Abstract
We have developed a noninvasive magnetic resonance imaging (MRI) assay to characterize human umbilical vein endothelial cell (HUVEC) motility, invasion, and network formation in response to the presence of cancer cells. HUVECs were labeled with a superparamagnetic iron oxide T2 contrast agent and cocultured with MDA-MB-231 breast cancer cells in the presence of an extracellular matrix (ECM) gel. Invasion into the ECM gel by HUVECs in response to paracrine factors secreted by MDA-MB-231 cancer cells, as well as network formation by HUVECs, was easily tracked with MRI. The invasive behavior of HUVECs was not observed in the absence of cancer cells. This noninvasive assay used to characterize the response of endothelial cells (ECs) can be used to understand the role of proangiogenic or antiangiogenic stimuli, and to study the interactions between ECs and other disease-specific cells in pathologies with aberrant angiogenesis, such as retinopathy and arthritis.
Keywords: Invasion, iron oxide, endothelial cells, magnetic resonance, cell tracking
Introduction
Evaluation of the role of angiogenesis in cancer, as well as the identification of suitable antiangiogenic therapies, requires a thorough understanding of endothelial cell (EC) response to angiogenic-promoting or angiogenic-inhibiting factors or to antiangiogenic drugs. The induction of angiogenesis is regulated by a balance of numerous proangiogenic and antiangiogenic factors [1,2]. Cytokines and growth factors secreted by tumor cells induce paracrine loops that result in EC activation. Nondestructive assays are necessary to further understand the role of these factors in EC survival, motility, invasive potential, and tubulogenesis in systems that can mimic, to some extent, cancer cell/EC interactions that occur in or around tumors, under carefully controlled conditions. Previous studies have reported the design of assays to characterize the relationship between cancer cells and ECs [3,4]. These assays can detect tubule formation and length, but cannot investigate invasion in a three-dimensional matrix. Traditionally, transwell chambers are used to investigate the role of chemokines and cytokines in EC invasion and migration, but these chambers have permeable membranes that restrict cancer cell/EC interactions to soluble factors and are not equipped for dynamic monitoring. We therefore designed a magnetic resonance (MR)-compatible assay that has an ability to image network-like structures, as well as cell motility, both noninvasively and longitudinally, without restricting cancer cell/EC interactions with a semipermeable membrane. Here, we have characterized this noninvasive MR assay by studying the time course of EC invasion, migration, and tubulogenesis in a coculture system containing breast cancer cells and ECs. Human umbilical vein endothelial cells (HUVECs) were labeled, by endocytosis, with an intracellular superparamagnetic iron oxide (SPIO) contrast agent. SPIO particles effectively shorten the transverse relaxation time (T2) of protons through susceptibility-induced local magnetic field inhomogeneities, generating hypointensity contrast in T2-weighted MR images [5]. Labeled HUVECs were cocultured in a chamber containing MDA-MB-231 breast cancer cells and an extracellular matrix (ECM) gel. We demonstrated the suitability of our assay to non-invasively track the invasion and network formation of HUVECs, in the presence or absence of MDA-MB-231 breast cancer cells. This assay can easily be adapted to study EC response to perturbations, such as hypoxia or vascular endothelial growth factor (VEGF) overexpression, and to study a range of conditions involving aberrant angiogenesis, such as cancer, diabetic retinopathy, and tissue ischemia.
Methods
Cells and Cell Culture
MDA-MB-231 cells were originally derived from the pleural effusion of a breast cancer patient [6]. Cells were maintained in monolayer culture in RPMI 1640 (Sigma, St. Louis, MO) supplemented with 9% fetal bovine serum, 90 U/ml penicillin, and 90 µg/ml streptomycin. HUVECs (Clonetics, Walkersville, MD), were cultured in endothelial growth medium (EGM-2; Clonetics). All cells were maintained in 5% CO2 and 90% humidity at 37°C, except for 30 minutes or less during MR experiments.
HUVEC Labeling with Feridex and Poly-L-Lysine (PLL)
Cells were labeled with a commercially available ferumoxide suspension Feridex, which has been shown to effectively label cells for T2-weighted MR detection. PLL was used as a transfection agent for Feridex labeling [7,8]. A labeling solution was prepared by mixing 2, 9, or 25 µg Feridex/ml EGM-2 in the presence of either 30, 125, or 375 ng/ml PLL, respectively. Then, the labeling solution was gently stirred at 50 rpm using a Glas-Col minirotator system (Glas-Col LLC, Terre Haute, IN). HUVECs were incubated in the labeling solution at 37°C and 5% CO2 for 24 hours. Cells were collected by trypsinization and resuspended in EGM-2 prior to seeding on the ECM gel.
