Abstract
Bacterial surface appendages called pili often are associated with DNA and/or protein transfer between cells. The exact function of pili in the transfer process is not understood and is a matter of considerable debate. The Hrp pilus is assembled by the Hrp type III protein secretion system of Pseudomonas syringae pv. tomato (Pst) strain DC3000. In this study, we show that the hrpA gene, which encodes the major subunit of the Hrp pilus, is required for secretion of putative virulence proteins, such as HrpW and AvrPto. In addition, the hrpA gene is required for full expression of genes that encode regulatory, secretion, and effector proteins of the type III secretion system. hrpA-mediated gene regulation apparently is through effect on the mRNA level of two previously characterized regulatory genes, hrpR and hrpS. Ectopic expression of the hrpRS gene operon restored gene expression, but not protein secretion, in the hrpA mutant. Three single amino acid mutations at the HrpA carboxyl terminus were identified that affect the secretion or regulatory function of the HrpA protein. These results define an essential role of the Hrp pilus structural gene in protein secretion and coordinate regulation of the type III secretion system in Pst DC3000.
Many Gram-negative bacterial pathogens, including Pseudomonas syringae pv. tomato (Pst) DC3000, possess a unique protein secretion system called the type III protein secretion system that transfers virulence proteins directly into the host cell (1–3). This system has been shown to play a critical role in bacterial infection of plants, animals, and humans. Genes involved in type III protein secretion have been characterized extensively in several human, animal, and plant bacteria (1–3). However, the actual mechanism by which virulence proteins are transferred into the host cell is poorly understood. Several general features of type III protein secretion have been revealed: (i) type III protein secretion appears to be activated fully upon contact with the host cells in vivo (4, 5); (ii) extracellular filamentous appendages often are associated with type III protein secretion (6–8); (iii) a secretion signal is localized in the 5′ region of the mRNA of the secreted protein (9–11); and (iv) the secretion apparatus is genetically and morphologically similar to the bacterial flagellum (12).
In plant pathogenic bacteria the type III protein secretion system (also called the Hrp secretion pathway or system) is encoded by hrp (for hypersensitive reaction and pathogenicity) genes (2, 13, 14). Nine hrp genes have been renamed hrc (for hrp genes conserved) because of their broad conservation among all bacteria that harbor type III protein secretion systems (15). The Hrp secretion system of Pseudomonas syringae has been shown to secrete two families of proteins that elicit host responses: harpins, such as HrpZ and HrpW (16–18), and Avr proteins (19, 20). The expression of P. syringae hrc/hrp genes is tightly controlled. Most hrp genes are expressed at a very low level in standard, nutrient-rich medium. The expression of hrc/hrp genes is induced in infected plant tissues or in artificial hrp-inducing minimal media that presumably mimic the in planta conditions (21–23). Three intracellular positive regulatory proteins are required for expression of hrc/hrp genes: HrpR and HrpS, which belong to the NtrC family of two-component regulatory proteins (24–26), and HrpL, a member of the ECF (extracytoplasmic factor) family of alternate σ factors (27). The HrpS, HrpR, and HrpL proteins appear to function as a regulatory cascade in which HrpS and HrpR activate the expression of HrpL in response to a signal in host tissue or in hrp-inducing minimal medium (25, 26). HrpL is presumed to activate all hrp and avr genes by recognizing a consensus sequence motif (“harp box”) present in the upstream regions of many hrp and avr genes (26, 27). Recently, a putative negative regulator encoded by the hrpV gene has been identified in P. syringae (28). In hrp-inducing minimal medium, overexpression of the hrpV gene down-regulates hrp/hrc gene expression, whereas a hrpV mutant is elevated in hrp/hrc gene expression (28).
In a previous study, we found that Pst strain DC3000 assembles a hrp-dependent pilus (the Hrp pilus) (6). We showed that the Hrp pilus structural protein, HrpA, is required for Pst DC3000 to cause disease in Arabidopsis and to elicit the hypersensitive response (HR) in tobacco and tomato (6). Pili also have been shown to be required for bacterial conjugation (29) and for transfer of T-DNA to plant cells by Agrobacterium tumefaciens (30). The conjugative F pilus plays a major role in mediating contact between donor and recipient bacteria during mating (29). However, whether pili have other functions in protein and/or DNA transfer is not clear and is a matter of considerable debate. In this study, we show that the hrpA gene plays a key role in secretion of Hrp and Avr proteins in culture.
Materials and Methods
Bacterial Strains and Culture Conditions.
Pst DC3000 and four hrp mutant derivatives (hrpA−, hrpS−, hrcC−, and hrcC− hrpT−V−) were used in this study. The hrpA, hrpS, and hrcChrpTV mutants were made in previous studies (17). The hrpA mutant does not make the major structural protein of the Hrp pilus (6). The hrpS regulatory mutant is defective in expression of hrc/hrp genes (17). The hrcChrpTV mutant is defective in protein secretion (17). The hrcChrpTV mutant was used as a secretion mutant control in early experiments (Fig. 1). Because the Tn5Cm insertion has a polar effect on the downstream negative regulatory gene hrpV in the hrcChrpTV mutant, a hrcC deletion mutant was constructed in this study and was used in most experiments presented here.
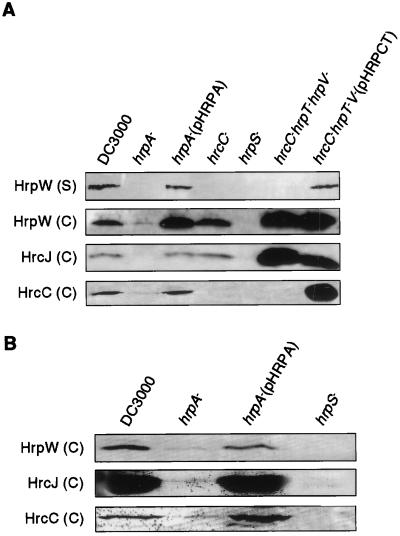
Figure 1.
