Abstract
Embryonic development requires generating cell types at the right place (spatial patterning) and the right time (temporal patterning). Drosophila neuroblasts undergo stem cell-like divisions to generate an ordered sequence of neuronal progeny, making them an attractive system to study temporal patterning. Embryonic neuroblasts sequentially express Hunchback, Krüppel, Pdm1/Pdm2 (Pdm), and Castor (Cas) transcription factors. Hunchback and Krüppel specify early-born temporal identity, but the role of Pdm and Cas in specifying temporal identity has never been addressed. Here we show that Pdm and Cas regulate late-born motor neuron identity within the NB7-1 lineage: Pdm specifies fourth-born U4 motor neuron identity, while Pdm/Cas together specify fifth-born U5 motor neuron identity. We conclude that Pdm and Cas specify late-born neuronal identity; that Pdm and Cas act combinatorially to specify a temporal identity distinct from either protein alone, and that Cas repression of pdm expression regulates the generation of neuronal diversity.
Keywords: [Drosophila, birth order, neuroblast, temporal, timing]
In the mammalian CNS, individual neural progenitors give rise to an ordered series of cell types in the cerebral cortex, retina, and spinal cord (for review, see Pearson and Doe 2004). The cerebral cortex is arranged in layers 1, 2/3, 4, 5, and 6—from superficial to deep—with each layer expressing unique molecular markers and having lamina-specific axon and dendrite projection patterns (McConnell 1992; Callaway 2002; Kubo and Nakajima 2003). Retroviral lineage analysis shows that individual progenitors can generate neurons of all layers, and birth-dating studies show that each layer is produced sequentially over time (Berry and Rogers 1965; Luskin and Shatz 1985; Walsh and Reid 1995; Reid et al. 1997). Layer 1 neurons are made first, followed by layers 6, 5, 4, and 2/3. Thus, the temporal identity of each neuron (i.e., its birth order) is tightly correlated with its laminar identity. Elegant transplantation studies show that extrinsic cues act on the progenitor to specify its temporal identity, and that this identity becomes fixed or cell autonomous around the final G2/M phase of the neuronal cell cycle (McConnell 1988; McConnell and Kaznowski 1991). These studies predict that extrinsic cues trigger an intrinsic developmental program in the newborn post-mitotic neurons. To date, only one intrinsic temporal identity factor has been identified: Foxg1, which is necessary and sufficient to repress early-born layer 1 neuronal identity (Hanashima et al. 2004). However, recent work has confirmed that single cortical progenitors cultured in vitro can recapitulate many aspects of their normal lineage (Shen et al. 2006), strongly implicating lineage-intrinsic cues in regulating temporal identity.
In addition to the cerebral cortex, temporal patterning is important for retinal development. Mammalian retinal progenitors divide to produce seven major cell types. Birthdating and retroviral lineage studies show that individual progenitors can generate many or all of these cell types, and that there is an evolutionarily conserved sequence of cell type production (Turner and Cepko 1987; Holt et al. 1988; Turner et al. 1990; Wetts and Fraser 1991; Fekete et al. 1994; Cepko et al. 1995; Chang and Harris 1998). Transplantation and in vitro culture studies reveal that both intrinsic and extrinsic cues regulate neuronal temporal identity in the retina (Ezzeddine et al. 1997; Morrow et al. 1998; Rapaport et al. 2001; Cayouette et al. 2003; Li et al. 2004; Shen et al. 2006), and that retinal progenitors undergo progressive restriction in their ability to respond to these cues (Belliveau et al. 2000).
Recently, the Drosophila embryonic CNS has emerged as an attractive system to study the specification of neuronal temporal identity and progressive restriction of progenitor competence. Drosophila neurogenesis begins with the delamination of neural stem cells, called neuroblasts, into the interior of the embryo while the cells remaining at the surface make ventral epidermis (Skeath 1999). Each neuroblast can be uniquely identified by its position, time of formation, and pattern of gene expression (Doe 1992; Broadus et al. 1995). Neuroblasts divide asymmetrically in a stem cell mode to bud off a series of smaller cells called ganglion mother cells (GMCs); each GMC typically generates a pair of neurons or glia. Early-born GMCs and neurons are passively displaced further inward by later-born GMCs, such that the first-born neurons are located in the deepest layer of the CNS and the last-born neurons lie in the most superficial layer of the CNS (Isshiki et al. 2001; Pearson and Doe 2003). Cell lineage studies show that neuroblast lineages are invariant, with each neuroblast contributing a characteristic family of neurons and glia to the CNS (Bossing et al. 1996; Schmidt et al. 1997; Schmid et al. 1999; Pearson and Doe 2003). For example, the NB in the seventh row, first column (NB7-1) sequentially generates the U1–U5 motor neurons and then ~30 interneurons (Pearson and Doe 2003).
Four transcription factors, Hunchback (Hb), Krüppel (Kr), Pdm1/Pdm2 (Pdm), Castor (Cas), are sequentially expressed in Drosophila neuroblasts and maintained in their neuronal progeny (summarized in Fig. 1A; Kambadur et al. 1998; Isshiki et al. 2001). Due to their sequential expression pattern and persistence in neurons based on their birth order, these transcription factors are excellent candidates for specifying temporal identity in the CNS. In fact, Hb and Kr are necessary and sufficient for specifying early-born temporal identity in multiple neuroblast lineages (Isshiki et al. 2001; Novotny et al. 2002; Pearson and Doe 2003; Kanai et al. 2005; Cleary and Doe 2006). In addition, experiments in which pulses of Hb or Kr are given at different points in the NB7-1 lineage reveal a single early competence window for responding to Hb or Kr, during which time NB7-1 undergoes progressive restriction (Isshiki et al. 2001; Novotny et al. 2002; Pearson and Doe 2003; Kanai et al. 2005; Cleary and Doe 2006).
Figure 1.
