Abstract
In Escherichia coli, the global regulatory protein CsrA (carbon store regulator A) binds to leader segments of target mRNAs, affecting their translation and stability. CsrA activity is regulated by two noncoding RNAs, CsrB and CsrC, which act by sequestering multiple CsrA dimers. Here, we describe a protein (CsrD) that controls the degradation of CsrB/C RNAs. The dramatic stabilization of CsrB/C RNAs in a csrD mutant altered the expression of CsrA-controlled genes in a manner predicted from the previously described Csr regulatory circuitry. A deficiency in RNase E, the primary endonuclease involved in mRNA decay, also stabilized CsrB/C, although the half-lives of other RNAs that are substrates for RNase E (rpsO, rpsT, and RyhB) were unaffected by csrD. Analysis of the decay of CsrB RNA, both in vitro and in vivo, suggested that CsrD is not a ribonuclease. Interestingly, the CsrD protein contains GGDEF and EAL domains, yet unlike typical proteins in this large superfamily, its activity in the regulation of CsrB/C decay does not involve cyclic di-GMP metabolism. The two predicted membrane-spanning regions are dispensable for CsrD activity, while HAMP-like, GGDEF, and EAL domains are required. Thus, these studies demonstrate a novel process for the selective targeting of RNA molecules for degradation by RNase E and a novel function for a GGDEF–EAL protein.
Keywords: RNA decay, biofilm formation, Hfq, polynucleotide phosphorylase, degradosome, GGDEF–EAL domain proteins
In many species of bacteria, the Csr (carbon storage regulator) and homologous Rsm (repressor of stationary phase metabolites) systems coordinate the expression of diverse genes that facilitate adaptation among major physiological phases of growth; e.g., exponential versus stationary phase, planktonic versus biofilm, and ostensibly acute versus chronic states of infection (Romeo 1998; Wei et al. 2001; Jackson et al. 2002; Goodman et al. 2004; Majdalani et al. 2005). These systems use the RNA-binding protein CsrA (Romeo et al. 1993) to regulate translation and mRNA stability by recognizing specific nucleotide sequences within mRNA leaders. Among the best-studied examples of these are the CsrA- repressed glgCAP, cstA, and pgaABCD mRNAs, which are involved in glycogen metabolism, peptide transport, and biofilm formation, respectively (Liu and Romeo 1997; Baker et al. 2002; X. Wang et al. 2005), as well as the CsrA-activated flhDC mRNA, which encodes the master regulator of motility and chemotaxis genes (Wei et al. 2001).
In Escherichia coli, CsrA protein activity is regulated by the CsrB and CsrC noncoding RNAs, which contain CsrA recognition sequences (18 and 9, respectively) primarily within the loops of predicted stem-loop structures. Interaction of CsrA with these sites leads to its sequestration (Liu et al. 1997; Weilbacher et al. 2003; Dubey et al. 2005). Thus, this system employs a mechanism distinct from that of other small RNAs such as OxyS and RyhB, which involve RNA–RNA base-pairing (Gottesman 2004). Furthermore, the Csr components in E. coli interact within an autoregulatory circuit that provides a homeostatic mechanism for control of CsrA activity (Fig. 1A). In this system, regulation of csrB/C transcription by CsrA requires the two-component signal transduction system (TCS) BarA/UvrY (Gudapaty et al. 2001; Suzuki et al. 2002; Weilbacher et al. 2003). Orthologous TCS regulates host–microbe interactions and quorum sensing (Hammer et al. 2002; Whistler and Ruby 2003; Altier 2005; Lenz et al. 2005), seemingly via Csr homologs.
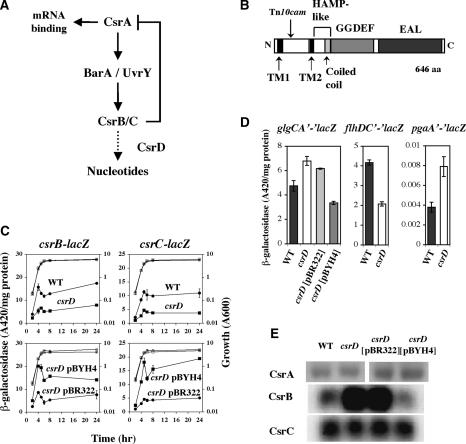
Figure 1.
(A) Csr regulatory circuitry (Suzuki et al. 2002; Weilbacher et al. 2003), with proposed CsrD function (broken line). (B) Domain structure of CsrD with predicted trans-membrane regions, HAMP-like domain, GGDEF and EAL domains, and Tn10 cam insertion site displayed. (C) Effect of csrD on β-galactosidase activity expressed from chromosomal csrB-lacZ and csrC-lacZ fusions. Strains containing csrB-lacZ and csrC-lacZ were KSB837 and GS1114, respectively. csrD strains contained the csrD[H11019]cam insertion. pBYH4 is a clone of csrD in pBR322. Closed and open symbols depict activity and growth, respectively. (D) Effect of csrD on gene expression required for glycogen synthesis (glgCA-lacZ), motility (flhDC-lacZ), and biofilm formation (pgaA-lacZ). (E) Effect of csrD disruption and complementation (pBYH4) on CsrA protein and CsrB/C RNA levels by Western and Northern analyses, respectively.
Although many aspects of this complex regulatory network are now understood, one exception relates to the stability of the CsrB and CsrC RNAs. Since the overall levels of these regulatory transcripts are determined by their relative synthesis and turnover rates, it is important to understand the factors that govern their degradation. In E. coli, bulk mRNA decay and many RNA processing reactions involve the essential enzyme RNase E, a single-strand-specific endoribonuclease (Kushner 2002). RNase E contains an N-terminal catalytic domain, an RNA-binding domain, and a C-terminal domain that serves as a scaffold for the association of the 3'- to-5' exonuclease polynucleotide phosphorylase (PNPase), the glycolytic enzyme enolase, and an RNA helicase (RhlB or CsdA) to form an RNA-degrading complex called the degradosome (Py et al. 1994, 1996; Carpousis 2002; Callaghan et al. 2004; Morita et al. 2004; Prud'homme-Généreux et al. 2004).
RNase E levels are autoregulated by a mechanism involving the degradation of its own transcript (Mudd and Higgins 1993; Jain and Belasco 1995; Diwa et al. 2000; Sousa et al. 2001; Ow et al. 2002). In addition, RNase E catalytic activity can be inhibited by binding to the RraA protein (Lee et al. 2003). Some mRNAs can be selectively targeted for turnover by RNase E by base-pairing with noncoding antisense RNAs, which undergo coincident decay during this process (for reviews, see Majdalani et al. 2005; Storz et al. 2005). The latter reactions require the Sm-like RNA chaperone, Hfq (Massé et al. 2003). It has been suggested recently that Hfq forms a complex with RNase E (Morita et al. 2005). Hfq also interacts with PNPase and poly(A) polymerase I (PAP I) to form a complex that stimulates polyadenylation of mRNAs containing intrinsic transcription terminators (Mohanty et al. 2004). Any or all of these proteins (RNase E,PNPase, PAP I, and Hfq) might participate in the turnover of the CsrB and CsrC RNAs.
Although regulation of CsrB and CsrC levels could simply be a function of transcription, a previous study has suggested the presence of at least one additional undefined regulator of csrB expression (Suzuki et al. 2002). Our search for this regulator led to the identification and characterization of the CsrD protein, a member of a large family of proteins that contain GGDEF and EAL signaling domains (for reviews, see D'Argenio and Miller 2004;
Jenal 2004; Römling et al. 2005). In various species, GGDEF and EAL proteins affect production of exopolysaccharides and surface proteins, and influence adhesion, motility, biofilm formation, and host–pathogen interactions (D'Argenio and Miller 2004; Hisert et al. 2005).
A number of GGDEF and EAL domain proteins are known to synthesize and hydrolyze, respectively, bis- (3'–5')-cyclic dimeric guanosine monophosphate (c-di- GMP), a secondary messenger (e.g., see Hickman et al. 2005; Hisert et al. 2005; Ryjenkov et al. 2005; Schmidt et al., 2005; Simm et al. 2005; Camilli and Bassler 2006). Amino acid residues required for c-di-GMP synthesis and hydrolysis have been defined by mutagenesis and other approaches (Chan et al. 2004; Kirillina et al. 2004; Paul et al. 2004; Simm et al. 2004; Tischler and Camilli 2004; Christen et al. 2005; Tamayo et al. 2005). In some dual domain proteins, the GGDEF or EAL domain may be inactive or assume an alternate activity (Christen et al. 2005; Schmidt et al. 2005). Although no GGDEF–EAL protein is known to function independently of c-di- GMP, metabolism of this nucleotide signal would seem to be inadequate to account for the sheer abundance of these proteins (e.g., E. coli has 19-GGDEF- and 17-EAL- containing proteins). Here we show that the CsrD protein is not involved in c-di-GMP metabolism, but rather appears to target the CsrB and CsrC RNAs for degradation by RNase E.
