Abstract
We report that mouse liver cells are highly resistant to extensive telomere dysfunction. In proliferating cells, telomere dysfunction results in chromosome end fusions, a DNA damage signal, and apoptosis or senescence. To determine the consequences of telomere dysfunction in noncycling cells, we used conditional deletion of the telomeric protein TRF2 in hepatocytes. TRF2 loss resulted in telomeric accumulation of γ-H2AX and frequent telomere fusions, indicating telomere deprotection. However, there was no induction of p53 or apoptosis, and liver function appeared unaffected. Furthermore, the loss of TRF2 did not compromise liver regeneration after partial hepatectomy. Remarkably, liver regeneration occurred without cell division involving endoreduplication and cell growth, thereby circumventing the chromosome segregation problems associated with telomere fusions. We conclude that nondividing hepatocytes can maintain and regenerate liver function despite substantial loss of telomere integrity.
Keywords: DNA damage, TRF2, endoreduplication, hepatocyte, telomere
Telomeres are essential DNA–protein structures that distinguish chromosome ends from sites of DNA damage. Interference with telomere function in proliferating cells has detrimental effects on cell viability and organ function. In mammalian cells, telomere dysfunction occurs after extensive replicative attrition of telomeric DNA or upon loss of shelterin, a six-subunit complex that binds to telomeric DNA and protects chromosome ends (for review, see de Lange 2005). Conditional deletion of the shelterin component TRF2 from proliferating mouse embryo fibroblasts (MEFs) results in the activation of a DNA damage signal, accumulation of DNA damage factors at telomeres, induction of senescence, and DNA ligase IV/Ku70-dependent nonhomologous end joining (NHEJ) of the telomeres (Celli and de Lange 2005; Celli et al. 2006). Similarly, extensive telomere attrition in the later generations of the telomerase-deficient (mTerc−/−) mouse strain results in chromosome end fusions, curbs the function of highly proliferative organs, and compromises self-renewal functions, including liver regeneration (Blasco et al. 1997; Lee et al. 1998; Rudolph et al. 2000; Hemann et al. 2001; Farazi et al. 2003). Here we use Cre-mediated deletion of TRF2 from adult mouse liver to determine whether telomeres are also essential in nondividing cells.
Results and Discussion
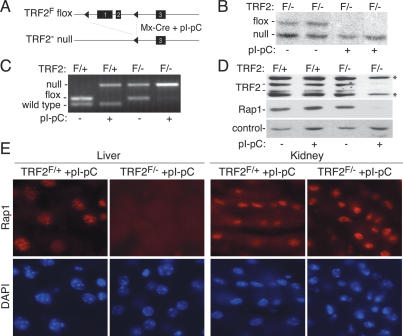
To induce telomere dysfunction in quiescent hepatocytes, we used TRF2 conditional knockout mice (TRF2F/−) (Celli and de Lange 2005) that also contained Mx-Cre transgene (see Fig. 1A for schematic; Kuhn et al. 1995). When induced with pI-pC, the Mx1 promoter directs high levels of expression of Cre-recombinase in the liver, whereas other organs express lower levels of Cre (Kuhn et al. 1995). Six days following pI-pC injection, Cre-mediated recombination resulted in disappearance of the “floxed” TRF2F allele as assessed by genomic blotting and PCR (Fig. 1B,C) and the concomitant loss of TRF2 protein as shown by immunoblotting (Fig. 1D). The deletion of TRF2 was also apparent from the disappearance of Rap1, a TRF2-interacting protein that is unstable in the absence of TRF2 (Celli and de Lange 2005). Immunoblots and in situ indirect immunofluorescence (IF) showed that the vast majority (>90%) of hepatocytes had lost Rap1 (Fig. 1D,E). In contrast, Rap1 remained detectable in kidney sections derived from the same mice (Fig. 1E) as expected from the lesser induction of Cre in this organ. These data indicate that the Mx-Cre TRF2F/− system affords efficient removal of TRF2 from the vast majority of quiescent hepatocytes.
Figure 1.