Chamber Preparation
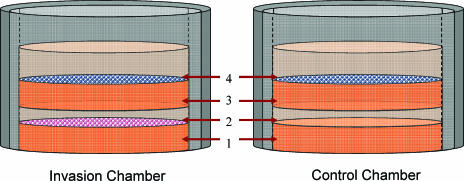
Cell invasion assays were housed in Millicell non-cell culture inserts (Millipore, Billerica, MA) with a 0.4-µm membrane pore. A schematic of chamber construction is shown in Figure 1. A total of 120 µl of the ECM gel (protein concentration, 8.8 mg/ml), thawed at 4°C, was placed on the membrane and allowed to polymerize for 30 minutes at 37°C in a CO2 incubator. MDA-MB-231 cells (1.5 x 105), suspended in 100 µl of EGM-2, were seeded in invasion chambers; 100 µl of EGM-2 was added to control chambers. After 4 hours of incubation to allow the cancer cells to adhere to the ECM gel, 100 µl of the ECM gel (diluted 1:2 in DMEM to prevent meniscus formation) was added to each chamber. The second layer of the ECM gel was then polymerized for 30 minutes at 37°C. Finally, 1.5 x 105 of labeled HUVECs in 300 µl of EGM-2 were seeded in all chambers, approximately 24 hours prior to magnetic resonance imaging (MRI), allowing for the attachment and formation of lumen-like structures. The chambers were maintained in a 24-well tissue culture plate with 300 µl/well EGM-2 added outside of the chamber. The chambers were maintained in an incubator at 37°C, 5% CO2, and 90% humidity, and removed only for brief periods of time during the MR experiments.
Figure 1.
Schematic of an invasion chamber. (1) A total of 120 µl of the ECM gel was added and incubated at 37°C for 30 minutes for polymerization. (2) A total of 1.5 x 105 MDA-MB-231 cells in 100 µ1 of EGM-2 were seeded on the surface of the ECM gel and incubated for 4 hours to allow cells to attach; 100 µ1 of EGM-2 alone was added to the control chambers. (3) A total of 200 µl of the ECM gel (diluted 1:2 in DMEM) was added and polymerized within 30 minutes at 37°C. (4) A total of 1.5 x 105 labeled HUVECs in 300 µl of EGM-2 were seeded and incubated at 37°C for 24 hours to allow for the attachment of HUVECs and the formation of lumen-like structures.
MRI
MR images were obtained on a 500-MHz (11.74-T) wide-bore MRI system with a Bruker Avance (Bruker BioSpin GmBH, Rheinstetten, Germany) spectrometer equipped with triple-axis gradients. The chambers were submerged, under sterile conditions, in a 15-mm nuclear magnetic resonance tube containing EGM-2 and then placed within a 23-mm birdcage coil for imaging. Coronal and axial images of the chambers were obtained at room temperature using a T2-weighted, multislice, spin-echo sequence. All chambers were imaged by MRI for at least two time points: 24 and 120 hours postseeding. After MRI, the chambers were returned to the 24-well plate, and the medium was refreshed in each well and chamber.
T2-weighted MR images were acquired in one scan with a field-of-view = 1.6 cm, acquisition matrix = 256 x 256, slice thickness = 0.5 mm, TE = 60 milliseconds, and TR = 617 milliseconds, unless stated otherwise. These T2-weighted MR images were used to calculate the percent fractional area of hypointensity. Gray-scale MR images were binarized using adaptive thresholding, a method by which the threshold operator dynamically changes the threshold over the image to accommodate intensity changes across the image, and the percent fractional area of hypointensity was calculated from these binarized images using Image J (National Institutes of Health, Bethesda, MD). The percent fractional area of hypointensity in MRI was considered to be proportional to the number of iron oxide-labeled HUVECs. An increase in the number of labeled HUVECs in a given layer relates to an increase in the percent fractional area of hypointensity, which was used as an index of labeled HUVECs within that layer.
Fluorescence Microscopy
Chambers were incubated in the dark for 30 minutes in the presence of EC-specific antibody CD31 (Serotec, Oxford, UK) at 1:10 dilution in phosphate-buffered saline (PBS). The chambers were washed thrice with PBS and observed under a fluorescent microscope.
Phase-Contrast Microscopy
Chambers were monitored with phase-contrast microscopy before and after each MRI experiment. All MR experiments were completed by the 120-hour time point. At the end of the MR experiments, the cells were stained with Prussian blue to detect the presence of iron. Chambers were washed in PBS and incubated for 30 minutes in 2% potassium ferrocyanide (Perls reagent) and 6% HCl, followed by a second wash in PBS. The chambers were then counterstained for 5 minutes with a nonspecific nuclei stain Nuclear Fast Red. Phase-contrast images of 15-µm sections of the chamber, sectioned using a cryostat, were also obtained.
Statistical Analysis
Statistical analysis was performed with JMP software (SAS Institute, Inc., Cary, NC) using a Wilcoxon rank-sum test. Unless otherwise noted, P < .05 was considered significant.