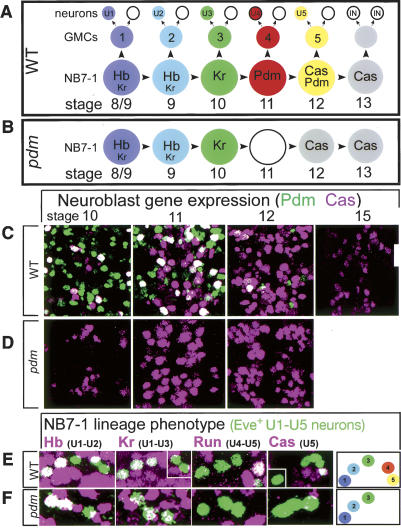
Pdm is required to generate late-born U4 and U5 neurons. (A) Schematic of the wild-type NB7-1 lineage and temporal identity gene expression profile (Schmid et al. 1999; Isshiki et al. 2001; Pearson and Doe 2003). Even-skipped (Eve) is detected in the first five GMCs and their progeny U1–U5 neurons, but not in their neuronal siblings or in the later-born GMCs and interneuronal progeny. Pdm is only transiently detected in U4. Embryonic stages (stage) according to Campos-Ortega and Hartenstein (1997). In this figure and below, neuroblast color reflect the neuronal identity produced at each point in the lineage. We infer that the first interneurons in the lineage arise from a Cas-positive Pdm-negative neuroblast based on the very transient window of Pdm/Cas coexpression, but the identification of markers for these interneurons will be necessary to resolve this point. (B) Schematic of NB7-1 gene expression profile in pdm mutants, based on the timing of gene expression in the entire neuroblast population at the indicated embryonic stages. (C,D) Wild-type and pdm mutant embryos showing Pdm and Cas expression in the neuroblast layer at the indicated embryonic stages. Ventral view of two segments; anterior is up. (E,F) Wild-type and pdm mutant embryos assayed for the indicated U1–U5 neuronal identity markers. (E) In wild type, U1–U5 were present. (F) In pdm mutants, U1–U3 were present but U4/ U5 neurons were often absent (74% both absent; n = 163 hemisegments). Each panel shows one hemisegment of a stage 16 CNS as a maximum intensity projection of the optical sections encompassing the U1–U5 neurons; anterior up, midline to left; neurons are shown as insets if they are obscured in the projection. (Hb) Hunchback; (Kr) Krüppel; (Run) Runt; (Cas) Castor. Summary of U1–U5 phenotype shown at right.
Although Pdm1/Pdm2 and Cas are excellent candidates for specifying late-born temporal identity based on their expression patterns, they have not been tested for a role in specifying temporal identity. pdm1 and pdm2 are adjacent genes that encode POU homeodomain proteins of the Oct1 family and are coexpressed within the embryonic CNS (Dick et al. 1991). In addition to being expressed in a temporal pattern between Kr and cas during neuroblast lineages, pdm1/pdm2 are also expressed in a small subset of GMCs and neurons, including the early-born RP2 neuron, where they act redundantly to regulate gene expression and axon pathfinding (Yang et al. 1993; Yeo et al. 1995; McDonald et al. 2003). For example, mutations in pdm1 alone (also called nubbin; FlyBase) or pdm2 alone show very weak defects in RP2 specification compared with mutations that remove both pdm1 and pdm2 (Yeo et al. 1995), and we observe a similar redundancy within the NB7-1 lineage (data not shown). Due to their common expression pattern and redundant function, we will refer to Pdm1/Pdm2 simply as Pdm. Previously, cas mutants were shown to have defects in neuronal gene expression and formation of specific CNS axon fascicles (Cui and Doe 1992; Mellerick et al. 1992), but due to the lack of markers and lineage data available in the early 1990s it was not possible to analyze temporal identity phenotypes. cas has also been shown to regulate axon pathfinding of post-embryonic neurons in the central complex and mushroom body (Hitier et al. 2001). Here we use loss-of-function and misexpression experiments to analyze the role of Pdm and Cas in temporal cell fate specification within the well-characterized NB7-1 lineage. We report that Pdm and Cas are required for development of late-born motor neuron identity within the NB7-1 lineage: Pdm specifies U4 motor neuron fate, whereas Pdm and Cas together specify U5 motor neuron fate. In addition, we confirm and extend previous results showing that NB7-1 has a single competence window to respond to Hb, Kr, Pdm, and Cas, and we provide additional information on the mechanisms regulating the precise timing of hb, Kr, pdm, and cas transcription in neuroblasts.
Results
To investigate the role of Pdm1/Pdm2 (henceforth Pdm) and Cas in specifying late-born neuronal identity we focus on the well-characterized NB7-1 lineage (Isshiki et al. 2001; Pearson and Doe 2003; Grosskortenhaus et al. 2005; Cleary and Doe 2006). In this lineage, the five Eve+ GMCs each produce one Eve+ neuron (U1–U5, based on birth order; Pearson and Doe 2003) and one Eve− sibling cell. The next ~15 GMCs generate different types of interneurons (Bossing et al. 1996; Schmid et al. 1999; Pearson and Doe 2003). Each of the U1–U5 motor neurons can be uniquely identified by molecular marker expression, cell position, and axon projections (Fig. 1A,E; Isshiki et al. 2001; Pearson and Doe 2003); there are no known molecular markers for identifying the U siblings or interneurons in the lineage.
Pdm is required to generate the late-born U4 and U5 neurons
During wild-type CNS development, neuroblasts express Hb at stage 8–9, Kr at stage 10, Pdm at stage 11, Pdm plus Cas at early stage 12, and Cas alone at stage 13 (Fig. 1A,C; Isshiki et al. 2001; Cleary and Doe 2006). The timing of Pdm neuroblast expression corresponds to the interval that NB7-1 is generating the U4 and U5 neurons, and consistent with this timing we can detect transient Pdm in the U4 and U5 neurons but not in earlier-born U1–U3 neurons (data not shown).
To investigate Pdm CNS function, we used the small deficiency Df(2L)ED773 that removes both pdm1 and pdm2 genes. In embryos homozygous for Df(2L)ED773 (subsequently called pdm mutant embryos), we found no change in the timing of Hb, Kr, or Cas in neuroblasts (Fig. 1B,D; data not shown), and conclude that Pdm is not required to down-regulate hb or Kr neuroblast expression or to activate cas neuroblast expression. We next examined the function of Pdm in specifying U1–U5 motor neuron identity. Whereas wild-type embryos always had five normally specified U1–U5 neurons (Fig. 1E), pdm mutant embryos had normal U1–U3 neuronal fates, but typically lacked the U4 and U5 neurons (Fig. 1F; quantified in legend). These neurons may have an alternate cell fate or undergo apoptosis (see below). We conclude that Pdm is required for the proper development of the late-born U4 and U5 neurons.
Pdm misexpression prolongs Pdm/Cas neuroblast coexpression and induces ectopic U5 neurons
Although Pdm is not required for proper expression of hb, Kr, or cas (preceding section), here we test whether misexpression of Pdm is sufficient to alter the timing of hb, Kr, or cas neuroblast expression. First, we extended the normal window of Pdm neuroblast expression using prospero-gal4 to express a haemagglutinin (HA)-tagged Pdm2 protein (UAS-HA:pdm2); this resulted in prolonged expression of Pdm from its normal initiation at stage 11 until late in embryogenesis (Fig. 2A,B). We found that cas expression persisted as long as Pdm was detectable, resulting in an extended window of Pdm/Cas coexpression within neuroblasts from stage 11 until late stage 16 (Fig. 2A,B; data not shown).
Figure 2.
