Abstract
Estrogen stimulates the proliferation of the most common type of human breast cancer that expresses estrogen receptor α (ERα) through the activation of the cyclin D1 (CCND1) oncogene. However, our knowledge of ERα transcriptional mechanisms remains limited. Hence, it is still elusive why ERα ectopically expressed in ER-negative breast cancer cells (BCC) is functional on ectopic reporter constructs but lacks activity on many endogenous target genes, including CCND1. Here, we show that estradiol (E2) stimulation of CCND1 expression in BCC depends on a novel cell-type-specific enhancer downstream from the CCND1 coding region, which is the primary ERα recruitment site in estrogen-responsive cells. The pioneer factor FoxA1 is specifically required for the active chromatin state of this enhancer and as such is crucial for both CCND1 expression and subsequent cell cycle progression. Interestingly, even in BCC, CCND1 levels and proliferation are tightly controlled by E2 through the establishment of a negative feedforward loop involving the induction of NFIC, a putative tumor suppressor capable of directly repressing CCND1 transcription. Taken together, our results reveal an estrogen-regulated combinatorial network including cell-specific cis- and trans-regulators of CCND1 expression where ERα collaborates with other transcription factors associated with the ER-positive breast cancer phenotype, including FoxA1 and NFIC.
Keywords: Breast cancer, estrogen receptor, cyclin D1, transcription, enhancer, chromatin
Cyclin D1 (CCND1) belongs to the family of D-type cyclins, which regulate G1–S cell cycle progression (Quelle et al. 1993). CCND1 acts through activation of cyclin-dependent kinases (cdks) that phosphorylate and inactivate the retinoblastoma protein. However, recent findings indicate that CCND1 could also promote cell cycle progression through cdk-independent mechanisms, such as interaction with and modulation of transcription factor activities (for reviews, see Ewen and Lamb 2004; Arnold and Papanikolaou 2005). CCND1 is a well recognized oncogene that is amplified and/or overexpressed in a substantial proportion of human cancers including parathyroid adenoma, colon cancer, lymphoma, melanoma, prostate cancer, and breast cancer (Ewen and Lamb 2004). More specifically, CCND1 is one of the most commonly overexpressed genes in human breast cancer (up to 50% of breast cancers) (Arnold and Papanikolaou 2005). CCND1 oncogenic activity in the mammary gland was confirmed by genetically modified mouse models where CCND1 overexpression leads to the development of mammary carcinoma (Wang et al. 1994), while CCND1 ablation results in mice resistant to cancer induced by several oncogenes (Yu et al. 2001). The CCND1-null mice also showed defective postnatal mammary development revealed by a lack of proliferation of alveolar epithelial cells in response to the sex steroid milieu of pregnancy (Fantl et al. 1995; Sicinski et al. 1995).
Indeed, CCND1 expression is induced in mammary epithelial cells by estrogen and progesterone (Sutherland et al. 1998), which act through transcription factors of the nuclear receptor superfamily; i.e., the estrogen receptors (ER) α and β and the progesterone receptors (PR) A and B. Over two-thirds of human breast cancers express ERα, which is positively correlated with high CCND1 expression levels as well as with the expression of a signature set of transcription factors, including FoxA1, GATA3, and XBP-1 (for reviews, see Lacroix and Leclercq 2004; Lacroix et al. 2004; Arnold and Papanikolaou 2005). Amplification of the CCND1 gene is present only in a minority of CCND1-overexpressing breast cancers, indicating that pathogenic transcriptional activation of this gene by factors such as ERα could be an important mechanism triggering its overexpression. Moreover, it has been shown that artificial induction of CCND1 mRNA expression in breast cancer cells (BCC) mimics estrogen effects and is sufficient to drive S-phase entry (Prall et al. 1998) but is also able to reverse the growth-inhibitory effect of antiestrogens (Wilcken et al. 1997). Accordingly, another study showed that the major antiproliferative effect of the antiestrogen tamoxifen could be related to its inhibition of CCND1 expression (Hodges et al. 2003). CCND1 silencing experiments in BCC further confirmed the correlation between CCND1 expression levels and cellular proliferation, emphasizing CCND1 as a potential therapeutic target for breast cancer (Arnold and Papanikolaou 2005; Grillo et al. 2006).
The mechanisms by which estrogen regulates CCND1 levels in BCC are primarily transcriptional. Transient transfection-based studies have mapped several potential estrogen-responsive sites in the CCND1 proximal promoter, including an atypical cyclic AMP response element (position −57), an AP1 site (position −954), and GC-rich sites (positions −110 and −143) (Altucci et al. 1996; Sabbah et al. 1999; Castro-Rivera et al. 2001; Liu et al. 2002; Park et al. 2005). Stable integration of reporter constructs in BCC was recently used by Cicatiello et al. (2004) to show that in this context estrogen stimulation required primarily the AP1 site. These authors proposed that ERα loading at the CCND1 promoter upon estrogen stimulation was mediated through interaction with AP1, resulting in the assembly of an activator complex responsible for CCND1 transcriptional induction.
While these studies suggested the presence of potential cis-regulatory elements involved in CCND1 expression, none explain the cell-type-specific regulation of CCND1 by ERα (Planas-Silva et al. 1999). Here, we describe the characterization of a novel estrogen-regulated cell-type-specific enhancer for CCND1 and the combinatorial transcription factor network that functions through this enhancer to regulate CCND1 expression in BCC.
Results
Identification of two DNase I-hypersensitive (HS) sites in the vicinity of the CCND1 gene that correspond to novel transcriptional enhancers
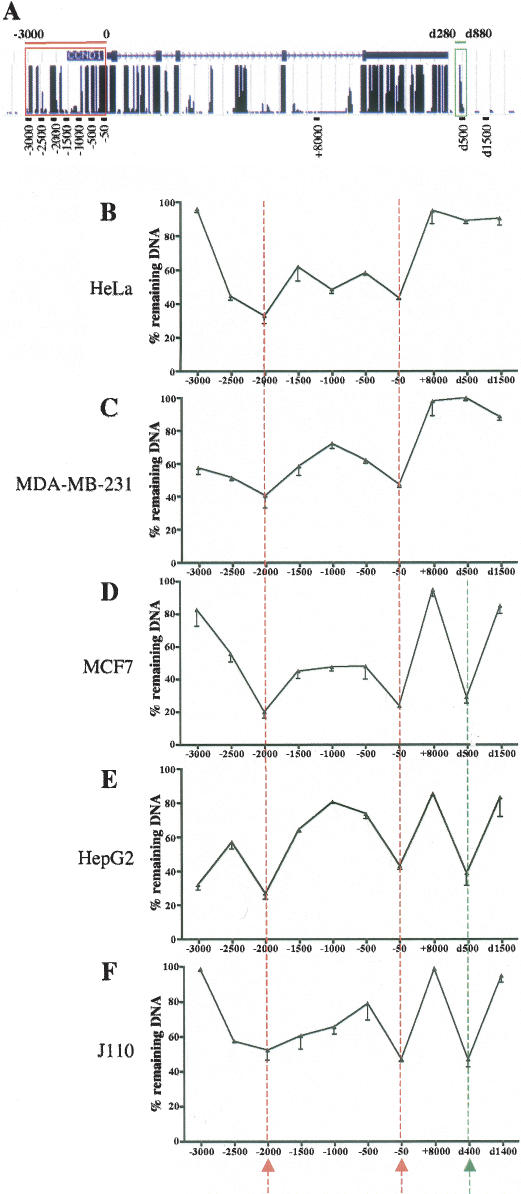
Although previous work defining CCND1 gene regulatory regions focused primarily on the proximal 1-kb promoter (cited above and Fukami-Kobayashi and Mitsui 1998; Eto 2000), analysis of sequence conservation across species indicated that additional regions ~2 and 3 kb upstream of the transcription start site were extremely well conserved (Fig. 1A, red frame). Moreover, a potential ERα-binding site was found downstream from the CCND1 gene by an unbiased approach combining chromatin immunoprecipitation (ChIP) and analysis of bound DNA by tiled microarrays covering the nonrepetitive sequences of the whole genome (J.S. Carroll, C.A. Meyer, J. Song, W. Li, T.R. Geistlinger, J. Eeckhoute, A.S. Brodsky, E.K. Keeton, K.C. Fertuck, G.F. Hall, et al., in prep.). This binding site spanning the region between 280 and 880 base pairs (bp) downstream from the human CCND1 coding sequence (noted d280–d880) was also highly conserved between species (Fig. 1A, green frame). Since DNase I hypersensitivity is a hallmark of active regulatory regions when they are analyzed in the in vivo chromatin context (Gross and Garrard 1988), we first analyzed the DNase I sensitivity of the highly conserved regions found in the vicinity of CCND1. To robustly quantify the DNase I sensitivity we used real-time PCR with primers designed to generate very small amplicons (<110 bp) tiled along these regions. This approach has been described and validated recently on a large-scale analysis (McArthur et al. 2001; Dorschner et al. 2004). DNase I HS site size ranges from 200 bp to 1 kb (Gross and Garrard 1988). We targeted sequences 500 bp apart spanning from −3000 to −50 bp of the CCND1 gene; one amplicon was centered on +8000 bp in intron 4, and two amplicons covered sequences 500 and 1500 bp downstream from the CCND1 coding sequence (Fig. 1A). The analysis was conducted in a variety of human cancer cell lines: the cervical adenocarcinoma cell line, HeLa (Fig. 1B), the ERα-negative mammary adenocarcinoma cell line MDA-MB-231 (Fig. 1C), the ERα-positive mammary adenocarcinoma cell line MCF7 (Fig. 1D), and the hepatocellular carcinoma cell line HepG2 (Fig. 1E). In all of these cell lines, the 3-kb region upstream of the CCND1 mRNA start site harbored two HS sites around −2000 and −50 bp (Fig. 1B–E, red dotted lines). The surrounding regions showed intermediate sensitivity when compared with the transcribed region (+8000 bp), which was relatively DNase I-resistant as expected. It has been reported that DNase I sensitivity can spread around HS sites (McArthur et al. 2001). Moreover, as mentioned earlier, transcription factor-binding sites exist in the region between −1000 and −500 bp that may induce intermediate DNase I sensitivity (McArthur et al. 2001). Interestingly, analysis of the conserved region downstream from CCND1 revealed a cell-type-specific HS site, which was present in MCF7 and HepG2 but not in HeLa or MDAMB-231 cells (Fig. 1B–D, green dotted line). Since these HS sites were within evolutionary conserved sequences, we analyzed whether they could also be found in the J110 mouse breast cancer cell line (low ERα-positive BCC derived from the tumor of an AIB1 transgenic mouse) (M.I. Torres-Arzayus, J. Yuan, J. Dellagatta, H. Lane, A. Kung, and M. Brown, in prep.). We observed that the HS sites found at −50 bp and downstream from CCND1 in the human cells were also present in this mouse cell line (Fig. 1F). The region at −2000 bp was DNAse I sensitive, but the difference in sensitivity with the neighboring sites was not as pronounced as in the human cell lines (Fig. 1F). The HS site at −50 bp is located near the transcription start site, which contains known CRE- and Sp1-binding sites that are conserved between human and mouse genomes (Eto 2000). In contrast, the HS sites at −2000 and d500 bp revealed new potentially important regulatory regions.