Results
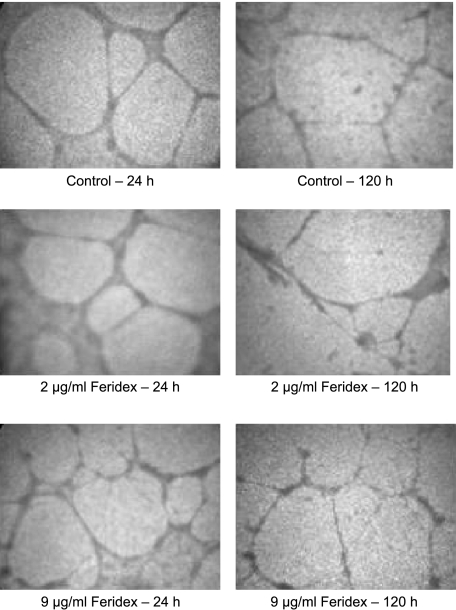
As shown in Figure 2, both labeled and unlabeled HUVECs formed nearly identical network structures at matched time points, demonstrating the absence of any deleterious effect of Feridex labeling on HUVEC network formation in the ECM gel.
Figure 2.
Light microscopy images of chambers with HUVECs, showing no discernable difference in network structure between unlabeled and labeled HUVECs at 24 and 120 hours postseeding.
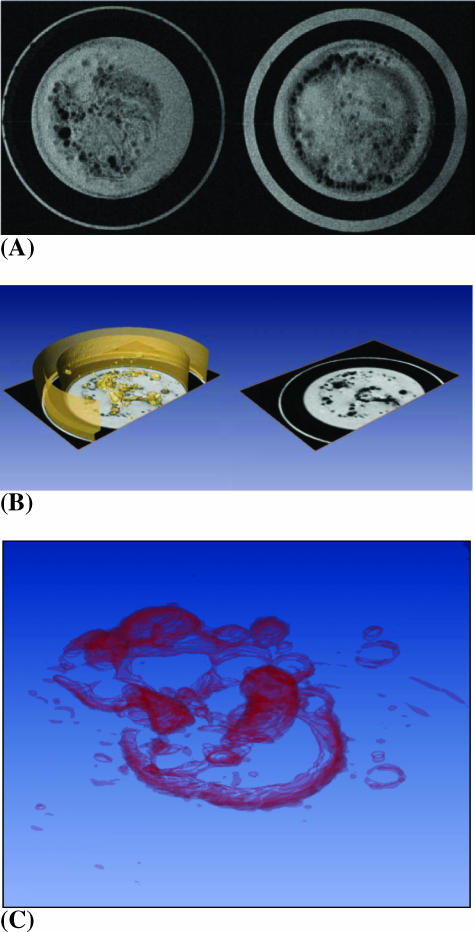
Figure 3A demonstrates the ability of T2-weighted MRI to detect Feridex-labeled HUVECs. Three-dimensional MRI revealed lumen-like structures of labeled HUVECs, as shown in Figure 3, B and C. MDA-MB-231 breast cancer cells formed a net-like pattern 24 hours postseeding, as observed by light microscopy (data not shown). In chambers containing MDA-MB-231 breast cancer cells, the network of HUVECs was well-preserved over the course of the 120-hour experiment, whereas the controls showed a diminished network structure.
Figure 3.
(A) Representative axial MR images of two contiguous 0.5-mm-thick slices containing HUVECs labeled with 9 µg/ml Feridex. A lumen-like network formation of ECs 24 hours postseeding is apparent in these images, which were obtained using a T2-weighted spin-echo sequence with a field-of-view = 1.6 cm, acquisition matrix = 256 x 256, TE = 60 milliseconds, TR = 617 milliseconds, and number of averages = 2. (B) A three-dimensional reconstructed image of multislice data showing a lumen-like HUVEC network in the ECM gel (left). Orthogonal slices in the axial direction provide the ability to dynamically follow migration, invasion, and network structure through the ECM gel (right). (C) A three-dimensional reconstructed image from a multislice MR data set, with volume rendering demonstrating an intricate HUVEC network.
Effective labeling of HUVECs was achieved at all Feridex concentrations (2, 9, and 25 µg/ml), as evident from the brown intracytoplasmic granules from Feridex or the presence of blue regions following Prussian blue staining for iron content.
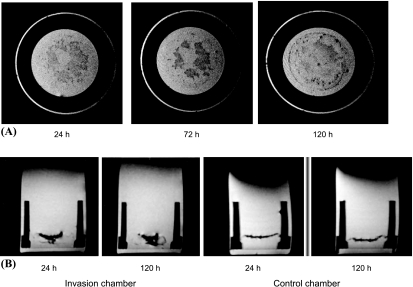
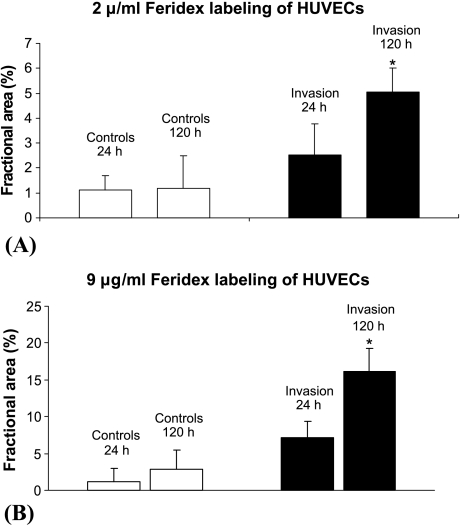
Representative axial (Figure 4A) and coronal (Figure 4B) MR images showed an increased presence of HUVECs in the cancer cell layer between the two time points, as observed by MRI, whereas in the control chambers without cancer cells, the HUVECs did not invade significantly through the ECM gel. The percent fractional area occupied by HUVECs in the cancer cell seed layer increased over the course of the experiment (Figure 5, A and B). For ECs labeled with 2 µg/ml Feridex, the fractional area of HUVECs in the cancer cell seed layer increased significantly, from 2.5 ± 1.3% at 24 hours to 5.0 ± 0.9% at 120 hours (mean ± SD; n = 6, P < .05), whereas no significant change was observed in controls from 24 hours (1.1 ± 0.6%) to 120 hours (1.2 ± 0.2%) (mean ± SD; n = 6). For cells labeled with 9 µg/ml Feridex, the fractional area of HUVECs in the cancer cell seed layer increased significantly, from 7.1 ± 2.6% at 24 hours to 16.2 ± 3.1% at 120 hours postseeding (mean ± SD; n = 5, P < .05). There were no significant changes in fractional area in the corresponding layer in the control chambers (1.2 ± 1.8% at 24 hours and 2.9 ± 2.3% at 120 hours postseeding) (mean ± SD; n = 4).