Immunoblot analysis of HrpW, HrcJ, and HrcC proteins in Pst DC3000 and hrp mutants in hrp-inducing medium (A) and in planta (B). Conditions for bacterial growth and immunoblot analysis are described in Materials and Methods. For analysis of HrpW expression in hrp-inducing cultures, the levels of HrpW in cell-associated (C) and supernatant (S) fractions were examined.
pHRPA (6) and pHRPCT (this study) contain the hrpA gene and the hrcC and hrpT genes, respectively, downstream of the lac promoter of pUCP18 (31). pHRPRS1 carries the P. syringae pv. syringae 61 hrpRS ORFs under the control of the salicylic acid-inducible PG promoter in pKMY299 (32), and it complemented the hrpS mutation in Pst DC3000 (Table 1). pHRPRS2 carries the Pst DC3000 hrpRS ORFs under the control of Plac in pUCP18, and it also complemented the hrpS mutation (Table 1).
Table 1.
Plant reactions to DC3000 and various hrp mutants
| Bacteria | Reaction
|
|
|---|---|---|
| Arabidopsis thaliana (ecotype Columbia) | Tobacco (cv. Samsun NN) | |
| DC3000 ± SA | D | HR |
| hrpS− | Null | Null |
| hrpA− | Null | Null |
| hrpA−/pHRPA | D | HR |
| hrpS−/pHRPRS1 + SA | D | HR |
| hrpS−/pHRPRS2 | D | HR |
| hrpA−/pHRPRS1 + SA | Null | Null |
| hrpA−/pHRPRS2 | Null | Null |
| hrpA−/pHRPAD95S, pHRPRS1 + SA | Null | Null |
HR, rapid, localized tissue collapse in the infiltrated area within 24 hr; D, disease symptoms (slowly developing necrosis and spreading tissue chlorosis) observed 3 days after infiltration; Null, no visible plant reactions; SA, 35 μM salicylic acid.
Pst strains were grown at 22–28°C in King's medium B (33) or LB (34). For induction of hrp genes, bacteria were grown at 20°C in hrp-inducing minimal liquid or agar medium (6). Escherichia coli DH5α was used for all cloning experiments; it was grown in LB at 37°C. Antibiotics used were 100 mg/liter rifampicin, 100 mg/liter ampicillin, 34 mg/liter chloramphenicol, and 50 mg/liter kanamycin.
Protein and RNA Analysis.
For immunoblot analysis of Hrp/Hrc proteins in culture, bacteria first were grown at 28°C to an OD600 of 0.5–0.8 in 5 ml of LB broth supplemented with appropriate antibiotics. Bacteria then were pelleted and resuspended in 5 ml of hrp-inducing broth or LB and incubated with shaking (250 rpm) at 20°C for 12 hr. Cultures then were separated into cell (C) and supernatant (S) fractions by centrifugation. The cell fraction was resuspended in 0.5 ml of sterile water, whereas the supernatant fraction was concentrated 10-fold with Microcon 10 microconcentrators (Amicon). Hrp/Hrc proteins in these fractions were analyzed by SDS/PAGE followed by immunoblotting with appropriate antibodies as described before (17). For immunoblot analysis of Hrp/Hrc proteins expressed in planta, bacteria were grown in LB at 28°C to an OD600 of 0.8. Bacterial suspensions (OD600 of 1.5) prepared in distilled water were infiltrated into leaves of tobacco by using needleless syringes. Six hours after infiltration, the infiltrated leaf tissue was excised and bacteria were expelled from the infiltrated leaf tissue by reinfiltrating the leaf tissue with an excess amount of distilled water. Bacteria were collected by centrifugation, and the levels of Hrp/Hrc proteins in the expelled bacteria were analyzed by immunoblotting with appropriate antibodies. Immunoblot analysis of AvrPto followed the protocol of van Dijk et al. (20). In all immunoblotting experiments, gel staining with Coomassie brilliant blue R-250 was used to ensure equal loading of samples. For determination of steady-state levels of hrp and avr transcripts, total RNA was isolated from bacteria grown in LB or hrp-inducing minimal medium for 4 hr at 20°C by following a standard protocol (34). Five micrograms of total RNA isolated from each bacterial strain was analyzed by Northern blot.
Mutagenesis of the hrpA Gene.
For site-directed mutagenesis, the hrpA gene from pHRPA (6) was cloned into pAlter1 (Promega). Six amino acid residues (i.e., G23, A54, K93, D95, I101, and I111) that are conserved between the HrpA proteins of P. syringae pv. tomato and Erwinia amylovora and/or among the HrpA proteins of P. syringae pvs. tomato, syringae, and glycinea (35, 36) were replaced individually by residues with different properties (A23, E54, I93, S95, T101, and Pro111). The mutagenized hrpA inserts then were recloned into pUCP18 (31) and examined for complementation of the hrpA deletion mutation (see ref. 6 for procedures). For random mutagenesis of the hrpA gene, pHRPA was transformed into the E. coli mutator strain XL1-Red (Stratagene). After being subcultured four times, each for 12 hr at 37°C, pHRPA was isolated from XL1-Red and introduced en masse into the Pst DC3000 hrpA nonpolar mutant by electroporation. Transformants were grown individually in LB in microtiter plate wells overnight at 28°C (OD600 > 1.0). The overnight bacterial cultures were diluted 10-fold in distilled water to an OD600 of 0.1–0.2, and the diluted cultures were infiltrated into tobacco and Arabidopsis leaves for HR and pathogenesis tests, respectively (see ref. 6 for procedures).