Figure 1.
Profiling of the DNase I sensitivity of the evolutionarily conserved regions in the vicinity of the CCND1 gene. (A) Schematic representation of the conserved regions at the CCND1 locus from the University of California at Santa Cruz genome browser. The CCND1 gene is shown at the top with blocks representing the exons. The track at the bottom of the gene shows a measure of evolutionary conservation between 17 vertebrate species based on a phylogenetic hidden Markov model. The conserved regions studied in this work upstream (−3000 to 0 bp; red frame) and downstream (d280–d880 bp; green frame) from the CCND1 gene are highlighted. The location of real-time PCR amplicons generated by PCR primer sets used throughout this work is indicated at the bottom part of the panel. (B–F) DNase I sensitivity assays were performed as indicated in Materials and Methods with intact nuclei from the indicated cells. The percent (%) of remaining DNA after partial digestion is indicated for every analyzed sequence throughout the CCND1 locus. Results are means ± SE from two independent experiments. The HS sites are indicated by the arrows at the bottom of the graphs and by the vertical dotted lines.
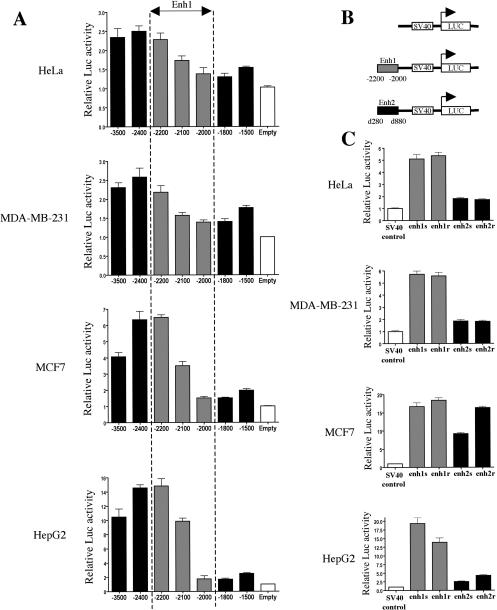
To test this hypothesis we cloned these conserved regions and assessed their transcriptional activities in luciferase reporter assays. First, we cloned the 3.5-kb upstream region of CCND1 and performed a serial deletion analysis ranging from −3.5 to −1.5 kb. The resultant reporter constructs were transfected in the four human cell lines: HeLa, MDA-MB-231, MCF7, and HepG2. The results clearly indicated that an enhancer element (called enh1) was located between −2.2 and −2 kb (Fig. 2A) in correlation with the HS site found at −2 kb. Next, we cloned enh1 and the region between d280 and d880 bp (thereafter called enh2) in both orientations in front of a minimal SV40 promoter (Fig. 2B). Enh1 was able to strongly increase the activity of the SV40 promoter in all cell lines (Fig. 2C). Transfection results indicated that enh2 only modestly increased the reporter activity in HeLa and MDA-MB-231 cells (less than twofold) but had a more significant enhancer activity in HepG2 (2.6- to 4.3-fold induction) and MCF7 cells (over ninefold induction) (Fig. 2C).
Figure 2.
The HS sites at −2000 and d500 bp correspond to transcriptional enhancers. (A) Reporter assays were performed in the indicated cell lines with various constructs spanning the upstream sequence of CCND1.(B) Schematic of the constructs used in C. The gray rectangles represent enh1 and the black rectangles represent enh2. (C) Reporter assays were performed in the indicated cell lines with constructs depicted in B. Each enhancer was cloned and analyzed in both orientations, which are noted sense (s) and reverse (r). Transcriptional activities are expressed relative to that of the respective control luciferase vector (white bar), which was set to 1. All results are means ± SE from at least three independent experiments performed in triplicate.
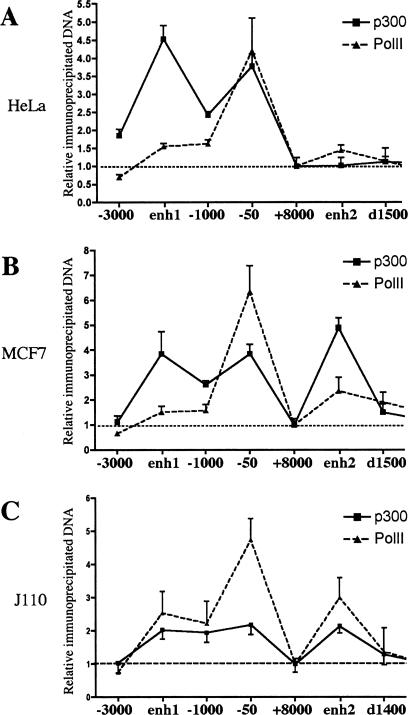
Enhancer elements are known to act in vivo through recruitment of multiprotein complexes involving transcription factors, cofactors, and potentially the RNA polymerase II (PolII) (Louie et al. 2003; Ogata et al. 2003). To test whether enh1 and enh2 were functional enhancers in an in vivo chromatin context, we analyzed recruitment of the cofactor p300 and PolII to the CCND1 regulatory regions by ChIP. Immunoprecipitated DNA was analyzed by real-time PCR with primer sets tiled every kilobase. ChIP experiments were performed in HeLa cells (Fig. 3A), where only enh1 was DNase I sensitive, and in MCF7 (Fig. 3B) and J110 cells, where both enhancers were sensitive (Fig. 3C). In all these cell lines, p300 was recruited to the proximal promoter (−50 bp) and to enh1 (−2000 bp) (Fig. 3A–C, solid line). Importantly, p300 was recruited to enh2 in MCF7 (Fig. 3B) and J110 cells (Fig. 3C) but not in Hela cells (Fig. 3A). PolII was primarily recruited to the proximal promoter as expected (Fig. 3A–C, dotted line) and was also significantly enriched at enh2 only in MCF7 and J110 cells (Fig. 3A–C). Control ChIP experiments with nonimmune IgG did not give any significant enrichment of the analyzed sequences (data not shown). Thus, results of ChIP assays confirmed the in vivo functionality of enh1 and enh2 and verified the cell-specific activity of enh2.
Figure 3.
Recruitment of p300 and PolII to the CCND1 gene regulatory regions. ChIP assays were used to analyze p300 (solid line) and PolII (dotted line) recruitment to the CCND1 regulatory regions in HeLa (A), MCF7 (B), and J110 (C) cells. Amounts of immunoprecipitated DNA were normalized to inputs and reported relative to the amount obtained at +8000 bp, which was set to one (indicated by the horizontal black dotted line). The +8000-bp site was used to normalize all the ChIP data in this study because it represented a negative control for transcription factor and coactivator binding (only low amounts of PolII were present at this site). Results are means ± SE from two to four independent experiments.
