DNA strand exchange is the molecular process that defines homologous recombination (Roca and Cox 1990; Eggleston and Kowalczykowski 1991; Kowalczykowski 1991; Folta-Stogniew et al. 2004). Members of the RecA family of recombinases promote DNA strand exchange by first assembling helical filaments on single-strand DNA (ssDNA). These filaments carry out a homology search of duplex DNAs. Recognition of homology is associated with DNA strand exchange, the process through which nucleotides on the incoming ssDNA strand form base pairs with nucleotides of the complementary strand of the target duplex.
Homologous recombination events involve formation and repair of DNA double-strand breaks (DSBs). In somatic cells, DSBs form spontaneously, while in meiotic cells, formation of these breaks is programmed (Paques and Haber 1999; Symington 2002). SPO11, a meiosis-specific transesterase, is required for formation of programmed meiotic breaks (Keeney et al. 1997). After breaks form, nucleases process DNA ends to yield 3′ ssDNA tails, which are the substrate for recombinase assembly into presynaptic filaments (Cox and Lehman 1982; Sung 1994; Sugawara et al. 2003; Wolner et al. 2003; Fukuda and Ohya 2006). DNA strand exchange then yields D-loops with a hybrid segment of DNA terminating with the 3′ end of the invading strand. Such ends serve as primers for the DNA repair synthesis needed to complete the recombination event (Paques and Haber 1999; Symington 2002).
Eukaryotes encode two major RecA-like recombinases, Rad51 and Dmc1 (Aboussekhra et al. 1992; Bishop et al. 1992; Shinohara et al. 1992). Rad51 is required for recombinational repair of DSBs in somatic cells. Dmc1 is a meiosis-specific recombinase found in a variety of organisms including budding and fission yeast, plants, and mammals (Villeneuve and Hillers 2001). In budding yeast, both Dmc1 and Rad51 play important roles in meiotic DNA strand exchange. The two recombinases have partially redundant and distinct functions; efficient, properly regulated meiotic recombination requires both proteins (Rockmill et al. 1995; Schwacha and Kleckner 1997; Shinohara et al. 1997a).
Rad51 and Dmc1 share several properties with bacterial RecA, including the abilities to form helical presynaptic filaments and to promote strand exchange in purified systems (Eggleston and Kowalczykowski 1991; Sehorn and Sung 2004). All RecA-like recombinases are DNA-dependent ATPases, and all require ATP as a cofactor during DNA strand exchange; however, hydrolysis of ATP is not essential for homology recognition and DNA strand exchange (Menetski et al. 1990; Sung and Stratton 1996; Campbell and Davis 1999a; Masson et al. 1999; Hong et al. 2001; Bugreev et al. 2005). The ability of recombinases to function without ATP hydrolysis led to two different explanations for the function of energy in RecA-mediated reactions. In one model, RecA helical filaments are rotary motors that use ATP hydrolysis to drive unidirectional branch migration to extend heteroduplexes even across regions of imperfect homology (Cox 2003; Cox et al. 2005). A second model is based largely on the finding that ATP hydrolysis converts a form of recombinase with high affinity for DNA (RecAATP) to a form of recombinase with low affinity (RecAADP). Thus, ATP hydrolysis promotes the dynamics of RecA–DNA interactions, allowing release of recombination products after strand exchange and/or making it possible to correct discontinuities that form during assembly of presynaptic helical filaments on ssDNA (Menetski et al. 1988; Kowalczykowski 1991).
Although the core DNA strand exchange reaction is likely to proceed by a mechanism conserved since the divergence of prokaryotes and eukaryotes, certain biochemical properties are significantly different (Eggleston and Kowalczykowski 1991). Three biochemical differences are particularly relevant to the present study. First, the ATPase activity of the eukaryotic recombinases is ∼80- to 200-fold weaker than that of RecA (discussed further below). Second, the eukaryotic recombinases show relatively little binding preference for ssDNA relative to double-strand DNA (dsDNA) (Ogawa et al. 1993; Sung 1994; Li et al. 1997; Hong et al. 2001). Third, although all recombinases use factors that promote their assembly on tracts of ssDNA (Kowalczykowski 2000; Gasior et al. 2001; Symington 2002; Hayase et al. 2004), optimal activity of the eukaryotic recombinases also requires Rad54 and Tid1/Rdh54, proteins that do not appear to have counterparts in prokaryotes (Petukhova et al. 1998, 2000).
Rad54 and Tid1 are members of the Swi2/Snf2 family of helicase-like proteins (Eisen et al. 1995; Klein 1997; Shinohara et al. 1997b). Members of this family are potent DNA-dependent ATPases that translocate along DNA in an ATP hydrolysis-dependent manner. Swi2/Snf2 and other members of the family have been shown to be capable of remodeling chromatin, thereby regulating gene expression at the transcriptional level (Eisen et al. 1995). Rad54 and Tid1 act in vivo to enhance the efficiency of Rad51- and Dmc1-dependent DSB repair in mitosis and meiosis (Klein 1997; Shinohara et al. 1997b). As mentioned above, Rad54 and Tid1 also stimulate DNA-strand exchange activity in purified systems (Petukhova et al. 1998, 1999, 2000). The mechanism underlying this stimulation is still the subject of study.
Several activities have been described for Rad54 and Tid1 that may contribute to its ability to stimulate DNA strand exchange in vivo. Both Rad54 and Tid1 locally unwind naked duplex DNA in an ATP hydrolysis-dependent manner (Petukhova et al. 1999, 2000), an activity that may enhance the efficiency of the homology search. Like other members of the Swi2/Snf2 family, Rad54 can displace nucleosomes, an activity that could also contribute to homology searching in vivo (Alexeev et al. 2003; Jaskelioff et al. 2003). Finally, physical assays of recombination intermediates in living cells suggest that Rad54 plays a post-synaptic role during Rad51-mediated recombination (Sugawara et al. 2003). The ability of Rad54 to promote dissociation of Rad51 from duplex DNA in vitro could account for such a late role (Solinger et al. 2002; Wesoly et al. 2006). Dissociation of Rad51 from duplex requires the ATPase activity of Rad54, indicating that this activity involves Rad54 translocation on dsDNA. Recent single-molecule studies show directly that Rad54 is a DNA translocase (Amitani et al. 2006). The biochemical properties of Tid1 have not been as extensively characterized as those of Rad54; however, the partial redundancy of the proteins in vivo and their similar activities in DNA strand exchange and in DNA unwinding reactions indicate that the two could share additional biochemical properties (Klein 1997; Shinohara et al. 1997b, 2000).
Tid1 and Rad54 have been shown to play a role in promoting the relative distribution of the two recombinases, Rad51 and Dmc1, during meiotic recombination. In wild-type cells, Rad51 and Dmc1 form DSB-dependent subnuclear foci (Bishop 1994). About 80% of Dmc1 foci localize at, or immediately adjacent to, Rad51 foci. The Swi2/Snf2 like recombination proteins control recombinase colocalization; Dmc1 and Rad51 colocalization is dramatically reduced in tid1Δ single mutants and eliminated in tid1Δ rad54Δ double mutants (Shinohara et al. 2000). These observations led to the proposal that Tid1 and Rad54 share an activity that promotes coordinated assembly of the recombinases at sites of DSBs.
In the present study we demonstrate that Tid1 functions to prevent the accumulation of Dmc1 on chromatin in the absence of DSBs. This function depends on the ATPase activity of Tid1. Chromatin immunoprecipitation (ChIP) experiments indicate that Rad54 and Tid1 share an activity required for Dmc1 to load at sites of DSBs, and that in the absence of this shared activity, Dmc1 is sequestered on duplex chromatin. These results suggest that Tid1 and Rad54 indirectly coordinate assembly of the recombinases at DSBs by promoting dissociation of recombinase from non-DSB sites.
On the basis of these observations, we propose that a major function of the energy expended during Dmc1-mediated recombination is to maintain a pool of unbound protein available for targeted assembly on ssDNA at sites of DSBs. The need for this energy expenditure is proposed to reflect a mechanism of DNA strand exchange, in which free energy is released as the ssDNA filament acquires a complementary DNA strand from the target duplex DNA.