Pathogenesis Assays.
Pathogenesis assay procedures were the same as described before (6).
Results
Initial Observations of the Effect of a hrpA Deletion Mutation on the Secretion of HrpW.
To examine a role of the Hrp pilus in secretion of proteins, we investigated the effect of a hrpA deletion mutation on the secretion of HrpW in culture, in which a possible involvement of the Hrp pilus in mediating bacterial attachment to host cells is excluded. The cellular distribution of HrpW in Pst DC3000 and hrpA, hrcC, hrcChrpTV, and hrpS mutants was determined. As shown in Fig. 1A, HrpW was produced and secreted in DC3000 and in the hrpA mutant complemented by pHRPA, which carries the wild-type hrpA gene (6); produced but not secreted in the hrcC and hrcChrpTV secretion mutants; and not detectable in the hrpS regulatory mutant. In the hrpA deletion mutant, HrpW was not secreted in the medium and only barely detectable in the cell fraction (Fig. 1A). However, the amount of HrpW was higher in the hrpA mutant than in the hrpS mutant. The same expression and secretion patterns were observed when bacteria were grown in liquid or agar medium. For all subsequent experiments, liquid cultures were used. The higher level of HrpW in the hrcChrpTV mutant, compared with that in the wild-type DC3000, is due to the polar effect of the Tn5Cm-induced mutation in the hrcC gene on the downstream hrpV gene, which was shown previously to encode a putative negative regulator in P. syringae pv. syringae (28). Consistent with this prediction, the HrpW was not overproduced in a nonpolar hrcC mutant, whereas it was overproduced in the hrcChrpTV mutant carrying pHRPCT, which is equivalent to the hrpV mutant (Fig. 1A). Overproduction of HrpW in the hrcChrpTV mutant carrying pHRPCT suggests that HrpV is also a negative regulator in Pst DC3000.
hrpA Is Required for Full Expression of Other Hrp Proteins.
The unexpected down-regulation of the secreted protein HrpW in the hrpA deletion mutant prompted us to examine a possible effect of the hrpA mutation on the expression of those Hrp proteins that are components of the type III secretion machinery. Specifically, we examined expression of HrcC and HrcJ (37, 38), for which we have antibodies. As expected, in hrp-inducing medium HrcJ was highly expressed in DC3000, the hrcC and hrcChrpTV mutants, and the hrpA mutant complemented by pHRPA, but not in the hrpS mutant. However, the expression of HrcJ was suppressed significantly in the hrpA mutant (Fig. 1A). The same expression and localization pattern was observed for HrcC, except that HrcC was not detected in the hrcC or hrcChrpTV mutant (Fig. 1A). To rule out the possibility that the effect of the hrpA mutation on expression of hrc/hrp genes is due to an artifact in hrp-inducing culture, we determined the expression patterns of HrpW, HrcJ, and HrcC in DC3000 and the hrp mutants in planta. We found an expression pattern similar to that in hrp-inducing culture (Fig. 1B). Specifically, HrpW, HrcJ, and HrcC were produced highly in DC3000, but not in the hrpS mutant. The amounts of these proteins were very low in the hrpA mutant, but higher than in the hrpS mutant. pHRPA restored the expression of these proteins in the hrpA mutant (Fig. 1B). Thus, we conclude that the hrpA gene is required for full expression of both secreted proteins (HrpW) and components of the Hrp secretion machinery (HrcC and HrcJ) in hrp-inducing minimal medium and in planta.
The hrpA Deletion Mutation Affects the Expression of hrp, hrc, and avr Genes at the RNA Level.
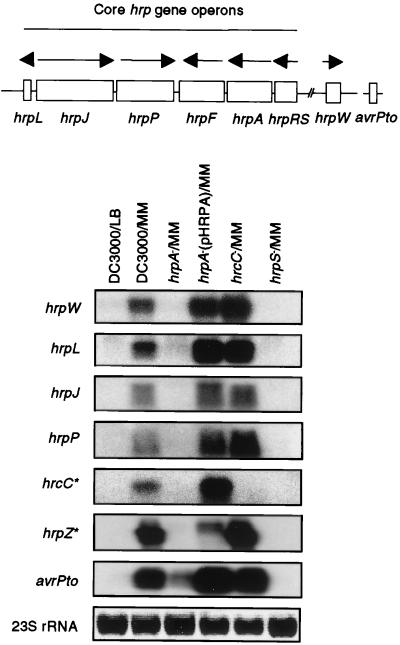
To determine whether the hrpA gene controls the expression of hrc/hrp genes at the RNA as well as the protein level, we directly measured the steady-state mRNA level of the hrpW operon in DC3000 and various mutants. Consistent with the immunoblot results, this operon was significantly down-regulated in the hrpA and hrpS mutants, as compared with DC3000, the hrpA deletion mutant complemented by pHRPA, and the hrcC mutant (Fig. 2). We also determined the effect of the hrpA mutation on the expression of five (i.e., hrpL, hrpJ, hrpP, hrpF, and hrpA; see Fig. 2) of the six core hrc/hrp gene operons and avrPto, which encodes a protein presumably delivered into the plant cell via the Hrp secretion system (40, 41). As shown in Fig. 2, the steady-state mRNA levels of all five hrc/hrp gene operons and avrPto were suppressed significantly in the hrpA and hrpS mutants, although the basal level of the avrPto transcript in the hrpA mutant was higher than those of other transcripts. Thus, the effect of the hrpA mutation is at the transcript level.