Figure 4.
(A) A panel of three axial MR images acquired 24, 72, and 120 hours postseeding. An increased number of HUVECs were detected with time in the layer containing MDA-MB-231 cancer cells, as evident from hypointense regions in this layer. HUVECs were labeled with 9 µg/ml Feridex. The MR images were obtained using a T2-weighted spin-echo sequence with field-of-view = 1.6 cm, acquisition matrix = 256 x 256, eight slices of slice thickness = 0.5 mm, TE = 60 milliseconds, TR = 617 milliseconds, and number of averages = 2. (B) Representative coronal MR images of a chamber containing MDA-MB-231 cells demonstrating the presence of Feridex-labeled HUVECs confined mainly to the upper seed layer 24 hours postseeding (i), with an increased presence of HUVECs in the cancer cell layer 120 hours postseeding (ii). By comparison, the control chambers showed no invasion of the ECM gel by the HUVECs, 24 hours (iii) and 120 hours postseeding (iv). The images were obtained in a single scan, using a T2-weighted spin-echo sequence with a field-of-view = 1.6 cm, eight slices of slice thickness = 1 mm, acquisition matrix = 256 x 256, TE = 60 milliseconds, and TR = 617 milliseconds.
Figure 5.
Fractional area occupied by HUVECs in the cancer cell seed layer for (A) HUVECs labeled with 2 µg/ml Feridex and (B) HUVECs labeled with 9 µg/ml Feridex. The fractional area was calculated for T2-weighted MR images acquired with TE = 60 milliseconds and TR = 617 milliseconds. A significant increase (P < .05) was detected in the fractional area (mean ± SD) occupied by HUVECs in the cancer cell layer for chambers containing MDA-MB-231 breast cancer cells (n = 6 for 2 µg/ml Feridex; n = 5 for 9 µg/ml Feridex), whereas no significant change (P < .05) was observed in the corresponding layer in the absence of cancer cells (n = 6 for 2 µg/ml Feridex; n = 4 for 9 µg/ml Feridex).
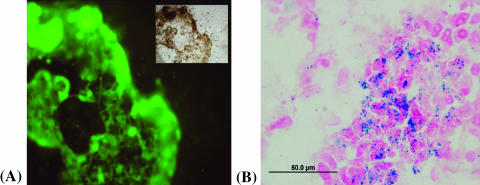
The presence of HUVECs in the cancer cell seed layer was evident from staining with endothelial cell-specific CD31 monoclonal antibody (Figure 6A) and with Prussian blue staining of HUVECs mixed with cancer cells (Figure 6B).
Figure 6.
The presence of HUVECs in the cancer cell layer detected by the fluorescence microscopy of immunohistochemistry staining with the CD31 monoclonal antibody (A). The layer in sharp focus is the layer containing MDA-MB-231 cancer cells which the HUVECs invaded. The inset shows a corresponding phase-contrast light micrograph. The presence of HUVECs in the cancer cell layer was also detected by Prussian blue staining for iron content (B). Staining with Prussian blue and Nuclear Fast Red (a nonspecific red stain for nuclei) shows the colocalization of iron-labeled ECs and unlabeled MDA-MB-231 cancer cells.
Discussion
Here, we have noninvasively and longitudinally characterized HUVEC invasion in response to cancer cells. HUVECs grown in the presence of cancer cells showed a marked increase in both network formation and cell invasion, compared to HUVECs grown without cancer cells.
We started with the published labeling concentration of 25 µg Feridex/ml medium [7–9]. We found that cells labeled with 40% or even 10% of the initial concentration were easily detected with our high-field microimaging system. A labeling concentration of 9 µg/ml Feridex was found to be the suitable labeling concentration for HUVECs in this study. The labeling showed no detrimental effect on HUVEC viability and tubulogenesis for Feridex concentrations at or below 9 µg/ml, as measured by trypan blue exclusion and as observed by MRI and light microscopy. This is in agreement with previous studies suggesting no adverse consequences of cell labeling with PLL Feridex [9,10], except for chondrogenic differentiation [9,11]. A Feridex labeling concentration of 25 µg/ml affected network formation, whereas a lower concentration (2 µg/ml) showed discernable contrast in MR images, which detected the presence of clusters of HUVECs, but not the finer structures of the HUVEC network.