Results
Formation of DSB-independent Dmc1 foci is enhanced in the absence of Tid1
The frequency of colocalization of Rad51 and Dmc1 foci is reduced in tid1Δ single and tid1Δ rad54Δ double mutants (Shinohara et al. 2000). These observations raised the possibility that Tid1 and Rad54 might be acting to restrict Dmc1 and/or Rad51 foci to sites of DSBs. If Tid1 acts to restrict recombinase assembly to DSBs, the reduction in Dmc1–Rad51 colocalization in the tid1Δ mutant could reflect an increased tendency for recombinase assembly at nonrecombinogenic sites on chromatin. To test this possibility, we assayed the effect of a tid1Δ mutation on Dmc1 focus formation in a spo11Δ mutant that does not make DSBs. Nuclear spreads were prepared from meiotic cultures of spo11Δ and spo11Δ tid1Δ cells at 4 h (the time at which the percentage of nuclei containing Dmc1 foci reaches a peak in wild type) and stained with anti-Dmc1 antibodies. Spreads were also prepared from wild-type, tid1Δ, and dmc1Δ cells for comparison.
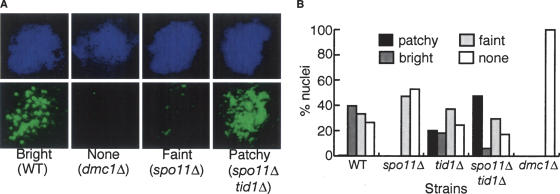
Stained nuclei were categorized as displaying one of four qualitatively different staining patterns as follows: “Bright”: those with >20 brightly staining foci of relatively uniform staining intensity. This pattern is typical of wild-type meiotic nuclei. “None”: those with a level of staining indistinguishable from that seen in dmc1Δ mutant controls. “Faint”: nuclei with only faintly staining foci. “Patchy”: those characterized by >20 brightly staining foci but with a wide range of staining intensities including at least one abnormally bright staining structure. Fifty random nuclei were scored for each strain (Fig. 1). In contrast to previous reports in which no Dmc1 staining was detected in a spo11Δ mutant (Bishop 1994; Shinohara et al. 1997a), a faint focal staining pattern was detected in 40% of spo11Δ nuclei using a more recently raised polyclonal anti-Dmc1 antibody (Fig. 1A,B). This result indicates that Dmc1 focus formation is not strictly dependent on DSBs. The spo11Δ tid1Δ mutant showed a strikingly different staining pattern as compared with the spo11Δ single mutant: Nearly 60% of spo11Δ tid1Δ double-mutant nuclei were scored as either bright or patchy, indicating a much higher level of focus formation than the spo11Δ single mutant.
Figure 1.
TID1 blocks Dmc1 focus formation in the absence of DSBs. (A) Examples of typical representatives of Dmc1 staining patterns. (Left) Wild -type nucleus displaying brightly staining Dmc1 foci. (Second from left) Background staining with anti-Dmc1 as observed in a dmc1Δ nucleus. (Second from right) spo11Δ nucleus displaying “faint” Dmc1 staining. (Right) spo11Δ tid1Δ nucleus showing the type of Dmc1 staining pattern scored as “patchy.” Note the large number of faintly staining foci combined with a predominant staining structure. (B) Fifty unselected nuclei from wild-type, tid1Δ, spo11Δ, spo11Δ tid1Δ, and dmc1Δ strains were scored with respect to having a Dmc1 staining pattern that was “bright,” “faint,” “patchy,” or “none” as described in the text.
Spo11-independent Dmc1 foci do not result from Dmc1 overexpression or DNA damage
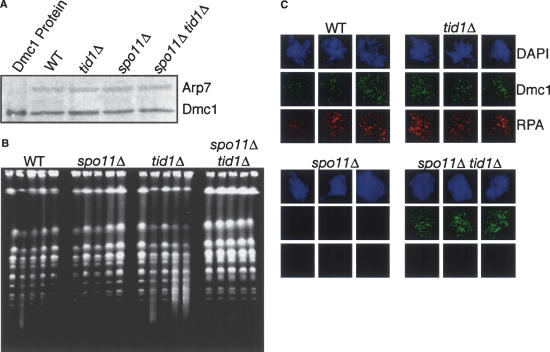
The difference between the spo11Δ and spo11Δ tid1Δ mutant phenotypes suggests that Tid1 directly opposes assembly of Dmc1 at undamaged sites on chromatin. Experiments were carried out to eliminate alternative interpretations. We considered the possibility that the high frequency of Dmc1 foci in spo11Δ tid1Δ double mutants compared with spo11Δ single mutants resulted from higher steady-state levels of Dmc1 protein, but no difference in Dmc1 levels was detected by Western blot analysis of the relevant cells (Fig. 2A). We next considered the possibility that the absence of Tid1 might result in the accumulation of Spo11-independent DSBs, perhaps as a result of errors during premeiotic replication. To test this, we examined the integrity of chromosomes by contour-clamped homogenous electric field (CHEF) gel electrophoresis (Fig. 2B). As reported previously (Zenvirth et al. 1992), formation and resection of meiotic DSBs results in a loss of chromosome integrity, indicated by smearing of the chromosome bands. Smearing disappears as DSBs are repaired during meiosis (Fig. 2B, cf. wild-type, 3.5 h vs. 8 h). Smearing is not observed in a spo11Δ mutant as expected for a mutant that does not form DSBs. Smearing is more pronounced in a tid1Δ single mutant than wild type, which is expected because repair of DSBs is slow and incomplete in the absence of Tid1 (Shinohara et al. 1997b). The spo11Δ tid1Δ double mutant was indistinguishable from the spo11Δ single mutant and showed no loss of chromosome integrity during meiosis. This analysis indicates that if DSBs form in spo11Δ tid1Δ mutants, their number is much less than that in wild type. This is in contrast to the number of DSB-independent Dmc1 foci in spo11Δ tid1Δ double mutant, which is similar to or greater than in wild type.
Figure 2.
Dmc1 foci in spo11Δ tid1Δ do not result from elevated Dmc1 levels, SPO11-independent chromosome breaks, or SPO11-independent ssDNA tracts. (A) Western blot analysis of Dmc1 protein levels from wild-type, tid1Δ, spo11Δ, and spo11Δ tid1Δ compared with Arp7 loading control. (B) Analysis of the integrity of meiotic chromosomes during a meiotic time course from wild-type, tid1Δ, spo11Δ, and spo11Δ tid1Δ strains by CHEF gel electrophoresis (per strain from left to right: 0, 3.5, 4, 6, and 8 h in meiosis). Note that in wild type, formation of DSBs at 3.5 h is evident, followed by restoration of chromatin integrity at 8 h. spo11Δ and spo11Δ tid1Δ mutants do not show loss of chromosome integrity. (C) Three representative spread nuclei prepared from cells collected 4 h after transfer to sporulation medium. The relevant genotype is indicated. Dmc1 foci are shown in green, and RPA foci are shown in red.
Finally, we considered the possibility that the absence of Tid1 function resulted in the accumulation of a Spo11-independent lesion associated with tracts of ssDNA that could not be detected by CHEF gel (e.g., ssDNA gaps). To test this possibility, we stained spread nuclei with antibodies against RPA, the ssDNA-specific binding protein. All wild-type nuclei containing Dmc1 foci also contained multiple RPA foci as predicted by previous studies (Bishop 1994; Gasior et al. 1998). In stark contrast to wild type, spread nuclei from the spo11Δ tid1Δ double-mutant nuclei containing Dmc1 foci contained very few, if any, RPA foci (Fig. 2C). This observation is not consistent with the possibility that the Dmc1 foci observed in the spo11Δ tid1Δ double mutant form on tracts of ssDNA, but it is consistent with the possibility that Dmc1 accumulates on chromatin in a DSB- and ssDNA-independent manner when Tid1 is absent.