Figure 2.
RNA blot analysis of hrp and avr transcripts in Pst DC3000 and hrp mutants. hrp gene operons, named by their first genes, are diagrammed at the top. The directions of transcription are indicated by arrows. The hrpW gene is linked to the core hrp cluster, whereas avrPto is not. Conditions for bacterial growth and RNA analysis are described in Materials and Methods. The genes indicated on the left were used as probes. The 23S rRNA visualized after ethidium bromide staining was used as loading control. RNA was isolated from bacteria grown in hrp-repressing LB or hrp-inducing minimal medium (MM). *, The hrcC and hrcZ genes are located within the hrpF and hrpA gene operons, respectively. See ref. 39 for a more detailed description of the P. syringae hrp gene cluster.
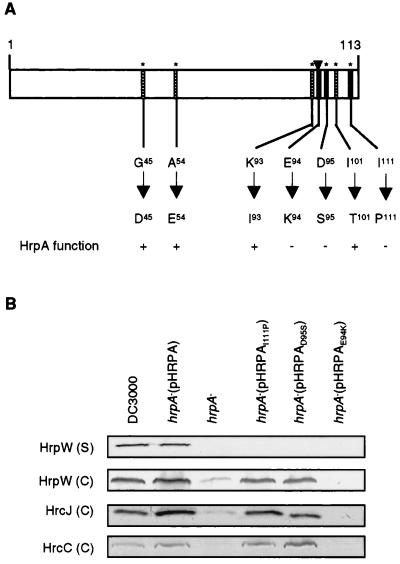
Three Amino Acid Residues Located at the Carboxyl Terminus Are Essential for the Secretion and/or Regulation Function of HrpA.
We next attempted to define amino acid substitution mutations that affect the secretion and/or regulatory function of the HrpA protein. An HR assay was used to screen for these mutations. As reported previously, the wild-type hrpA gene cloned in pHRPA restored the ability of a hrpA deletion mutant to elicit an HR in tobacco (6). We reasoned that a mutation that affects the regulatory or secretion function of the HrpA protein would eliminate the ability of the hrpA gene (pHRPA) to complement the genomic hrpA deletion mutation. We used both site-directed and random mutagenesis procedures in the screen. Site-directed mutagenesis was facilitated by the relatively few residues (six were substituted in this study) conserved among the hypervariable HrpA proteins of P. syringae pathovars and E. amylovora (35, 36). Residue substitutions in two positions (D95 to S95 and I111 to P111) were found to eliminate the ability of the hrpA gene to complement the hrpA mutation for the elicitation of HR in tobacco (Fig. 3A). Random mutagenesis enabled us to identify a third mutant hrpA gene that fails to complement the hrpA mutation. Sequence analysis revealed an amino acid substitution at position 94 (from E94 to K94) in the HrpA protein (Fig. 3A). Thus, all three identified mutations are at the carboxyl terminus of the HrpA protein. The E94K mutation appears primarily to affect the regulatory function of HrpA, whereas the D95S and I111P mutations primarily affect the secretion function (Fig. 3B). However, the absolute amounts of Hrc and Hrp proteins in these mutant strains varied somewhat from experiment to experiment.
Figure 3.
(A) A diagram of the P. syringae pv. tomato DC3000 HrpA protein (113 aa in length). Six amino acid residues (*) conserved among HrpA proteins of P. syringae pvs. tomato, syringae, and glycinea and E. amylovora were mutated by site-directed mutagenesis. The E94-to-K94 mutation, indicated by an arrowhead, was obtained by random mutagenesis. Amino acid residue substitutions that did not affect the HrpA function are indicated by hatched bars. Those substitutions that eliminated the ability of pHRPA to complement the genomic hrpA mutation are indicated by solid bars. The HrpA function was assayed by the ability (+) or inability (−) of the corresponding pHRPA derivatives to complement the genomic hrpA deletion mutation for HR elicitation in tobacco leaves and disease causation in Arabidopsis thaliana leaves. (B) Immunoblot analysis of the effect of single amino acid mutations of HrpA on the production of HrpW, HrcJ, and HrcC in hrp-inducing medium. Both cell-associated (C) and supernatant (S) fractions were analyzed for HrpW. Conditions for bacterial growth and immunoblot analysis are described in Materials and Methods.
The hrpA Mutation Affects the Transcript Level of the hrpRS Operon.