Efficient TRF2 deletion in quiescent hepatocytes. (A) Strategy for conditional deletion of TRF2 from mouse liver. (B) Genomic blot showing deletion of the TRF2F allele upon pI-pC induction of Cre. Liver DNA from TRF2F/− Mx-CRE mice (F/−) isolated 6 d after pI-pC injection (+) was digested with HindIII and probed with a probe 3′ of the third exon (Celli and de Lange 2005). (C) PCR analysis showing Cre-mediated deletion of TRF2. PCR for the indicated alleles was performed on liver DNA from Mx-Cre TRF2F/+ (F/+) or Mx-Cre TRF2F/−(F/−) mice treated with pI-pC 6 d before DNA isolation as indicated. (D) Immunoblot for TRF2 (1254) and Rap1 (1252) in liver extracts of Mx-Cre TRF2F/+ (F/+) or Mx-Cre TRF2F/− (F/−) mice treated with pI-pC 6 d prior to harvest as indicated. Asterisks represent nonspecific bands detected by the TRF2 antibody. (E) In situ IF for Rap1 (1252) in the liver and kidney of Mx-Cre TRF2F/+ and Mx-Cre TRF2F/− mice treated with pI-pC.
Telomere fusions in quiescent hepatocytes lacking TRF2
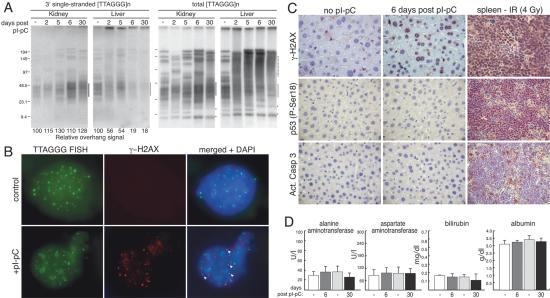
A prominent outcome of telomere dysfunction in proliferating cells is the generation of dicentric and multicentric chromosomes, formed through NHEJ of the deprotected telomeres (van Steensel et al. 1998; Smogorzewska et al. 2002; Celli and de Lange 2005). To test whether NHEJ is active in quiescent cells, we analyzed the telomere restriction pattern in liver and kidney of TRF2F/− mice before and after induction of Cre (Fig. 2A). As expected, the telomeric restriction fragment pattern is identical in the liver and kidney of each mouse when Cre was not induced. In contrast, in Cre-induced TRF2F/− mice, the telomeric pattern differed between liver and kidney. Liver DNA showed a marked increase in the apparent molecular weight of the telomeric restriction fragments, a pattern that was previously shown to be indicative of NHEJ-mediated telomere fusions (van Steensel et al. 1998; Celli and de Lange 2005). Furthermore, the new telomeric fragments in the TRF2-deficient livers lacked the single-stranded TTAGGG repeats that normally protrude 3′ from the chromosome end (Fig. 2A, left panels). Removal of this overhang has been shown to occur when telomeres are processed by NHEJ (van Steensel et al. 1998; Celli and de Lange 2005). Thus, upon TRF2 deletion in the liver, telomeres undergo endto-end fusions, indicating that the NHEJ DNA repair pathway is active in quiescent cells and threatens telomeres as it does in proliferating cells. We verified that in these experiments the hepatocytes remained in G0 by measuring BrdU incorporation after pI-pC injection. The results showed that there was no active DNA replication under these conditions (Supplementary Fig. 1B).
Figure 2.
Consequences of telomere deprotection in quiescent hepatocytes. (A) Telomeric restriction fragment analysis on DNA isolated from either kidney or liver of Mx-Cre TRF2F/− mice untreated or treated with pI-pC and collected at the indicated times following injection. In the panels on the right, the single-stranded telomeric overhang DNA was detected under native conditions using a C-strand probe. After in situ denaturation of the DNA, the total telomeric DNA was detected in the same gel using the same probe (left panels). The position of the bulk of the telomeric restriction fragments is highlighted by the line on the right of the gels. The dotted line indicates the position of fragments resulting from telomere fusions. Chromosome internal telomeric DNA fragments are indicated with an asterisk. The overhang signal was quantified with ImageQuant software and normalized to the total TTAGGG signal in the same lane. These ratios were compared between the samples from one organ. The relative overhang value is given, with the value for the DNA from the uninduced mouse set at 100. (B) γ-H2AX at telomeres 6 d after Cre-mediated TRF2 deletion. IF for γ-H2AX (Upstate Biotechnology) in combination with FISH for telomeric DNA (PNA-TelC probe) on liver cells derived from Mx-Cre TRF2F/− mice before (control) and after induction with pI-pC (6 d). Arrowheads in the merged panels highlight TIFs. (C) Immunhistochemistry for the detection of the indicated proteins in liver sections of Mx-Cre TRF2F/− mice either untreated or 6 d following pI-pC administration. Spleen sections derived from an irradiated (4 Gy) mouse were used as a positive control. (D) Serum levels of the indicated markers for liver function and liver damage in Mx-Cre TRF2F/− mice 6 or 30 d following pI-pC induction and of untreated matching controls.