Dmc1 associates with recombination hotspots in a DSB-dependent manner
Given that Dmc1 forms only a few faint foci in a spo11Δ single mutant, it seemed likely that the intense staining patterns seen in spo11Δ tid1Δ double mutants resulted from the association of Dmc1 at random loci rather than from the specific assembly of Dmc1 at sites that normally give rise to recombination intermediates. In order to distinguish between these possibilities, we used a ChIP assay capable of detecting specific association of Dmc1 at recombination sites. This assay detects the relative amount of DNA precipitated by anti-Dmc1 antibodies from chromosomal regions that are recombination hotspots versus recombination coldspots. Specific loci examined for Dmc1 association include the particularly strong HIS4::LEU2 recombination hotspot (Symington 2002) and CIT2, a well-characterized coldspot (Baudat and Nicolas 1997). Quantitation of PCR-amplified fragments from the anti-Dmc1 precipitates showed a three- to fourfold excess of DNA from the HIS4 ::LEU2 recombination hotspot as compared with that from the CIT2 recombination coldspot (hotspot:coldspot ratio = 3.5 ± 0.2; Fig. 3A). Similar results were obtained when fragments from two other pairs of hotspots (ARG4, HIS2) and coldspots (SUL1, STF2) were examined (data not shown). The preferential association of Dmc1 with recombination sites depends on DSB formation, as equal amounts of hotspot and coldspot DNA are precipitated by anti-Dmc1 antibodies in spo11Δ and spo11Δ tid1Δ mutants (hotspot:coldspot ratio = 1.1 ± 0.1 and 1.1 ± 0.2, respectively; Fig. 3A). We did not detect any signal in this assay using primers for hot or coldspot regions when immunoprecipitation was carried out using extracts from dmc1Δ mutants. Therefore, ChIP coldspot signals detected in DMC1+ strains reflect significant Dmc1– coldspot association. Nonetheless, preferential association of Dmc1 with hotspots is readily and reproducibly detected in wild-type strains. The difference in hotspot/ coldspot ratios in wild type and spo11Δ tid1Δ provides biochemical evidence that Dmc1 does not preferentially associate with hotspot DNA in the absence of DSBs and supports the view that the Dmc1 foci observed in spo11Δ tid1Δ mutants do not mark recombination sites.
Figure 3.
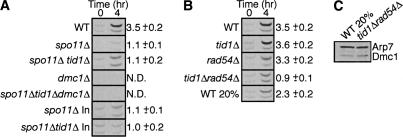
Dmc1 ChIP. The average hotspot:coldspot ratios corrected to input DNA are shown for each strain (n = 3). (A) Preferential association of Dmc1 at the HIS4::LEU2 hotspot depends on SPO11. Dmc1 enrichment was not detected in control experiments without cross-linking and without antibody (data not shown). Hotspot and coldspot signal is not detectable in strains lacking DMC1. Dmc1 is not enriched at hotspots in spo11Δ tid1Δ mutants, but twofold more DNA is immunoprecipitated at both the hotspot and the coldspot after 4 h in sporulation medium vs. the spo11Δ single mutant. (B) Dmc1 does not preferentially associate with DSBs at HIS4 ::LEU2 in a tid1Δ rad54Δ double mutant. “WT 20%” represents a mixture of cells consisting of 20% wild-type cells from 4 h in meiosis with 80% wild-type cells from a mitotic culture. This mixture was designed to mimic the low efficiency of meiotic induction in the tid1Δ rad54Δ mutant in order to demonstrate that the ChIP assay is capable of detecting preferential hotspot association, even when meiotic induction is inefficient. (C) Dmc1 Western blot showing accuracy of mixing in the “WT 20%” sample compared with tid1Δ rad54Δ.
Tid1 reduces random association of Dmc1 with unbroken chromatin
When the levels of DNA isolated by ChIP are normalized to total input DNA (Fig. 3A), spo11Δ tid1Δ double mutants yield twofold more DNA from both hotspots and coldspots than DNA from ChIP of a spo11Δ single mutant. Furthermore, this signal is not detectable in the triple mutant strain lacking Dmc1. This result provides a second line of evidence supporting the conclusion that Tid1 functions to prevent nonspecific DSB-independent association of Dmc1 with chromatin.
Tid1 ATPase activity is required for suppression of DSB-independent Dmc1 focus formation
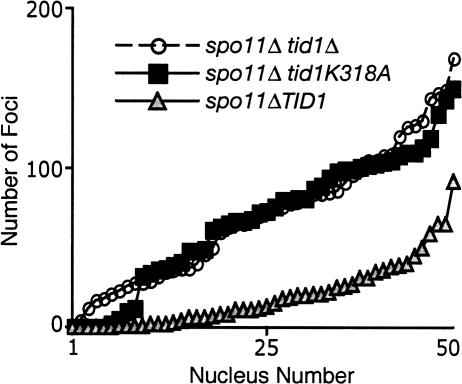
Biochemical analysis demonstrated that Tid1 is a DNA-dependent ATPase and that the ATPase activity is required for its ability to stimulate DNA strand exchange in vitro (Petukhova et al. 1998). Phenotypic analysis supports the contention that the ATPase activity of Tid1 is critical for its function in vivo (Petukhova et al. 2000). In order to determine whether the ability of Tid1 to block DSB-independent association of Dmc1 with chromosomes requires its ATPase activity, the conserved lysine residue 318 of the Tid1 Walker ATPase motif was mutated to alanine (tid1-K318A). Western analysis showed strains carrying this mutation display normal steady-state levels of Tid1 protein and two-hybrid analysis showed that the mutant protein retains the ability to bind Dmc1 (data not shown). These control experiments support the view that any phenotype detected for the tid1-K318A mutant reflects a defect in binding or hydrolysis of ATP. We compared the spo11Δ tid1Δ double mutant to spo11Δ tid1-K318A with respect to the total number of Dmc1 foci as well as average focus intensity. The spo11Δ single mutant served as a control. We did not detect a difference in the distribution of focus counts between the two double mutants (p = 0.936, Mann-Whitney U-test), although such a difference was readily detected by comparison of each double mutant with the spo11Δ single (p < 10−4, Fig. 4). The average staining intensities of Dmc1 foci in the spo11Δ tid1Δ and of spo11Δ tid1-K318A double mutants were essentially identical (106 ± 11 and 139 ± 39, respectively) and these numbers are nearly five times higher than the average staining intensity obtained for the spo11Δ single mutant (22 ± 13). Thus, the tid1-K318A mutation confers a phenotype quantitatively indistinguishable from that of the tid1Δ in a spo11Δ mutant background. Together, the results suggest that the DNA-dependent ATPase activity of Tid1 acts to prevent the accumulation of Dmc1 in DSB-independent foci.
Figure 4.
Tid1 ATPase activity is required for dissociation of Dmc1 from dsDNA. Distribution of Dmc1 focus counts from spo11Δ and spo11Δ tid1 mutants after 4 h of meiosis. Focus counts from 50 unselected nuclei are plotted in ascending order.
Tid1 and Rad54 share a function required for association of Dmc1 with recombination hotspots
Having concluded that Tid1 acts to prevent accumulation of Dmc1 at nonrecombinogenic sites, we were interested to determine whether this activity maintains a pool of Dmc1 available for assembly at DSBs by preventing its sequestration at non-DSB sites. Hotspot-specific association of Dmc1 by ChIP was carried out as before, except that the strains examined were SPO11+ and therefore formed DSBs. In a tid1Δ single mutant, Dmc1 localized preferentially to recombination hotspots (Fig. 3B, hotspot:coldspot ratio = 3.6 ± 0.2), suggesting that even in the absence of Tid1, a sufficient pool of free Dmc1 is still available to localize to recombination hotspots. Previous studies have shown that Rad54, the paralog of Tid1, can partially substitute for the role of Tid1 in both meiotic DSB repair, and exit from the meiotic stage at which Dmc1 foci are present (Shinohara et al. 1997b, 2000). We therefore tested hotspot-specific localization of Dmc1 in a rad54Δ single mutant as well as a tid1Δ rad54Δ double mutant. As in the tid1Δ single mutant, the rad54Δ single mutant was capable of loading Dmc1 preferentially at recombination hotspots (Fig. 3B, hotspot:coldspot ratio = 3.3 ± 0.2). Importantly, Dmc1 showed no preferential association with hotspots in a tid1Δ rad54Δ double mutant (Fig. 3B; hotspot:coldspot ratio = 0.9 ± 0.1).