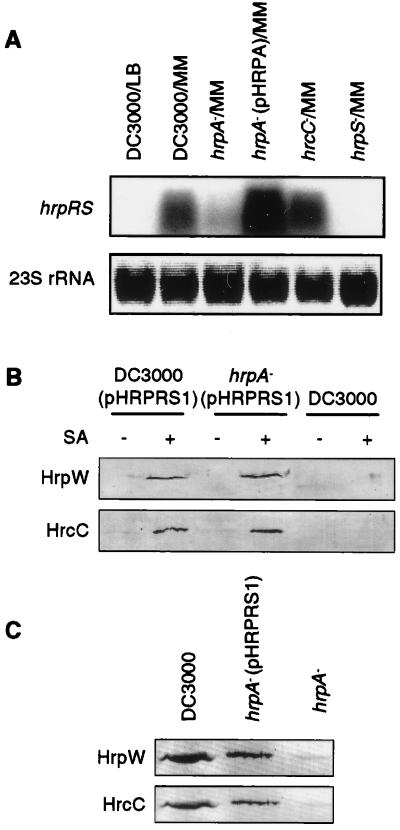
To examine further the effect of the hrpA deletion mutation on gene expression, we determined the steady-state message levels of the hrpRS positive regulatory operon in DC3000 and the hrpA mutant. As expected, the expression of the hrpRS operon was barely detectable in DC3000 grown in LB or in the hrpS mutant grown in the hrp-inducing medium, but was induced when DC3000 and the hrc mutant were grown in hrp-inducing minimal medium. The expression of hrpRS operon was significantly down-regulated in the hrpA mutant, whereas overexpression of the hrpA gene from pHRPA elevated the expression of the hrpRS operon (Fig. 4A). Furthermore, when the hrpRS operon was expressed ectopically from a plasmid (pHRPRS1), the repression of hrc/hrp gene expression in the hrpA mutant was completely relieved. Specifically, HrpW and HrcC were produced in both DC3000 and the hrpA mutant when hrpRS genes were ectopically expressed in the otherwise hrp-repressing LB (Fig. 4B). Similarly, ectopic expression of the hrpRS gene operon allowed expression of HrpW and HrcC proteins in the hrpA mutant in planta (Fig. 4C). The same results were obtained for the HrcJ protein (data not shown). These experiments suggest that the hrpA mutation affects expression of hrc, hrp, and avr genes through a reduction of the transcript level of the hrpRS operon.
Figure 4.
(A) Effect of the hrpA mutation on the steady-state message abundance of hrpRS. The procedures for bacterial growth and RNA preparation and blotting are the same as in the legend to Fig. 2, except that the RNA gel blot was hybridized to the hrpRS gene probe. (B) Effects of ectopic expression of hrpRS on the accumulation of Hrp/Hrc proteins in the hrpA mutant. Bacteria were grown in LB supplemented with (+) or without (−) 35 μM salicylic acid (SA). The original culture was used directly for analysis by SDS/PAGE followed by immunoblot analysis with antibodies against HrpW and HrcC, respectively, without further fractionation. (C) Effects of SA-induced expression of hrpRS from pHRPRS1 on the accumulation of Hrp/Hrc proteins in the hrpA mutant in planta. Bacteria were supplemented with 35 μM SA before infiltration. Constitutive expression of Pst DC3000 hrpRS genes from pHRPRS2 also restored the expression of HrpW in the hrpA mutant (see Fig. 5).
Secretion of HrpW and AvrPto Is Blocked in the hrpA Deletion Mutant Even When Gene Expression Is Restored.
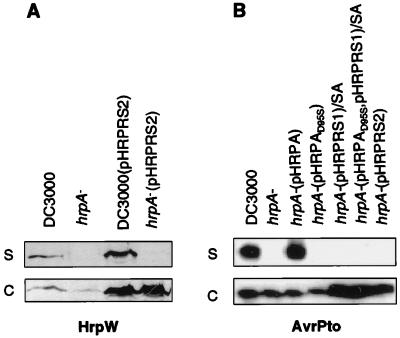
The complete restoration of gene expression in the hrpA deletion mutant by the ectopically expressed hrpRS gene operon enabled the testing of a direct role of HrpA in protein secretion again. As shown in Fig. 5A, when grown in the hrp-inducing medium, DC3000 produced and secreted HrpW to the medium with or without constitutive expression of the hrpRS gene operon from pHRPRS2. However, HrpW was not detected in the culture medium of the hrpA mutants even when the hrpRS gene operon was constitutively expressed. Instead, the expressed HrpW was recovered exclusively in the cell fraction. Similarly, secretion of AvrPto was blocked by both the hrpA deletion mutation and the hrpAD95S point mutation (Fig. 5B). pHRPA restored the secretion of AvrPto. Ectopic expression of the hrpRS operon from pHRPRS1 and pHRPRS2 increased the level of AvrPto, but did not result in the secretion of AvrPto in the hrpA mutant. Consistent with a defect in protein secretion, the hrpA mutant ectopically expressing the hrpRS genes from pHRPRS1 or pHRPRS2 did not elicit HR in tobacco or cause disease in Arabidopsis (Table 1). Thus, in addition to the involvement of the hrpA gene in the regulation of the Hrp secretion system, the Hrp pilus structural protein is required for the secretion of HrpW and AvrPto in culture.
Figure 5.
Effect of ectopic expression of the hrpRS operon in the hrpA mutant on HrpW and AvrPto secretion. Bacteria were grown in hrp-inducing minimal medium. Bacterial supernatant (S) and cell (C) fractions were analyzed by SDS/PAGE followed by immunoblotting with HrpW (A) or AvrPto antibody (B).
Discussion
In this study, we have attempted to define the function of the hrpA gene, which encodes the Hrp pilus structural protein, in type III protein secretion in P. syringae pv. tomato DC3000. We show that a functional HrpA protein is required for secretion of HrpW and AvrPto in culture. In addition, we found that the hrpA mutation affects the full expression of all six core hrc/hrp gene operons as well as hrpW and avrPto that reside outside the core hrc/hrp gene cluster. We identified 3 aa residues at the carboxyl terminus that affect the secretion and/or regulatory function of the HrpA protein. Finally, we show that the hrpA mutation affects the transcript level of the two positive regulatory genes hrpR and hrpS and that ectopic expression of the hrpRS gene operon can completely restore gene expression in the hrpA mutant. We suggest that the Hrp pilus is an integral component of a supramolecular protein secretion structure that enables Pst DC3000 to deliver virulence proteins at the right place and time during bacterial infection of plants.