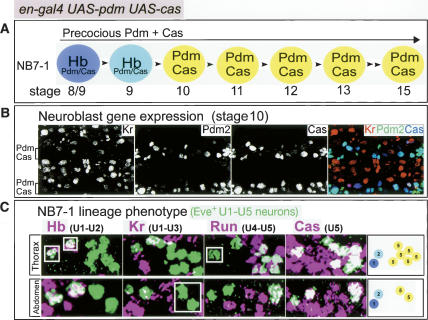
Prolonged Pdm expression can extend the Pdm/Cas coexpression window and generate ectopic U5 neurons (A) Schematic of NB7-1 gene expression profile in embryos with prolonged Pdm expression (prospero-gal4 UAS-HA:pdm2). (B) prospero-gal4 UAS-HA:pdm2 embryos showing Pdm and Cas in the neuroblast layer at the indicated embryonic stages. Ventral view of two segments; anterior is up. (C) prospero-gal4 UASHA:pdm2 embryos assayed for U1–U5 neuronal identities. Wild-type embryos had U1–U5 neurons (Fig. 1E), whereas prospero-gal4 UASHA:pdm2 embryos typically had four extra U5 neurons in the thorax (n = 58; range 2–5) and at most one extra U5 neuron in the abdomen (n = 68; 12% one extra). Panels show one thoracic or abdominal hemisegment at stage 16; anterior up, midline to left; summary of the most common U1–U5 phenotype is shown at right.
To determine the consequences of prolonged Pdm misexpression on neuronal temporal identity, we assayed U1–U5 cell fates within the NB7-1 lineage. We found that prolonged Pdm expression resulted in normal U1–U3 neuronal identity (Fig. 2C), as expected because Pdm is not present in this part of the lineage. We also observed normal formation of the Pdm-dependent U4 neuron (Fig. 2C). Strikingly, we observed four or more super-numerary U5 neurons within thoracic segments, and a similar but weaker phenotype in abdominal segments (Fig. 2C). The ectopic U5 neurons express both Pdm and Cas, consistent with the extended window of Pdm/Cas expression seen in neuroblasts. We conclude that extended Pdm expression can prolong cas expression and induce ectopic U5 neurons.
In addition, we induced Pdm expression prior to its normal time of expression in the NB7-1 lineage using the engrailed-gal4 driver (Fig. 3A). We observed that precocious Pdm resulted in strong repression of Kr expression, weak early activation of cas expression, and prolonged Pdm/Cas coexpression for the duration of the Pdm misexpression window (Fig. 3B; quantified in Supplementary Table 1; data not shown). Assaying older embryos of this genotype for U1–U5 neuronal identity revealed the expected supernumerary U5 neurons as well as a partial loss of U4 and U3 neurons (Fig. 3C). We propose that the extra U5 neurons are due to the extended period of Pdm/Cas coexpression, and that the loss of U4 may be due to the precocious activation of Cas, which transforms the normally Pdm+ Cas− U4 neuron into a Pdm+ Cas+ U5 neuron. In contrast, the loss of U3 is likely due to Pdm repression of Kr expression, because Kr is required for U3 development (Isshiki et al. 2001).
Figure 3.
Precocious Pdm expression can repress Kr, activate cas, and generate ectopic U5 neurons at the expense of U3/U4 neurons. (A) Schematic of NB7-1 gene expression profile in embryos with precocious Pdm expression (engrailed-gal4 UAS-HA:pdm2). (B) engrailed-gal4 UAS-HA:pdm2 embryos stained for Kr and HA:Pdm2 (using anti-HA antibody) at the indicated stages. An optical section through the neuroblast layer is shown. The domain of high HA:Pdm2 in the posterior of each segment (brackets) results in strong repression of Kr and weak activation of Cas; the wild-type pattern of Kr and Cas can be seen in the anterior of each segment where there is no HA:Pdm2 misexpression. Ventral view of two segments; anterior is up. (C) engrailed-gal4 UAS-HA:pdm2 embryos assayed for U1–U5 neuronal identities. Wild-type embryos had U1–U5 neurons (Fig. 1E), whereas engrailed-gal4 UAS-HA:pdm2 embryos had a variable loss of U3/U4 neurons and extra U5 neurons (thoracic segments: 5.2 extra, range 4–9 extra, n = 128; abdominal segments 0–1 extra, n = 359). Panels show one hemisegment of a stage 16 CNS; anterior up, midline to left; summary of the most common phenotypes shown at right.
Taken together, our Pdm experiments allow us to draw several conclusions and make one prediction. First, we conclude that Pdm is expressed during the formation of the U4 neuron and is required for U4 neuron survival and/or specification. Second, we conclude that Pdm and Cas are coexpressed during the formation of the U5 neuron, and at least Pdm is required for U5 survival and/or specification. Third, we conclude that misexpression of Pdm can extend the window of Pdm/Cas coexpression and induce ectopic U5 neurons. Finally, we predict that Pdm/Cas together specify the identity of the late-born U5 neuron, and we test this prediction below.
Castor is required to suppress U4 and promote U5 neuronal identity
In wild-type embryos, cas is expressed at the time of U5 formation (stage 12) and into the subsequent window of interneuron production (Fig. 1A,C). To determine the role of cas in U1–U5 neuron specification we assayed embryos homozygous for cas24 or cas39, both of which act as strong cas alleles (see Materials and Methods; Cui and Doe 1992). We found that cas mutant embryos show the proper timing of hb and Kr neuroblast expression (data not shown), whereas pdm expression persists as late as stage 15 (Fig. 4B). We conclude that Cas is required to repress pdm neuroblast expression, confirming previous results (Kambadur et al. 1998). In addition, neuronal temporal identity assays revealed that cas mutants have supernumerary U4 neurons and a lack of U5 neurons (Fig. 4C). These results suggest that prolonged Pdm expression induces U4 neuronal identity and the lack of Pdm/Cas coexpression prevents specification of U5 neuronal identity.
Figure 4.
Castor is required to suppress U4 and promote U5 neuronal identity. (A) Schematic of NB7-1 gene expression profile in cas mutants. (B) cas24 homozygous mutant embryos showing Pdm in the neuroblast layer at the indicated embryonic stages. Note that Pdm persists until stage 15, which is not observed in wild-type embryos (Fig. 1C). Ventral view of two segments; anterior is up. The neuroblast layer dips out of the focal plane at the segment border (*). (C) cas39 homozygous mutant embryos assayed for U1–U5 neuronal identities. Wild-type embryos had U1– U5 neurons (Fig. 1E), whereas cas mutant embryos had ectopic U4 neurons in both thoracic (6.6 extra; n = 20) and abdominal hemisegments (3.5 extra; n = 29). In this experiment, U5 neurons were identified by expression of a lacZ gene at the cas39 locus, which is expressed in the normal cas pattern despite lack of functional Cas protein (see Materials and Methods for details; see Supplementary Fig. 1A for lacZ expression in the wild-type U5 neuron). Panels show one hemisegment of a stage 16 CNS; anterior up, midline to left; summary of the most common U1–U5 phenotype shown at right. (D) Schematic of NB7-1 gene expression profile in pdm cas24 double mutants. (E) pdm cas24 double-mutant embryos assayed for U1–U5 neuronal identities. Panels show one abdominal hemisegment of a stage 16 CNS; anterior up, midline to left; summary of U1–U5 phenotype shown at right.