Results
Identification of csrD and determination of its role in the Csr regulatory circuitry
CsrA indirectly activates csrB/C transcription, possibly via effects on BarA sensor-kinase activity. To identify novel regulators of csrB, we screened for transposon insertions that alter expression of a csrB-lacZ transcriptional fusion in strain KSB837 (Table 1). A mutation that decreased csrB-lacZ expression was isolated within a gene, csrD (formerly yhdA), which is predicted to encode a membrane-bound signaling protein (Fig. 1B) containing GGDEF and EAL domains (http://www.sanger.ac.uk/ Software/Pfam). Analysis of the effects of the csrD transposon insertion on genes and phenotypes regulated by CsrA—including csrB, csrC (Fig. 1C), and genes for motility (flhDC), biofilm formation (pgaABCD), and glycogen biosynthesis (glgCAP)—suggested that CsrA activity might be decreased by the csrD mutation (Fig. 1D). However, CsrA protein levels and csrA-lacZexpression were unaltered in this mutant (Fig. 1E; data not shown, respectively). In addition, expression of uvrY-lacZ and barA- lacZ fusions showed minimal or no effects of csrD (data not shown). Surprisingly, while csrB-lacZ and csrC-lacZ fusions exhibited decreased expression in the mutant (Fig. 1C), CsrB RNA levels were elevated (2.4-fold) and CsrC levels were essentially unchanged (Fig. 1E). Effects of the csrD[H11019]cam mutation were complemented by the cloned csrD gene (pBYH4) (e.g., see Fig. 1C). Furthermore, deletion of the csrD coding region in the genome reproduced the mutant phenotype (data not shown), confirming that it is caused by inactivation of csrD.
Table 1.
Bacterial strains, plasmids, and phages used in this study
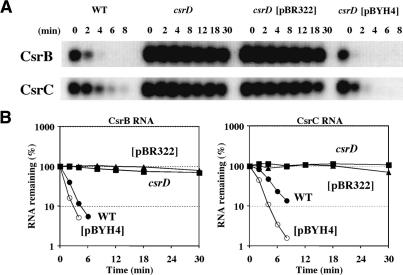
Based on the Csr autoregulatory circuitry (Fig. 1A), the above observations suggested that csrD might be required for functional inactivation of CsrB and/or CsrC, since the csrD mutation would be predicted to increase CsrA sequestration. To test this hypothesis, we determined the effects of csrD on the degradation of CsrB and CsrC RNAs in rifampicin-treated cultures. CsrB and CsrC decay rates were drastically decreased in the csrD mutant strain, a phenotype that was complemented by a plasmid carrying the wild-type csrD gene (Fig. 2). These dramatic effects of csrD on CsrB/C decay were consistently observed in mid-exponential, transition, and early stationary phases of growth under both gluconeogenic (LB medium) and glycolytic (Kornberg medium) conditions (data not shown).
Figure 2.
Effect of csrD on CsrB and CsrC decay. (A) Northern blot of CsrB and CsrC RNAs, following rifampicin addition to 37°C cultures of MG1655 (wild type), or DCMG (csrD) with or without plasmids pBR322 and pBYH4 (csrD++) at the transition from exponential to stationary phase of growth. RNA was separated by electrophoresis on 1.5% agarose gels containing form- aldehyde. (B) Quantitative PhosphorImager data obtained from the experiment shown in A. The RNA half-lives were determined from the linear portions of the decay curves of B. The CsrB half-life in isogenic strains MG1655 (wild type) and csrD mutant (csrD, csrD[pBR322]) and overexpressing (csrD[pBYH4]) strains was 1.4, >30, >30, and 0.9 min, respectively. The CsrC half-life in the same strains was 2.2, >30, >30, and 1.1 min, respectively.
Epistasis experiments were conducted to determine whether the effects of csrD on bacterial gene expression were mediated through the Csr regulatory circuitry (Table 2). We observed that regulation of all genes and phenotypes that were tested by csrD—including csrB- lacZ expression (Table 2, lines 1–4, 15–19), glycogen synthesis (data not shown), and biofilm formation (Table 2, lines 5–8, 11–14)—required functional csrB/C and csrA genes, respectively. Our model (Fig. 1A) predicts that in the absence of decay, CsrB/C RNAs should accumulate and sequester CsrA, leading to decreased transcription of csrB/C. We tested this model by constructing a plasmid from which csrB transcription was driven by a heterologous promoter (pCB44), thereby permitting CsrB to be overproduced. This plasmid clone repressed chromosomal csrB-lacZ expression, as predicted (Table 2, lines 9–10). Previous analyses revealed that activation of csrB expression by CsrA was completely dependent on the response regulator UvrY, but only partly dependent on the sensor-kinase BarA. Furthermore, ectopic expression of csrA restored csrB-lacZ expression in csrA and barA mutants, but not in a uvrY mutant, and ectopic expression of barA was unable to restore a csrA mutation (Suzuki et al. 2002). Similarly, ectopic expression of csrD restored csrB-lacZ expression in csrD and barA mutants (Table 2, lines 15–17, 20–21), but not in csrA or uvrY mutants (Table 2, lines 18–19, 22–23). In addition, csrB- lacZ expression in a csrD mutant was restored by ectopic expression of csrA but not by barA (Table 2, cf. lines 16 and 24–25). These studies fully supported the regulatory circuitry depicted in Figure 1A.
Table 2.
Epistasis analyses of csrD effects on csrB-lacZ expression and biofilm formation
CsrD-mediated RNA decay requires RNase E
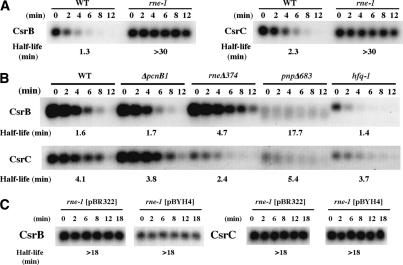
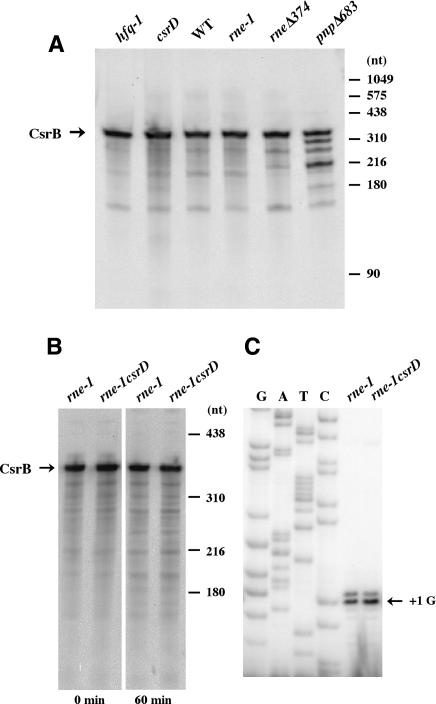
To determine which enzymes participated in the turnover of CsrB/C RNAs, we measured their half-lives in a series of mutants that affect mRNA turnover. Strikingly, the half-lives of both CsrB and CsrC increased >10-fold in an rne-1 mutant at the nonpermissive temperature (Fig. 3A). This immediate (<2 min) cessation of decay and large increase in half-life suggested that RNase E may be directly involved in the decay of both regulatory RNAs. Ectopic expression of csrD from a multicopy plasmid did not cause CsrB/C turnover under RNase E nonpermissive conditions (Fig. 3C). Furthermore, CsrB decay over an extended period of time (90 min) was identical in a strain lacking RNase E activity versus one lacking both RNase E and CsrD (Supplementary Fig. S1), confirming that these two factors function within a single decay pathway. Decay of CsrB was also dependent to a lesser extent upon PNPase, a 3'-to-5' exonuclease that is also a component of the RNase E-based degradosome (Fig. 3B). In fact, CsrB decay intermediates accumulated in the pnp[H9004]683 mutant (Figs. 3B, 4A). Interestingly, the half- life of CsrB RNA increased almost threefold in the absence of degradosome assembly in rne[H9004]374 (Fig. 3B), but was unaffected by loss of PAP I ([H9004]pcnB1) or Hfq (Fig. 3B). While the decay of the CsrC RNA also required RNase E activity, loss of PNPase activity did not substantially affect the half-life of its products (Fig. 3B). In addition, CsrC half-life decreased modestly in the absence of degradosome assembly (Fig. 3B). Further evidence that the decay of CsrB and CsrC RNAs employed slightly different mechanisms was derived from an analysis of a csrA csrD double mutant. This experiment determined if the formation of CsrA/CsrB or CsrA/CsrC complexes required CsrD for their turnover. In the case of CsrB, turnover was still dependent on CsrD, while CsrC decay became CsrD independent (Supplementary Fig. S2). CsrB and CsrC did not appreciably influence each other's decay (Supplementary Fig. S2).