Enh2 is an estradiol (E2)-responsive enhancer corresponding to the primary ERα recruitment site of the CCND1 gene in ERα-positive BCC
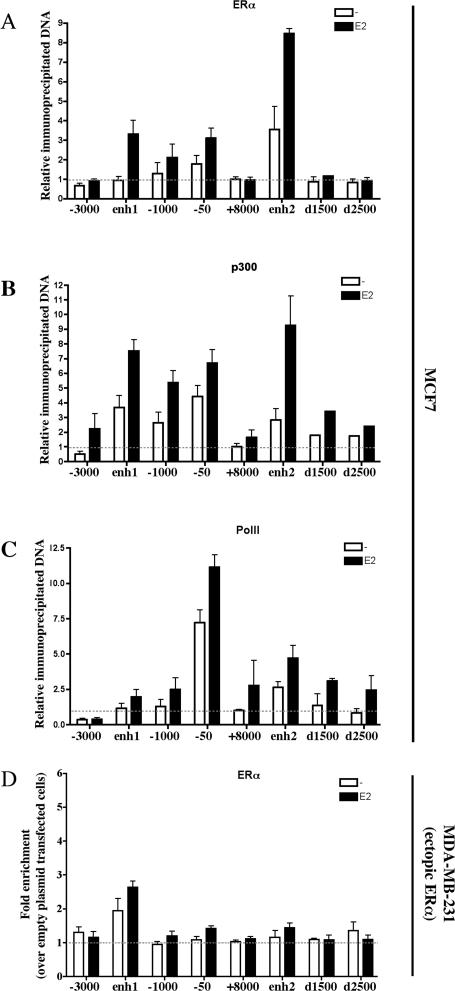
As noted earlier, CCND1 overexpression is particularly important in human breast cancer and is highly associated with the expression of ERα. The novel enhancer elements described here are devoid of estrogen response element (ERE) or ERE half-sites (Supplementary Fig. 1). However, ERα has the ability to regulate target genes through interaction with other transcription factors that tether ERα to the DNA (Kushner et al. 2000; Safe 2001). In keeping with the relatively low E2 sensitivity of indirect regulatory sites reported in reporter assays (Xie et al. 1999; Stoner et al. 2004), transient transfections performed in MCF7 cells showed that enh2 activity could be modestly enhanced by E2 (Supplementary Fig. 2). In light of these results, we decided to reassess ERα recruitment to CCND1 regulatory regions. As such, we performed ChIP assays in MCF7 cells, which were hormone starved for 3 d and then stimulated with E2 for 45 min (Shang et al. 2000; Metivier et al. 2003; Carroll et al. 2005). We found that ERα was recruited at the upstream regulatory sites following E2 stimulation, including the 1-kb proximal promoter as previously reported (Castro-Rivera et al. 2001; Cicatiello et al. 2004) and enh1 (Fig. 4A). However, upon E2 treatment ERα was predominantly loaded at enh2 downstream from the CCND1 gene (Fig. 4A). A significant recruitment was also observed in the absence of hormone at enh2 (Fig. 4A). The unliganded ERα has previously been suggested to bind the promoter of another target gene (TFF-1) in MCF7 cells, where it plays an active transcriptional role (Metivier et al. 2004). ERα recruitment to enh2 was verified in three other human ERα-positive breast cancer cell lines (T47D, ZR75-1, and BT474) as well as in the mouse J110 breast cancer cell line (Supplementary Fig. 3), but was not observed in MDA-MB-231 cells expressing exogenous ERα, as expected from the cell-type-specific activity of this enhancer (Fig. 4D). Moreover, additional ChIP assays in MCF7 cells indicated that E2 increased p300 recruitment to the CCND1 regulatory regions, with the strongest effect at enh2 (Fig. 4B). As expected, E2 induced PolII loading at the proximal promoter with also a modest enrichment at enh2 (Fig. 4C).Thus, enh2 corresponds to the primary site of E2-induced ERα and p300 recruitment, strongly arguing for a crucial role of this enhancer in hormonal regulation of CCND1 gene expression in human BCC.
Figure 4.
Primary recruitment of ERα to the E2-responsive enh2 in MCF7 cells. ChIP assays were performed to analyze ERα (A), p300 (B), and PolII (C) recruitment to the CCND1 regulatory regions upon E2 stimulation of MCF7 cells. Amounts of immunoprecipitated DNA were normalized to inputs and reported relative to the amount obtained at +8000 bp in the absence of ligand, which was set to 1 (indicated by the horizontal black dotted line). (D) ChIP of ERα ectopically expressed in MDA-MB-231 was used to verify the cell-type-specific activity of enh2. Data were expressed as fold chromatin enrichment over empty control plasmid-transfected cells. All results are means ± SE from two to four independent experiments.
Identification and pattern of transcription factor recruitment to the CCND1 gene regulatory regions
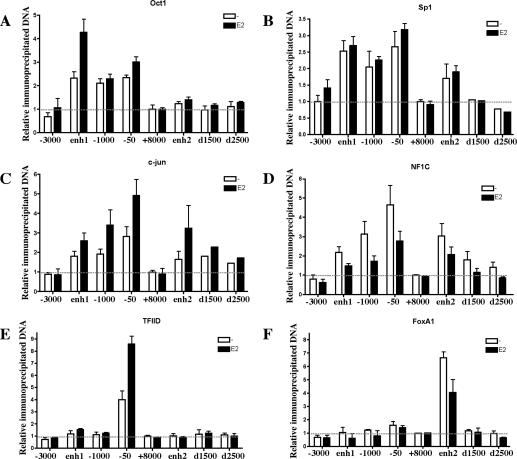
In order to further characterize the newly identified enhancer elements, we screened for evolutionarily conserved potential transcription factor-binding sites (Supplementary Fig. 1). Although a number of motifs were identified, we focused on specific transcription factors that were expressed in MCF7 cells (GNF SymAtlas) (Su et al. 2002), including Oct1, Sp1, c-jun, NFIC, and FoxA1. ChIP analysis of transcription factor recruitment showed various patterns on the different CCND1 regulatory regions. Oct1 was recruited only to the upstream region of CCND1; its recruitment was induced by E2 primarily at enh1 (Fig. 5A). Sp1, c-jun, and NFIC were all recruited at both the upstream and downstream regulatory sites (Fig. 5B–D); c-jun recruitment was increased by E2 (Fig. 5C), while Sp1 binding was independent of E2 (Fig. 5B). Interestingly, NFIC recruitment was reduced by E2 treatment (Fig. 5D). The E2 effects on Oct1 and c-jun recruitment were similar to those reported in previous studies (Cicatiello et al. 2004). Additional transcription factors, namely C/EBPβ, SRF, and PR, were also recruited to the CCND1 regulatory sites, including, most importantly, enh2, suggesting this element could also be involved in regulation of CCND1 expression by other stimuli (data not shown). All of these transcription factors were also recruited to the CCND1 upstream regulatory sites in HeLa cells, but none was found at enh2, which is not active in this cell line (Supplementary Fig. 4). The general transcription factor TFIID was specifically recruited to the proximal promoter as expected (Fig. 5G). In contrast, FoxA1 was found recruited only to enh2 with reduced binding upon E2 stimulation (Fig. 5G), a result consistent with our previous report (Carroll et al. 2005). FoxA1 loading to enh2 was also observed in each one of the analyzed ERα-positive breast cancer cell lines, including T47D, ZR75-1, BT474, and J110 (Supplementary Fig. 3), but not in HeLa or MDA-MB-231 cells where this factor is not expressed (data not shown). Thus, FoxA1 is specifically recruited to enh2 prior to ERα and could mark this region as one competent for ERα binding. Taken together, our ChIP experiments define an E2-orchestrated transcription factor recruitment pattern to the discrete CCND1 regulatory sites.
Figure 5.
Pattern of transcription factor recruitment to the CCND1 gene regulatory regions in BCC. Recruitment of the indicated transcription factors was analyzed by ChIP in MCF7 cells. Data were analyzed as in Figure 4. Results are means ± SE from two to five independent experiments.
FoxA1 activates while NFIC represses both CCND1 expression and MCF7 cell cycle progression
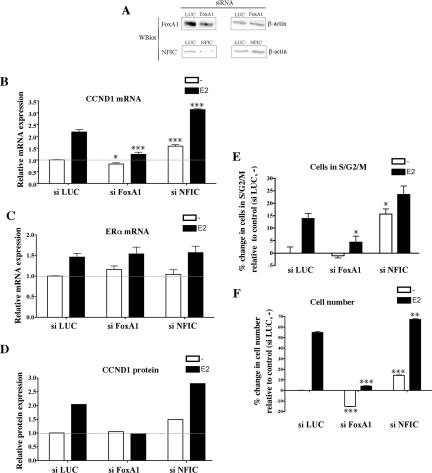
FoxA1 expression correlates with ERα in human breast cancers (Lacroix and Leclercq 2004; Williamson et al. 2005), and we and others have shown FoxA1 to be important for the activation of a significant subset of ERα target genes in BCC by allowing ERα to bind to regulatory sequences (Carroll et al. 2005; Laganiere et al. 2005). Interestingly, among the other transcription factors recruited to the CCND1 regulatory sites, we noticed that NFIC expression correlated with ERα expression in primary human breast cancers (van de Vijver et al. 2002). Moreover, we found that NFIC was up-regulated by E2 treatment in MCF7 cells (Supplementary Fig. 5), a result reminiscent of FoxA1 regulation (Laganiere et al. 2005). Hence, we decided to further analyze the functional role of FoxA1 and NFIC in CCND1 transcriptional regulation. With this aim, we monitored CCND1 mRNA and protein expression levels after silencing each of these factors by RNA interference (RNAi) in MCF7 cells. Small interfering RNA (siRNA) against luciferase (LUC) was used as a control. As shown in Figure 6A, RNAi against FoxA1 and NFIC was able to significantly reduce the protein expression level of each one of these transcription factors. Interestingly, real-time RT–PCR assays indicated that FoxA1 silencing inhibited E2 induction of CCND1 mRNA levels without significantly affecting the basal expression level (Fig. 6B). Surprisingly, RNAi against NFIC significantly increased CCND1 expression in both the basal and E2-treated conditions (Fig. 6B). These effects were not due to modulation of ERα expression (Fig. 6C; Carroll et al. 2005). Western blot assays confirmed the effect of FoxA1 and NFIC silencing on CCND1 expression at the protein level (Fig. 6D). In similar experiments, Oct1 silencing slightly reduced CCND1 levels, while Sp1 silencing had a very strong negative effect (Supplementary Fig. 7). It has been widely reported that CCND1 expression levels are tightly linked to BCC growth rates and that small variations in CCND1 expression (within the range of twofold observed in our data) robustly modulate BCC proliferation (Musgrove et al. 1993, 1994; Dupont et al. 2000; Castro-Rivera et al. 2001; Rubio et al. 2006).