To determine if the prolonged Pdm expression in cas mutants is responsible for the formation of ectopic U4 neurons, we generated pdm cas double mutants and analyzed neuronal temporal identity. We found that pdm cas double mutants lacked all extra U4 fates found in cas single mutants, without affecting the early U1–U3 fates (Fig. 4E). Thus, Pdm is required for U4 specification in wild-type embryos (Fig. 1F) as well as for specification of the ectopic U4 neurons in cas mutant embryos (Fig. 4E). We conclude that Pdm alone specifies U4 neuronal identity; both Pdm and Cas are required to specify U5 neuronal identity; and Cas is required to repress pdm expression and limit its window of expression within the NB7-1 lineage.
To test whether precocious Cas expression could repress endogenous Pdm expression and thereby prevent U4 neuron specification, we used the engrailed-gal4 driver to express UAS-cas from the time of neuroblast formation (Fig. 5A). We detected no change in the timing or levels of Hb and Kr expression despite high levels of Cas protein (data not shown), but Pdm levels were strongly reduced within the domain of Cas misexpression (Fig. 5B; quantified in Supplementary Table 1). Consistent with the loss of Pdm in neuroblasts, we observed absence of the U4 and U5 neurons (Fig. 5C), both of which require Pdm for their specification (see above). The same loss of U4/U5 phenotype was observed when we used prospero-gal4 to misexpress Cas (data not shown). Coexpression of Cas and the cell death inhibitor p35 (engrailed-gal4 UAS-cas UA-p35) also resulted in loss of U4/U5 neurons (data not shown), suggesting that ectopic Cas does not kill the U4/U5 neurons but may transform them into another neuronal identity (e.g., later-born interneurons) (see Discussion). We conclude that misexpression of Cas is sufficient to repress Pdm and block specification of U4/U5 neuronal identity, and that Cas alone is insufficient to specify U5 fate.
Figure 5.
Pdm is required for ectopic U4 neurons in cas mutant embryos. (A) Schematic of NB7-1 gene expression profile in embryos with precocious cas expression (engrailed-gal4 UAS-cas). (B) engrailed-gal4 UAS-cas embryos showing Pdm and Cas in the neuroblast layer at the indicated embryonic stages. Ventral view of two segments; anterior is up. Single channels are shown in rows 1 and 2, which then merge in row 3. Domain of ectopic Cas indicated by brackets. Pdm is down-regulated in the neuroblasts that express ectopic Cas (cf. Fig. 1C). Cas expression can significantly repress Pdm protein levels (quantified in Supplementary Table 1). (C) engrailed-gal4 UAS-cas embryos assayed for U1– U5 neuronal identities. In wild type, we observe U1–U5 neurons (Fig. 1E). In engrailed-gal4 UAS-cas embryos, the U1–U3 fates are normal, but U4 and U5 neurons are invariably missing. Panels show one abdominal hemisegment of a stage 16 CNS; anterior up, midline to left; summary of U1–U5 phenotype shown at right.
Pdm/Cas together specify U5 neuronal identity
The mutant and misexpression data described above support a model in which Pdm alone specifies U4 neuronal identity, and Pdm/Cas together specify U5 neuronal identity. Thus, we tested whether coexpressing Pdm/Cas together would be sufficient to induce U5 neuronal identity, at least during the NB7-1 competence window. We used engrailed-gal4 to misexpress UAS-HA:pdm2 and UAS-cas from the time of neuroblast formation (Fig. 6A). We observed normal Hb neuroblast expression but strong repression of Kr neuroblast expression (Fig. 6B), similar to the effect of misexpressing Pdm alone (see Fig. 3A,B). Furthermore, there was prolonged coexpression of Pdm/Cas within neuroblasts of the misexpression domain (data not shown). Within the NB7-1 lineage, we observed normal Hb+ U1/U2 neurons, a loss of the U3/ U4 neurons, and extra U5 neurons (Fig. 6C). To further test the hypothesis that Pdm and Cas together specify U5 identity, we misexpressed Pdm in a cas mutant background. We find that prospero-gal4 UAS-HA:pdm2; cas39/cas39 embryos have ectopic U4 neurons (Supplementary Fig. 1), whereas prospero-gal4 UAS-HA:pdm2 alone have ectopic U5 neurons (Fig. 2). We conclude that Pdm and Cas function together to induce U5 neuronal identity, and a similar competence window exists for generating ectopic U5 neurons as previously observed for Hb-, Kr-, or Pdm-induced earlier-born neuron identities (Fig. 4C; Pearson and Doe 2003; Cleary and Doe 2006).
Figure 6.
Pdm/Cas specify U5 neuronal identity. (A) Schematic of NB7-1 gene expression profile in embryos with Pdm/Cas misexpression (engrailed-gal4 UASHA:pdm2 UAS-cas). (B) engrailed-gal4 UAS-HA:pdm2 UAS-cas embryos showing Kr, Pdm, and Cas in the neuroblast layer at the indicated embryonic stages. Ventral view of two segments; anterior is up. Kr is repressed in the Pdm/Cas misexpression domain. (C) engrailed-gal4 UASHA:pdm2 UAS-cas embryos assayed for U1–U5 neuronal identities. In wild type, we observe U1–U5 neurons (Fig. 1E). In engrailed-gal4 UAS-HA:pdm2 UAS-cas embryos, we observe loss of Kr+ U3 neurons (79% lost; n = 110) and ectopic U5 neurons (approximately six in thoracic hemisegments, n = 35; ~2.5 in abdominal hemisegments, n = 75). We suggest these extra neurons have a U5 fate based on their Pdm− Run+ Cas+ marker expression, but we cannot rule out a U4 identity or a mixed U4/U5 identity without additional markers. Panels show one hemisegment of a stage 16 CNS; anterior up, midline to left; summary of U1–U5 phenotype shown at right.
Discussion
Hb, Kr, Pdm, and Cas are sequentially expressed in neuroblasts, but only Hb and Kr have been shown to have a function in specifying temporal identity (Isshiki et al. 2001; Novotny et al. 2002; Pearson and Doe 2003; Grosskortenhaus et al. 2005; Kanai et al. 2005; Cleary and Doe 2006). Here we show that Pdm and Cas are required for specifying the late-born U4 and U5 neuron fates within the NB7-1 lineage. Thus, all four transcription factors are required to specify sequential temporal identities within the NB7-1 lineage: High Hb gives U1 fate, low Hb gives U2 fate, Kr gives U3 fate, Pdm gives U4 fate, and Pdm/Cas gives U5 fate. Cas is then transiently expressed in the lineage during the window of interneuron production, and it remains possible that Cas alone specifies one or more interneuron identities later in the lineage.
Pdm without Cas specifies U4 neuronal identity
Pdm is detected in neuroblasts during the window that NB7-1 is generating the GMC progenitors of the U4/U5 neurons, and pdm mutant embryos lack the U4/U5 neurons. What happens in the NB7-1 lineage following production of the U3 progenitor in pdm mutant embryos? It is unlikely that NB7-1 dies or is cell cycle arrested, because Cas+ neuroblasts can be observed well after the time of pdm expression (see Fig. 1B,C). It is more likely that the U4/U5 neurons undergo cell death or that NB7-1 “skips” production of U4/U5 neurons and goes directly to the interneuron phase of its lineage. Independent of the mechanism used, it is clear that Pdm is required for the proper development of the late-born U4 and U5 neurons.