Figure 3.
Effects of RNase E, PNPase, Hfq, PAP I, and the RNase E degradosome scaffold on CsrB/C turnover. (A) Northern blot of CsrB and CsrC RNAs from MG1693 (wild type) and SK5665 (rne-1), following temperature shift of cultures (at transition to stationary phase) from 30°C to 44°C and rifampicin addition, or from (B) MG1693 (wild type), SK7988 ([H9004]pcnB), SK9971 (rne[H9004]374), SK10019 (pnp[H9004]683), and SK10023 (hfq-1) grown at 37°C followed by rifampicin addition. (C) Northern blot of CsrB and CsrC RNAs following temperature shift of SK5665 (rne-1) strains containing pBR322 vector or pBYH4 (csrD +). RNA half-lives were determined as in Figure 2.
Figure 4.
CsrB decay patterns. (A) Northern blot of CsrB RNA from MG1693 (wild type), SK5665 (rne-1; grown at 30°C), SK9971 (rne[H9004]374), SK10019 (pnp[H9004]683), SK10023 (hfq-1), and DCMG (csrD). Steady-state RNA was prepared from cultures harvested at the transition to stationary phase growth. (B) Northern blot of CsrB RNA from SK5665 (rne-1) and DCSK5665 (rne-1 csrD) at T = 0 and 60 min following temperature shift from 30°C to 44°C and rifampicin addition. In this experiment, RNA was prepared from cultures harvested 0 and 60 min after temperature shift and rifampicin addition. (C) Primer extension analysis of RNA preparations (60 min) shown in B. The major 5' end of CsrB RNA is indicated by +1, and is identical to the CsrB transcription initiation site reported previously (Gudapaty et al. 2001).
CsrD is a specificity factor, not a regulator of bulk RNase E activity
Although CsrB/C decay was very defective in the csrD mutant (Fig. 2), its growth was unaffected (e.g., see Fig. 1C). Thus, we suspected that CsrD was not required for maintenance of bulk RNase E activity because loss of this enzyme would lead to either a significant growth defect (Ow et al. 2002) or the loss of cell viability. To test this idea directly, we examined the effect of the csrD[H11019]cam allele on the half-lives of three transcripts whose turnover depends on RNase E: the mRNAs rpsO and rpsT, and the noncoding antisense RNA RyhB. Half- lives of the mRNAs were determined after the addition rifampicin of to cultures, while RyhB transcription was induced by chelation of iron with 2,2'-dipyridyl and inhibited by addition of excess FeSO4, as described (Massé et al. 2003). The decay of these RNAs was unaltered in the csrD[H11019]cam mutant (Supplementary Fig. S3). In addition, Western blot analysis showed that RNase E protein levels were, within experimental error, identical in csrD mutant and wild-type control (M. Stead and S.R. Kushner, unpubl.).
CsrD does not appear to be a ribonuclease
The possibility that CsrD might initiate CsrB decay by modifying or cleaving this RNA was suggested by the fact that the bacterial GGDEF domain belongs to the ancestral palm domain family, which includes nucleotidyl transferases and RNA-binding proteins (Pei and Grishin 2001; Li et al. 2002). Thus, the effects of CsrD and other factors on the pattern of CsrB RNA decay products were examined. Steady-state RNA was prepared from wild-type, csrD[H11019]cam, hfq-1, rne-1, rne[H9004]374, and pnp[H9004]683 strains, separated on 6% urea PAGE gels, and analyzed by Northern blotting (Fig. 4A). In this analysis, the wild type, csrD[H11019]cam, hfq-1, and rne-1 yielded very similar patterns that contained few decay intermediates (Fig. 4A). In contrast, in the pnp[H9004]683 mutant there were a substantial number of apparent decay intermediates (Fig. 4A). Interestingly, some of these intermediates were observed in the rne[H9004]374 mutant, where degradosome assembly could not occur.
Subsequently, an experiment was conducted to determine whether CsrD was a nuclease that might facilitate attack by RNase E. rne-1 and csrD[H11019]cam rne-1 mutants were grown into transition phase, at which time rifampicin was added and the strains were transferred to non- permissive temperature. Immediately, and 60 min thereafter, RNA was isolated and examined by Northern blot analysis. As shown in Figure 4B, there was a single major transcript with the mobility of full-length CsrB and a ladder of minor decay products that were identical in both strains (Fig. 4B). Primer extension analysis revealed identical 5' ends for CsrB RNAs of both strains (Fig. 4C), which corresponded to the initiating nucleotide of this transcript (Gudapaty et al. 2001). These results indicate that CsrD does not cleave CsrB in order to permit prototypical 5' end recognition and turnover of this transcript by RNase E (Kushner 2002).
CsrD activity does not involve c-di-GMP synthesis or turnover
CsrD is predicted to be a membrane-bound signaling protein containing GGDEF and EAL domains. Proteins containing GGDEF and EAL domains have been proposed to synthesize and degrade c-di-GMP, respectively (Ryjenkov et al. 2005; Schmidt et al. 2005). Since domain deletions and plasmid complementation tests determined that both of these domains were required for CsrD activity (Supplementary Fig. S4), we conducted experiments to assess the possibility that CsrD activity is based on c-di-GMP. Isogenic wild-type, mutant, or csrD-overexpressing strains were grown in limiting phosphate and labeled with 32Pi, and guanine nucleotides were extracted and analyzed by 2D-TLC, an approach that is highly sensitive for detection of c-di-GMP (Tischler and Camilli 2004; Hickman et al. 2005). While the other gua- nine nucleotides were readily identified from these strains, c-di-GMP was not detected (data not shown).
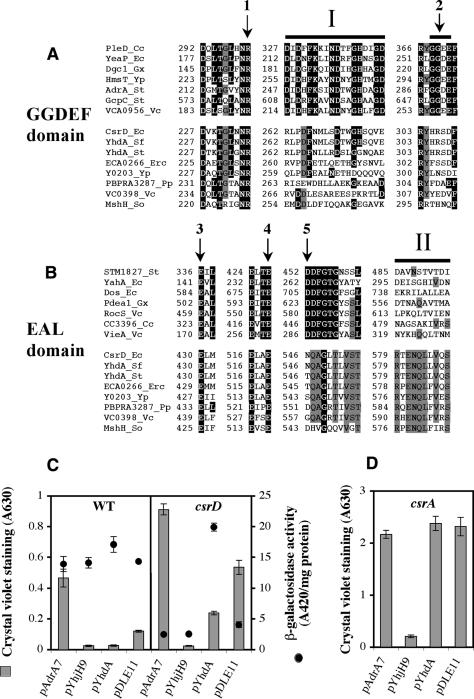
Site-directed mutations in the GGDEF signature sequence have been found to disrupt the function of this domain (García et al. 2004; Kirillina et al. 2004; Paul et al. 2004; Simm et al. 2004). Furthermore, studies of the three-dimensional (3D) structure of PleD (Chan et al. 2004) revealed that the GGDEF motif contains residues involved in substrate binding (G368, G369, and E371) and catalysis (E370), and that residue R300 is also involved in substrate binding. In addition, the GGDEF domain of CsrD contains HRSDF in place of the conserved GG(D/E)EF motif (Chan et al. 2004), suggesting that CsrD might not synthesize c-di-GMP (Fig. 5A). To obtain direct evidence concerning the role of the HRSDF sequence in csrD function, single or double amino acid residue changes of HRSD and R235 (corresponding to R300 of PleD) residues were introduced into CsrD by site-directed mutagenesis and tested for their effects on csrB-lacZ expression and biofilm formation (Fig. 5A; Supplementary Table S4). None of the residues examined were required for CsrD to regulate these activities.
Figure 5.