Because the efficiency of induction of meiosis in the tid1Δ rad54Δ double mutant was only 20% of that in the wild-type control, it was important to demonstrate that the failure to detect specific association of Dmc1 with DSB sites was not a consequence of inefficient meiosis. To eliminate this possibility, a reconstruction experiment was carried out using a mixture of meiotic and mitotic wild-type cells, designed to mimic the level of meiotic induction in tid1Δ rad54Δ mutants. The accuracy of mixing was confirmed by comparing the level of Dmc1 protein, which is induced in meiosis, in the mixed wild-type sample with that in the tid1Δ rad54Δ double mutant (Fig. 3C). Immunostaining of meiotic nuclei also confirmed that the fraction of tid1Δ rad54Δ double-mutant nuclei displaying Dmc1 foci was ∼20% of that in wild type (data not shown). This control was designed to eliminate the possibility that tid1Δ rad54Δ double-mutant cells entered meiosis with normal efficiency, but went on to express only 20% of the normal level of Dmc1. The wild-type mixture displayed significant and reproducible preferential association of Dmc1 with hotspot DNA (hotspot:coldspot ratio = 2.3 ± 0.2; Fig. 3B), whereas no hotspot preference was detected for the tid1Δ rad54Δ double mutant. Fifty percent to 75% less total DNA is immunoprecipitated by anti-Dmc1 starting with lysates from the tid1Δ rad54Δ double mutant or the mitotic–meiotic control mixture as compared with the amount precipitated from the pure meiotic control. This result is as expected if the total yield depends on Dmc1 binding. These data indicate that the reduction in the hotspot to coldspot ratio observed in the tid1Δ rad54Δ double mutant reflects abnormal distribution of Dmc1 on chromatin and not simply a reduction in the fraction of cells undergoing meiosis. These results indicate that Tid1 and Rad54 share a function required for assembly of Dmc1 at sites of DSBs.
Discussion
Dmc1 forms DSB-independent foci
Previous studies showed that Dmc1 focus formation is strongly Spo11 dependent and, therefore, strongly DSB dependent (Bishop 1994; Shinohara et al. 1997a; Gasior et al. 1998; Romanienko and Camerini-Otero 2000). Here we present cytological and physical evidence that Dmc1 can associate with chromatin in the absence of DSBs. Faint focal Dmc1 staining was detected in spo11Δ nuclei. The DSB-independent Dmc1 foci could indicate that a small number of breaks form in the absence of Spo11, either during meiosis or during the spreading procedure; however, the dramatic effect on DSB-independent Dmc1 foci by mutating TID1 in a spo11Δ background suggests that the faint foci seen in the spo11Δ single mutant are likely to mark sites of Dmc1 assembly on unbroken chromatin. Given previous studies showing that Dmc1, like Rad51, binds to dsDNA with similar affinity to ssDNA (Li et al. 1997; Hong et al. 2001), it is not entirely surprising that Dmc1 focus formation is not strictly DSB dependent.
Does Dmc1 have DSB-independent function?
The description of Dmc1 activity presented thus far has been restricted to the repair of DSBs. Considering that Dmc1 can associate with chromatin independently of DSBs, it is reasonable to speculate that Dmc1 may have a DSB-independent function. Recent work from Dresser and colleagues, using a different strain background than that used here, suggested that Tid1 may have a role in dissolution of sister-chromatid cohesion (Kateneva et al. 2005). Deletion of TID1 in spo11Δ strains caused an anaphase arrest of meiotic cells, which has not been observed in the SK-1 strain background used in this study (data not shown). Importantly, this phenotype was alleviated upon the deletion of DMC1 (Kateneva and Dresser 2006). These results were interpreted to mean that Dmc1 plays a role in sister-chromatid cohesion and that Tid1 antagonizes this activity. Consistent with the proposed role in cohesion, we do observe DSB-independent Dmc1 foci in our strains. However, our observations in spo11Δ tid1Δ nuclei present the possibility that the anaphase arrest observed by Dresser and colleagues could be a consequence of the aberrant accumulation of Dmc1 on chromatin in this double-mutant background. Demonstration of a cohesion defect in a dmc1Δ single mutant is needed to support the hypothesis that Dmc1 plays a role in this process.
Tid1 and Rad54 control distribution of Dmc1 on chromatin
The data presented support a model in which Tid1 has an activity that prevents accumulation of Dmc1 at non-DSB sites. This finding is consistent with a recent report of increased association of Dmc1 with telomeric sequences in a tid1Δ mutant (Fukuda and Ohya 2006). The results presented here suggest that the redistribution observed is not limited to increases at telomeres but rather is more general, being driven in large part by loss of restriction of Dmc1 to DSB-proximal regions.
Double-mutant analysis shows that Tid1 and Rad54 share an activity required for the preferential association of Dmc1 at sites of DSBs. Given our finding that Tid1 is required to block DSB-independent association of Dmc1, results of the ChIP analysis suggest that Rad54 can partially substitute for the ability of Tid1 to promote dissociation of Dmc1 from non-DSB sites. The Dmc1 dissociation function of Rad54 may only make a significant contribution in the absence of Tid1, as a spo11Δ rad54Δ double mutant does not display the high level of Dmc1 foci observed in spo11Δ tid1Δ (data not shown). This difference is consistent with several previous results showing that the meiotic phenotypes conferred by a rad54Δ mutation are modest unless TID1Δ is also mutated (Shinohara et al. 1997b, 2000). The prior demonstration that Rad54 can promote dissociation of Rad51 from duplex DNA in a purified system is strongly consistent with our proposal that Rad54 can also promote dissociation of Dmc1 (Solinger et al. 2002). Efforts to detect an analogous biochemical activity for Tid1 on Dmc1 are underway.
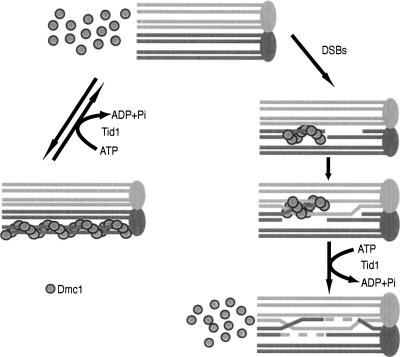
An important recent study showed that the Mei5 and Sae3 proteins also promote Dmc1 assembly at DSBs (Hayase et al. 2004). The role of Tid1 and Rad54 in controling Dmc1 distribution provides an interesting comparison to Mei5 and Sae3. Dmc1 fails to form foci in the absence of either Mei5 or Sae3, indicating that these proteins are positive regulators of Dmc1 assembly at DSB sites. In contrast, Dmc1 foci form and accumulate to greater than normal numbers in the absence of Tid1 and Rad54 (Shinohara et al. 2000; Solinger et al. 2002). This finding, together with ChIP results, leads us to a model in which Tid1 and Rad54 indirectly promote assembly of Dmc1 at DSB sites by preventing sequestration of the protein at non-DSB sites on chromatin (Fig. 5).
Figure 5.
Model for Tid1 activity. Tid1 has an ATP-dependent function that opposes Dmc1–dsDNA interaction. This can occur at two steps during meiosis. First, Dmc1 is removed from nonspecific sites on duplex DNA prior to DSB formation, creating a pool of Dmc1 available for binding when DSBs do form. After DSB formation and strand exchange, Tid1 then removes Dmc1 from the duplex product of strand exchange to allow recombination to be completed.
ChIP results indicate that the ability of Dmc1 to preferentially associate at DSBs does not depend on Tid1 so long as Rad54 is present. This is true even though Dmc1 has a dramatically increased tendency to accumulate on chromatin when DSBs are absent. These findings can be reconciled if preferential association of Dmc1 at DSB sites is determined by a lower off rate from the ssDNA tracts at DSBs as compared with non-DSB sites. Alternatively, the similarity in the degree of hotspot preference in wild-type and tid1Δ mutants may reflect a balance of opposing effects, with reduced efficiency of recruitment to DSBs compensated for by increased residency time (as discussed further below).