Figure 6.
Analysis of CCND1 expression and cell cycle progression after silencing of transcription factors recruited to the CCND1 regulatory regions. (A) Western blot (Wblot) assays were performed on whole-cell extracts of cells transfected with si LUC, si FoxA1, or si NFIC to analyze expression of the specific transcription factors silenced (left) and of β-actin (right). CCND1 (B) and ERα (C) mRNA expression levels were determined by real-time RT–PCR in MCF7 cells transfected with the indicated siRNA and challenged with E2 or vehicle alone for 3 h. CCND1 and ERα mRNA expression levels were expressed relative to expression in si LUC-transfected cells treated with vehicle, which was set to 1. Results are means ± SE from at least three independent experiments. (*) p < 0.05, (***) p < 0.001 versus si LUC-transfected cells similarly treated. (D) CCND1 expression was analyzed by Western blot using whole-cell extracts from MCF7 cells transfected with the indicated siRNA and challenged with E2 or vehicle alone for 6 h. CCND1 expression was quantified by densitometry and normalized using calnexin as a loading control. CCND1 expression levels were expressed relative to expression in si LUC-transfected cells treated with vehicle, which was set to 1. Images of the Western blots are presented in Supplementary Figure 6. (E) MCF7 cells transfected with the indicated siRNAs were challenged with E2 or vehicle alone for 20–24 h. DNA contents were analyzed by propidium iodide staining and flow cytometry. Results are expressed as percent change in the fraction of cells in the S, G2, and M phases of the cell cycle relative to control (si LUC-transfected cells treated with vehicle) and are means ± SE from three separate assays. (*) p < 0.05 versus si LUC-transfected cells similarly treated. For si NFIC-transfected cells challenged with E2, p = 0.069. (F) Cell viability assays were performed as described in Materials and Methods using MCF7 cells transfected with the indicated siRNAs. Relative cell numbers were determined after 3 d of E2 stimulation and are expressed as percent change relative to control (si LUC-transfected cells treated with vehicle). Results are means ± SE from one representative experiment performed in sextuplicate. p < 0.01 (**) and p < 0.001 (***) versus si LUC-transfected cells similarly treated.
Hence, we analyzed MCF7 cell growth after silencing of FoxA1 and NFIC. MCF7 cells transfected with the various siRNAs were hormone starved for 3 d and then stimulated with E2 for 20–24 h before analysis for cell cycle distribution. Silencing of FoxA1 significantly blunted the E2-mediated increase in the fraction of cells in S/G2/M phases of the cell cycle (Fig. 6E). In contrast, NFIC silencing increased the fraction of proliferating cells in both basal and E2-induced conditions (Fig. 6E). Analysis of the sub-G1 fraction of cells revealed only slight differences in apoptosis between conditions (data not shown). In a cell growth assay, we found, as expected, that FoxA1 was required for MCF7 proliferation predominantly upon E2 stimulation (Fig. 6F). In contrast, NFIC silencing increased the number of both vehicle and E2-treated MCF7 cells (Fig. 6F).
These results suggest that FoxA1 is predominantly required for the E2-mediated stimulation of CCND1 expression and cell cycle progression, while NFIC (as well as Oct1 and Sp1) (Supplementary Fig. 7) had a broader role in the regulation of both the basal and E2-stimulated conditions.
Functional competency of enh2 is defined by FoxA1 and is crucial for E2-mediated induction of PolII recruitment to the CCND1 promoter in MCF7 cells
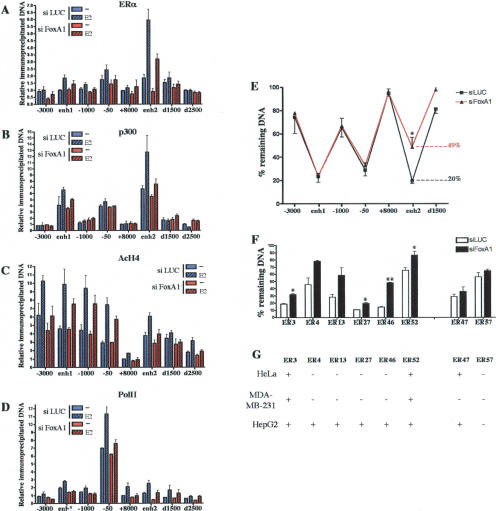
To better characterize the functional role of FoxA1 in E2 induction of CCND1 expression, we assessed by ChIP the consequences of FoxA1 silencing on recruitment of ERα, p300, and PolII as well as histone H4 acetylation at the CCND1 regulatory sequences. MCF7 cells were transfected with siRNA against FoxA1 or LUC as a control and processed for ChIP assays. FoxA1 silencing dramatically impaired ERα (Fig. 7A) and p300 (Fig. 7B) recruitment as well as H4 acetylation (Fig. 7C) at enh2 in E2-stimulated cells. Note that ERα loading at enh2 was also significantly reduced in the absence of hormone, while only a small reduction in p300 recruitment and H4 acetylation was observed, presumably because p300 is primarily recruited through other transcription factors in the absence of E2 stimulation. Interestingly, FoxA1 silencing also tended to slightly reduce ERα and p300 recruitment, as well as H4 acetylation to the upstream CCND1 regulatory region in E2-treated MCF7 cells (Fig. 7A–C, respectively), suggesting a potential communication between the downstream enh2 and the upstream sites upon hormone treatment. In agreement with this hypothesis, siRNA against FoxA1 strongly reduced E2-induced recruitment of PolII to the transcriptional start site (Fig. 7D). These results indicate that FoxA1 is required for activity of enh2 that mediates E2 induction to the proximal promoter of the CCND1 gene.
Figure 7.
FoxA1 regulates CCND1 expression by defining the functionality of enh2 in BCC. MCF7 cells transfected with si LUC or si FoxA1 were used in ChIP experiments analyzing recruitment of ERα (A), p300 (B), and PolII (D) as well as H4 acetylation (AcH4) (C) to the CCND1 regulatory regions. Amounts of immunoprecipitated DNA were normalized to inputs and reported relative to the amount obtained at +8000 bp with si LUC-transfected cells treated with vehicle alone, which was set to 1. Results are means ± SE from two or three independent experiments. (E) Intact nuclei from MCF7 cells transfected with si LUC or si FoxA1 were used in DNase I sensitivity assays performed as in Figure 1. The percent of remaining DNA corresponding to the enh2 (d500 bp) after partial digestion is indicated. (F) DNase I sensitivity assays were performed using primers that spanned ERα-binding sites previously identified (Carroll et al. 2005). p < 0.05 (*) and p < 0.01 (**) versus si LUC-transfected cells. For ER4 and ER13, p values were 0.076 and 0.053, respectively. (G) DNase I sensitivity assays were performed in HeLa, MDA-MB-231, and HepG2 cells as indicated. DNase I sensitivity (+) and insensitivity (−) were determined using the DNase I insensitive control rhodopsin locus as a reference. Results of all DNase I sensitivity assays are from at least two independent experiments.
FoxA1 is a tissue-specific transcription factor that has the ability to bind and remodel compacted chromatin (Cirillo et al. 2002; Holmqvist et al. 2005). We noticed that FoxA1 was expressed in MCF7 (as well as in the other ERα-positive breast cancer cell lines analyzed) and HepG2 but not in HeLa and MDA-MB-231 cells (Crowe et al. 1999; Williamson et al. 2005), paralleling the cell-specific functional activity of enh2. As such we tested the hypothesis that FoxA1 could be important in determining an active chromatin structure at enh2 by performing DNase I sensitivity assays with nuclei isolated from MCF7 cells transfected with si LUC or si FoxA1. As shown in Figure 7E, FoxA1 silencing significantly and specifically reduced the DNase I sensitivity of the HS site corresponding to enh2. Thus, FoxA1 is required to produce a chromatin environment that allows binding and formation of a multiprotein complex at enh2.