Amino acid sequence comparisons among putative CsrD orthologs and other GGDEF and EAL domain proteins, and effects of c-di-GMP metabolism genes on csrB-lacZ expression and biofilm formation. (A,B) Conserved regions of c-di-GMP metabolizing GGDEF and EAL domain proteins (upper sets of sequences) and CsrD orthologs (lower sets). Residues identical in >80% of the c-di-GMP metabolizing EAL or GGDEF domains are indicated by a black background. Residues identical in >80% of the CsrD orthologs are indicated by a gray background. The positions shown by arrows (1–5) depict amino acid(s) that have been substituted by alanine or other amino acids in one or more c-di-GMP metabolizing GGDEF or EAL domain proteins or CsrD (see Supplementary Table S4). Region I is highly conserved in c-di-GMP biosynthetic GGDEF domains and region II is one that is highly conserved in the EAL domain of CsrD orthologs, but not in other EAL domain proteins. (Cc) Caulobacter crescentus; (Ec) E. coli; (Gx) Gluconacetobacter xylinus; (Yp) Yersinia pestis; (St) Salmonella enterica Typhimurium; (Vc) Vibrio cholerae; (Sf) Shigella flexneri; (Erc) Erwinia cartovora; (Pp) Photobacterium profundum; (So) Shewanella oneidensis. (C,D) Effects of ectopic expression of adrA (pAdrA7), yhjH (pYhjH9), and csrD (pYhdA) vs. empty vector (pDLE11) on expression of csrB-lacZ and biofilm formation in wild type or csrD mutant of KSB837 (C) and on biofilm formation in acsrA mutant of MG1655 (D). Strains were cultured in the presence of IPTG (1 mM) to maintain expression of the cloned genes. The vector control was pDLE11 in each case. Crystal violet staining for monitoring biofilm formation and β-galactosidase activity are shown as bars and closed circles, respectively. Values are reported as the average ± standard deviation.
In the case of the EAL domain, the 3D structure has not been solved and active site residues have not been clearly defined. Nevertheless, single-residue changes in the conserved EAL signature sequence inactivate EAL domain proteins (Kirillina et al. 2004; Simm et al. 2004; Tischler and Camilli 2004; Tamayo et al. 2005). However, E430A substitution in the EML (corresponding to EAL) sequence of CsrD did not affect CsrD activity (Fig. 5B; Supplementary Table S4). Another substitution, E519A, in a highly conserved amino acid residue of EAL proteins, resulted in partial loss of activity (Fig. 5B; Supplementary Table S4). In contrast, YhjH lost all activity by E136A substitution, which corresponds to E519A of CsrD (Simm et al. 2004). These results suggested that the EAL domain of CsrD activity does not hydrolyze c-di-GMP.
In addition, we determined the effects of genes involved in the synthesis or degradation of c-di-GMP on csrB-lacZ expression and biofilm formation. These experiments were based on observations that genes for c-di- GMP-metabolizing GGDEF proteins, as well as EAL proteins, often cross-complement, especially when the heterologous gene is overexpressed (García et al. 2004; Simm et al. 2004, 2005; Tischler and Camilli 2004). In Salmonella species, AdrA (GGDEF) and YhjH (EAL) proteins possess diguanylate cyclase and c-di-GMP phosphodiesterase (PDE-A) activity, respectively, through which they activate and repress biofilm formation (Simm et al. 2004). E. coli K-12 has closely related orthologs of AdrA and YhjH (75% and 79% identical, respectively), which we expected to possess the same activities. Plasmid clones of these genes were active in vivo. adrA stimulated biofilm formation and yhjH repressed biofilm formation, via unknown target(s), in both wild-type and csrD[H11019]cam mutant strains. However, unlike csrD, neither adrA nor yhjH affected csrB-lacZ expression (Fig. 5C). While CsrD had no effect on biofilm formation in the csrA mutant background (Table 2, 11– 14), biofilm formation was repressed by yhjH in this background (Fig. 5D), indicating that the latter effect is not mediated through the Csr system.
Phylogenetic distribution of csrD
BLAST searches (http://www.ncbi.nlm.nih.gov/BLAST) and Clustal X analyses (Thompson et al. 1997) were conducted to assess the phylogenetic distribution of CsrD (data not shown). CsrD orthologs were apparent in the sequenced genomes of Enterobacteriaceae (50%–99% amino acid identity), Vibrionaceae ([H11350]31%), and Shewanellaceae ([H11350]28%) species (Fig. 5). These proposed orthologs were identified based on (1) identical domain structure and high sequence similarity with respect to CsrD, (2) divergence of the GGDEF signature and the conserved EAL and DDFGTG sequences of the EAL domain (Schmidt et al. 2005), and (3) the presence of amino acid sequences that are conserved in the CsrD orthologs (e.g., region II in Fig. 5B) but not in c-di-GMP metabolizing proteins. Site-directed replacement of six residues of the later type in CsrD by alanine revealed a change, L584A, which partially eliminated activity (71% inactivation based on csrB-lacZ expression) without affecting protein accumulation (Supplementary Table S5), suggesting an important function for this leucine. The uniform presence of rne and csrA genes in the genomes of these species (data not shown) is also consistent with a common function for their CsrD-like proteins. While csrB/C orthologs are challenging to identify by comparative sequence analyses (Weilbacher et al. 2003), functional homologs of these RNAs are known from other Enterobacteriaceae (Ma et al. 2001; Altier 2005), Vibrionaceae (Lenz et al. 2005), and Pseudomonadaceae (Heurlier et al. 2004).
Discussion
The results presented here demonstrate that in E. coli the global regulatory RNAs CsrB and CsrC require a specificity factor, CsrD, for their decay through an RNase E-mediated pathway. To our knowledge, this is the first report of RNA turnover being selectively controlled by a predicted modular signaling protein containing GGDEF and EAL domains. Of equal significance is that the CsrD protein does not function either in the synthesis or degradation of c-di-GMP. It is likely that the CsrD–RNase E-mediated decay pathway operates in many Gram-negative bacteria and broadly influences metabolism, motility/sessility, quorum sensing, host– microbe interactions, and virulence factor expression.
While our studies have not yet clearly defined the mechanism of CsrD action, analysis of decay products (Fig. 4) suggests that CsrD is not a nuclease. In fact, we have isolated a recombinant CsrD protein that lacks the membrane-spanning regions and contains an N-terminal His6 tag (see pQDY3, Supplementary Fig. S4). This purified protein binds to CsrB and CsrC RNAs with high affinity, but without specificity (Supplementary Fig. S5), and does not degrade CsrB or hydrolyze c-di-GMP (data not shown). These observations are of particular interest since bioinformatics analyses have suggested that the GGDEF domain is homologous to eukaryotic adenylyl cyclases and the palm domains of DNA polymerase β, CCA-adding enzyme, and a number of other proteins that interact with RNA (Pei and Grishin 2001; Li et al. 2002, and references therein).
We therefore hypothesize that CsrD functions by binding to the CsrB and CsrC RNAs, converting them into substrates for RNase E degradation. We predict that the binding of CsrD to CsrB and CsrC may change their structures in such a way as to make them accessible to RNase E. We also hypothesize that CsrD activity is not constant under all conditions, and by modulating CsrB/C decay, it helps to determine when CsrA is active. Consistent with this idea, expression of a chromosomal csrD-lacZ translational fusion was modestly repressed (twofold) by CsrA (data not shown). This observation also indicates that CsrD is part of an additional autoregulatory loop within the Csr system. CsrB/C decay rates vary significantly over the course of the growth curve (data not shown), although the precise role of CsrD in this response remains to be determined.
This model predicts that CsrD should be an RNA- binding protein. In fact, in vitro experiments have shown that CsrD binds to both CsrB and CsrC RNAs with high affinity (~25 nM), but that the binding was not specific (Supplementary Fig. S5). Thus, it is possible that there is an additional specificity factor that we have yet to identify. Alternatively, CsrD could be a generalized RNA- binding protein that in vivo is prevented from associating with most RNAs either by proteins (e.g., ribosomes, Hfq) or due to their conformations.
Because the mechanism of action of CsrB/C is fundamentally different from other noncoding regulatory RNAs, it should not be surprising that their decay pathways are distinct. The latter RNAs (with the exception of 6S RNA) (Wassarman and Storz 2000) are antisense RNAs that require Hfq to mediate base-pairing with mRNA targets. Hfq binding typically stabilizes antisense RNAs until they interact with a cognate mRNA, and thereafter targets both RNAs for turnover by RNase E. It has been suggested that this depends on the fact that RNase E and Hfq have similar (AU-rich) target sequences that permit Hfq to protect antisense RNA from RNase E attack until base-pairing has occurred (Massé et al. 2003). Since the decay of RyhB antisense RNA is not affected by CsrD (Supplementary Fig. S4), while CsrB/C degradation is not affected by Hfq (Fig. 3), we hypothesize that CsrD– RNA interactions are necessary for turnover of CsrB/C because they do not contain any obvious RNase E (or Hfq) recognition regions.