Pathogenic bacteria devote a large number of genes to type III protein secretion. In the P. syringae hrp gene cluster alone, for example, about 27 genes are coregulated and they encode either regulatory, secretion, or effector proteins (42). In addition, several hrp-regulated genes that encode effector (Avr) proteins have been shown to be unlinked to the hrp gene cluster (43, 44). Thus, turning on the type III protein secretion system is an energy-consuming process. Because the final outcome of turning on the type III secretion system is delivery of some virulence proteins into the host cell, it would be beneficial for bacteria to prevent full induction of type III-secretion-associated genes until host cells are available for protein injection. This prediction is consistent with the observed “contact-dependent activation” (transcriptionally or posttranscriptionally) of type III protein secretion in Yersinia pseudotuberculosis, Salmonella typhimurium, Shigella flexneri, and E. coli (7, 8, 45–47). Expression of hrp genes also has been shown to be induced in infected plant tissues (21–23). Host–bacterium contact appears to be important for bacterial elicitation of an HR (48) and for expression of hrp genes in Ralstonia solanacearum (5), although evidence for contact-dependent activation of the P. syringae Hrp system has yet to be obtained. The molecular basis of bacterial sensing of host–bacterium contact is not understood in any bacterium. In Yersinia, the current hypothesis for contact-dependent activation assumes that host–bacterium contact removes bacterial surface sensors, such as YopN (49), LcrG (50, 51), and TyeA (52), that normally prevent secretion (53). However, an involvement of secreted proteins (e.g., LcrV) as part of an extracellular secretion appendage in the positive regulation of Yop secretion is a formal possibility (53). In R. solanacearum, host cell-dependent hrp gene expression is controlled by a novel signaling pathway involving prhA and prhJ (54, 55).
The significant down-regulation of hrp, hrc, and avr genes in the hrpA deletion mutant in vivo suggests that assembly of the Hrp pilus, secretion of effector proteins, and expression of type III-secretion-associated genes are coordinately regulated in Pst DC3000 and that the HrpA protein, as part of a supramolecular secretion structure, may participate in sensing host cells by Pst DC3000. In support of a role of a pilus protein in contact-based gene regulation, the adhesive P pilus of E. coli has been shown to sense the host–bacterium contact signal, resulting in the induction of bacterial iron starvation-response genes (56). In Yersinia spp., an artificial minimal medium has been shown to mimic the host–bacterium contact signal (by presumably changing the conformation of surface sensors) to induce the Ysc type III secretion system (45, 53). It is therefore likely that the composition of the hrp-inducing minimal medium and growth conditions in vitro mimic the plant–bacterium contact signal in vivo.
Whether pili are involved directly in the transfer of DNA or protein or indirectly in mediating cell–cell contact has been a long-standing and unresolved question. We show here that the HrpA protein is necessary for secretion of HrpW and AvrPto in culture, in which the possible function of pilus-mediated bacterial attachment to the host cell is excluded. The demonstrated role of HrpA in the secretion of AvrPto and HrpW therefore is consistent with a hypothesis that the Hrp pilus is an integral component of the Hrp secretion structure. The Hrp pilus could be the functional equivalent of the extracellular part (called the short-needle extension) of the S. typhimurium type III secretion supramolecular structure (12), presumably providing a conduit for protein secretion. Assembly of the longer pilus in Pst DC3000 may reflect a need for this bacterium to deliver proteins through the thick plant cell wall, which is lacking in animal cells. Alternatively, HrpA, as part of the secretion structure, is required for maintaining the integrity of the Hrp secretion apparatus. Finally, the HrpA protein may function as a chaperone protein that pilots HrpW, AvrPto, and other secreted proteins through the Hrp secretion machinery (and along the Hrp pilus).
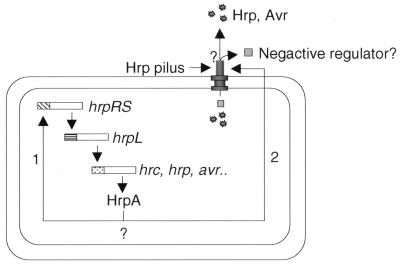
The observed dual function of HrpA in gene regulation and protein secretion suggests that HrpA-mediated gene regulation may be linked to the HrpA function in protein secretion. The simplest explanation would be that HrpA, as a component of the Hrp secretion structure, is required for secretion of a negative regulator, such as HrpV. Secretion of such a negative regulator would derepress the hrc, hrp, and avr genes through activation of the hrpRS/hrpL regulatory cascade (Fig. 6). Gene regulation based on export of a negative regulator (i.e., LcrQ) via the type III protein secretion system, but not a pilus, has been shown in Y. pseudotuberculosis (46). However, our isolation of mutant hrpA genes that primarily affect protein secretion without significantly affecting gene expression (Fig. 3B) does not appear to support this hypothesis, unless the mutant HrpA proteins can counteract the action of a negative regulator in the bacterial cell. It therefore is possible that the regulatory function of the hrpA gene is independent of its role in protein secretion. The hrpA mutation may directly or indirectly affect the transcription and/or RNA stability of the hrpRS operon (Fig. 6). Our future research is aimed at resolving the exact mechanism by which the hrpA gene controls type III protein secretion and affects gene regulation.
Figure 6.
A hypothetical model depicting hrpA-mediated coupling of protein secretion and gene regulation. Arrow 1 indicates that hrpA directly affects the transcription or RNA stability of the hrpRS operon independent of its effects on secretion of Hrp and Avr proteins. Arrow 2 indicates that hrpA indirectly regulates the hrpRS operon through its involvement in secretion of a negative regulator (HrpV?) as well as other Hrp and Avr proteins. The question marks indicate uncertainties in the model regarding whether (i) hrpA affects hrpRS expression directly and/or indirectly through its effects on secretion, (ii) Hrp and Avr proteins are secreted through the pilus, and (iii) a negative regulator (e.g., HrpV) is secreted.