Telomere dysfunction-induced foci (TIFs) but not p53 activation or apoptosis after telomere deprotection
A second prominent outcome of telomere deprotection in proliferating cells is the accumulation of DNA damage factors at chromosome ends, forming TIFs (d'Adda di Fagagna et al. 2003; Takai et al. 2003). Similarly, phosphorylation of histone H2AX in the chromatin at or near telomeres is observed in the quiescent hepatocytes that have lost TRF2 (Fig. 2B). Most of the TRF2-deficient hepatocytes showed a level of γ-H2AX staining similar to that observed in splenocytes of irradiatiated mice (Fig. 2C), suggesting a strong DNA damage response. In contrast, pI-pC treatment of control (TRF2F/+) animals did not induce formation of γ-H2AX TIFs in the liver (data not shown). These data show that deprotected telomeres are recognized as sites of DNA damage in these noncycling cells.
In proliferating human and mouse cells, telomere damage invariably leads to stabilization of p53, phosphorylation of p53 on Ser15 (Ser18 in the mouse), induction of p21, and a G1 arrest accompanied by senescence or apoptosis. However, we failed to find evidence for upregulation/activation of the p53 pathway in TRF2-deficient livers based on IHC for p53 phosphorylated on Ser18 (Fig. 2C) or immunoblotting for p53 (Supplementary Fig. 1D). Moreover, IHC for activated caspase 3 suggested that there was no prominent induction of apoptosis (Fig. 2C), and staining for SA-β-galactosidase failed to provide evidence for senescent cells (data not shown). Apoptotic or senescent cells were also undetectable at earlier time points (2–5 d following pI-pC induction) (data not shown).
Our finding that acute telomere deprotection in quiescent hepatocytes does not result in senescence or apoptosis contrasts with the results obtained with Adenoviral delivery of a dominant-negative allele of TRF2 (AdTRF2ΔBΔM), which induced both senescence and apoptosis in the mouse liver (Lechel et al. 2005). We consider it unlikely that the telomere damage induced in our study was insufficient, since the effect of AdTRF2ΔBΔM on telomeres is minor compared with the effect of TRF2 deletion (Celli and de Lange 2005). The more likely explanation for the lack of detectable levels of senescence and apoptosis after TRF2 deletion is that the hepatocytes remained in G0 in our experimental setting, whereas adenoviral infection induced cell cycle entry, as evidenced by a sharp increase in mitotic index (Lechel et al. 2005).
TRF2-mediated telomere protection is dispensable for liver function
TRF2F/− mice were fully viable up to 1 mo after deletion of TRF2. At this time point we did not detect increased levels of γ-H2AX in hepatocytes compared with untreated controls, contrary to what was observed at 6 d post-pI-pC (Supplementary Fig. 1A). The disappearance of γ-H2AX is consistent with the NHEJ-mediated repair of the deprotected telomeres. In order to assess the function of TRF2-deficient livers, we measured the levels of serum alanine-and aspartate-aminotransferase, liver enzymes whose release into the circulation is indicative of liver damage. Both markers were in the normal range and indistinguishable from the controls (Fig. 2D). Furthermore, the circulating levels of bilirubin and albumin, markers for liver function, were indistinguishable in Cre-induced and control mice (Fig. 2D). It is unlikely that liver function is maintained by TRF2-proficient hepatocytes, since PCR for the TRF2F allele, immunoblotting, and IF for Rap1 (Supplementary Fig. 2A–C), showed that the livers were TRF2 deficient. Furthermore, genomic blotting showed that at day 30 after deletion of TRF2, the telomeric restriction fragments continued to represent a pattern consistent with fused telomeres (Fig. 2A; data not shown). Hence we conclude that the extensive loss of telomere integrity resulting from TRF2 deletion does not compromise liver function.