Although CsrD is predicted to be anchored in the plasma membrane and it has been suggested that RNase E is associated with the inner membrane (Liou et al. 2001), the presence of the predicted N-terminal membrane anchor was not essential for CsrD activity when the protein was ectopically expressed (Supplementary Fig. S4). Nevertheless, this finding does not exclude a possible role of the membrane anchor in subsubcellular localization or signal sensing. In contrast, the HAMP- like domain (Appleman and Stewart 2003) of CsrD was required for activity. We suspect that this region, including its predicted coiled-coil, may be needed for protein– protein interactions, but have not examined this possibility. Previous proteomic analyses of E. coli cytoplasmic membrane proteins did not identify CsrD (YhdA) (Fountoulakis and Gasser 2003), and attempts to prepare clones expressing CsrD fusion proteins for membrane topology analyses failed (Daley et al. 2005). These results may be explained by the fact that csrD is expressed at extremely low levels, and the full-length CsrD protein, containing the membrane-spanning regions, causes cell lysis upon overexpression (data not shown). Because csrB/C expression is relatively strong and these RNAs are abundant (Gudapaty et al. 2001; Weilbacher et al. 2003), the high rates of turnover mediated by CsrD further imply that this protein is active on these RNAs at substoichiometric concentrations.
Interestingly, the conundrum posed by the abundance of GGDEF–EAL proteins in many species has been partially resolved by this study. Clearly, all of these proteins are not dedicated to c-di-GMP metabolism. Based on criteria defined above, we tentatively identified GGDEF– EAL proteins of other species that are CsrD orthologs (Fig. 5) and likely function in the decay of Csr (Rsm) RNAs. There are also GGDEF and EAL proteins in various species that do not fit our criteria for CsrD orthologs, but nevertheless lack amino acid sequences that should be required for c-di-GMP metabolism (data not shown). Thus, the strategies taken here should be useful for establishing which of these proteins possesses novel mechanisms.
The role of CsrA is not simply to switch genes on or off, but to fine-tune expression; e.g., for governing relative fluxes of competing metabolic pathways (Sabnis et al. 1995; Pernestig et al. 2003). Consequently, CsrA activity is not regulated by covalent modification or small ligand binding, but rather by RNA antagonists whose levels can be rapidly adjusted to offer continuous high- fidelity control of CsrA activity. While CsrB and CsrC RNAs are functionally related and expressed via the same regulatory circuitry, they differ quantitatively in both respects (Suzuki et al. 2002; Weilbacher et al. 2003). Likewise, decay of both CsrB and CsrC utilizes a CsrD– RNase E pathway, but differs in response to ancillary decay factors (PNPase and degradosome assembly) (Fig. 4; data not shown) and CsrA (Supplementary Fig. S2). The presence of orthologs of these RNAs in enteric species suggests that these subtle distinctions, while not fully understood, are likely to be biologically important.
Due to the explosive growth of research on post-transcriptional regulation and RNA decay, common features of eukaryotic and bacterial processes are emerging (e.g., see Gottesman 2005). Antisense RNAs dominate both worlds, no doubt, because base-pairing reactions can mediate highly specific targeting interactions. Csr homologs are not apparent in Archaea or Eukarya. Nevertheless, BC1 RNA of neurons and germ cells is analogous to CsrB/C, in that it binds to and antagonizes RNA-binding proteins involved in translation control and forms ribonucleoprotein complexes (H. Wang et al. 2005, and references therein). Since we have only begun to understand RNA regulatory mechanisms and networks in eukaryotes (Mattick and Makunin 2005), it would not be surprising to find Csr-like regulatory systems in eukaryotes as well.
Materials and methods
Bacterial strains, plasmids, bacteriophage, and growth conditions
All E. coli K-12 strains, plasmids, and bacteriophage used in this study are listed in Table 1 or in Supplemental Material, as appropriate. The various mutant alleles of this study were moved among strains by bacteriophage P1vir transduction, as described previously (Miller 1972). Luria-Bertani (LB) growth medium (Miller 1972) was used for routine cultures, flhDC-lacZ and pgaA-lacZ gene expression assays, and biofilm formation as- says. Thymine (50 μg/mL) was added to LB medium for growth of strains containing the thyA715 allele. Kornberg growth medium (1.1% K2HPO4, 0.85% KH2PO4, 0.6% yeast extract containing 0.5% glucose for liquid or 1% for solid medium) was used for other gene expression assays, selection of transposon mutants, glycogen phenotype determination, and Western and Northern blot analyses, with the exception of Northern analyses of thyA715 strains, which used LB plus thymine. The following antibiotics were added, as required, at the indicated concentrations: 20 μg/mL chloramphenicol, 50 μg/mL kanamycin, 100 μg/mL ampicillin, 10 μg/mL streptomycin, 10 μg/mL tetracycline, and 200 μg/mL rifampicin, except that kanamycin was used at 100 μg/mL for the selection of csrA[H11019]kan strains.
Isolation of transposon mutants and identification of insertion sites
KSB837 (csrB-lacZ) strain was infected with [H9261]NK1324 containing mini-Tn10cam at a multiplicity of infection of 0.15, as described previously (Kleckner et al. 1991). Mutants with altered β-galactosidase activity on Kornberg agar containing X-gal (40 μg/mL) and sodium pyrophosphate (2.5 mM) were isolated and then examined for glycogen accumulation. Mutants with altered csrB-lacZ expression and glycogen production were retained. Amplification of chromosomal DNA flanking the transposon insertions by arbitrarily primed PCR and sequencing of the PCR products were conducted as described previously (Wang et al. 2004).
Glycogen, β -galactosidase, motility, and quantitative biofilm assay
Glycogen accumulation was examined by staining colonies with iodine vapor (Liu et al. 1997). β-Galactosidase activity was assayed as described previously (Suzuki et al. 2002). Assays for flhDC-lacZ and pgaA-lacZ expression were conducted as described (Wei et al. 2001; X. Wang et al. 2005). Single time point β-galactosidase assays were conducted at 6 h of growth. Biofilm formation by cultures at 24 h of growth was assayed by crystal violet staining, as described previously (Jackson et al. 2002).
Cloning of csrD, adrA, and yhjH genes
Plasmid pYhdA encoding the csrD gene, including 222 base pairs (bp) upstream of and 266 bp downstream from the csrD ORF, was constructed by amplifying the E. coli csrD gene with primers yhdAF and yhdAR and ligating it into pCR2.1-TOPO by TA cloning. The orientation of csrD was the same as that of the lac promoter of the vector, which was confirmed by PCR. The EcoRI fragment of pYhdA was subcloned into the EcoRI site of pBR322 to generate pBYH4. Plasmids pAdrA7 and pYhjH9, which express adrA and yhjH under control of the lac promoter, respectively, were constructed by amplifying the E. coli adrA and yhjH genes with primer pairs adrA-F/adrA-R and yhjH-F/ yhjH-R and ligating them into pCR2.1-TOPO by TA cloning. Each forward primer contains its own ribosome-binding site and ATG start codon. The orientation of each gene was confirmed by PCR. Nucleotide sequences of all plasmid inserts were determined to avoid PCR-mediated mutations. The oligonucleo- tide primers used in this study are listed in Supplementary Table S2.
Site-directed mutations and domain deletions of csrD
Site-directed mutations and domain deletions of the csrD gene were constructed using the plasmids pBYH4 and pNC-His. DNA fragments upstream of and downstream from each mutation/deletion were amplified by PCR using pBYH4 as a template. Primers containing the mutation were complementary in sequence with each other (Supplementary Table S2). The two resulting PCR fragments were annealed together and amplified using the primer pair for the ends of the gene. The final PCR product was digested with appropriate restriction enzymes, cloned into pBYH4 or pNC-His (Supplementary Table S2), and confirmed by nucleotide sequencing. The restriction enzymes and primers are listed in Supplementary Tables S2 and S3.