An increasing presence of HUVECs was observed in the MDA-MB-231 cancer cell seed layer at a later time point, demonstrating the invasion and migration of HUVECs through the ECM gel toward the cancer cells. Cell migration requires proteolytic ECM remodeling for the lowering of matrix organization barriers to migration and then for tissue remodeling [12]. Proteolytic enzymes, such as urokinase-type plasminogen activator and matrix metalloproteinase, secreted by cancer cells are known to degrade the surrounding ECM, leading to directional migration of ECs [13,14]. The presence of a large population of HUVECs in the cancer cell seed layer, along with ECM gel architecture, indicated that cancer cells stimulated the HUVECs to invade through the ECM and to migrate toward the cancer cells, whereas there was no significant movement of HUVECs through the ECM gel in the absence of cancer cells. These results were confirmed by staining with the endothelial-specific monoclonal antibody CD31 and with Prussian blue staining for iron content, which detected HUVECs in the cancer cell seed layer. Khodarev et al. [15] have observed an increase in the migration of HUVECs toward U87 human glioma cells, compared to controls, using a collagen I transwell migration assay. Gene expression profiling of HUVECs, cultured in the presence of cancer cells, revealed the expression of several growth factors (such as transforming growth factor β3 and FGF7) and their receptors. A panel of experiments in their study suggests that growth factors released by cancer cells induce autocrine loops in HUVECs, thereby generating an angiogenic response with increased migration toward the cancer cells. The data also suggest a role for cancer cells in the activation of ECs through the formation of paracrine loops with soluble ligands secreted by the cancer cells. Although paracrine factors secreted by cancer cells most likely stimulated the invasion and migration of HUVECs in our study, it is also possible that local hypoxic environments created within the cancer cells may have acted as a stimulus for EC migration and invasion. This possibility is currently under investigation. We are also currently determining if the migration and invasion of ECs is cancer cell-specific in this system, or if it occurs for nonmalignant cells as well.
In all chambers, HUVECs formed a lumen-like network 24 hours postseeding, in agreement with previous observations [16,17]. Our data are consistent with previous studies, which have shown that cell/cell and cell/ECM interactions stimulate both HUVEC survival and network formation [18–22]. At the end of 120 hours, network structures were well-preserved in chambers with cancer cells, compared to the control chambers where diminished HUVEC survival and network structure were observed. One possible explanation is the presence of antiapoptotic signals derived from the cancer cells, which increased HUVEC survival [23–28]. The presence of a small population of HUVECs in the cancer cell layer in the control chambers was probably attributable to migration and the baseline invasive ability of ECs due to the presence of some growth factors in the ECM gel.
In addition, although HUVECs were used as a model of ECs to validate the assay described in the present study, the human microvascular endothelium is not well-represented by HUVECs [29,30]. For instance, Anderson et al. [31] used iron oxide labeling to show that a population of mouse bone marrow-derived cells is enriched in endothelial precursor cells, which incorporate and differentiate into endothelial-like cells in the tumor vasculature in a glioma model. Our future studies will use cells that are more representative of the human tumor neovasculature, such as VEGF-R2-overexpressing ECs [32–36] and endothelial precursor cells [37,38].
In conclusion, cell labeling with SPIO, combined with high-resolution MRI, allowed a noninvasive and dynamic three-dimensional characterization of EC networks formed in a three-dimensional ECM gel. Vessel formation, maturation [39], quiescence [40], and regression depend on the competing effects of growth [41–43] and inhibitory factors, with survival and proliferation on one hand and apoptosis on the other hand [2]. Therefore, this assay can be expanded to study either EC responses to the synergistic effects of these competing factors, or the roles of a single perturbation such as hypoxia [13,14,44–48], a chemokine, or a cytokine. We are currently studying endothelial response to hypoxia and VEGF overexpression to understand the precise role of each factor in angiogenesis. Additionally, our ability to obtain high-resolution three-dimensional MR images may provide further insights into the remodeling of basement membranes in response to various factors and conditions, such as upregulation of growth factors and receptor activity [49]. Our assay serves as a model system for a range of conditions involving aberrant angiogenesis, such as cancer, rheumatoid arthritis, diabetic retinopathy, and tissue ischemia. This novel assay may prove useful in longitudinal high-throughput studies to determine the efficacy of proangiogenic and antiangiogenic drugs.
Acknowledgement
The authors thank Dmitri Artemov for useful discussions and technical advice.
Footnotes
Work from the authors' program was supported through funding by National Institutes of Health grants 2RO1 CA82337, 1P50 CA 103175, and 1RO1 NS045062.