Liver regeneration in the absence of TRF2 through endoreduplication and cell growth
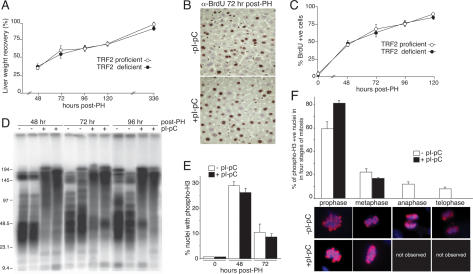
Cells with frequent telomere fusions show diminished proliferation, presumably because anaphase bridges generated by dicentric and multicentric chromosomes hamper the completion of mitosis. Therefore, we asked whether the telomere fusions induced by TRF2 deletion impaired liver regeneration after partial hepatectomy. In order to circumvent potential inhibition of cell cycle progression arising from an ongoing DNA damage response, partial hepatectomy was performed 1 mo after Cre-mediated deletion of TRF2, allowing sufficient time for repair of the deprotected telomeres and dissipation of the telomere damage signal. In parallel, partial hepatectomy was performed on control mice that were TRF2 proficient (TRF2F/+ mice treated with pI-pC and untreated TRF2F/− mice). All 15 mice lacking TRF2 in the liver survived the procedure, as did all of the control mice. The gain in liver mass was indistinguishable between the TRF2-deficient and TRF2-proficient settings (Fig. 3A) and liver function recovered to the same extent (Supplementary Fig. 4C). In agreement with this, we did not detect apoptotic or senescent cells in the regenerating TRF2-deficient livers throughout this experiment (data not shown). Furthermore, BrdU uptake was identical in the two groups at 24–122 h post-partial hepatectomy (Fig. 3B,C; data not shown), indicating that TRF2 status did not affect entry into the cell cycle or the extent of DNA replication. The regenerated TRF2-deficient livers retained the telomeric restriction pattern typical of fused chromosome ends (Fig. 3D) and continued to show absence of the TRF2 gene and Rap1 protein (Supplementary Fig. 4A,B).
Figure 3.
Liver regeneration despite telomere dysfunction. (A) Recovery in liver weight at the indicated time points following partial hepatectomy in Mx-Cre TRF2F/− mice induced with pI-pC (TRF2 proficient) and in control mice (TRF2 deficient). Partial hepatectomy was performed 30 d after induction of Cre with pI-pC. Liver weight recovery is expressed as the percentage of the remnant liver mass relative to the liver mass before partial hepatectomy. This value was obtained by dividing the wet weight of the regenerated liver normalized to the original liver/body weight ratio. Represented are the averages and standard deviations (error bars) obtained by analyzing three independent mice per time point. (B) IHC for BrdU at 72 h post-partial hepatectomy in Mx-Cre TRF2F/− mice. BrdU was administered continuously following partial hepatectomy at 12-h intervals; pI-pC treatment is as in A. (C) Quantification of percentage of BdrU-positive cells following partial hepatectomy at the indicated time points. At least 200 nuclei were counted for each mouse analyzed (n = 3 for the treated and untreated cohorts). Methods are as in B. (D) Telomeric restriction fragments from livers isolated from Mx-Cre TRF2F/− mice; pI-pC treatment and partial hepatectomy are as above. (E) Quantification cells containing phosphorylated H3 in livers from Mx-Cre TRF2F/− mice; pI-pC treatment and partial hepatectomy are as above. (F) Occurrence of four stages of mitosis in phospho-H3-positive liver nuclei (n = 100) of Mx-Cre TRF2F/− mice at 48 h post-partial hepatectomy. Staging was based on morphological appearance as shown by the examples from control mice below. No examples of anaphase or telophase were found in the pI-pC-treated mice. No distinction could be made between prophase and failed anaphase in the +pI-pC setting
The absence of TRF2 did not affect the ability of hepatocytes to enter mitosis as shown by the percentage of nuclei containing a phosphorylated from of histone H3 associated with late G2 and M phase (Fig. 3E). However, in TRF2-deficient settings the ability of cells to complete mitosis was severely impaired (Fig. 3F). In control livers, 20% of the phospho-H3-positive hepatocytes were in the late stages of mitosis (12% in anaphase and 8% in telophase) at 48 h post-surgery, while in the TRF2-deficient setting anaphase and telophase were not observed (Fig. 3F). Concomitant with the disappearance of cells in anaphase and telophase, there was a proportional increase of cells in prophase (83% vs. 60% in controls). Given that the percentage of cells in metaphase remained largely unaffected in TRF2-deficient hepatocytes relative to controls (17% and 22%, respectively), it seems unlikely that the accumulation of cells in prophase is due to an arrest in this mitotic phase. A more likely explanation is that a fraction of the nuclei that appear to be in prophase are actually attempting progression through anaphase. The numerous end-to-end fusions in these cells may create a structure that is morphologically indistinguishable from prophase.