Construction of csrD-null mutant
The chromosomal csrD gene was deleted by targeted gene substitution, as described (Datsenko and Wanner 2000). The kan
gene was amplified from pKD13 by PCR using primers yhP1 and yhP4, and introduced by electroporation into arabinose-treated BW25113[pKD46]. Transformants were selected on kanamycin, and their insertion sites were confirmed by PCR.
Isolation of total RNA
Bacterial cultures were mixed with 2 vol of RNAprotect Bacterial Reagent (Qiagen) and incubated for 5 min at room temperature. Total cellular RNA was subsequently prepared and treated with DNase I using the MasterPure RNA Purification Kit as recommended (Epicentre).
Northern and Western blotting
Total cellular RNA was separated by electrophoresis on 1.5% agarose gels containing formaldehyde or 6% polyacrylamide gels containing 7 M urea. The RNA in agarose gels was then transferred overnight to positively charged nylon membranes (Roche) by capillary action in 20× SSC. The RNA in polyacryl- amide gels was electroblotted onto the same membranes using a Trans-Blot SD semidry transfer cell (Bio-Rad) according to the manufacturer's directions. The blotted membranes were then baked for 30 min at 120°C. DIG-labeled riboprobes were hybridized to RNA on the blots and detected using DIG luminescent detection kit (Roche). Chemiluminescent signals were visualized with ChemiDoc or VersaDoc system (Bio-Rad), and band intensities were quantified using Quantity One software (Bio- Rad). DIG-labeled riboprobes for detection of CsrB, CsrC, RyhB, and rpsO and rpsT transcripts were synthesized from PCR products containing a T7 promoter using a DIG RNA labeling kit (Roche). The primer pairs (Supplementary Table S2) csrBT7- csrBR, csrCT7-csrCR, ryhBT7-ryhBR, rpsOT7-rpsOR, and rpsTT7-rpsTR were used for synthesis of the templates for csrB, csrC, ryhB, rpsO, and rpsT DIG-labeled riboprobes, respectively.
Western blotting of CsrA was performed as described (Gudapaty et al. 2001) on cultures at 4 h of growth or as otherwise indicated.
Primer extension of CsrB RNA
Cells were grown in LB supplemented with thymine at 30°C to the transition to stationary phase of the growth and shifted to 44°C, and rifampicin was added to inhibit transcription. Total RNA was prepared 60 min after the addition of rifampicin. Primer PEX1 that anneals at position +12 to +35 relative to the transcription start site of CsrB was 5'-end-labeled with [[H9253]-32P]ATP (3000 Ci mmol−1, NEN Life Science Products) using T4 polynucleotide kinase (Promega). Unincorporated [[H9253]-32P]-ATP was removed using a MicroSpinTM G-25 Column (Amersham Biosciences). Approximately 3 pmol of labeled primer was added to 5 μg of total RNA. Subsequent cDNA synthesis was performed using the ThermoScriptTM RT–PCR system (Invitrogen). The same labeled primer and pCSRBSF were used to generate a corresponding DNA sequencing ladder using the SequiTherm EXCELTM II DNA Sequencing Kit (Epicentre). The primer extension products were separated alongside the sequencing ladder on an 8% polyacrylamide sequencing gel containing 6 M urea. The gel was dried and subjected to autoradiography using a PhosphorImager (Storm Gel and Blot Imaging system, Amersham Bioscience).
Analysis of guanine nucleotides
Bacterial growth and nucleotide labeling were conducted as described previously, with some modification (Bochner and Ames1982; Tischler and Camilli 2004). Overnight cultures were grown at 37°C in supplemented MOPS medium (Wanner et al. 1977) and used to inoculate the same medium. These cultures were grown at 37°C with aeration until an OD600 = 0.6 was reached. Cells were collected by centrifugation, resuspended in medium plus 100 μCi mL−1 32Pi (PerkinElmer), and incubated for 1–4 h at 37°C to label nucleotides. Following the labeling, nucleotides were extracted and 2D-TLC was conducted as described previously (Tischler and Camilli 2004). The TLC plate was dried and subjected to autoradiography using a PhosphorImager (Storm Gel and Blot Imaging system, Amer- sham Bioscience).
ACKNOWLEDGMENTS
We are grateful to Mark Gomelsky for the generous gifts of cyclic-di-GMP and linear-di-GMP. These studies were funded in part by the National Institutes of Health (GM059969; GM066794). Kane Biotech, Inc., may develop applications related to the findings herein. T.R. serves as Chief Scientific Advisor for, owns equity in, and may receive royalties from this company. The terms of this arrangement have been reviewed and approved by Emory University in accordance with its conflict of interest policies.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1461606.
References
- Altier, C. Genetic and environmental control of Salmonella invasion. J. Microbiol. 2005;43:85–92. [PubMed] [Google Scholar]
- Appleman, J.A., Stewart, V. Mutational analysis of a conserved signal-transducing element: The HAMP linker of the Escherichia coli nitrate sensor NarX. J. Bacteriol. 2003;185:89–97. doi: 10.1128/JB.185.1.89-97.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arraiano, C.M., Yancey, S.D., Kushner, S.R. Stabilization of discrete mRNA breakdown products in ams pnp rnb multiple mutants of Escherichia coli K-12. J. Bacteriol. 1988;170:4625–4633. doi: 10.1128/jb.170.10.4625-4633.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, C.S., Morozov, I., Suzuki, K., Romeo, T., Babitzke, P. CsrA regulates glycogen biosynthesis by preventing translation of glgC in Escherichia coli . Mol. Microbiol. 2002;44:1599–1610. doi: 10.1046/j.1365-2958.2002.02982.x. [DOI] [PubMed] [Google Scholar]
- Bochner, B.R., Ames, B.N. Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. J. Biol. Chem. 1982;257:9759–9769. [PubMed] [Google Scholar]
- Callaghan, A.J., Aurikko, J.P., Ilag, L.L., Grossmann, J.G., Chandran, V., Kühnel, K., Poljak, L., Carpousis, A.J., Robinson, C.V., Symmons, M.F., et al. Studies of the RNA degradosome-organizing domain of the Escherichia coli ribonuclease RNase E. J. Mol. Biol. 2004;340:965–979. doi: 10.1016/j.jmb.2004.05.046. [DOI] [PubMed] [Google Scholar]
- Camilli, A., Bassler, B.L. Bacterial small-molecule signaling pathways. Science. 2006;311:1113–1116. doi: 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpousis, A.J. The Escherichia coli RNA degradosome: Structure, function and relationship in other ribonucleolytic multienzyme complexes. Biochem. Soc. Trans. 2002;30:150–155. [PubMed] [Google Scholar]
- Chan, C., Paul, R., Samoray, D., Amiot, N.C., Giese, B., Jenal, U., Schirmer, T. Structural basis of activity and allosteric control of diguanylate cyclase. Proc. Natl. Acad. Sci. 2004;101:17084–17089. doi: 10.1073/pnas.0406134101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen, M., Christen, B., Folcher, M., Schauerte, A., Jenal, U. Identification and characterization of a cyclic di- GMP-specific phosphodiesterase and its allosteric control by GTP. J. Biol. Chem. 2005;280:30829–30837. doi: 10.1074/jbc.M504429200. [DOI] [PubMed] [Google Scholar]
- D'Argenio, D.A., Miller, S.I. Cyclic di-GMP as a bacterial second messenger. Microbiol. 2004;150:2497–2502. doi: 10.1099/mic.0.27099-0. [DOI] [PubMed] [Google Scholar]
- Daley, D.O., Rapp, M., Granseth, E., Melén, K., von Heijne, G. Global topology analysis of the Escherichia coli inner membrane proteome. Science. 2005;308:1321–1323. doi: 10.1126/science.1109730. [DOI] [PubMed] [Google Scholar]
- Datsenko, K.A., Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diwa, A., Bricker, A.L., Jain, C., Belasco, J.G. An evolutionarily conserved RNA stem-loop functions as a sensor that directs feedback regulation of RNase E gene expression. Genes & Dev. 2000;14:1249–1260. [PMC free article] [PubMed] [Google Scholar]