An alternative explanation for the dramatic effect of Tid1 mutation on DSB-independent Dmc1 foci is that Tid1 might prevent aggregation of Dmc1, independent of any DNA interaction. Such aggregates could be trapped by the nuclear matrix during preparation of spread nuclei. Although this possibility cannot be excluded for all Spo11-independent foci, particularly the very large structures that help define the “patchy” class, the ChIP experiments indicate that the absence of Tid1 results in a global increase in Dmc1–DNA association. Furthermore, the ATPase activity of Tid1 is DNA dependent, and the finding that the same number of DSB-independent foci form in the ATPase defective point mutant as in the tid1Δ mutant suggests that interaction of Tid1 with DNA is critical for preventing DSB-independent Dmc1 assembly.
Energetics of DNA strand exchange
The phenotypic similarity of the tid1 ATPase mutant and a tid1Δ mutant strongly suggests that the ability of Tid1 and Rad54 to prevent accumulation of Dmc1 at non-DSB sites depends on the ATPase activity of these proteins. This model proposes that much of the energy necessary for Dmc1-dependent recombination is expended prior to DNA strand exchange in order to assemble recombinase at DSBs. How might this seemingly wasteful energy expenditure contribute to the recombination process? One possibility is that Tid1- and Rad54-mediated energy expenditure allows for the assembly of Dmc1–ssDNA filaments that are less stable than Dmc1 filaments bound to the heteroduplex DNA (hDNA) products of DNA strand exchange. If Dmc1–hDNA is structurally similar to Dmc1–dsDNA that forms de novo, the requirement for a recycling factor is easily explained by the fact that DSB-associated tracts of ssDNA are estimated to represent <3% of total DNA during meiotic recombination (T.M. Holzen and D.K. Bishop, unpubl.).
The idea that DNA strand exchange could be driven by the relative stability of recombinase–dsDNA filaments is consistent with the properties of the best-studied recombinase, Escherichia coli RecA. Each protomer of RecA has two DNA-binding sites: Site I is a high-affinity DNA-binding site, and site II is a lower-affinity site (Takahashi et al. 1989; Muller et al. 1990; Zlotnick et al. 1993; Mazin and Kowalczykowski 1996, 1998). RecA binds ssDNA via site I in assembling the presynaptic filament, and these initial protomer–ssDNA binding interactions are not disrupted during DNA-strand exchange. Rather, the complementary strand of the target duplex is transferred onto the RecA–siteI–ssDNA structure. Thus, the duplex DNA strand exchange product is associated with RecA protomers via site I by virtue of the original interaction between site I and the invading ssDNA strand. The displaced strand from the original duplex briefly occupies site II before being displaced from the RecA–hDNA filament. As with the eukaryotic proteins, RecA-mediated DNA strand exchange occurs without hydrolysis of ATP, suggesting that this process is driven by the relative stability of the RecA–heteroduplex filament compared with that of the RecA–ssDNA filament (Menetski et al. 1990; Rosselli and Stasiak 1990). Furthermore, ATP hydrolysis is required for release of RecA from hDNA (Gumbs and Shaner 1998). In the absence of ATP hydrolysis, RecA filaments on heteroduplex products are very stable, as are RecA–dsDNA filaments formed in the absence of DNA strand exchange and hydrolysis (Menetski et al. 1990; Zaitsev and Kowalczykowski 1998). Taken together, these observations are consistent with the proposal that RecA-mediated DNA strand exchange is driven by energy stored in the RecA–ssDNA filament relative to that of the product RecA–hDNA filament.
If these considerations are correct, how does bacterial RecA manage to avoid sequestration on duplex DNA without the help of a Tid1- or Rad54-like recycling factor? Two functional differences between RecA and the two eukaryotic recombinases are likely to be relevant. First, the intrinsic DNA-dependent ATPase activity of RecA is nearly 80- to 200-fold greater than that of the eukaryotic proteins (Sung 1994; Li et al. 1997; Hong et al. 2001). Because RecA-ADP has relatively low affinity for dsDNA, the ATPase activity of RecA may play a role analogous to that proposed for the Swi2/Snf2 like proteins in eukaryotes. As discussed in the introduction, the idea that RecA ATPase functions to promote filament dynamics in vivo is a long-standing one (Kowalczykowski 1991). Another property of RecA that may allow it to function without a specialized recycling factor is its strong preference for binding ssDNA. This is in marked contrast to eukaryotic recombinases, which readily bind to dsDNA (Ogawa et al. 1993; Li et al. 1997; Hong et al. 2001). The preference of RecA for ssDNA results from a kinetic barrier to RecA filament nucleation on dsDNA rather than a thermodynamic impediment to dsDNA association (Pugh and Cox 1987a, b; Zaitsev and Kowalczykowski 1998). Thus, bacterial RecA and its eukaryotic counterparts may have evolved different approaches to solving the same requirement for driving DNA strand exchange reactions forward.
A possible role for Tid1 and Rad54 after DNA strand exchange
Little is known about how eukaryotic recombinases release heteroduplex DNA after DNA strand exchange occurs in vivo. The evidence that Tid1 and Rad54 act to promote Dmc1 dissociation from duplex DNA raises the possibility that these factors may also function following DNA strand exchange to dissociate Dmc1 from hDNA. This hypothesis has already been proposed as the biologically relevant mechanism behind the ability of Rad54 to dissociate Rad51 from dsDNA in vitro (Solinger et al. 2002). Also in support of this view, ChIP experiments have shown that Rad54 is not required for Rad51-mediated capture of homologous targets but is required for DNA synthesis from the invading filament after strand invasion has occurred (Sugawara et al. 2003). One interpretation of this result was that Rad54 is required to remove Rad51 from the DNA to allow for loading of PCNA and DNA polymerase and for DNA repair synthesis to occur (Solinger et al. 2002; Sugawara et al. 2003). Abnormally extensive Rad51 staining on spermatocyte chromosome spreads from mRad54 −/− and mRad54 −/− mRad54B −/− mice may also indicate a role for these proteins in releasing recombinase after recombination (Wesoly et al. 2006). It will be interesting to determine whether, in addition to preventing sequestration of Dmc1 at nonrecombining sites, Tid1 and Rad54 also promote dissociation of Dmc1 from DNA strand exchange products in vitro and in vivo.
Given our findings with Dmc1, it is interesting to speculate that disruption of unproductive recombinase–DNA interactions is a feature shared by divergent homologous recombination systems. An obvious candidate for this is Rad51. Thus far, we have not detected Rad51 foci in spo11Δ rad54Δ or spo11Δ tid1Δ double mutants. However, we have not been able to induce meiosis in a spo11Δ tid1Δ rad54Δ triple mutant, leaving open the possibility that there is a higher level of redundancy for Tid1 and Rad54 activity on Rad51 as compared with that on Dmc1. Thus, it remains to be determined whether Rad51 avoids assembly at non-DSB sites by a mechanism equivalent to that used by Dmc1.
A study of certain alleles of E. coli recA could indicate that disruption of unproductive recombinase interactions occurs in bacteria. It is possible that activation of the intrinsic ATPase activity of RecA by its binding to dsDNA is sufficient to disassemble the protein from dsDNA substrates, hence preventing its accumulation at non-DSB sites. In this view, Tid1 and Rad54 are required to assist eukaryotic recombinases to achieve a level of dynamics functionally equivalent to that achieved by the intrinsic activity of RecA. This idea is consistent with the isolation of alleles of recA that kill E. coli cells, but nonetheless promote ∼25% the normal level of recombination. Biochemical studies of these mutant proteins show that they are profoundly defective in ATP hydrolysis and have markedly increased dsDNA binding (Campbell and Davis 1999a, b). The toxicity of these alleles was attributed to the slow dissociation of RecA from hDNA following strand exchange. In light of the work presented here, it is interesting to consider that the toxicity of these forms of RecA results from their association at nonrecombinogenic sites.
Materials and methods
Yeast strains and sporulation conditions
Yeast strains used in all experiments were derived by transformation of SK-1 and/or by crosses with other such SK-1 derivatives (Table 1). All sporulation conditions for yeast cultures were as described previously (Bishop 1994).
Table 1.