To determine if this could be a general mode of action for FoxA1, we surveyed the effect of its silencing on the DNase I sensitivity of ERα-binding sites from chromosomes 21 and 22 that we previously reported to recruit FoxA1 (named ER3, ER4, ER13, ER27, ER46, and ER52; Carroll et al. 2005). Interestingly, RNAi against FoxA1 also significantly reduced the DNase I sensitivity of all those sites while having no significant effect on two ERα-binding sites (ER47 and ER57) that did not bind FoxA1 (Fig. 7F). Moreover, most of the ERα-binding sites whose DNase I sensitivity was reduced by FoxA1 silencing in MCF7 displayed cell-specific DNase I sensitivity (Fig. 7G) identical to the one observed for the CCND1 gene enh2 (Fig. 1).
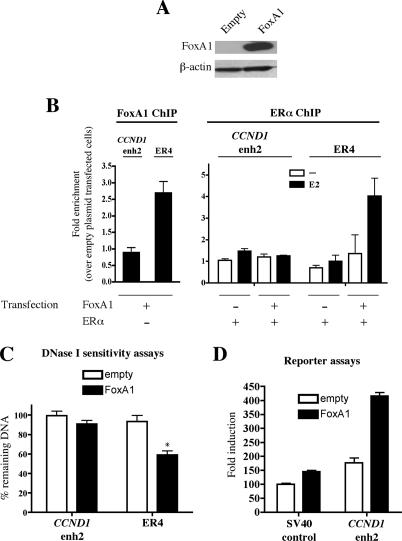
Hence, we investigated whether FoxA1 was sufficient to induce DNaseI sensitivity of cell-type-specific regulatory regions such as CCND1 enh2 by ectopically expressing FoxA1 in MDA-MB-231 cells (Fig. 8A). In these conditions, FoxA1 was not recruited to CCND1 enh2 but could be detected at another cell-specific ERα regulatory site (ER4) by ChIP (Fig. 8B). Accordingly, when coexpressed with FoxA1 in MDA-MB-231 cells, ERα was recruited to ER4 but not enh2 (Fig. 8B). Consistent with these results, FoxA1 expression did not modify CCND1 enh2 DNaseI sensitivity, but importantly was able to induce that of ER4 (Fig. 8C). Thus, results on ER4 confirmed that FoxA1 recruitment was sufficient to induce chromatin remodeling and DNaseI sensitivity of a cell-specific regulatory region allowing ERα recruitment. The lack of FoxA1 binding to CCND1 enh2 may be due to inhibitory chromatin-bound factors or specific histone modifications, since FoxA1 was able to induce enh2 activity in MDA-MB-231 cells in the context of a nonchromatinized transiently transfected reporter construct (Fig. 8D). Together with previous reports, these results indicate that despite being necessary for ERα recruitment, FoxA1 expression alone is not sufficient to predict its own recruitment to chromatin, which is regulated by other factors that remain to be determined (Tan et al. 2001; Long and Spear 2004).
Figure 8.
Functional activities of FoxA1 ectopically expressed in MDA-MB-231 cells. (A) Western blot performed with extracts from MDA-MB-231 cells transfected with a FoxA1 expression vector or the empty control plasmid. (B) FoxA1 and ERα recruitment to CCND1 enh2 and ER4 in transfected MDA-MB-231 cells was analyzed by ChIP experiments. Results are means ± SE from two independent experiments and are expressed as fold chromatin enrichment over empty control plasmid transfected cells. (C) DNase I sensitivity assays were performed using nuclei from transfected MDA-MB-231 cells. Results are means ± SE from two independent experiments. (*) p < 0.05 versus cells transfected with the empty control vector. (D) MDAMB-231 cells were transfected with 25 ng of the pGL3-SV40 promoter construct (control) or pGL3-SV40 enh2 together with 1 ng of empty pcDNA3 (white bars) or pcDNA3-FoxA1 (black bars). Transcriptional activities were expressed relative to the activity obtained with control plasmids only, which was set to 100. Results are means ± SE from three independent experiments performed in triplicate.
Discussion
ERα is the master transcriptional regulator of the behavior of over two-thirds of human breast cancers. The ability of estrogen to stimulate the growth of these tumors depends on the ability of ERα to promote cell cycle progression through the induction of CCND1. However, the ERα-positive breast cancer phenotype is characterized by low growth rates and low S-phase fractions. In these studies we have defined the critical cis-regulatory elements and associated transcription factors that allow for both the estrogen induction of CCND1 and its tight regulation. We have defined two enhancers in the CCND1 gene removed from the previously described proximal promoter region (Altucci et al. 1996; Fukami-Kobayashi and Mitsui 1998; Sabbah et al. 1999; Eto 2000;
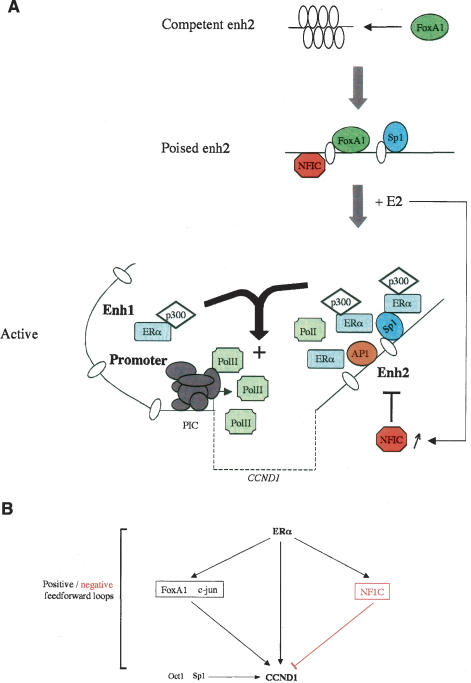
Castro-Rivera et al. 2001; Liu et al. 2002; Cicatiello et al. 2004). Specifically, we show that E2 induction of CCND1 expression in BCC depends on a cell-type-specific enhancer downstream from the CCND1 coding region (enh2) corresponding to the primary ERα recruitment site. Most interestingly, we find that E2 functions through a combinatorial network mediated by ERα acting directly at the CCND1 gene through a FoxA1-dependent interaction with enh2 and collaborating with other transcription factors to achieve controlled induction of CCND1 expression in BCC (Fig. 9A).
Figure 9.
Model of the combinatorial transcription factor network regulating CCND1 expression in ERα-positive BCC. (A) The recruitment pattern of the key transcription factors involved in E2 regulation of CCND1 expression is shown. Competent enh2 is recognized by FoxA1 that triggers chromatin remodeling, allowing recruitment of additional transcription factors including Sp1 that make enh2 sensitive to E2 induction (poised). Upon E2 stimulation, ERα is predominantly loaded at enh2 and mediates E2 transcriptional induction through communication with the upstream regulatory sites. NFIC is a repressor that enables E2 to balance its regulation of CCND1 transcription. The CCND1 transcribed sequence is represented as a dotted line, andnucleosomes are represented as ellipses. (B) Transcription factor network involved in estrogen modulation of CCND1 expression in human BCC. Arrows indicate transcriptional activations. NFIC that serves as a transcriptional repressor is displayed in red.
Enhancers represent DNA regions containing clusters of transcription factor-binding sites where recruitment of multiprotein complexes occurs to transmit signals to the basal transcription machinery (Ogata et al. 2003; Arnosti and Kulkarni 2005). Recruitment of transcription factors to regulatory sequences can be mediated by direct recognition of the DNA or indirectly through protein–protein interactions (Ogata et al. 2003; Arnosti and Kulkarni 2005). Using ChIP we determined the recruitment pattern of several cooperating transcription factors to the CCND1 enhancers and promoter region. Although numerous potential binding sites exist in the regulatory regions bound by the studied factors (Supplementary Fig. 1; Eto 2000), the exact in vivo recruitment mechanisms remain to be established. Moreover, the recruitment of transcriptional complexes at the CCND1 gene upon E2 stimulation may be modulated by both regulation of direct ERα-mediated genomic function as well as indirect regulation of the activity of collaborating factors through E2-induced nongenomic effects (Marino et al. 2002). Interestingly, no ERE or ERE half-site is present within the CCND1 gene regulatory regions (Supplementary Fig. 1; Sabbah et al. 1999). Indirect loading of ERα to transcriptional regulatory sites has already been documented and could represent an important pathway through which this factor acts (Hall et al. 2001). Interestingly, several transcription factors recruited to the CCND1 regulatory regions have been shown to physically interact with hormone-activated ERα and could therefore tether it to these regions: C/EBPβ (Chang et al. 2005), c-jun (Kushner et al. 2000), and Sp1 (Safe 2001; Kim et al. 2005). ERα ChIP experiments following Sp1 silencing revealed a decrease in ERα recruitment at enh2 (Supplementary Fig. 8). Thus, several factors including Sp1 collaborate to recruit ERα to the CCND1 regulatory sites (Fig. 9A).