If Pdm is required for both U4 and U5 fates, what distinguishes these neuronal identities? The Cas transcription factor is detected in U5 but not U4, leading to a model in which Pdm alone specifies U4 identity and Pdm/Cas specifies U5 identity. Our data fully support such a model. First, cas mutant embryos have an extended window of Pdm only expression, and we observe the formation of supernumerary U4 neurons during this window of Pdm expression. Furthermore, pdm cas double mutants lack these ectopic U4 neurons, showing that the extended window of Pdm is required for specifying the ectopic U4 neurons. These data provide strong support for the conclusion that Pdm without Cas specifies U4 neuronal identity.
If Pdm specifies U4 neuron identity, then why are ectopic U4 neurons not observed following Pdm misexpression? The answer to this apparent paradox is that Pdm misexpression induces Cas expression, resulting in the Pdm/Cas double-positive state that specifies U5 identity (see below). Precocious expression of Pdm also results in repression of Kr, and the occasional loss of the U3 neuron. Finally, misexpression of Pdm can result in the absence of U5 at very low frequency. One possible explanation is that in these segments, Pdm induces sufficiently high levels of Cas to trigger production of the Cas+ interneuron identity that normally occurs after U5 production.
Pdm and Cas together specify U5 neuronal identity
The proposal that Pdm and Cas together specify U5 neuronal identity is supported by several observations: (1) Both pdm and cas single mutants lack U5 neuronal identity; (2) misexpression of Pdm can extend the window of Pdm/Cas coexpression and generate ectopic U5 neurons; (3) misexpression of Pdm in a cas mutant background generates U4 neurons but not U5 neurons; and (4) misexpression of Pdm and Cas together results in ectopic U5 neurons. How might the combination of Pdm and Cas specify a unique neuronal identity, different from either factor alone? It is possible that Pdm and Cas form a heterodimer with a different set of target genes than either factor alone; POU domain proteins such as Pdm are known to heterodimerize with a wide range of transcription factors, including zinc finger transcription factors (Kakizawa et al. 1999; Prefontaine et al. 1999). However, there are no reported Pdm1/Cas or Pdm2/Cas interactions in a genome-wide yeast two-hybrid screen (Giot et al. 2003), and we have been unable to coimmunoprecipitate HA:Pdm2/Cas after co-overexpression (K. Robinson, unpubl.). It is also possible that genes differ in the composition of Pdm- and Cas-binding sites, some genes having sites for Pdm, others having Cas sites, and yet others having coclusters of both sites. Testing this hypothesis using bioinformatics is currently not possible due to the low information content of the Pdm DNA-binding motif (Neumann and Cohen 1998).
Are Pdm and Cas temporal identity genes?
It is clear that Pdm and Cas specify late-born U4/U5 motor neuron fates within the NB7-1 lineage. If they specify late-born neuronal fates in other lineages, they would be temporal identity genes; if they only have this function in the NB7-1 lineage, then they would be better defined as having a U4/U5 cell type specification function. Currently, not enough information exists to distinguish these two possibilities. Besides NB7-1, the only other neuroblast lineage where birth-order data exists is NB7-3, but that lineage is short—just three GMCs—and it does not express cas (Isshiki et al. 2001; Novotny et al. 2002; Karcavich and Doe 2005). In the future, it will be important to determine birth-order relationships in additional embryonic neuroblast lineages, and then test Pdm and Cas for a role in specifying late-born neuronal identity in these lineages. Pdm is known to specify the first-born GMC in the NB4-2 lineage (Yang et al. 1993; Yeo et al. 1995; McDonald et al. 2003), which shows Pdm is not restricted to specifying late-born temporal identity, but this does not preclude it from specifying late-born cell fates in NB4-2 or other neuroblast lineages.
The NB7-1 competence window
NB7-1 has the longest embryonic lineage of any neuroblast, producing ~40 neurons (U1–U5 motor neurons, five U siblings, and 30 interneurons). We have previously shown that pulses of low levels of Hb or Kr can induce one to three extra Eve+ early-born neurons only during the first five cell cycles of the NB7-1 lineage, and misexpression of high levels of Hb or Kr can only generate an average of 4.1 and 4.8 extra Eve+ U neurons, respectively (Pearson and Doe 2003; Cleary and Doe 2006). Thus, NB7-1 has a single early competence window to respond to Hb and Kr. Interestingly, in this study we found that high levels of Pdm or Pdm/Cas also induced approximately four extra Eve+ U neurons. Thus, NB7-1 may lose competence to respond to all four temporal identity factors at the same time—after nine to 10 cell cycles. These data support the conclusion that NB7-1 has a single competence window for all four temporal identity factors. Alternatively, Pdm or Pdm/Cas may induce levels of Cas that exceed a threshold for inducing Eve− interneuron identity.
Differences in thoracic and abdominal phenotypes
We find that prolonged Pdm or Pdm/Cas coexpression generates more U4 or U5 neurons in thoracic segments than in abdominal segments (Figs. 2,3,4,6). One explanation might be the effect of homeotic gene expression on the NB7-1 lineage. Homeotic genes are known to regulate the length of neuroblast lineages (Prokop et al. 1998), the type of neurons generated within neuroblast lineages (Prokop and Technau 1994; Berger et al. 2005), and the timing of neuroblast apoptosis (Bello et al. 2003). Thus, it is possible that homeotic genes also regulate the ability of Pdm or Cas to induce late-born neuronal identity, the length of the competence window, or the survival/proliferation state of NB7-1.
The neuroblast gene expression ‘timer’
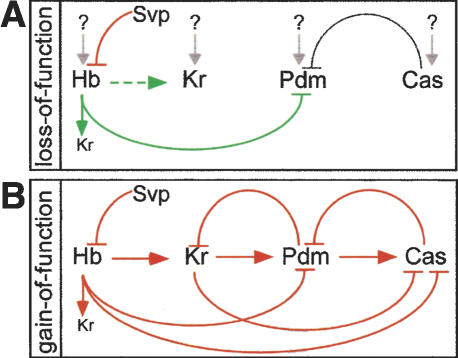
Our results provide new information on the “gene expression timer” that regulates sequential hb, Kr, pdm, and cas expression in embryonic neuroblasts. Previous studies showed that loss of Hb or Kr in neuroblasts did not significantly alter the timing of hb, Kr, pdm or cas neuroblast expression (Isshiki et al. 2001; Grosskortenhaus et al. 2005; Kanai et al. 2005); our studies confirm and extend these conclusions. We find that pdm or cas mutants have no effect on the timing of transcriptional initiation of hb, Kr, pdm, or cas within neuroblast lineages. Thus, expression of hb, Kr, pdm, cas must be induced by one or more unknown transcriptional activators (Fig. 7A). This highlights the importance of identifying the relevant cis-regulatory region controlling the timing of hb, Kr, pdm, and cas expression, and characterizing the trans-acting factors that initiate temporally accurate neuroblast gene expression.
Figure 7.