Strains
Spreading and immunofluorescent staining of meiotic nuclei
Preparation of slides of spread meiotic nuclei and immunostaining of the slides were as described previously (Bishop 1994; Gasior et al. 1998; Rua et al. 2004). To stain meiotic nuclei for Dmc1, a 1:1000 dilution of rabbit anti-Dmc1 serum was used (Covance; DB Ab #113). For RPA, Dmc1 double-staining experiments, a 1:500 dilution of guinea pig anti-RPA serum was also used. Fluorescein-conjugated secondary antibodies detected and amplified signal from the primary antibodies. Alexa-fluor 488-conjugated goat anti-rabbit IgG (Molecular Probes) was used at a dilution of 2 μg/mL for Dmc1 staining and Alexa-fluor 594-conjugated goat anti-guinea pig IgG (Molecular Probes) was used at a dilution of 1 μg/mL for RPA staining. Focus counts were done without the aid of image analysis software. Average focus-staining intensity was measured at a set exposure time chosen to be subsaturating for the brightest foci. The total fluorescence signal from an area containing a nucleus was divided by the number of foci in that area.
Microscopy
Microscopy was performed as described previously (Bishop 1994; Gasior et al. 1998, 2001). Images from a Hamamatsu C-4742 digital CCD camera (Hamamatsu Photonics, K.K.) were acquired using QED Imaging or ImagePro Express software (Media Cybernetics).
Pulsed-field gel electrophoresis
Plugs were prepared from 5 mL of sporulating cells using previously described methods (Borde et al. 1999). Plugs were loaded into a 1% pulsed-field certified agarose gel in 0.5× TBE. The gel was run in prechilled 0.5× TBE buffer at 6 V/cm, 120° switch angle, 90-sec initial, and final switch time for 23 h at 14°C. DNA was visualized using SYBR Green DNA stain (Molecular Probes).
Extraction of whole-yeast cellular proteins and Western blotting
Five milliliters of a sporulating culture was harvested at the indicated times and fixed by addition of 1 mL of 50% trichloroacetic acid. Samples were incubated for 5 min at 60°C, then chilled on ice for 5 min. Samples were centrifuged, and the pellet was resuspended in 250 μL of SDS-PAGE loading buffer (58 mM Tris-Cl/0.05% SDS at pH 6.8, 5% glycerol, 0.02% SDS, 0.1 M DTT, 20 μg/mL bromphenol blue, 50 mM Na+PIPES at pH 7.5) and boiled for 10 min. Protein samples were run in standard 8% SDS–polyacrylamide gels. Western blots were prepared on Immobilon-P PVDF transfer membrane (Millipore). A 1:5000 dilution of rabbit anti-Dmc1 serum (Covance) and 0.4 μg/mL goat anti-Arp7 IgG (Santa Cruz Biotechnology) were used as primary antibodies. Primary antibody signal was detected with horseradish peroxidase-conjugated secondary antibodies (Amersham Biosciences and Santa Cruz Biotechnology) and chemiluminesence detection agents (PerkinElmer Life Sciences). Blots were visualized with a Storm PhosphorImager (Molecular Dynamics) and analyzed using ImageQuant 4.0 (Molecular Dynamics).
ChIP
Twenty-five milliliters of sporulating cells were harvested at the times indicated and cross-linked by addition of formalde-hyde to a final concentration of 1%. Cross-linking was quenched by addition of glycine to 125 mM. For the mixing experiment in Figure 3B, wild-type 0-h cross-linked culture that had not yet entered meiosis was mixed with wild-type culture that had been cross-linked after 4 h in sporulation medium to a final concentration of 20% 4-h culture. Cells were lysed with glass beads in 1 M NaCl lysis buffer (50 mM HEPES/HOH at pH
7.5, 1 M NaCl, 5 mM EDTA, 1% Triton X-100, 0.1% Na Deoxycholate, 0.1 mg/mL leupeptin, 1 mM PMSF, 0.1 mg/mL pepstatin). Lysate was sonicated for 20 pulses, three times, using a Branson Sonifier 450 at output control 1 and 50% duty cycle. Samples were incubated for 5 min on ice between sonications. All samples were adjusted to the same input protein concentration and an amount of sonicated lysate, equivalent to 2 mg of protein, was incubated with 1 μg of guinea pig polyclonal anti-Dmc1 serum (Covance) and rotated overnight at 4°C. Protein G magnetic beads (New England Biolabs) were washed and resuspended in 1 M NaCl lysis buffer. Twenty-five microliters beads were added to the lysate and BSA was added to a final concentration of 11 mM. Samples were incubated for 2 h at 4°C with rotation. Beads were washed five times in 1 mL of 1 M NaCl lysis buffer, three times in 0.5 M NaCl buffer (50 mM HEPES/ KOH at pH 7.5, 0.5 M NaCl, 5 mM EDTA, 1% Triton X-100, 0.1% Na Deoxycholate), once in 1 mL of LiCl buffer (10 mM Tris-HCl at pH 8.0, 250 mM LiCl, 0.5% NP-40, 0.5% Na Deoxycholate, 5 mM EDTA), and once in 1 mL TE; all washes were performed at room temperature for 5 min on a rotator. Beads were resuspended in 100 μL of TE and incubated overnight at 65°C to reverse cross-linking. Samples were heated for 30 min at 95°C and the supernatant was removed for PCR analysis. Multiplex PCR was performed with 200 μM dNTPs and 2 μL of ChIP extract using primers in LEU2 (forward: 5′-GT TAAGAACCCAACCCACCTAAATGG-3′, reverse: 5′-CAT CATTGCAGCAGACAAGATAGTGG-3′) and CIT2 (forward: 5′-TGGACCCAAATGCCGATTATG-3′, reverse: 5′-AGCCAA CCCGTTCAAACCTG-3′). PCR products were separated on a 1.5 mM, 8% acrylamide gel, and detected with SYBR Green DNA stain (Molecular Probes). Images were obtained using a Storm PhosphorImager (Molecular Dynamics), and the amount of signal from each band was analyzed with ImageQuant 4.0 software (Molecular Dynamics). The relative enrichment of Dmc1 at hotspots was determined by calculating the ratio of hotspot signal to coldspot signal for each sample, and the ratios were corrected to input levels of DNA.
Site-directed mutagenesis of TID1
Lysine residue 318 in the putative Walker A ATP-binding motif of TID1 (DxxxxGKT) was mutated to alanine (K318A) using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). pRS304-TID1 was mutagenized using forward primer 5′-GGC TGATGATATGGGTTTAGGTGCAACACTAATGAGTATAA CTTT GAT-3′ and reverse primer 5′-ATC AA AG TTATA CC A TTAGTGTTGCACCTAAACCCATATCATCAGCC-3′. tid1-K318A as well a wild-type TID1 control were introduced at the TID1 locus by transformation.
Acknowledgments
We thank Steve Kowalczykowski, Joe Piccirilli, Molly Przeworski, and Phoebe Rice for helpful discussions, and Rob Shroff for help in developing the ChIP assay. We also thank Akio Sugino and Akira Shinohara for antibodies, as well as Lily Yeh and Ting-Fang Wang for two-hybrid constructs. We are grateful to Wolf-Dietrich Heyer and Jon Staley for critical reading of the manuscript. This work was supported by N.I.H. GM50936 to D.K.B.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1447106.