- Dubey, A.K., Baker, C., Romeo, T., Babitzke, P. RNA sequence and secondary structure participate in high-affinity CsrA–RNA interaction. RNA. 2005;11:1579–1587. doi: 10.1261/rna.2990205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fountoulakis, M., Gasser, R. Proteomic analysis of the cell envelope fraction of Escherichia coli . Amino Acids. 2003;24:19–41. doi: 10.1007/s00726-002-0339-z. [DOI] [PubMed] [Google Scholar]
- García, B., Latasa, C., Solano, C., García-del Portillo, F., Gamazo, C., Lasa, I. Role of the GGDEF protein family in Salmonella cellulose biosynthesis and biofilm formation. Mol. Microbiol. 2004;54:264–277. doi: 10.1111/j.1365-2958.2004.04269.x. [DOI] [PubMed] [Google Scholar]
- Goodman, A.L., Kulasekara, B., Rietsch, A., Boyd, D., Smith, R.S., Lory, S. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa . Dev. Cell. 2004;7:745–754. doi: 10.1016/j.devcel.2004.08.020. [DOI] [PubMed] [Google Scholar]
- Gottesman, S. The small RNA regulators of Escherichia coli: Roles and mechanisms. Annu. Rev. Microbiol. 2004;58:303–328. doi: 10.1146/annurev.micro.58.030603.123841. [DOI] [PubMed] [Google Scholar]
- Gottesman, S. Micros for microbes: Non-coding regulatory RNAs in bacteria. Trends Genet. 2005;21:399–404. doi: 10.1016/j.tig.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Gudapaty, S., Suzuki, K., Wang, X., Babitzke, P., Romeo, T. Regulatory interactions of Csr components: The RNA binding protein CsrA activates csrB transcription in Escherichia coli . J. Bacteriol. 2001;183:6017–6027. doi: 10.1128/JB.183.20.6017-6027.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer, B.K., Tateda, E.S., Swanson, M.S. A two- component regulator induces the transmission phenotype of stationary-phase Legionella pneumophila . Mol. Microbiol. 2002;44:107–118. doi: 10.1046/j.1365-2958.2002.02884.x. [DOI] [PubMed] [Google Scholar]
- Heurlier, K., Williams, F., Heeb, S., Dormond, C., Pessi, G., Singer, D., Cámara, M., Williams, P., Haas, D. Positive control of swarming, rhamnolipid synthesis, and lipase production by the posttranscriptional RsmA/RsmZ system in Pseudomonas aeruginosa PA01. J. Bacteriol. 2004;156:2936–2945. doi: 10.1128/JB.186.10.2936-2945.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman, J.W., Tifrea, D.F., Harwood, C.S. A chemo- sensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. 2005;102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisert, K.B., MacCoss, M., Shiloh, M.U., Darwin, K.H., Singh, S., Jones, R.A., Ehrt, S., Zhang, Z., Gaffney, B.L., Gandotra, S., et al. A glutamate–alanine–leucine (EAL) domain protein of Salmonella controls bacterial survival in mice, antioxidant defence and killing of macrophages: Role of cyclic diGMP. Mol. Microbiol. 2005;56:1234–1245. doi: 10.1111/j.1365-2958.2005.04632.x. [DOI] [PubMed] [Google Scholar]
- Jackson, D.W., Suzuki, K., Oakford, L., Simecka, J.W., Hart, M.E., Romeo, T. Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli . J. Bacteriol. 2002;184:290–301. doi: 10.1128/JB.184.1.290-301.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain, C., Belasco, J.G. RNase E autoregulates its synthesis by controlling the degradation rate of its own mRNA in Escherichia coli Unusual sensitivity of the rnetranscript to RNase E activity. Genes& Dev. 1995;9:84–96. doi: 10.1101/gad.9.1.84. [DOI] [PubMed] [Google Scholar]
- Jenal, U. Cyclic di-guanosine-monophosphate comes of age: A novel secondary messenger involved in modulating cell surface structures in bacteria? Curr. Opin. Microbiol. 2004;7:185–191. doi: 10.1016/j.mib.2004.02.007. [DOI] [PubMed] [Google Scholar]
- Kirillina, O., Fetherston, J.D., Bobrov, A.G., Abney, J., Perry, R.D. HmsP, a putative phosphodiesterase, and HmsT, a putative diguanylate cyclase, control Hms-dependent bio- film formation in Yersinia pestis . Mol. Microbiol. 2004;54:75–88. doi: 10.1111/j.1365-2958.2004.04253.x. [DOI] [PubMed] [Google Scholar]
- Kleckner, N., Bender, J., Gottesman, S. Uses of transposons with emphasis on Tn10 . Methods Enzymol. 1991;204:139–180. doi: 10.1016/0076-6879(91)04009-d. [DOI] [PubMed] [Google Scholar]
- Kushner, S.R. mRNA decay in Escherichia colicomes of age. J.Bacteriol discussion 4657. 2002;184:4658–4665. doi: 10.1128/JB.184.17.4658-4665.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K., Zhan, X., Gao, J., Qiu, J., Feng, Y., Meganathan, R., Cohen, S.N., Georgiou, G. RraA. a protein inhibitor of RNase E activity that globally modulates RNA abundance in E. coli . Cell. 2003;114:623–634. [PubMed] [Google Scholar]
- Lenz, D.H., Miller, M.B., Zhu, J., Kulkarni, R.V., Bassler, B.L. CsrA and three redundant small RNAs regulate quorum sensing in Vibrio cholerae . Mol. Microbiol. 2005;58:1186–1202. doi: 10.1111/j.1365-2958.2005.04902.x. [DOI] [PubMed] [Google Scholar]
- Li, F., Xiong, Y., Wang, J., Cho, H.D., Tomita, K., Weiner, A.M., Steitz, T.A. Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and its complexes with ATP or CTP. Cell. 2002;111:815–824. doi: 10.1016/s0092-8674(02)01115-7. [DOI] [PubMed] [Google Scholar]
- Liou, G.G., Jane, W.N., Cohen, S.N., Lin, N.S., Lin-Chao, S. RNA degradosomes exist in vivo in Escherichia coli as multicomponent complexes associated with the cytoplasmic membrane via N-terminal region of ribonuclease E. Proc. Natl. Acad. Sci. 2001;98:63–68. doi: 10.1073/pnas.011535498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, M.Y., Romeo, T. The global regulator CsrA of Escherichia coli is a specific mRNA-binding protein. J. Bacteriol. 1997;179:4639–4642. doi: 10.1128/jb.179.14.4639-4642.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, M.Y., Gui, G., Wei, B., Preston, J.F., III, Oakford, L., Yüksel, U., Giedroc, D.P., Romeo, T. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli . J. Biol. Chem. 1997;272:17502–17510. doi: 10.1074/jbc.272.28.17502. [DOI] [PubMed] [Google Scholar]
- Ma, W., Cui, Y., Liu, Y., Dumenyo, C.K., Mukherjee, A., Chatterjee, A.K. Molecular characterization of global regulatory RNA species that control pathogenicity factors in Erwinia amylovora and Erwinia herbicola pv. gypsophilae. J. Bacteriol. 2001;183:1870–1880. doi: 10.1128/JB.183.6.1870-1880.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdalani, N., Vanderpool, C.K., Gottesman, S. Bacterial small RNA regulators. Crit. Rev. Biochem. Mol. Biol. 2005;40:93–113. doi: 10.1080/10409230590918702. [DOI] [PubMed] [Google Scholar]
- Massé, E., Escorcia, F.E., Gottesman, S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli . Genes & Dev. 2003;17:2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick, J.S., Makunin, I.V. Small regulatory RNAs in mammals. Hum. Mol. Genet. 2005;14:R121–R132. doi: 10.1093/hmg/ddi101. [DOI] [PubMed] [Google Scholar]
- Miller, J.H. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor; NY: 1972. [Google Scholar]
- Mohanty, B.K., Kushner, S.R. Genomic analysis in Escherichia coli demonstrates differential roles for poly- nucleotide phosphorylase and RNase II in mRNA abundance and decay. Mol.Microbiol. 2003;50:645–658. doi: 10.1046/j.1365-2958.2003.03724.x. [DOI] [PubMed] [Google Scholar]
- Mohanty, B.K., Maples, V.F., Kushner, S.R. The Sm- like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli . Mol. Microbiol. 2004;54:905–920. doi: 10.1111/j.1365-2958.2004.04337.x. [DOI] [PubMed] [Google Scholar]
- Morita, T., Kawamoto, H., Mizota, T., Inada, T., Aiba, H. Enolase in the RNA degradosome plays a crucial role in the rapid decay of glucose transporter mRNA in the response to phosphosugar stress in Escherichia coli . Mol. Microbiol. 2004;54:1063–1075. doi: 10.1111/j.1365-2958.2004.04329.x. [DOI] [PubMed] [Google Scholar]
- Morita, T., Maki, K., Aiba, H. RNase E-based ribonucleoprotein complexes: Mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes & Dev. 2005;19:2176–2186. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudd, E.A., Higgins, C.F. Escherichia coli endoribonuclease RNase E: Autoregulation of expression and site- specific cleavage of mRNA. Mol. Microbiol. 1993;9:557–568. doi: 10.1111/j.1365-2958.1993.tb01716.x. [DOI] [PubMed] [Google Scholar]
