Wang S, Sharov V, Saeed AI, White J, Li J, et al. Within the fold: assessing differential expression measures and reproducibility in microarray assays. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-11-research0062. research0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar-Sinha C, Shah RB, Laxman B, Tomlins SA, Harwood J, Schmitz W, Conzelmann E, Sanda MG, Wei JT, Rubin MA, et al. Elevated alpha-methylacyl-CoA racemase enzymatic activity in prostate cancer. Am J Pathol. 2004;164:787–793. doi: 10.1016/s0002-9440(10)63167-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci USA. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rubin MA, Varambally S, Beroukhim R, Tomlins SA, Rhodes DR, Paris PL, Hofer MD, Storz-Schweizer M, Kuefer R, Fletcher JA, et al. Overexpression, amplification, and androgen regulation of TPD52 in prostate cancer. Cancer Res. 2004;64:3814–3822. doi: 10.1158/0008-5472.CAN-03-3881. [DOI] [PubMed] [Google Scholar]
- 24.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiang CC, Chen M, Ma L, Phan QN, Inman JM, Kozhich OA, Brownstein MJ. A new strategy to amplify degraded RNA from small tissue samples for microarray studies. Nucleic Acids Res. 2003;31:e53. doi: 10.1093/nar/gng053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 28.Young RH Armed Forces Institute of Pathology (US) RH, author; Universities Associated for Research and Education in Pathology RH, author. Tumors of the Prostate Gland, Seminal Vesicles, Male Urethra, and Penis American Registry of Pathology. Washington, DC: Armed Forces Institute of Pathology; 2000. [Google Scholar]
- 29.Stuart RO, Wachsman W, Berry CC, Wang-Rodriguez J, Wasserman L, Klacansky I, Masys D, Arden K, Goodison S, McClelland M, et al. In silico dissection of cell-type-associated patterns of gene expression in prostate cancer. Proc Natl Acad Sci USA. 2004;101:615–620. doi: 10.1073/pnas.2536479100. [DOI] [PMC free article] [PubMed] [Google Scholar]