References
- 1.Liotta LA, Steeg PS, Stetlet-Stevenson WG. Cancer metastasis and angiogenesis: an imbalance of positive and negative regulation. Cell. 1991;64:327–336. doi: 10.1016/0092-8674(91)90642-c. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Folkman J. Patterns and emerging mechanisms of the angiogenic switch during tumorigenesis. Cell. 1996;86:353–364. doi: 10.1016/s0092-8674(00)80108-7. [DOI] [PubMed] [Google Scholar]
- 3.Bishop ET, Bell GT, Bloor S, Broom IJ, Hendry NF, Wheatley DN. An in vitro model of angiogenesis: basic features. Angiogenesis. 1999;3:335–344. doi: 10.1023/a:1026546219962. [DOI] [PubMed] [Google Scholar]
- 4.Donovan D, Brown NJ, Bishop ET, Lewis CE. Comparison of three in vitro human “angiogenesis” assays with capillaries formed in vivo. Angiogenesis. 2001;4:113–121. doi: 10.1023/a:1012218401036. [DOI] [PubMed] [Google Scholar]
- 5.Bulte JW, Kraitchman DL. Iron oxide MR contrast agents for molecular and cellular imaging. NMR Biomed. 2004;17:484–499. doi: 10.1002/nbm.924. [DOI] [PubMed] [Google Scholar]
- 6.Cailleau R, Young R, Olive M, Reeves WJ., Jr Breast tumor cell lines from pleural effusions. J Natl Cancer Inst. 1974;53:661–674. doi: 10.1093/jnci/53.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frank JA, Zywicke H, Jordan EK, Mitchell J, Lewis BK, Miller B, Bryant LH, Jr, Bulte JW. Magnetic intracellular labeling of mammalian cells by combining (FDA-approved) superparamagnetic iron oxide MR contrast agents and commonly used transfection agents. Acad Radiol. 2002;9(Suppl 2):S484–S487. doi: 10.1016/s1076-6332(03)80271-4. [DOI] [PubMed] [Google Scholar]
- 8.Kraitchman DL, Heldman AW, Atalar E, Amado LC, Martin BJ, Pittenger MF, Hare JM, Bulte JW. In vivo magnetic resonance imaging of mesenchymal stem cells in myocardial infarction. Circulation. 2003;107:2290–2293. doi: 10.1161/01.CIR.0000070931.62772.4E. [DOI] [PubMed] [Google Scholar]
- 9.Kostura L, Kraitchman DL, Mackay AM, Pittenger MF, Bulte JW. Feridex labeling of mesenchymal stem cells inhibits chondrogenesis but not adipogenesis or osteogenesis. NMR Biomed. 2004;17:513–517. doi: 10.1002/nbm.925. [DOI] [PubMed] [Google Scholar]
- 10.Frank JA, Miller BR, Arbab AS, Zywicke HA, Jordan EK, Lewis BK, Bryant LH, Jr, Bulte JW. Clinically applicable labeling of mammalian and stem cells by combining superparamagnetic iron oxides and transfection agents. Radiology. 2003;228:480–487. doi: 10.1148/radiol.2281020638. [DOI] [PubMed] [Google Scholar]
- 11.Bulte JW, Kraitchman DL, Mackay AM, Pittenger MF. Chondrogenic differentiation of mesenchymal stem cells is inhibited after magnetic labeling with ferumoxides. Blood. 2004;104:3410–3412. doi: 10.1182/blood-2004-06-2117. (author reply, 3412–3413) [DOI] [PubMed] [Google Scholar]
- 12.Friedl P, Brocker EB. The biology of cell locomotion within three-dimensional extracellular matrix. Cell Mol Life Sci. 2000;57:41–64. doi: 10.1007/s000180050498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeClerck YA, Mercurio AM, Stack MS, Chapman HA, Zutter MM, Muschel RJ, Raz A, Matrisian LM, Sloane BF, Noel A, et al. Proteases, extracellular matrix, and cancer: a workshop of the path B study section. Am J Pathol. 2004;164:1131–1139. doi: 10.1016/S0002-9440(10)63200-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borg TK. It's the matrix! ECM, proteases, and cancer. Am J Pathol. 2004;164:1141–1142. doi: 10.1016/S0002-9440(10)63201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khodarev NN, Yu J, Labay E, Darga T, Brown CK, Mauceri HJ, Yassari R, Gupta N, Weichselbaum RR. Tumour-endothelium interactions in co-culture: coordinated changes of gene expression profiles and phenotypic properties of endothelial cells. J Cell Sci. 2003;116:1013–1022. doi: 10.1242/jcs.00281. [DOI] [PubMed] [Google Scholar]
- 16.Pollman MJ, Naumovski L, Gibbons GH. Endothelial cell apoptosis in capillary network remodeling. J Cell Physiol. 1999;178:359–370. doi: 10.1002/(SICI)1097-4652(199903)178:3<359::AID-JCP10>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 17.Kubota Y, Kleinman HK, Martin GR, Lawley TJ. Role of laminin and basement membrane in the morphological differentiation of human endothelial cells into capillary-like structures. J Cell Biol. 1988;107:1589–1598. doi: 10.1083/jcb.107.4.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grant DS, Kleinman HK, Martin GR. The role of basement membranes in vascular development. Ann NY Acad Sci. 1990;588:61–72. doi: 10.1111/j.1749-6632.1990.tb13197.x. [DOI] [PubMed] [Google Scholar]