Acknowledgments
We thank Wenqi Hu for preparation of media, our lab members for useful discussions, Karen Bird for help in preparation of this paper, and Dr. Jonathan Walton for critical reading of the paper. This work was supported by grants from the United States Department of Agriculture and Department of Energy (S.Y.H.) and Grant MCB-9631530 from the National Science Foundation (A.C.).
Abbreviations
- Pst, Pseudomonas syringae pv. tomato
hrp, hypersensitive reaction and pathogenicity
- HR
hypersensitive response
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.040570097.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.040570097
References
- 1.Galan J E, Collmer A. Science. 1999;284:1322–1328. doi: 10.1126/science.284.5418.1322. [DOI] [PubMed] [Google Scholar]
- 2.He S Y. Annu Rev Phytopathol. 1998;36:363–392. doi: 10.1146/annurev.phyto.36.1.363. [DOI] [PubMed] [Google Scholar]
- 3.Hueck C J. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pettersson J, Nordfelth R, Dubinina E, Bergman T, Gustafsson M. Science. 1996;273:1231–1233. doi: 10.1126/science.273.5279.1231. [DOI] [PubMed] [Google Scholar]
- 5.Aldon D, Guéneron M, Brito B, Arlat M, Genin S, Vasse J, Van Gijsegem F, Barberis P, Boucher C. In: Biology of Plant–Microbe Interactions. de Wit P J G M, Bisseling T, Stiekema W J, editors. Vol. 2. St. Paul, MN: Int. Soc. Molecular Plant–Microbe Interactions; 2000. , in press. [Google Scholar]
- 6.Roine E, Wei W, Yuan J, Nurmiaho-Lassila E-L, Kalkkinen N, Romantschuk M, He S Y. Proc Natl Acad Sci USA. 1997;94:3459–3464. doi: 10.1073/pnas.94.7.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ginocchio C, Olmsted S B, Wells C L, Galan J E. Cell. 1994;76:717–724. doi: 10.1016/0092-8674(94)90510-x. [DOI] [PubMed] [Google Scholar]
- 8.Knutton S, Rosenshine I, Pallen M J, Nisan I, Neves B C, Bain C, Wolff C, Dougan G, Frankel G. EMBO J. 1998;17:2166–2176. doi: 10.1093/emboj/17.8.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson D M, Schneewind O. Science. 1997;278:1140–1143. doi: 10.1126/science.278.5340.1140. [DOI] [PubMed] [Google Scholar]
- 10.Anderson D M, Schneewind O. Curr Opin Microbiol. 1999;2:18–24. doi: 10.1016/s1369-5274(99)80003-4. [DOI] [PubMed] [Google Scholar]
- 11.Anderson D M, Fouts D E, Collmer A, Schneewind O. Proc Natl Acad Sci USA. 1999;96:12839–12843. doi: 10.1073/pnas.96.22.12839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, Sukhan A, Galan J E, Aizawa S I. Science. 1998;280:602–605. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- 13.Lindgren P B. Annu Rev Phytopathol. 1997;35:129–152. doi: 10.1146/annurev.phyto.35.1.129. [DOI] [PubMed] [Google Scholar]
- 14.Lindgren P B, Peet R C, Panopoulos N J. J Bacteriol. 1986;168:512–522. doi: 10.1128/jb.168.2.512-522.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bogdanove A J, Beer S V, Bonas U, Boucher C A, Collmer A, Coplin D L, Cornelis G R, Huang H-C, Hutcheson S W, Panopoulos N J, et al. Mol Microbiol. 1996;20:681–683. doi: 10.1046/j.1365-2958.1996.5731077.x. [DOI] [PubMed] [Google Scholar]
- 16.He S, Huang H, Collmer A. Cell. 1993;73:1255–1266. doi: 10.1016/0092-8674(93)90354-s. [DOI] [PubMed] [Google Scholar]
- 17.Yuan J, He S Y. J Bacteriol. 1996;178:6399–6402. doi: 10.1128/jb.178.21.6399-6402.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Charkowski A O, Alfano J R, Preston G, Yuan J, He S Y, Collmer A. J Bacteriol. 1998;180:5211–5217. doi: 10.1128/jb.180.19.5211-5217.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mudgett M B, Staskawicz B J. Mol Microbiol. 1999;32:927–941. doi: 10.1046/j.1365-2958.1999.01403.x. [DOI] [PubMed] [Google Scholar]
- 20.van Dijk K, Fouts D E, Rehm A H, Hill A R, Collmer A, Alfano J R. J Bacteriol. 1999;181:4790–4797. doi: 10.1128/jb.181.16.4790-4797.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huynh T V, Dahlbeck D, Staskawicz B J. Science. 1989;245:1374–1377. doi: 10.1126/science.2781284. [DOI] [PubMed] [Google Scholar]
- 22.Rahme L G, Mindrinos M N, Panopoulos N J. J Bacteriol. 1992;174:3499–3507. doi: 10.1128/jb.174.11.3499-3507.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao Y, Lu Y, Heu S, Hutcheson S W. J Bacteriol. 1992;174:1734–1741. doi: 10.1128/jb.174.6.1734-1741.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grimm C, Panopoulos N J. J Bacteriol. 1989;171:5031–5038. doi: 10.1128/jb.171.9.5031-5038.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grimm C, Aufsatz W, Panopoulos N J. Mol Microbiol. 1995;15:155–165. doi: 10.1111/j.1365-2958.1995.tb02230.x. [DOI] [PubMed] [Google Scholar]