Expression profiling analyses have revealed profound differences between ERα-positive and ERα-negative breast tumors (for review, see Lacroix et al. 2004). ERα expression is positively associated with that of several E2 target genes (TFF-1 or CCND1), including the transcription factors FoxA1 (Laganiere et al. 2005) and NFIC (this study). However, the functional connection that may exist between ERα and these factors remains largely unknown (Lacroix and Leclercq 2004). It has been reported recently that FoxA1 could be important for ERα recruitment to a subset of its target gene regulatory sequences (Carroll et al. 2005; Laganiere et al. 2005). Here, we describe a functional connection between these factors in induction of a critical E2 target in BCC, namely the oncogene CCND1. Consequently, FoxA1 is required for E2 subsequent induction of cell cycle progression and MCF7 cell growth. The role of FoxA1 is specifically associated with the activity of the CCND1 cell-specific enh2, which provides a link with the promoter to mediate E2 induction (Fig. 9A). Thus, this novel downstream enhancer establishes a functional connection between ERα and FoxA1 expression and CCND1 levels in breast cancer cells. Importantly, we provide a mechanistic explanation for FoxA1 role in ERα activity. We showed that FoxA1 expression in MCF7 cells controlled enh2 DNase I sensitivity, indicating that FoxA1 chromatin remodeling activities play a central role. The same results were obtained with several other ERα-binding sites recruiting FoxA1. Indeed, FoxA1 recruitment is able to induce remodeling of compacted chromatin structures both in vitro (Cirillo et al. 2002) and in vivo (Fig. 8). Reduced DNase I sensitivity upon FoxA1 silencing in MCF7 cells was accompanied by a decrease in transcriptional activator loading to enh2. Thus, FoxA1 is required for activity of a subset of the regulatory sequences used by ERα in BCC by defining a chromatin state that allows formation of an ERα-bound multiprotein transcriptional complex (Fig. 9A). Another important observation regarding our understanding of ERα activity in BCC is that at least some of these regulatory sites used by ERα, including CCND1 enh2, harbor a cell-type-specific activity. Hence, their lack of functionality may explain why ERα ectopic expression in various cell lines including MDA-MB-231 does not restore hormonal sensitivity to many ERα target genes, and in particular was found insufficient to mediate estrogen induction of CCND1 (Planas-Silva et al. 1999).
NFIC belongs to a family of transcription factors thought to play key roles in differentiation and development (Gronostajski 2000). In the mammary gland, NFIC is involved in the transcription of genes associated with lactation and involution (Murtagh et al. 2003). A few observations have also suggested a role for NFIC in cell growth control (Gronostajski 2000), including its over-expression in chick embryo fibroblasts, which prevents transformation by several oncogenes (Schuur et al. 1995). Here we show that NFIC represses MCF7 cell growth in two- and three-dimensional culture systems (Fig. 6; data not shown). Together with the fact that NFIC expression correlates with primary nonaggressive tumors in a number of human malignancies (Ramaswamy et al. 2001), these data emphasize NFIC as a potential tumor suppressor gene. The negative effect of NFIC on cell cycle progression is associated with a direct repression of CCND1 transcription. Indeed, NFIC is recruited to both the upstream and downstream CCND1 regulatory sites and dissociates upon E2 stimulation of MCF7 cells, suggesting that release of NFIC is part of the molecular mechanisms involved in E2 activation of CCND1 transcription. NFIC transcriptional repression activities could involve as yet poorly characterized repression domains within the protein (Gronostajski 2000). Interestingly, we found that NFIC expression is induced by E2 in MCF7 cells, revealing a potential negative feedforward loop (Fig. 9B). This mechanism is reminiscent of c-myc promotion of cell cycle progression through induction of E2F1 expression, where c-myc directly activates E2F1 but concomitantly induces specific microRNAs that negatively regulate E2F1 levels (O'Donnell et al. 2005). These findings suggest that even in some cancer cells negative growth regulatory loops may be intact and might be targets for therapeutic augmentation.
Materials and methods
Reagents and antibodies
E2 and DNase I (bovine, recombinant) were purchased from Sigma. The following antibodies were used: ERα (Ab-10) and CCND1 (Ab-3) from Neomarkers; AcH4 and β-actin from Cell Signaling; calnexin from Stressgen; FoxA1 (Ab5089) from Abcam; ERα (HC-20), PolII (H-224), p300 (C-20), Sp1 (PEP 2), Oct1 (C-21), TFIID (SI-1), c-jun (N) and FoxA1 (H-120) from Santa Cruz Biotechnology; and NFIC antisera 8199 and 2092 kindly provided by Dr N. Tanese (New York University, School of Medicine, New York).
Transfection assays
Cell culture and DNA constructs are described in the Supplemental Material. For transfection assays, cells were grown in 96-well plates and transfected using Lipofectamine2000 (Invitrogen) in OptiMEM for 5 h. Luciferase activities were assayed 16–24 h later using the Bright-Glo luciferase assay system (Promega). In experiments analyzing E2 stimulatory effects, MCF7 cells were grown in phenol red-free DMEM containing 1% char-coal/dextran-treated FBS (CDT-FBS) (Hyclone) and were stimulated for 5–6 h before harvesting. As recommended recently, use of an internal control was avoided and experiments were performed at least three times in triplicate and with independent plasmid preparations to ensure reproducibility of the results (Ishikawa et al. 2004).
ChIP assays
Cells were hormone-deprived by culture for 3–4 d in phenol red-free medium (Invitrogen) supplemented with 10% CDTFBS. Cells were challenged with hormone for 45 min and cross-linked using 1% formaldehyde. Cells were lyzed and sonicated three times for 10 sec each at 12% amplitude (Fisher Sonic desmembrator, model 500). Immunoprecipitation, reverse cross-linking, and DNA purification were performed as described in Carroll et al. (2005). Immunoprecipitated DNA was quantified by real-time PCR.
Real-time PCR
Real-time PCR was performed using Sybr Green PCR Master Mix (Applied Biosystems) and the primers listed in Supplementary Table 1. The presence of a single amplicon was systemically verified by dissociation curve analysis.
RNA silencing
Cells were transfected with siRNA at a final concentration of 75 nM using Lipofectamine2000. Forty-eight hours after transfection, cells were stimulated and harvested for analysis. Cells were hormone-deprived for 3 d at the moment of stimulation. siRNA oligonucleotide duplexes were purchased from Dharmacon. The targeted sequences for LUC and FoxA1 were described previously (Carroll et al. 2005). The SMARTpool siRNA was used to silence Sp1, NFIC, and Oct1.
Real-time RT–PCR
RNA isolation and real-time RT–PCR were performed as in Keeton and Brown (2005). Expression was normalized to the 40S ribosomal protein S28 (RPS28).
DNase I sensitivity assays
Nuclei were prepared by incubating cells for 5 min on ice in lysis buffer (10 nM Tris at pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.3% NP40). Nuclei were pelleted and washed in the same buffer without detergent. Cellular lysis was checked by visualization under a microscope. Two micrograms of DNA were then partially digested for 3 min at 37°C in a total volume of 200 μL with2Uof DNase I. The reaction was stopped by addition of 50 μL of 0.5 M EDTA. The DNA was purified using the DNeasy genomic DNA preparation kit (Qiagen) and analyzed by real-time PCR as described above. DNA amounts were normalized using PCR against a DNase-insensitive region corresponding to the rhodopsin locus (Dorschner et al. 2004). Amplicons were kept very small (51–119 bp), and we checked that in those conditions differences in amplicon size did not modulate significantly the DNase I sensitivity of the target sequences.
Western blotting
Western blot assays were performed as in Keeton and Brown (2005). Images were acquired using a FluorChem 5500 chemiluminescence imager, and bands were quantified by densitometry with the AlphaEase software (AlphaInnotec Corp.) according to the manufacturer's instructions.
Cell cycle distribution analysis and cell growth assays
MCF-7 cell cycle distribution was analyzed 20–24 h after stimulation with E2 or vehicle alone using propidium iodide staining and flow cytometry as in Keeton and Brown (2005). For cell viability assays, immediately after transfection with the various siRNAs MCF7 cells (6 × 103 cells per well) were seeded in a 96-well plate and stimulated 48 h later with E2 or vehicle alone. Relative cell number was determined before (time 0 used for normalization) and after stimulation (3 d) using the CellTiter-Glo luminescent assay (Promega).
Data analysis
Statistical analyses were performed using Prism software. Statistical significance was determined using Student's t-test comparison for unpaired data and was indicated as follows: (*) p < 0.05; (**) p < 0.01; (***) p < 0.001.
Acknowledgments
We thank Dr. N. Tanese (New York University School of Medicine) for the NFIC antisera. Laboratory members are acknowledged for helpful discussions. This work was supported by grants from the Claudia Adams Barr Program in Innovative Basic Cancer Research (to M.B.), the DF/HCC Breast Cancer SPORE Grant (to M.B.), and NIH grants P01 CA80111 (to M.B.) and R56 DK074967 (to M.B.), and by fellowships from the Fondation Recherche Medicale (to J.E.), the NIH (to T.R.G), and the DOD Breast Cancer Research Program (to J.S.C.).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1446006.