- 19.Grant DS, Lelkes PI, Fukuda K, Kleinman HK. Intracellular mechanisms involved in basement membrane induced blood vessel differentiation in vitro. In Vitro Cell Dev Biol. 1991;27A:327–336. doi: 10.1007/BF02630910. [DOI] [PubMed] [Google Scholar]
- 20.Eccles SA. Parallels in invasion and angiogenesis provide pivotal points for therapeutic intervention. Int J Dev Biol. 2004;48:583–598. doi: 10.1387/ijdb.041820se. [DOI] [PubMed] [Google Scholar]
- 21.Brooks PC, Clark RA, Cheresh DA. Requirement of vascular integrin alpha v beta 3 for angiogenesis. Science. 1994;264:569–571. doi: 10.1126/science.7512751. [DOI] [PubMed] [Google Scholar]
- 22.Friedlander M, Brooks PC, Shaffer RW, Kincaid CM, Varner JA, Cheresh DA. Definition of two angiogenic pathways by distinct alpha v integrins. Science. 1995;270:1500–1502. doi: 10.1126/science.270.5241.1500. [DOI] [PubMed] [Google Scholar]
- 23.Gerber HP, McMurtrey A, Kowalski J, Yan M, Keyt BA, Dixit V, Ferrara N. Vascular endothelial growth factor regulates endothelial cell survival through the phosphatidylinositol 3′-kinase/Akt signal transduction pathway. Requirement for Flk-1/KDR activation. J Biol Chem. 1998;273:30336–30343. doi: 10.1074/jbc.273.46.30336. [DOI] [PubMed] [Google Scholar]
- 24.Fujio Y, Walsh K. Akt mediates cytoprotection of endothelial cells by vascular endothelial growth factor in an anchorage-dependent manner. J Biol Chem. 1999;274:16349–16354. doi: 10.1074/jbc.274.23.16349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spyridopoulos I, Brogi E, Kearney M, Sullivan AB, Cetrulo C, Isner JM, Losordo DW. Vascular endothelial growth factor inhibits endothelial cell apoptosis induced by tumor necrosis factor-alpha: balance between growth and death signals. J Mol Cell Cardiol. 1997;29:1321–1330. doi: 10.1006/jmcc.1996.0365. [DOI] [PubMed] [Google Scholar]
- 26.Nor JE, Christensen J, Mooney DJ, Polverini PJ. Vascular endothelial growth factor (VEGF)-mediated angiogenesis is associated with enhanced endothelial cell survival and induction of Bcl-2 expression. Am J Pathol. 1999;154:375–384. doi: 10.1016/S0002-9440(10)65284-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tran J, Rak J, Sheehan C, Saibil SD, LaCasse E, Korneluk RG, Kerbel RS. Marked induction of the IAP family antiapoptotic proteins survivin and XIAP by VEGF in vascular endothelial cells. Biochem Biophys Res Commun. 1999;264:781–788. doi: 10.1006/bbrc.1999.1589. [DOI] [PubMed] [Google Scholar]
- 28.Mesri M, Morales-Ruiz M, Ackermann EJ, Bennett CF, Pober JS, Sessa WC, Altieri DC. Suppression of vascular endothelial growth factor-mediated endothelial cell protection by survivin targeting. Am J Pathol. 2001;158:1757–1765. doi: 10.1016/S0002-9440(10)64131-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Asahara T, Murohara T, Sullivan A, Silver M, van der Zee R, Li T, Witzenbichler B, Schatteman G, Isner JM. Isolation of putative progenitor endothelial cells for angiogenesis. Science. 1997;275:964–967. doi: 10.1126/science.275.5302.964. [DOI] [PubMed] [Google Scholar]
- 30.Lin Y, Weisdorf DJ, Solovey A, Hebbel RP. Origins of circulating endothelial cells and endothelial outgrowth from blood. J Clin Invest. 2000;105:71–77. doi: 10.1172/JCI8071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anderson SA, Glod J, Arbab AS, Noel M, Ashari P, Fine HA, Frank JA. Noninvasive MR imaging of magnetically labeled stem cells to directly identify neovasculature in a glioma model. Blood. 2005;105:420–425. doi: 10.1182/blood-2004-06-2222. [DOI] [PubMed] [Google Scholar]
- 32.Shalaby F, Rossant J, Yamaguchi TP, Gertsenstein M, Wu XF, Breitman ML, Schuh AC. Failure of blood-island formation and vasculogenesis in Flk-1-deficient mice. Nature. 1995;376:62–66. doi: 10.1038/376062a0. [DOI] [PubMed] [Google Scholar]