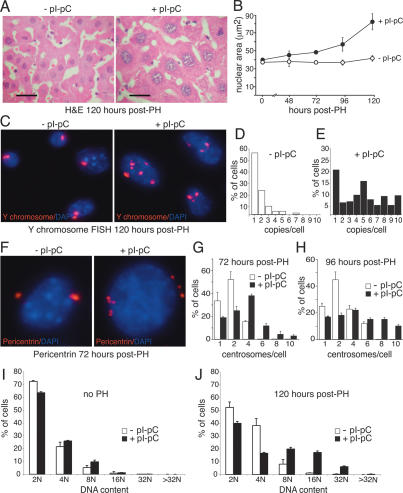
The combination of normal levels of DNA replication and diminished cell division suggested that cells underwent endoreduplication. Consistent with this hypothesis, the size of both the nuclei and the cells was significantly increased in regenerating TRF2-deficient livers compared with regenerating TRF2-proficient livers or nonregenerating liver samples (Fig. 4A,B). To determine the ploidy of the cells, we performed FISH for the Y chromosome. Before partial hepatectomy, the average number of Y chromosomes per cell was similar in control and induced mice (median 1.4 and 1.6, respectively). In contrast, 5 d after partial hepatectomy, the majority of TRF2-deficient hepatocytes contained multiple copies of the Y chromosome (TRF2F/− + pI-pC: average 4.4 ± 1.0 copies/cell, n = 202; control: average 1 ± 2.8 copies per cell, n = 208) with some cells containing >10 copies (Fig. 4C–E). Furthermore, the TRF2-deficient cells showed centrosome amplification (Fig. 4F–H) and their ploidy increased compared with the controls resulting in >40% of the cells having DNA content ≥8 N (Fig. 4I,J). Interestingly, in regenerating TRF2-deficient livers we detected induction of p53 and a concomitant rise in p21 levels (Supplementary Fig. 3). This is reminiscent of the induction of the so-called tetraploidy checkpoint induced by aberrant mitotic exit without concomitant chromosome segregation (Margolis et al. 2003).
Figure 4.
Endoreduplication and cell growth in regenerating livers lacking TRF2. (A) H&E-stained sections of livers from Mx-Cre TRF2F/− mice treated with pI-pC as indicated and subject to partial hepatectomy 30 d later. The two images are shown at the same magnification. Bar, 10 μm. (B) Average nuclear area (in square micrometers) in regenerating livers. At least 300 nuclei were measured for each mouse. Values represent the average and the standard deviation (error bar) as measured from three independent mice for each time point. Genotypes and treatment are as in A (three mice per time point). (C) Y chromosome FISH on liver sections of the mice in A. (D,E) Quantification of Y chromosome copy number per cell based on data as shown in C. At least 200 nuclei of two independent mice were analyzed. (F) Supernumerary centrosomes in regenerating TRF2-deficient mouse liver cells. Mice and treatment are as in A. Centrosomes were detected with a polyclonal anti-pericentrin antibody. (G,H) Quantification of percentage of cells with the indicated number of centrosomes based on data as in F. For every data point, at least 100 nuclei of two independent mice were analyzed. (I,J) Single-cell DNA measurement of Feulgen-stained hepatocytes before (no PH) and 120 h after partial hepatectomy. Mice and treatment are as in A. DNA content was measured on three independent mice for each time point for both untreated and pI-pC-treated animals.