Regulation of hb, Kr, pdm, and cas neuroblast gene expression. (A) Regulatory hierarchy based on loss-of-function experiments (Isshiki et al. 2001; Kanai et al. 2005; this study). The small Kr below Hb represents low-level Kr that is always coexpressed with Hb; in contrast, the large Kr to the right of Hb represents the high-level Kr expression that occurs after Hb expression is down-regulated. (B) Regulatory hierarchy based on misexpression experiments. In both panels, arrows indicate positive regulation, “T” indicates negative regulation, and “?” indicates one or more unknown transcriptional activators.
Although mutant analysis reveals the presence of unknown transcriptional activators of hb, Kr, pdm, and cas neuroblast expression, misexpression experiments reveal regulatory interactions between each of these genes. We confirm and extend previous studies (Kambadur et al. 1998; Isshiki et al. 2001), showing that each temporal identity factor is capable of activating transcription of the next gene in the pathway (Fig. 7B). The ability of each transcription factor to activate the next gene in the cascade may act redundantly with the unknown transcriptional activators to maintain the linear cascade of gene expression. In addition, we observe many repressive interactions between the temporal identity genes (Fig. 7B), which may serve to maintain distinct temporal windows of expression. It is unknown whether these regulatory interactions are direct or indirect; this is a question we are actively investigating.
Materials and methods
Fly stocks and transgene construction
pdm mutant phenotype was analyzed using Df (2L)ED773 (removes both pdm1 and pdm2). The cas mutant phenotype was analyzed using cas24 and cas39 (Cui and Doe 1992), balanced over TM3 ftz-lacZ. Both cas alleles are strong alleles for their U1–U5 phenotype (same phenotype in trans to a deficiency or to each other) and were made by imprecise excision of the cas2092 lacZ enhancer trap line (Cui and Doe 1992). The cas2092 transposon is inserted at base 1,542,578 of the BDGP release 4.3 genome sequence, within castor 5′ untranslated region (UTR). The cas39 allele has a small deletion that strongly reduces cas expression, but retains a functional lacZ gene that is expressed in the normal cas CNS expression pattern, allowing lacZ expression to be used as a U5 marker in cas mutant embryos (Fig. 4C). The cas24 allele is a deletion that removes lacZ-containing transposon and eliminates cas expression in the NB7-1 lineage, although a few Cas+ cells are detected scattered throughout the CNS. For overexpression experiments, we used prospero-gal4 on chromosome III (F. Matsuzaki, Kobe, Japan), engrailed-gal4 on chromosome II (A. Brand, Cambridge, UK), UAS-cas on chromosome III (W. Odenwald, National Institutes of Health, Washington, DC), UAS-HA:pdm2 on chromosome II and III, and UAS-p35 (Hay et al. 1995). The UAS-HA:pdm2 transgene was made by cloning the ORF from EST RE34565 following the haemagglutinin (HA) sequence into pUAST, and standard methods were used to generate the transgenic flies.
Antibodies, immunofluorescence, and semiquantitation of protein levels
The Pdm2 antibody was made by PCR amplification using the primer pair GCGACACACGAATTCATGCGGCACAT and GCGCTTCTCGAGTCTGCGTTACAGGA from a pdm2 EST RE49429 template, digesting with EcoRI/XhoI, cloning into the EcoRI/XhoI sites of pET-28a, and sequencing for confirmation. Standard methods were used to bacterially express and purify the protein, and inject it into rats to generate polyclonal sera.
Antibody staining was performed according to Grosskortenhaus et al. (2005). Primary antibodies, dilutions, and sources are rabbit anti-β-galactosidase, 1:1000, Cappel; mouse anti-β-galactosidase, 1:500, Promega; guinea pig anti-Hb, 1:400, East Asian Distribution Center for Segmentation Antibodies (EADC); guinea pig anti-Kr, 1:400, EADC; rat anti-Pdm2, 1:10, this work; rabbit anti-Cas, 1:1000, W. Odenwald; guinea pig anti-Eve, 1:400, EADC; mouse anti-Eve, 1:10, Developmental Studies Hybridoma Bank; guinea pig anti-Runt, 1:400, EADC; rat anti-HA, 1:100, Roche. Species-specific secondary antibodies were conjugated to Alexa 488, RhodamineRedX, Cy5 (Jackson), or Biotin (Vector Labs) and were used at 1:400.
Images were collected as confocal image stacks using a Leica SP2 confocal microscope, processed in ImageJ (NIH) and shown as two-dimensional projections. U1–U5 neurons are shown as insets in their approximate spatial position if they would be obscured in the projection.
Semiquantitation of protein levels was done by staining wild-type and misexpression embryos in the same tube, collecting confocal images with the same settings, ensuring all data is within the linear range of 1–255 pixel intensity, outlining each neuroblast using ImageJ (Polygon tool), and quantifying average pixel intensity of each neuroblast using ImageJ (Measure tool).
Acknowledgments
We thank Mike Cleary and Bret Pearson for comments on the manuscript; Takako Isshiki for sharing unpublished data; and Ward Odenwald, Fumio Matsuzaki, Andrea Brand, Steve Cohen, and Bloomington stock center for antibodies and/or fly stocks. This work was supported by a post-doctoral fellowship from the Deutsche Forschungsgemeinschaft to R.G. and an NIH grant HD27056 to C.Q.D., who is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1445306.