- O'Hara, E.B., Chekanova, J.A., Ingle, C.A., Kushner, Z.R., Peters, E., Kushner, S.R. Polyadenylylation helps regulate mRNA decay in Escherichia coli . Proc. Natl. Acad. Sci. 1995;92:1807–1811. doi: 10.1073/pnas.92.6.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ow, M.C., Liu, Q., Kushner, S.R. Analysis of mRNA decay and rRNA processing in Escherichia coli in the absence of RNase E-based degradosome assembly. Mol. Micro- biol. 2000;38:854–866. doi: 10.1046/j.1365-2958.2000.02186.x. [DOI] [PubMed] [Google Scholar]
- Ow, M.C., Liu, Q., Mohanty, B.K., Andrew, M.E., Maples, V.F., Kushner, S.R. RNase E levels in Escherichia coli are controlled by a complex regulatory system that involves transcription of the rne gene from three promoters. Mol. Microbiol. 2002;43:159–171. doi: 10.1046/j.1365-2958.2002.02726.x. [DOI] [PubMed] [Google Scholar]
- Paul, R., Weiser, S., Amiot, N.C., Chan, C., Schirmer, T., Giese, B., Jenal, U. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes & Dev. 2004;18:715–727. doi: 10.1101/gad.289504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei, J., Grishin, N.V. GGDEF domain is homologous to adenylyl cyclase. Proteins. 2001;42:210–216. doi: 10.1002/1097-0134(20010201)42:2<210::aid-prot80>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Pernestig, A.K., Georgellis, D., Romeo, T., Suzuki, K., Tomenius, H., Normark, S., Melefors, Ö. The Escherichia coli BarA–UvrY two-component system is needed for efficient switching between glycolytic and gluconeogenic carbon sources. J. Bacteriol. 2003;185:843–853. doi: 10.1128/JB.185.3.843-853.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prud'homme-Généreux, A., Beran, R.K., Iost, I., Ramey, C.S., Mackie, G.A., Simons, R.W. Physical and functional interactions among RNase E, polynucleotide phosphorylase and the cold-shock protein, CsdA: Evidence for a ‘cold shock degradosome.’. Mol. Microbiol. 2004;54:1409–1421. doi: 10.1111/j.1365-2958.2004.04360.x. [DOI] [PubMed] [Google Scholar]
- Py, B., Causton, H., Mudd, E.A., Higgins, C.F. A protein complex mediating mRNA degradation in Escherichia coli . Mol. Microbiol. 1994;14:717–729. doi: 10.1111/j.1365-2958.1994.tb01309.x. [DOI] [PubMed] [Google Scholar]
- Py, B., Higgins, C.F., Carpousis, A.J., Higgins, C.F. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Mol. Microbiol. 1996;26:387–398. [Google Scholar]
- Romeo, T. Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol. Microbiol. 1998;29:1321–1330. doi: 10.1046/j.1365-2958.1998.01021.x. [DOI] [PubMed] [Google Scholar]
- Romeo, T., Gong, M., Liu, M.Y., Brun-Zinkernagel, A.M. Identification and molecular characterization of csrA, a pleiotropic gene from Escherichia coli that affects glycogen biosynthesis, gluconeogenesis, cell size, and surface properties. J. Bacteriol. 1993;175:4744–4755. doi: 10.1128/jb.175.15.4744-4755.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling, U., Gomelsky, M., Galperin, M.Y. C-di- GMP: The dawning of a novel bacterial signalling system. Mol. Microbiol. 2005;57:629–639. doi: 10.1111/j.1365-2958.2005.04697.x. [DOI] [PubMed] [Google Scholar]
- Ryjenkov, D.A., Tarutina, M., Moskvin, O.V., Gomelsky, M. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: Insights into biochemistry of the GGDEF protein domain. J. Bacteriol. 2005;187:1792–1798. doi: 10.1128/JB.187.5.1792-1798.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabnis, N.A., Yang, H., Romeo, T. Pleiotropic regulation of central carbohydrate metabolism in Escherichia coli via the gene. csrA.J. Biol. Chem. 1995;270:29096–29104. doi: 10.1074/jbc.270.49.29096. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Russell, D.W. 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor; NY: 2001. Molecular cloning: A laboratory manual. [Google Scholar]
- Schmidt, A.J., Ryjenkov, D.A., Gomelsky, M. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: Enzymatically active and inactive EAL domains. J. Bacteriol. 2005;187:4774–4781. doi: 10.1128/JB.187.14.4774-4781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simm, R., Morr, M., Kader, A., Nimtz, M., Römling, U. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 2004;53:1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- Simm, R., Fetherston, J.D., Kader, A., Römling, U., Perry, R.D. Phenotypic convergence mediated by GGDEF- domain-containing proteins. J. Bacteriol. 2005;187:6816–6823. doi: 10.1128/JB.187.19.6816-6823.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa, S., Marchand, I., Dreyfus, M. Autoregulation allows Escherichia coliRNase E to adjust continuously its synthesis to that of its substrates. Mol. Microbiol. 2001;42:867–878. doi: 10.1046/j.1365-2958.2001.02687.x. [DOI] [PubMed] [Google Scholar]
- Storz, G., Altuvia, S., Wassarman, K.M. An abundance of RNA regulators. Annu. Rev. Biochem. 2005;74:199–217. doi: 10.1146/annurev.biochem.74.082803.133136. [DOI] [PubMed] [Google Scholar]
- Suzuki, K., Wang, X., Weilbacher, T., Pernestig, A.K., Melefors, Ö, Georgellis, D., Babitzke, P., Romeo, T. Regulatory circuitry of the CsrA/CsrB and BarA/UvrY systems of Escherichia coli. J. Bacteriol. 2002;184:5130–5140. doi: 10.1128/JB.184.18.5130-5140.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo, R., Tischler, A.D., Camilli, A. The EAL domain protein VieA is a cyclic diguanylate phosphodiesterase. J. Biol. Chem. 2005;280:33324–33330. doi: 10.1074/jbc.M506500200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischler, A.D., Camilli, A. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol. Microbiol. 2004;53:857–869. doi: 10.1111/j.1365-2958.2004.04155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., Preston, J.F., III, Romeo, T. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J. Bacteriol. 2004;186:2724–2734. doi: 10.1128/JB.186.9.2724-2734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Iacoangeli, A., Lin, D., Williams, K., Denman, R.B., Hellen, C.U.T., Tiedge, H. Dentritic BC 1 RNA in translational control mechanisms. J. Cell Biol. 2005;171:811–821. doi: 10.1083/jcb.200506006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., Dubey, A.K., Suzuki, K., Baker, C.S., Babitzke, P., Romeo, T. CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccha- ride adhesin of Escherichia coli . Mol. Microbiol. 2005;56:1648–1663. doi: 10.1111/j.1365-2958.2005.04648.x. [DOI] [PubMed] [Google Scholar]
- Wanner, B.L., Kodaira, R., Neidhart, F.C. Physiological regulation of a decontrolled lac operon. J. Bacteriol. 1977;130:212–222. doi: 10.1128/jb.130.1.212-222.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman, K.M., Storz, G. RNA regulates E. coliRNA polymerase activity. Cell. 2000;101:6S. 613–623. doi: 10.1016/s0092-8674(00)80873-9. [DOI] [PubMed] [Google Scholar]
- Wei, B.L., Brun-Zinkernagel, A.M., Simecka, J.W., Prüß, B.M., Babitzke, P., Romeo, T. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli . Mol. Microbiol. 2001;40:245–256. doi: 10.1046/j.1365-2958.2001.02380.x. [DOI] [PubMed] [Google Scholar]
- Weilbacher, T., Suzuki, K., Dubey, A.K., Wang, X., Gudapaty, S., Morozov, I., Baker, C.S., Georgellis, D., Babitzke, P., Romeo, T. A novel sRNA component of the carbon storage regulatory system of Escherichia coli . Mol. Micro- biol. 2003;48:657–670. doi: 10.1046/j.1365-2958.2003.03459.x. [DOI] [PubMed] [Google Scholar]
- Whistler, C.A., Ruby, E.G. GacA regulates symbiotic colonization traits of Vibrio fischeri and facilitates a beneficial association with an animal host. J. Bacteriol. 2003;185:7202–7212. doi: 10.1128/JB.185.24.7202-7212.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]