References
- Aboussekhra, A., Chanet, R., Adjiri, A., Fabre, F. Semidominant suppressors of Srs2 helicase mutations of Saccharomyces cerevisiae map in the RAD51 gene, whose sequence predicts a protein with similarities to procaryotic RecA proteins. Mol. Cell. Biol. 1992;12:3224–3234. doi: 10.1128/mcb.12.7.3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexeev, A., Mazin, A., Kowalczykowski, S.C. Rad54 protein possesses chromatin-remodeling activity stimulated by the Rad51–ssDNA nucleoprotein filament. Nat. Struct. Biol. 2003;10:182–186. doi: 10.1038/nsb901. [DOI] [PubMed] [Google Scholar]
- Amitani, I., Baskin, R.J., Kowalczykowski, S.C. Visualization of Rad54, a chromatin remodeling protein, translocating on single DNA molecules. Mol. Cell. 2006;23:143–148. doi: 10.1016/j.molcel.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Baudat, F., Nicolas, A. Clustering of meiotic double-strand breaks on yeast chromosome III. Proc. Natl. Acad. Sci. 1997;94:5213–5218. doi: 10.1073/pnas.94.10.5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, D.K. RecA homologs Dmc1 and Rad51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell. 1994;79:1081–1092. doi: 10.1016/0092-8674(94)90038-8. [DOI] [PubMed] [Google Scholar]
- Bishop, D.K., Park, D., Xu, L., Kleckner, N. DMC1: A meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- Borde, V., Wu, T.C., Lichten, M. Use of a recombination reporter insert to define meiotic recombination domains on chromosome III of Saccharomyces cerevisiae . Mol. Cell. Biol. 1999;19:4832–4842. doi: 10.1128/mcb.19.7.4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugreev, D.V., Golub, E.I., Stasiak, A.Z., Stasiak, A., Mazin, A.V. Activation of human meiosis-specific recombinase Dmc1 by Ca2+ . J. Biol. Chem. 2005;280:26886–26895. doi: 10.1074/jbc.M502248200. [DOI] [PubMed] [Google Scholar]
- Campbell, M.J., Davis, R.W. On the in vivo function of the RecA ATPase. J. Mol. Biol. 1999a;286:437–445. doi: 10.1006/jmbi.1998.2457. [DOI] [PubMed] [Google Scholar]
- Campbell, M.J., Davis, R.W. Toxic mutations in the recA gene of E. coli prevent proper chromosome segregation. J. Mol. Biol. 1999b;286:417–435. doi: 10.1006/jmbi.1998.2456. [DOI] [PubMed] [Google Scholar]
- Cox, M.M. The bacterial RecA protein as a motor protein. Annu. Rev. Microbiol. 2003;57:551–577. doi: 10.1146/annurev.micro.57.030502.090953. [DOI] [PubMed] [Google Scholar]
- Cox, M.M., Lehman, I.R. recA protein-promoted DNA strand exchangeStable complexes of recA protein and single-stranded DNA formed in the presence of ATP and single-stranded DNA binding protein. J. Biol. Chem. 1982;257:8523–8532. [PubMed] [Google Scholar]
- Cox, J.M., Tsodikov, O.V., Cox, M.M. Organized unidirectional waves of ATP hydrolysis within a RecA filament. PLoS Biol. 2005;3:e52. doi: 10.1371/journal.pbio.0030052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggleston, A.K., Kowalczykowski, S.C. An overview of homologous pairing and DNA strand exchange proteins. Biochimie. 1991;73:163–176. doi: 10.1016/0300-9084(91)90199-b. [DOI] [PubMed] [Google Scholar]
- Eisen, J.A., Sweder, K.S., Hanawalt, P.C. Evolution of the SNF2 family of proteins: Subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folta-Stogniew, E., O'Malley, S., Gupta, R., Anderson, K.S., Radding, C.M. Exchange of DNA base pairs that coincides with recognition of homology promoted by E. coli RecA protein. Mol. Cell. 2004;15:965–975. doi: 10.1016/j.molcel.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Fukuda, T., Ohya, Y. Recruitment of RecA homologs Dmc1p and Rad51p to the double-strand break repair site initiated by meiosis-specific endonuclease VDE (PI-SceI) Mol. Genet. Genomics. 2006;275:204–214. doi: 10.1007/s00438-005-0078-4. [DOI] [PubMed] [Google Scholar]
- Gasior, S.L., Wong, A.K., Kora, Y., Shinohara, A., Bishop, D.K. Rad52 associates with RPA and functions with rad55 and rad57 to assemble meiotic recombination complexes. Genes & Dev. 1998;12:2208–2221. doi: 10.1101/gad.12.14.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasior, S.L., Olivares, H., Ear, U., Hari, D.M., Weichselbaum, R., Bishop, D.K. Assembly of RecA-like recombinases: Distinct roles for mediator proteins in mitosis and meiosis. Proc. Natl. Acad. Sci. 2001;98:8411–8418. doi: 10.1073/pnas.121046198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbs, O.H., Shaner, S.L. Three mechanistic steps detected by FRET after presynaptic filament formation in homologous recombination. ATP hydrolysis required for release of oligonucleotide heteroduplex product from RecA. Biochemistry. 1998;37:11692–11706. doi: 10.1021/bi980646s. [DOI] [PubMed] [Google Scholar]
- Hayase, A., Takagi, M., Miyazaki, T., Oshiumi, H., Shinohara, M., Shinohara, A. A protein complex containing Mei5 and Sae3 promotes the assembly of the meiosis-specific RecA homolog Dmc1. Cell. 2004;119:927–940. doi: 10.1016/j.cell.2004.10.031. [DOI] [PubMed] [Google Scholar]
- Hong, E.L., Shinohara, A., Bishop, D.K. Saccharomyces cerevisiae Dmc1 protein promotes renaturation of single-strand DNA (ssDNA) and assimilation of ssDNA into homologous super-coiled duplex DNA. J. Biol. Chem. 2001;276:41906–41912. doi: 10.1074/jbc.M105563200. [DOI] [PubMed] [Google Scholar]
- Jaskelioff, M., Van Komen, S., Krebs, J.E., Sung, P., Peterson, C.L. Rad54p is a chromatin remodeling enzyme required for heteroduplex DNA joint formation with chromatin. J. Biol. Chem. 2003;278:9212–9218. doi: 10.1074/jbc.M211545200. [DOI] [PubMed] [Google Scholar]
- Kateneva, A.V., Dresser, M.E. Sister chromatid cohesion remodeling and meiotic recombination. Cell Cycle. 2006;5:467–471. doi: 10.4161/cc.5.5.2498. [DOI] [PubMed] [Google Scholar]
- Kateneva, A.V., Konovchenko, A.A., Guacci, V., Dresser, M.E. Recombination protein Tid1p controls resolution of cohesin-dependent linkages in meiosis in Saccharomyces cerevisiae . J. Cell Biol. 2005;171:241–253. doi: 10.1083/jcb.200505020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney, S., Giroux, C.N., Kleckner, N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- Klein, H.L. RDH54, a RAD54 homologue in Saccharomyces cerevisiae, is required for mitotic diploid-specific recombination and repair and for meiosis. Genetics. 1997;147:1533–1543. doi: 10.1093/genetics/147.4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczykowski, S.C. Biochemistry of genetic recombination: Energetics and mechanism of DNA strand exchange. Annu. Rev. Biophys. Biophys. Chem. 1991;20:539–575. doi: 10.1146/annurev.bb.20.060191.002543. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski, S.C. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 2000;25:156–165. doi: 10.1016/s0968-0004(00)01569-3. [DOI] [PubMed] [Google Scholar]
- Li, Z., Golub, E.I., Gupta, R., Radding, C.M. Recombination activities of HsDmc1 protein, the meiotic human homolog of RecA protein. Proc. Natl. Acad. Sci. 1997;94:11221–11226. doi: 10.1073/pnas.94.21.11221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson, J.Y., Davies, A.A., Hajibagheri, N., Van Dyck, E., Benson, F.E., Stasiak, A.Z., Stasiak, A., West, S.C. The meiosis-specific recombinase hDmc1 forms ring structures and interacts with hRad51. EMBO J. 1999;18:6552–6560. doi: 10.1093/emboj/18.22.6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazin, A.V., Kowalczykowski, S.C. The specificity of the secondary DNA binding site of RecA protein defines its role in DNA strand exchange. Proc. Natl. Acad. Sci. 1996;93:10673–10678. doi: 10.1073/pnas.93.20.10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazin, A.V., Kowalczykowski, S.C. The function of the secondary DNA-binding site of RecA protein during DNA strand exchange. EMBO J. 1998;17:1161–1168. doi: 10.1093/emboj/17.4.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menetski, J.P., Varghese, A., Kowalczykowski, S.C. Properties of the high-affinity single-stranded DNA binding state of the Escherichia coli recA protein. Biochemistry. 1988;27:1205–1212. doi: 10.1021/bi00404a021. [DOI] [PubMed] [Google Scholar]