References
- Belliveau, M.J., Young, T.L., Cepko, C.L. Late retinal progenitor cells show intrinsic limitations in the production of cell types and the kinetics of opsin synthesis. J. Neurosci. 2000;20:2247–2254. doi: 10.1523/JNEUROSCI.20-06-02247.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello, B.C., Hirth, F., Gould, A.P. A pulse of the Drosophila Hox protein Abdominal-A schedules the end of neural proliferation via neuroblast apoptosis. Neuron. 2003;37:209–219. doi: 10.1016/s0896-6273(02)01181-9. [DOI] [PubMed] [Google Scholar]
- Berger, C., Pallavi, S.K., Prasad, M., Shashidhara, L.S., Technau, G.M. A critical role for cyclin E in cell fate determination in the central nervous system of Drosophila melanogaster . Nat. Cell Biol. 2005;7:56–62. doi: 10.1038/ncb1203. [DOI] [PubMed] [Google Scholar]
- Berry, M., Rogers, A.W. The migration of neuroblasts in the developing cerebral cortex. J. Anat. 1965;99:691–709. [PMC free article] [PubMed] [Google Scholar]
- Bossing, T., Udolph, G., Doe, C.Q., Technau, G.M. The embryonic central nervous system lineages of Drosophila melanogaster. I.Neuroblast lineages derived from the ventral half of the neuroectoderm. Dev. Biol. 1996;179:41–64. doi: 10.1006/dbio.1996.0240. [DOI] [PubMed] [Google Scholar]
- Broadus, J., Skeath, J.B., Spana, E.P., Bossing, T., Technau, G., Doe, C.Q. New neuroblast markers and the origin of the aCC/pCC neurons in the Drosophila central nervous system. Mech. Dev. 1995;53:393–402. doi: 10.1016/0925-4773(95)00454-8. [DOI] [PubMed] [Google Scholar]
- Callaway, E.M. Cell type specificity of local cortical connections. J. Neurocytol. 2002;31:231–237. doi: 10.1023/a:1024165824469. [DOI] [PubMed] [Google Scholar]
- Campos-Ortega, J.A., Hartenstein, V. The embryonic development of Drosophila melanogaster. Springer; Berlin: 1997. [Google Scholar]
- Cayouette, M., Barres, B.A., Raff, M. Importance of intrinsic mechanisms in cell fate decisions in the developing rat retina. Neuron. 2003;40:897–904. doi: 10.1016/s0896-6273(03)00756-6. [DOI] [PubMed] [Google Scholar]
- Cepko, C., Ryder, E.F., Austin, C.P., Walsh, C., Fekete, D.M. Lineage analysis using retrovirus vectors. Methods Enzymol. 1995;254:387–419. doi: 10.1016/0076-6879(95)54027-x. [DOI] [PubMed] [Google Scholar]
- Chang, W.S., Harris, W.A. Sequential genesis and determination of cone and rod photoreceptors in Xenopus . J. Neurobiol. 1998;35:227–244. [PubMed] [Google Scholar]
- Cleary, M.D., Doe, C.Q. Regulation of neuroblast competence: Multiple temporal identity factors specify distinct neuronal fates within a single early competence window. Genes & Dev. 2006;20:429–434. doi: 10.1101/gad.1382206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, X., Doe, C.Q. ming is expressed in neuroblast sublineages and regulates gene expression in the Drosophila central nervous system. Development. 1992;116:943–952. doi: 10.1242/dev.116.4.943. [DOI] [PubMed] [Google Scholar]
- Dick, T., Yang, X.H., Yeo, S.L., Chia, W. Two closely linked Drosophila POU domain genes are expressed in neuroblasts and sensory elements. Proc. Natl. Acad. Sci. 1991;88:7645–7649. doi: 10.1073/pnas.88.17.7645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe, C.Q. Molecular markers for identified neuroblasts and ganglion mother cells in the Drosophila central nervous system. Development. 1992;116:855–863. doi: 10.1242/dev.116.4.855. [DOI] [PubMed] [Google Scholar]
- Ezzeddine, Z.D., Yang, X., DeChiara, T., Yancopoulos, G., Cepko, C.L. Postmitotic cells fated to become rod photoreceptors can be respecified by CNTF treatment of the retina. Development. 1997;124:1055–1067. doi: 10.1242/dev.124.5.1055. [DOI] [PubMed] [Google Scholar]
- Fekete, D.M., Perez-Miguelsanz, J., Ryder, E.F., Cepko, C.L. Clonal analysis in the chicken retina reveals tangential dispersion of clonally related cells. Dev. Biol. 1994;166:666–682. doi: 10.1006/dbio.1994.1346. [DOI] [PubMed] [Google Scholar]
- Giot, L., Bader, J.S., Brouwer, C., Chaudhuri, A., Kuang, B., Li, Y., Hao, Y.L., Ooi, C.E., Godwin, B., Vitols, E., et al. A protein interaction map of Drosophila melanogaster . Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- Grosskortenhaus, R., Pearson, B.J., Marusich, A., Doe, C.Q. Regulation of temporal identity transitions in Drosophila neuroblasts. Dev. Cell. 2005;8:193–202. doi: 10.1016/j.devcel.2004.11.019. [DOI] [PubMed] [Google Scholar]
- Hanashima, C., Li, S.C., Shen, L., Lai, E., Fishell, G. Foxg1 suppresses early cortical cell fate. Science. 2004;303:56–59. doi: 10.1126/science.1090674. [DOI] [PubMed] [Google Scholar]
- Hay, B.A., Wassarman, D.A., Rubin, G.M. Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell. 1995;83:1253–1262. doi: 10.1016/0092-8674(95)90150-7. [DOI] [PubMed] [Google Scholar]
- Hitier, R., Chaminade, M., Preat, T. The Drosophila castor gene is involved in postembryonic brain development. Mech. Dev. 2001;103:3–11. doi: 10.1016/s0925-4773(01)00312-4. [DOI] [PubMed] [Google Scholar]
- Holt, C.E., Bertsch, T.W., Ellis, H.M., Harris, W.A. Cellular determination in the Xenopus retina is independent of lineage and birth date. Neuron. 1988;1:15–26. doi: 10.1016/0896-6273(88)90205-x. [DOI] [PubMed] [Google Scholar]
- Isshiki, T., Pearson, B., Holbrook, S., Doe, C.Q. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell. 2001;106:511–521. doi: 10.1016/s0092-8674(01)00465-2. [DOI] [PubMed] [Google Scholar]
- Kakizawa, T., Miyamoto, T., Ichikawa, K., Kaneko, A., Suzuki, S., Hara, M., Nagasawa, T., Takeda, T., Mori, J., Kumagai, M., et al. Functional interaction between Oct-1 and retinoid X receptor. J. Biol. Chem. 1999;274:19103–19108. doi: 10.1074/jbc.274.27.19103. [DOI] [PubMed] [Google Scholar]
- Kambadur, R., Koizumi, K., Stivers, C., Nagle, J., Poole, S.J., Odenwald, W.F. Regulation of POU genes by castor and hunchback establishes layered compartments in the Drosophila CNS. Genes & Dev. 1998;12:246–260. doi: 10.1101/gad.12.2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai, M.I., Okabe, M., Hiromi, Y. seven-up Controls switching of transcription factors that specify temporal identities of Drosophilaneuroblasts. Dev. Cell. 2005;8:203–213. doi: 10.1016/j.devcel.2004.12.014. [DOI] [PubMed] [Google Scholar]
- Karcavich, R., Doe, C.Q. Drosophila neuroblast 7–3 cell lineage: A model system for studying programmed cell death, Notch/Numb signaling, and sequential specification of ganglion mother cell identity. J. Comp. Neurol. 2005;481:240–251. doi: 10.1002/cne.20371. [DOI] [PubMed] [Google Scholar]