References
- Altucci, L., Addeo, R., Cicatiello, L., Dauvois, S., Parker, M.G., Truss, M., Beato, M., Sica, V., Bresciani, F., Weisz, A. 17β-Estradiol induces cyclin D1 gene transcription, p36D1–p34cdk4 complex activation and p105Rb phosphorylation during mitogenic stimulation of G(1)-arrested human breast cancer cells. Oncogene. 1996;12:2315–2324. [PubMed] [Google Scholar]
- Arnold, A., Papanikolaou, A. Cyclin D1 in breast cancer pathogenesis. J. Clin. Oncol. 2005;23:4215–4224. doi: 10.1200/JCO.2005.05.064. [DOI] [PubMed] [Google Scholar]
- Arnosti, D.N., Kulkarni, M.M. Transcriptional enhancers: Intelligent enhanceosomes or flexible billboards? J. Cell. Biochem. 2005;94:890–898. doi: 10.1002/jcb.20352. [DOI] [PubMed] [Google Scholar]
- Carroll, J.S., Liu, X.S., Brodsky, A.S., Li, W., Meyer, C.A., Szary, A.J., Eeckhoute, J., Shao, W., Hestermann, E.V., Geistlinger, T.R., et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Castro-Rivera, E., Samudio, I., Safe, S. Estrogen regulation of cyclin D1 gene expression in ZR-75 breast cancer cells involves multiple enhancer elements. J. Biol. Chem. 2001;276:30853–30861. doi: 10.1074/jbc.M103339200. [DOI] [PubMed] [Google Scholar]
- Chang, W., Parra, M., Centrella, M., McCarthy, T.L. Interactions between CCAAT enhancer binding protein δ and estrogen receptor α control insulin-like growth factor I (igf1) and estrogen receptor-dependent gene expression in osteoblasts. Gene. 2005;345:225–235. doi: 10.1016/j.gene.2004.11.017. [DOI] [PubMed] [Google Scholar]
- Cicatiello, L., Addeo, R., Sasso, A., Altucci, L., Petrizzi, V.B., Borgo, R., Cancemi, M., Caporali, S., Caristi, S., Scafoglio, C., et al. Estrogens and progesterone promote persistent CCND1 gene activation during G1 by inducing transcriptional derepression via c-Jun/c-Fos/estrogen receptor (progesterone receptor) complex assembly to a distal regulatory element and recruitment of cyclin D1 to its own gene promoter. Mol. Cell. Biol. 2004;24:7260–7274. doi: 10.1128/MCB.24.16.7260-7274.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo, L.A., Lin, F.R., Cuesta, I., Friedman, D., Jarnik, M., Zaret, K.S. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol. Cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- Crowe, A.J., Sang, L., Li, K.K., Lee, K.C., Spear, B.T., Barton, M.C. Hepatocyte nuclear factor 3 relieves chromatin-mediated repression of the α-fetoprotein gene. J. Biol. Chem. 1999;274:25113–25120. doi: 10.1074/jbc.274.35.25113. [DOI] [PubMed] [Google Scholar]
- Dorschner, M.O., Hawrylycz, M., Humbert, R., Wallace, J.C., Shafer, A., Kawamoto, J., Mack, J., Hall, R., Goldy, J., Sabo, P.J., et al. High-throughput localization of functional elements by quantitative chromatin profiling. Nat. Methods. 2004;1:219–225. doi: 10.1038/nmeth721. [DOI] [PubMed] [Google Scholar]
- Dupont, J., Karas, M., LeRoith, D. The potentiation of estrogen on insulin-like growth factor I action in MCF-7 human breast cancer cells includes cell cycle components. J. Biol. Chem. 2000;275:35893–35901. doi: 10.1074/jbc.M006741200. [DOI] [PubMed] [Google Scholar]
- Eto, I. Molecular cloning and sequence analysis of the promoter region of mouse cyclin D1 gene: Implication in phorbol ester-induced tumor promotion. Cell Prolif. 2000;33:167–187. doi: 10.1046/j.1365-2184.2000.00176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewen, M.E., Lamb, J. The activities of cyclin D1 that drive tumorigenesis. Trends Mol. Med. 2004;10:158–162. doi: 10.1016/j.molmed.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Fantl, V., Stamp, G., Andrews, A., Rosewell, I., Dickson, C. Mice lacking cyclin D1 are small and show defects in eye and mammary gland development. Genes & Dev. 1995;9:2364–2372. doi: 10.1101/gad.9.19.2364. [DOI] [PubMed] [Google Scholar]
- Fukami-Kobayashi, J., Mitsui, Y. The regulation of cyclin D1 expression in senescent human fibroblasts. Exp. Cell Res. 1998;241:435–444. doi: 10.1006/excr.1998.4079. [DOI] [PubMed] [Google Scholar]
- Grillo, M., Bott, M.J., Khandke, N., McGinnis, J.P., Miranda, M., Meyyappan, M., Rosfjord, E.C., Rabindran, S.K. Validation of cyclin D1/CDK4 as an anticancer drug target in MCF-7 breast cancer cells: Effect of regulated overexpression of cyclin D1 and siRNA-mediated inhibition of endogenous cyclin D1 and CDK4 expression. Breast Cancer Res. Treat. 2006;95:185–194. doi: 10.1007/s10549-005-9066-y. [DOI] [PubMed] [Google Scholar]
- Gronostajski, R.M. Roles of the NFI/CTF gene family in transcription and development. Gene. 2000;249:31–45. doi: 10.1016/s0378-1119(00)00140-2. [DOI] [PubMed] [Google Scholar]
- Gross, D.S., Garrard, W.T. Nuclease hypersensitive sites in chromatin. Annu. Rev. Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- Hall, J.M., Couse, J.F., Korach, K.S. The multifaceted mechanisms of estradiol and estrogen receptor signaling. J. Biol. Chem. 2001;276:36869–36872. doi: 10.1074/jbc.R100029200. [DOI] [PubMed] [Google Scholar]
- Hodges, L.C., Cook, J.D., Lobenhofer, E.K., Li, L., Bennett, L., Bushel, P.R., Aldaz, C.M., Afshari, C.A., Walker, C.L. Tamoxifen functions as a molecular agonist inducing cell cycle-associated genes in breast cancer cells. Mol. Cancer Res. 2003;1:300–311. [PubMed] [Google Scholar]
- Holmqvist, P.H., Belikov, S., Zaret, K.S., Wrange, O. FoxA1 binding to the MMTV LTR modulates chromatin structure and transcription. Exp. Cell Res. 2005;304:593–603. doi: 10.1016/j.yexcr.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Ishikawa, T., Lee, E.J., Jameson, J.L. Nonhomologous end-joining ligation transfers DNA regulatory elements between cointroduced plasmids. Mol. Cell. Biol. 2004;24:8323–8331. doi: 10.1128/MCB.24.19.8323-8331.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeton, E.K., Brown, M. Cell cycle progression stimulated by tamoxifen-bound estrogen receptor-α and promoter-specific effects in breast cancer cells deficient in N-CoR and SMRT. Mol. Endocrinol. 2005;19:1543–1554. doi: 10.1210/me.2004-0395. [DOI] [PubMed] [Google Scholar]
- Kim, K., Barhoumi, R., Burghardt, R., Safe, S. Analysis of estrogen receptor α–Sp1 interactions in breast cancer cells by fluorescence resonance energy transfer. Mol. Endocrinol. 2005;19:843–854. doi: 10.1210/me.2004-0326. [DOI] [PubMed] [Google Scholar]
- Kushner, P.J., Agard, D.A., Greene, G.L., Scanlan, T.S., Shiau, A.K., Uht, R.M., Webb, P. Estrogen receptor pathways to AP-1. J. Steroid Biochem. Mol. Biol. 2000;74:311–317. doi: 10.1016/s0960-0760(00)00108-4. [DOI] [PubMed] [Google Scholar]
- Lacroix, M., Leclercq, G. About GATA3, HNF3A, and XBP1, three genes co-expressed with the oestrogen receptor-α gene (ESR1) in breast cancer. Mol. Cell. Endocrinol. 2004;219:1–7. doi: 10.1016/j.mce.2004.02.021. [DOI] [PubMed] [Google Scholar]
- Lacroix, M., Toillon, R.A., Leclercq, G. Stable ‘portrait’ of breast tumors during progression: Data from biology, pathology and genetics. Endocr. Relat. Cancer. 2004;11:497–522. doi: 10.1677/erc.1.00758. [DOI] [PubMed] [Google Scholar]
- Laganiere, J., Deblois, G., Lefebvre, C., Bataille, A.R., Robert, F., Giguere, V. Location analysis of estrogen receptor α target promoters reveals that FOXA1 defines a domain of the estrogen response. Proc. Natl. Acad. Sci. 2005;102:11651–11656. doi: 10.1073/pnas.0505575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, M.M., Albanese, C., Anderson, C.M., Hilty, K., Webb, P., Uht, R.M., Price, R.H., Jr., Pestell, R.G., Kushner, P.J. Opposing action of estrogen receptors α and β on cyclin D1 gene expression. J. Biol. Chem. 2002;277:24353–24360. doi: 10.1074/jbc.M201829200. [DOI] [PubMed] [Google Scholar]