- Menetski, J.P., Bear, D.G., Kowalczykowski, S.C. Stable DNA heteroduplex formation catalyzed by the Escherichia coli RecA protein in the absence of ATP hydrolysis. Proc. Natl. Acad. Sci. 1990;87:21–25. doi: 10.1073/pnas.87.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, B., Koller, T., Stasiak, A. Characterization of the DNA binding activity of stable RecA–DNA complexes. Interaction between the two DNA binding sites within RecA helical filaments. J. Mol. Biol. 1990;212:97–112. doi: 10.1016/0022-2836(90)90307-8. [DOI] [PubMed] [Google Scholar]
- Ogawa, T., Yu, X., Shinohara, A., Egelman, E.H. Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science. 1993;259:1896–1899. doi: 10.1126/science.8456314. [DOI] [PubMed] [Google Scholar]
- Paques, F., Haber, J.E. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae . Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petukhova, G., Stratton, S., Sung, P. Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature. 1998;393:91–94. doi: 10.1038/30037. [DOI] [PubMed] [Google Scholar]
- Petukhova, G., Van Komen, S., Vergano, S., Klein, H., Sung, P. Yeast Rad54 promotes Rad51-dependent homologous DNA pairing via ATP hydrolysis-driven change in DNA double helix conformation. J. Biol. Chem. 1999;274:29453–29462. doi: 10.1074/jbc.274.41.29453. [DOI] [PubMed] [Google Scholar]
- Petukhova, G., Sung, P., Klein, H. Promotion of Rad51-dependent D-loop formation by yeast recombination factor Rdh54/Tid1. Genes & Dev. 2000;14:2206–2215. doi: 10.1101/gad.826100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh, B.F., Cox, M.M.1987a.recA protein binding to the heteroduplex product of DNA strand exchange J. Biol Chem. 2621337–1343. [PubMed] [Google Scholar]
- Pugh, B.F., Cox, M.M. Stable binding of recA protein to duplex DNA. Unraveling a paradox. J. Biol. Chem. 1987b;262:1326–1336. [PubMed] [Google Scholar]
- Roca, A.I., Cox, M.M. The RecA protein: Structure and function. Crit. Rev. Biochem. Mol. Biol. 1990;25:415–456. doi: 10.3109/10409239009090617. [DOI] [PubMed] [Google Scholar]
- Rockmill, B., Sym, M., Scherthan, H., Roeder, G.S. Roles for two RecA homologs in promoting meiotic chromosome synapsis. Genes & Dev. 1995;9:2684–2695. doi: 10.1101/gad.9.21.2684. [DOI] [PubMed] [Google Scholar]
- Romanienko, P.J., Camerini-Otero, R.D. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol. Cell. 2000;6:975–987. doi: 10.1016/s1097-2765(00)00097-6. [DOI] [PubMed] [Google Scholar]
- Rosselli, W., Stasiak, A. Energetics of RecA-mediated recombination reactions. Without ATP hydrolysis RecA can mediate polar strand exchange but is unable to recycle. J. Mol. Biol. 1990;216:335–352. doi: 10.1016/S0022-2836(05)80325-0. [DOI] [PubMed] [Google Scholar]
- Rua, D., Holzen, T., Glick, B.S., Kron, S.J., Bishop, D.K. Monitoring changes in the subcellular location of proteins in S. cerevisiae . Methods Mol. Biol. 2004;241:299–311. doi: 10.1385/1-59259-646-0:299. [DOI] [PubMed] [Google Scholar]
- Schwacha, A., Kleckner, N. Interhomolog bias during meiotic recombination: Meiotic functions promote a highly differentiated interhomolog-only pathway. Cell. 1997;90:1123–1135. doi: 10.1016/s0092-8674(00)80378-5. [DOI] [PubMed] [Google Scholar]
- Sehorn, M.G., Sung, P. Meiotic recombination: An affair of two recombinases. Cell Cycle. 2004;3:1375–1377. doi: 10.4161/cc.3.11.1364. [DOI] [PubMed] [Google Scholar]
- Shinohara, A., Ogawa, H., Ogawa, T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- Shinohara, A., Gasior, S., Ogawa, T., Kleckner, N., Bishop, D.K. Saccharomyces cerevisiae recA homologues RAD51 and DMC1 have both distinct and overlapping roles in meiotic recombination. Genes Cells. 1997a;2:615–629. doi: 10.1046/j.1365-2443.1997.1480347.x. [DOI] [PubMed] [Google Scholar]
- Shinohara, M., Shita-Yamaguchi, E., Buerstedde, J.M., Shinagawa, H., Ogawa, H., Shinohara, A. Characterization of the roles of the Saccharomyces cerevisiae RAD54 gene and a homologue of RAD54, RDH54/TID1, in mitosis and meiosis. Genetics. 1997b;147:1545–1556. doi: 10.1093/genetics/147.4.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara, M., Gasior, S.L., Bishop, D.K., Shinohara, A. Tid1/Rdh54 promotes colocalization of rad51 and dmc1 during meiotic recombination. Proc. Natl. Acad. Sci. 2000;97:10814–10819. doi: 10.1073/pnas.97.20.10814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solinger, J.A., Kiianitsa, K., Heyer, W.D. Rad54, a Swi2/Snf2-like recombinational repair protein, disassembles Rad51:dsDNA filaments. Mol. Cell. 2002;10:1175–1188. doi: 10.1016/s1097-2765(02)00743-8. [DOI] [PubMed] [Google Scholar]
- Sugawara, N., Wang, X., Haber, J.E. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol. Cell. 2003;12:209–219. doi: 10.1016/s1097-2765(03)00269-7. [DOI] [PubMed] [Google Scholar]
- Sung, P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- Sung, P., Stratton, S.A. Yeast Rad51 recombinase mediates polar DNA strand exchange in the absence of ATP hydrolysis. J. Biol. Chem. 1996;271:27983–27986. doi: 10.1074/jbc.271.45.27983. [DOI] [PubMed] [Google Scholar]
- Symington, L.S. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, M., Kubista, M., Norden, B. Binding stoichiometry and structure of RecA–DNA complexes studied by flow linear dichroism and fluorescence spectroscopy. Evidence for multiple heterogeneous DNA co-ordination. J. Mol. Biol. 1989;205:137–147. doi: 10.1016/0022-2836(89)90371-9. [DOI] [PubMed] [Google Scholar]
- Villeneuve, A.M., Hillers, K.J. Whence meiosis? Cell. 2001;106:647–650. doi: 10.1016/s0092-8674(01)00500-1. [DOI] [PubMed] [Google Scholar]
- Wesoly, J., Agarwal, S., Sigurdsson, S., Bussen, W., Van Komen, S., Qin, J., van Steeg, H., van Benthem, J., Wassenaar, E., Baarends, W.M., et al. Differential contributions of mammalian Rad54 paralogs to recombination, DNA damage repair, and meiosis. Mol. Cell. Biol. 2006;26:976–989. doi: 10.1128/MCB.26.3.976-989.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolner, B., van Komen, S., Sung, P., Peterson, C.L. Recruitment of the recombinational repair machinery to a DNA double-strand break in yeast. Mol. Cell. 2003;12:221–232. doi: 10.1016/s1097-2765(03)00242-9. [DOI] [PubMed] [Google Scholar]
- Zaitsev, E.N., Kowalczykowski, S.C. Binding of double-stranded DNA by Escherichia coli RecA protein monitored by a fluorescent dye displacement assay. Nucleic Acids Res. 1998;26:650–654. doi: 10.1093/nar/26.2.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenvirth, D., Arbel, T., Sherman, A., Goldway, M., Klein, S., Simchen, G. Multiple sites for double-strand breaks in whole meiotic chromosomes of Saccharomyces cerevisiae . EMBO J. 1992;11:3441–3447. doi: 10.1002/j.1460-2075.1992.tb05423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotnick, A., Mitchell, R.S., Steed, R.K., Brenner, S.L. Analysis of two distinct single-stranded DNA binding sites on the recA nucleoprotein filament. J. Biol. Chem. 1993;268:22525–22530. [PubMed] [Google Scholar]