- 26.Xiao Y, Heu S, Yi J, Lu L, Hutcheson S. J Bacteriol. 1994;176:1025–1036. doi: 10.1128/jb.176.4.1025-1036.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao Y, Hutcheson S. J Bacteriol. 1994;176:3089–3091. doi: 10.1128/jb.176.10.3089-3091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Preston G, Deng W L, Huang H C, Collmer A. J Bacteriol. 1998;180:4532–4537. doi: 10.1128/jb.180.17.4532-4537.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Willetts N, Skurray R. Annu Rev Genet. 1980;14:41–76. doi: 10.1146/annurev.ge.14.120180.000353. [DOI] [PubMed] [Google Scholar]
- 30.Fullner K J, Lara J C, Nester E W. Science. 1996;273:1107–1109. doi: 10.1126/science.273.5278.1107. [DOI] [PubMed] [Google Scholar]
- 31.Schweizer H P. Gene. 1991;97:109–112. doi: 10.1016/0378-1119(91)90016-5. [DOI] [PubMed] [Google Scholar]
- 32.Yen K. J Bacteriol. 1991;173:5328–5335. doi: 10.1128/jb.173.17.5328-5335.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.King E O, Ward M K, Raney D E. J Lab Med. 1954;22:301–307. [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 35.Preston G, Huang H C, He S Y, Collmer A. Mol Plant–Microbe Interact. 1995;8:717–732. doi: 10.1094/mpmi-8-0717. [DOI] [PubMed] [Google Scholar]
- 36.Kim J F, Wei Z M, Beer S V. J Bacteriol. 1997;179:1690–1697. doi: 10.1128/jb.179.5.1690-1697.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang H-C, He S Y, Bauer D W, Collmer A. J Bacteriol. 1992;174:6878–6885. doi: 10.1128/jb.174.21.6878-6885.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deng W L, Huang H C. J Bacteriol. 1999;181:2298–2301. doi: 10.1128/jb.181.7.2298-2301.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He S. Trends Microbiol. 1997;5:489–495. doi: 10.1016/S0966-842X(97)01163-3. [DOI] [PubMed] [Google Scholar]
- 40.Tang X, Frederick R D, Zhou J, Halterman D A, Jia Y, Martin G B. Science. 1996;274:2060–2063. doi: 10.1126/science.274.5295.2060. [DOI] [PubMed] [Google Scholar]
- 41.Scofield S R, Tobias C M, Rathjen J P, Chang J H, Lavelle D T, Michelmore R W, Staskawicz B J. Science. 1996;274:2063–2065. doi: 10.1126/science.274.5295.2063. [DOI] [PubMed] [Google Scholar]
- 42.Huang H, Lin R, Chang C, Collmer A, Deng W. Mol Plant–Microbe Interact. 1995;8:733–746. doi: 10.1094/mpmi-8-0733. [DOI] [PubMed] [Google Scholar]
- 43.Collmer A. Curr Opin Plant Biol. 1998;1:329–335. doi: 10.1016/1369-5266(88)80055-4. [DOI] [PubMed] [Google Scholar]
- 44.Leach J E, White F F. Annu Rev Phytopathol. 1996;34:153–179. doi: 10.1146/annurev.phyto.34.1.153. [DOI] [PubMed] [Google Scholar]
- 45.Rosqvist R, Magnusson K, Wolf-Watz H. EMBO J. 1994;13:964–972. doi: 10.1002/j.1460-2075.1994.tb06341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pettersson J, Nordfelth R, Dubinina E, Bergman T, Gustafsson M, Magnusson K E, Wolf-Watz H. Science. 1996;273:1231–1233. doi: 10.1126/science.273.5279.1231. [DOI] [PubMed] [Google Scholar]
- 47.Menard R, Sansonetti P J, Parsot C. EMBO J. 1994;13:5293–5302. doi: 10.1002/j.1460-2075.1994.tb06863.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stall R E, Cook A A. Physiol Plant Pathol. 1979;14:77–84. [Google Scholar]
- 49.Forsberg A, Viitanen A-M, Skurnik M, Wolf-Watz H. Mol Microbiol. 1991;5:977–986. doi: 10.1111/j.1365-2958.1991.tb00773.x. [DOI] [PubMed] [Google Scholar]
- 50.Sarker M R, Sory M P, Boyd A P, Iriarte M, Cornelis G R. Infect Immunol. 1998;66:2976–2979. doi: 10.1128/iai.66.6.2976-2979.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Skrzypek E, Straley S C. J Bacteriol. 1993;175:3520–3528. doi: 10.1128/jb.175.11.3520-3528.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iriarte M, Sory M P, Boland A, Boyd A P, Mills S D, Lambermont I, Cornelis G R. EMBO J. 1998;17:1907–1918. doi: 10.1093/emboj/17.7.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cornelis G R. J Bacteriol. 1998;180:5495–5504. doi: 10.1128/jb.180.21.5495-5504.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brito B, Marenda M, Barberis P, Boucher C, Genin S. Mol Microbiol. 1999;31:237–251. doi: 10.1046/j.1365-2958.1999.01165.x. [DOI] [PubMed] [Google Scholar]
- 55.Marenda M, Brito B, Callard D, Genin S, Barberis P, Boucher C, Arlat M. Mol Microbiol. 1998;27:437–453. doi: 10.1046/j.1365-2958.1998.00692.x. [DOI] [PubMed] [Google Scholar]
- 56.Zhang J P, Normark S. Science. 1996;273:1234–1236. doi: 10.1126/science.273.5279.1234. [DOI] [PubMed] [Google Scholar]