- 33.Ferrara N, Carver-Moore K, Chen H, Dowd M, Lu L, O'Shea KS, Powell-Braxton L, Hillan KJ, Moore MW. Heterozygous embryonic lethality induced by targeted inactivation of the VEGF gene. Nature. 1996;380:439–442. doi: 10.1038/380439a0. [DOI] [PubMed] [Google Scholar]
- 34.Carmeliet P, Ferreira V, Breier G, Pollefeyt S, Kieckens L, Gertsenstein M, Fahrig M, Vandenhoeck A, Harpal K, Eberhardt C, et al. Abnormal blood vessel development and lethality in embryos lacking a single VEGF allele. Nature. 1996;380:435–439. doi: 10.1038/380435a0. [DOI] [PubMed] [Google Scholar]
- 35.Waltenberger J, Claesson-Welsh L, Siegbahn A, Shibuya M, Heldin CH. Different signal transduction properties of KDR and Flt1, two receptors for vascular endothelial growth factor. J Biol Chem. 1994;269:26988–26995. [PubMed] [Google Scholar]
- 36.Niethammer AG, Xiang R, Becker JC, Wodrich H, Pertl U, Karsten G, Eliceiri BP, Reisfeld RA. A DNA vaccine against VEGF receptor 2 prevents effective angiogenesis and inhibits tumor growth. Nat Med. 2002;8:1369–1375. doi: 10.1038/nm1202-794. [DOI] [PubMed] [Google Scholar]
- 37.Bagley RG, Walter-Yohrling J, Cao X, Weber W, Simons B, Cook BP, Chartrand SD, Wang C, Madden SL, Teicher BA. Endothelial precursor cells as a model of tumor endothelium: characterization and comparison with mature endothelial cells. Cancer Res. 2003;63:5866–5873. [PubMed] [Google Scholar]
- 38.Gill M, Dias S, Hattori K, Rivera ML, Hicklin D, Witte L, Girardi L, Yurt R, Himel H, Rafii S. Vascular trauma induces rapid but transient mobilization of VEGFR2(+)AC133(+) endothelial precursor cells. Circ Res. 2001;88:167–174. doi: 10.1161/01.res.88.2.167. [DOI] [PubMed] [Google Scholar]
- 39.Benjamin LE, Hemo I, Keshet E. A plasticity window for blood vessel remodelling is defined by pericyte coverage of the preformed endothelial network and is regulated by PDGF-B and VEGF. Development. 1998;125:1591–1598. doi: 10.1242/dev.125.9.1591. [DOI] [PubMed] [Google Scholar]
- 40.Denekamp J. Review article: angiogenesis, neovascular proliferation and vascular pathophysiology as targets for cancer therapy. Br J Radiol. 1993;66:181–196. doi: 10.1259/0007-1285-66-783-181. [DOI] [PubMed] [Google Scholar]
- 41.Mukhopadhyay D, Tsiokas L, Sukhatme VP. Wild-type p53 and v-Src exert opposing influences on human vascular endothelial growth factor gene expression. Cancer Res. 1995;55:6161–6165. [PubMed] [Google Scholar]
- 42.Rak J, Mitsuhashi Y, Bayko L, Filmus J, Shirasawa S, Sasazuki T, Kerbel RS. Mutant ras oncogenes upregulate VEGF/VPF expression: implications for induction and inhibition of tumor angiogenesis. Cancer Res. 1995;55:4575–4580. [PubMed] [Google Scholar]
- 43.Rak J, Filmus J, Finkenzeller G, Grugel S, Marme D, Kerbel RS. Oncogenes as inducers of tumor angiogenesis. Cancer Metastasis Rev. 1995;14:263–277. doi: 10.1007/BF00690598. [DOI] [PubMed] [Google Scholar]
- 44.Shweiki D, Itin A, Soffer D, Keshet E. Vascular endothelial growth factor induced by hypoxia may mediate hypoxia-initiated angiogenesis. Nature. 1992;359:843–845. doi: 10.1038/359843a0. [DOI] [PubMed] [Google Scholar]
- 45.Shima DT, Deutsch U, D'Amore PA. Hypoxic induction of vascular endothelial growth factor (VEGF) in human epithelial cells is mediated by increases in mRNA stability. FEBS Lett. 1995;370:203–208. doi: 10.1016/0014-5793(95)00831-s. [DOI] [PubMed] [Google Scholar]
- 46.Ikeda E, Achen MG, Breier G, Risau W. Hypoxia-inducedtran-scriptional activation and increased mRNA stability of vascular endothelial growth factor in C6 glioma cells. J Biol Chem. 1995;270:19761–19766. doi: 10.1074/jbc.270.34.19761. [DOI] [PubMed] [Google Scholar]
- 47.Levy AP, Levy NS, Wegner S, Goldberg MA. Transcriptional regulation of the rat vascular endothelial growth factor gene by hypoxia. J Biol Chem. 1995;270:13333–13340. doi: 10.1074/jbc.270.22.13333. [DOI] [PubMed] [Google Scholar]
- 48.Krishnamachary B, Berg-Dixon S, Kelly B, Agani F, Feldser D, Ferreira G, Iyer N, LaRusch J, Pak B, Taghavi P, et al. Regulation of colon carcinoma cell invasion by hypoxia-inducible factor 1. Cancer Res. 2003;63:1138–1143. [PubMed] [Google Scholar]
- 49.Moses MA. The regulation of neovascularization of matrix metalloproteinases and their inhibitors. Stem Cells. 1997;15:180–189. doi: 10.1002/stem.150180. [DOI] [PubMed] [Google Scholar]