- Long, L., Spear, B.T. FoxA proteins regulate H19 endoderm enhancer E1 and exhibit developmental changes in enhancer binding in vivo. Mol. Cell. Biol. 2004;24:9601–9609. doi: 10.1128/MCB.24.21.9601-9609.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie, M.C., Yang, H.Q., Ma, A.H., Xu, W., Zou, J.X., Kung, H.J., Chen, H.W. Androgen-induced recruitment of RNA polymerase II to a nuclear receptor–p160 coactivator complex. Proc. Natl. Acad. Sci. 2003;100:2226–2230. doi: 10.1073/pnas.0437824100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino, M., Acconcia, F., Bresciani, F., Weisz, A., Trentalance, A. Distinct nongenomic signal transduction pathways controlled by 17β-estradiol regulate DNA synthesis and cyclin D(1) gene transcription in HepG2 cells. Mol. Biol. Cell. 2002;13:3720–3729. doi: 10.1091/mbc.E02-03-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur, M., Gerum, S., Stamatoyannopoulos, G. Quantification of DNaseI-sensitivity by real-time PCR: Quantitative analysis of DNaseI-hypersensitivity of the mouse β-globin LCR. J. Mol. Biol. 2001;313:27–34. doi: 10.1006/jmbi.2001.4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metivier, R., Penot, G., Hubner, M.R., Reid, G., Brand, H., Kos, M., Gannon, F. Estrogen receptor-α directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- Metivier, R., Penot, G., Carmouche, R.P., Hubner, M.R., Reid, G., Denger, S., Manu, D., Brand, H., Kos, M., Benes, V., et al. Transcriptional complexes engaged by apo-estrogen receptor-α isoforms have divergent outcomes. EMBO J. 2004;23:3653–3666. doi: 10.1038/sj.emboj.7600377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtagh, J., Martin, F., Gronostajski, R.M. The Nuclear Factor I (NFI) gene family in mammary gland development and function. J. Mammary Gland Biol. Neoplasia. 2003;8:241–254. doi: 10.1023/a:1025909109843. [DOI] [PubMed] [Google Scholar]
- Musgrove, E.A., Hamilton, J.A., Lee, C.S., Sweeney, K.J., Watts, C.K., Sutherland, R.L. Growth factor, steroid, and steroid antagonist regulation of cyclin gene expression associated with changes in T-47D human breast cancer cell cycle progression. Mol. Cell. Biol. 1993;13:3577–3587. doi: 10.1128/mcb.13.6.3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musgrove, E.A., Lee, C.S., Buckley, M.F., Sutherland, R.L. Cyclin D1 induction in breast cancer cells shortens G1 and is sufficient for cells arrested in G1 to complete the cell cycle. Proc. Natl. Acad. Sci. 1994;91:8022–8026. doi: 10.1073/pnas.91.17.8022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell, K.A., Wentzel, E.A., Zeller, K.I., Dang, C.V., Mendell, J.T. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- Ogata, K., Sato, K., Tahirov, T.H. Eukaryotic transcriptional regulatory complexes: Cooperativity from near and afar. Curr. Opin. Struct. Biol. 2003;13:40–48. doi: 10.1016/s0959-440x(03)00012-5. [DOI] [PubMed] [Google Scholar]
- Park, K.J., Krishnan, V., O'Malley, B.W., Yamamoto, Y., Gaynor, R.B. Formation of an IKKα-dependent transcription complex is required for estrogen receptor-mediated gene activation. Mol. Cell. 2005;18:71–82. doi: 10.1016/j.molcel.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Planas-Silva, M.D., Donaher, J.L., Weinberg, R.A. Functional activity of ectopically expressed estrogen receptor is not sufficient for estrogen-mediated cyclin D1 expression. Cancer Res. 1999;59:4788–4792. [PubMed] [Google Scholar]
- Prall, O.W., Rogan, E.M., Musgrove, E.A., Watts, C.K., Sutherland, R.L. c-Myc or cyclin D1 mimics estrogen effects on cyclin E–Cdk2 activation and cell cycle reentry. Mol. Cell. Biol. 1998;18:4499–4508. doi: 10.1128/mcb.18.8.4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quelle, D.E., Ashmun, R.A., Shurtleff, S.A., Kato, J.Y., Bar-Sagi, D., Roussel, M.F., Sherr, C.J. Overexpression of mouse D-type cyclins accelerates G1 phase in rodent fibro-blasts. Genes & Dev. 1993;7:1559–1571. doi: 10.1101/gad.7.8.1559. [DOI] [PubMed] [Google Scholar]
- Ramaswamy, S., Tamayo, P., Rifkin, R., Mukherjee, S., Yeang, C.H., Angelo, M., Ladd, C., Reich, M., Latulippe, E., Mesirov, J.P., et al. Multiclass cancer diagnosis using tumor gene expression signatures. Proc. Natl. Acad. Sci. 2001;98:15149–15154. doi: 10.1073/pnas.211566398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio, M.F., Werbajh, S., Cafferata, E.G., Quaglino, A., Colo, G.P., Nojek, I.M., Kordon, E.C., Nahmod, V.E., Costas, M.A. TNF-α enhances estrogen-induced cell proliferation of estrogen-dependent breast tumor cells through a complex containing nuclear factor-κ B. Oncogene. 2006;25:1367–1377. doi: 10.1038/sj.onc.1209176. [DOI] [PubMed] [Google Scholar]
- Sabbah, M., Courilleau, D., Mester, J., Redeuilh, G. Estrogen induction of the cyclin D1 promoter: Involvement of a cAMP response-like element. Proc. Natl. Acad. Sci. 1999;96:11217–11222. doi: 10.1073/pnas.96.20.11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safe, S. Transcriptional activation of genes by 17 β-estradiol through estrogen receptor–Sp1 interactions. Vitam. Horm. 2001;62:231–252. doi: 10.1016/s0083-6729(01)62006-5. [DOI] [PubMed] [Google Scholar]
- Schuur, E.R., Kruse, U., Iacovoni, J.S., Vogt, P.K. Nuclear factor I interferes with transformation induced by nuclear oncogenes. Cell Growth Differ. 1995;6:219–227. [PubMed] [Google Scholar]
- Shang, Y., Hu, X., DiRenzo, J., Lazar, M.A., Brown, M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- Sicinski, P., Donaher, J.L., Parker, S.B., Li, T., Fazeli, A., Gardner, H., Haslam, S.Z., Bronson, R.T., Elledge, S.J., Weinberg, R.A. Cyclin D1 provides a link between development and oncogenesis in the retina and breast. Cell. 1995;82:621–630. doi: 10.1016/0092-8674(95)90034-9. [DOI] [PubMed] [Google Scholar]
- Stoner, M., Wormke, M., Saville, B., Samudio, I., Qin, C., Abdelrahim, M., Safe, S. Estrogen regulation of vascular endothelial growth factor gene expression in ZR-75 breast cancer cells through interaction of estrogen receptor α and SP proteins. Oncogene. 2004;23:1052–1063. doi: 10.1038/sj.onc.1207201. [DOI] [PubMed] [Google Scholar]
- Su, A.I., Cooke, M.P., Ching, K.A., Hakak, Y., Walker, J.R., Wiltshire, T., Orth, A.P., Vega, R.G., Sapinoso, L.M., Moqrich, A., et al. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl. Acad. Sci. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland, R.L., Prall, O.W., Watts, C.K., Musgrove, E.A. Estrogen and progestin regulation of cell cycle progression. J. Mammary Gland Biol. Neoplasia. 1998;3:63–72. doi: 10.1023/a:1018774302092. [DOI] [PubMed] [Google Scholar]
- Tan, Y., Costa, R.H., Kovesdi, I., Reichel, R.R. Adenovirus-mediated increase of HNF-3 levels stimulates expression of transthyretin and sonic hedgehog, which is associated with F9 cell differentiation toward the visceral endoderm lineage. Gene Expr. 2001;9:237–248. doi: 10.3727/000000001783992542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Vijver, M.J., He, Y.D., van't Veer, L.J., Dai, H., Hart, A.A., Voskuil, D.W., Schreiber, G.J., Peterse, J.L., Roberts, C., Marton, M.J., et al. A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- Wang, T.C., Cardiff, R.D., Zukerberg, L., Lees, E., Arnold, A., Schmidt, E.V. Mammary hyperplasia and carcinoma in MMTV–cyclin D1 transgenic mice. Nature. 1994;369:669–671. doi: 10.1038/369669a0. [DOI] [PubMed] [Google Scholar]
- Wilcken, N.R., Prall, O.W., Musgrove, E.A., Sutherland, R.L. Inducible overexpression of cyclin D1 in breast cancer cells reverses the growth-inhibitory effects of antiestrogens. Clin. Cancer Res. 1997;3:849–854. [PubMed] [Google Scholar]
- Williamson, E.A., Wolf, I., O'Kelly, J., Bose, S., Tanosaki, S., Koeffler, H.P. BRCA1 and FOXA1 proteins coregulate the expression of the cell cycle-dependent kinase inhibitor p27(Kip1) Oncogene. 2005;25:1391–1399. doi: 10.1038/sj.onc.1209170. [DOI] [PubMed] [Google Scholar]
- Xie, W., Duan, R., Safe, S. Estrogen induces adenosine deaminase gene expression in MCF-7 human breast cancer cells: Role of estrogen receptor–Sp1 interactions. Endocrinology. 1999;140:219–227. doi: 10.1210/endo.140.1.6394. [DOI] [PubMed] [Google Scholar]
- Yu, Q., Geng, Y., Sicinski, P. Specific protection against breast cancers by cyclin D1 ablation. Nature. 2001;411:1017–1021. doi: 10.1038/35082500. [DOI] [PubMed] [Google Scholar]