- Kubo, K., Nakajima, K. Cell and molecular mechanisms that control cortical layer formation in the brain. Keio J. Med. 2003;52:8–20. doi: 10.2302/kjm.52.8. [DOI] [PubMed] [Google Scholar]
- Li, S., Mo, Z., Yang, X., Price, S.M., Shen, M.M., Xiang, M. Foxn4 controls the genesis of amacrine and horizontal cells by retinal progenitors. Neuron. 2004;43:795–807. doi: 10.1016/j.neuron.2004.08.041. [DOI] [PubMed] [Google Scholar]
- Luskin, M.B., Shatz, C.J. Neurogenesis of the cat's primary visual cortex. J. Comp. Neurol. 1985;242:611–631. doi: 10.1002/cne.902420409. [DOI] [PubMed] [Google Scholar]
- McConnell, S.K. Fates of visual cortical neurons in the ferret after isochronic and heterochronic transplantation. J. Neurosci. 1988;8:945–974. doi: 10.1523/JNEUROSCI.08-03-00945.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The control of neuronal identity in the developing cerebral cortex. Curr. Opin. Neurobiol. 1992;2:23–27. doi: 10.1016/0959-4388(92)90156-f. ———. [DOI] [PubMed] [Google Scholar]
- McConnell, S.K., Kaznowski, C.E. Cell cycle dependence of laminar determination in developing neocortex. Science. 1991;254:282–285. doi: 10.1126/science.254.5029.282. [DOI] [PubMed] [Google Scholar]
- McDonald, J.A., Fujioka, M., Odden, J.P., Jaynes, J.B., Doe, C.Q. Specification of motoneuron fate in Drosophila: Integration of positive and negative transcription factor inputs by a minimal eve enhancer. J. Neurobiol. 2003;57:193–203. doi: 10.1002/neu.10264. [DOI] [PubMed] [Google Scholar]
- Mellerick, D.M., Kassis, J.A., Zhang, S.D., Odenwald, W.F. castor encodes a novel zinc finger protein required for the development of a subset of CNS neurons in Drosophila . Neuron. 1992;9:789–803. doi: 10.1016/0896-6273(92)90234-5. [DOI] [PubMed] [Google Scholar]
- Morrow, E.M., Belliveau, M.J., Cepko, C.L. Two phases of rod photoreceptor differentiation during rat retinal development. J. Neurosci. 1998;18:3738–3748. doi: 10.1523/JNEUROSCI.18-10-03738.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann, C.J., Cohen, S.M. Boundary formation in Drosophila wing: Notch activity attenuated by the POU protein Nubbin. Science. 1998;281:409–413. doi: 10.1126/science.281.5375.409. [DOI] [PubMed] [Google Scholar]
- Novotny, T., Eiselt, R., Urban, J. Hunchback is required for the specification of the early sublineage of neuroblast 7-3 in the Drosophila central nervous system. Development. 2002;129:1027–1036. doi: 10.1242/dev.129.4.1027. [DOI] [PubMed] [Google Scholar]
- Pearson, B.J., Doe, C.Q. Regulation of neuroblast competence in Drosophila . Nature. 2003;425:624–628. doi: 10.1038/nature01910. [DOI] [PubMed] [Google Scholar]
- Specification of temporal identity in the developing nervous system. Annu. Rev. Cell Dev. Biol. 2004;20:619–647. doi: 10.1146/annurev.cellbio.19.111301.115142. ———. [DOI] [PubMed] [Google Scholar]
- Prefontaine, G.G., Walther, R., Giffin, W., Lemieux, M.E., Pope, L., Hache, R.J. Selective binding of steroid hormone receptors to octamer transcription factors determines transcriptional synergism at the mouse mammary tumor virus promoter. J. Biol. Chem. 1999;274:26713–26719. doi: 10.1074/jbc.274.38.26713. [DOI] [PubMed] [Google Scholar]
- Prokop, A., Technau, G.M. Early tagma-specific commitment of Drosophila CNS progenitor NB1-1. Development. 1994;120:2567–2578. doi: 10.1242/dev.120.9.2567. [DOI] [PubMed] [Google Scholar]
- Prokop, A., Bray, S., Harrison, E., Technau, G.M. Homeotic regulation of segment-specific differences in neuroblast numbers and proliferation in the Drosophila central nervous system. Mech. Dev. 1998;74:99–110. doi: 10.1016/s0925-4773(98)00068-9. [DOI] [PubMed] [Google Scholar]
- Rapaport, D.H., Patheal, S.L., Harris, W.A. Cellular competence plays a role in photoreceptor differentiation in the developing Xenopus retina. J. Neurobiol. 2001;49:129–141. doi: 10.1002/neu.1070. [DOI] [PubMed] [Google Scholar]
- Reid, C.B., Tavazoie, S.F., Walsh, C.A. Clonal dispersion and evidence for asymmetric cell division in ferret cortex. Development. 1997;124:2441–2450. doi: 10.1242/dev.124.12.2441. [DOI] [PubMed] [Google Scholar]
- Schmid, A., Chiba, A., Doe, C.Q. Clonal analysis of Drosophila embryonic neuroblasts: Neural cell types, axon projections and muscle targets. Development. 1999;126:4653–4689. doi: 10.1242/dev.126.21.4653. [DOI] [PubMed] [Google Scholar]
- Schmidt, H., Rickert, C., Bossing, T., Vef, O., Urban, J., Technau, G.M. The embryonic central nervous system lineages ofDrosophila melanogaster. II. Neuroblast lineages derived from the dorsal part of the neuroectoderm. Dev. Biol. 1997;189:186–204. doi: 10.1006/dbio.1997.8660. [DOI] [PubMed] [Google Scholar]
- Shen, Q., Wang, Y., Dimos, J.T., Fasano, C.A., Phoenix, T.N., Lemischka, I.R., Ivanova, N.B., Stifani, S., Morrisey, E.E., Temple, S. The timing of cortical neurogenesis is encoded within lineages of individual progenitor cells. Nat. Neurosci. 2006;9:743–751. doi: 10.1038/nn1694. [DOI] [PubMed] [Google Scholar]
- Skeath, J.B. At the nexus between pattern formation and cell-type specification: The generation of individual neuroblast fates in the Drosophila embryonic central nervous system. Bioessays. 1999;21:922–931. doi: 10.1002/(SICI)1521-1878(199911)21:11<922::AID-BIES4>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Turner, D.L., Cepko, C.L. A common progenitor for neurons and glia persists in rat retina late in development. Nature. 1987;328:131–136. doi: 10.1038/328131a0. [DOI] [PubMed] [Google Scholar]
- Turner, D.L., Snyder, E.Y., Cepko, C.L. Lineage-independent determination of cell type in the embryonic mouse retina. Neuron. 1990;4:833–845. doi: 10.1016/0896-6273(90)90136-4. [DOI] [PubMed] [Google Scholar]
- Walsh, C., Reid, C. Cell lineage and patterns of migration in the developing cortex. Ciba Found. Symp. 1995;193:21–40. doi: 10.1002/9780470514795.ch2. [DOI] [PubMed] [Google Scholar]
- Wetts, R., Fraser, S.E. Microinjection of fluorescent tracers to study neural cell lineages. Development. 1991;2(Suppl.):1–8. [PubMed] [Google Scholar]
- Yang, X., Yeo, S., Dick, T., Chia, W. The role of a Drosophila POU homeo domain gene in the specification of neural precursor cell identity in the developing embryonic central nervous system. Genes & Dev. 1993;7:504–516. doi: 10.1101/gad.7.3.504. [DOI] [PubMed] [Google Scholar]
- Yeo, S.L., Lloyd, A., Kozak, K., Dinh, A., Dick, T., Yang, X., Sakonju, S., Chia, W. On the functional overlap between two Drosophila POU homeo domain genes and the cell fate specification of a CNS neural precursor. Genes & Dev. 1995;9:1223–1236. doi: 10.1101/gad.9.10.1223. [DOI] [PubMed] [Google Scholar]