Conclusions
This report demonstrates that quiescent hepatocytes are resistant to the DNA damage signal elicited by dysfunctional telomeres and are fully functional in the presence of chromosome end fusions. The viability of hepatocytes with dysfunctional telomeres contrasts with the response in cycling cells that either undergo apoptosis or senescence. Previous work has shown that cycling cells can arrest due to telomere deprotection even if telomere fusions and their associated breakage fusion bridge (BFB) cycles do not occur (Karlseder et al. 1999). Thus, the secondary DNA damage associated with BFB cycles is not a likely source of the difference in cellular outcome in cycling cells and quiescent hepatocytes with telomere dysfunction. Hepatocytes have the ability to detect telomere deprotection, as shown by the presence of γ-H2AX at telomeres. Yet, hepatocytes survive this damage, suggesting that the effector pathway of the telomere DNA damage response is different in these cells. This impaired response can be due to an intrinsic property of hepatocytes or alternatively to a feature of quiescent hepatocytes. An impaired DNA damage response in quiescent cells has been reported in muscle cells, which can sense DNA damage when proliferating (myoblasts) and when quiescent (myotubes), yet the activation of p53 and its effector pathway only occurs in myoblasts (Latella et al. 2004).
Whereas TRF2 deletion does not curb the viability of hepatocytes, the cells appear incapable of dividing when challenged to proliferate by partial hepatectomy. Although cells attempt mitosis, anaphase and telophase are virtually absent and cell numbers do not increase. However, entry into S phase is not impeded, and as a result, the TRF2-deficient hepatocytes become highly polyploid. The increase in cell size associated with polyploidy accounts for the regeneration of liver mass and liver function after partial hepatectomy. Similar results were recently reported for mouse hepatocytes lacking Separase, the protease that removes cohesin from sister chromatids, thereby allowing their separation in anaphase. Upon partial hepatectomy, Separase-deficient hepatocytes appear to have the ability to regenerate normal liver function through endoreduplication and cell growth (Wirth et al. 2006). Hepatocytes without Separase attempt to exit mitosis, leading to aberrant nuclei and multinucleated cells when chromosome segregation fails. Thus, when chromatid segregation fails in mitosis, either because cohesin persists or because the telomeres are joined, hepatocytes can remain viable and regenerate the required liver mass through repeated DNA replication, resulting in polyploid nuclei and large cells. The findings on hepatocytes without TRF2 or Separase reveal the remarkable tolerance of these cells for aberrant cell cycles and predict that loss of other factors required for progression through mitosis may be tolerated in the liver.
Our results pertain to the role of telomere dynamics in human malignancy. Human hepatocellular carcinoma (HCC) is caused by chronic liver damage, which results in extensive telomere erosion (Plentz et al. 2004). Such telomere erosion is thought to have two opposing effects on tumorigenesis (Maser and DePinho 2002). On the one hand, it can curb cell proliferation through the induction of apoptosis or senescence. On the other hand, loss of telomere protection can promote chromosomal rearrangements and thus enhance malignant transformation. The balance of these two forces is in part determined by the extent to which telomere deprotection leads to proliferative arrest and cell death in the incipient tumor cells. Our finding that quiescent hepatocytes are resistant to the cell-lethal effects of TRF2 deletion predicts that telomere erosion might be well tolerated in hepatocytes that experience telomere deprotection before entering quiescence. These cells represent a permissive setting for telomere-related chromosome instability since upon a subsequent induction of proliferation the cells might initiate BFB cycles or become polyploid, as observed in TRF2-deficient hepatocytes. Therefore, our data would predict that in the liver, telomere attrition promotes the early stages of tumorigenesis and that the resulting HCC contain aneuploid cells. Consistent with this prediction, late-generation telomerase-deficient mice, which have extensively shortened telomeres, were found to have an increase in the frequency of the earliest stages of neoplastic transformation (hyperplastic nodules and initiation foci) after chronic liver damage (Farazi et al. 2003). Moreover, the analysis of human HCC revealed that tumors characterized by the presence of short telomeres also display a significant increase in polyploid cells relative to HCC with longer telomeres (Plentz et al. 2005). If human hepatocytes are similarly resistant to the lethal aspects of telomere dysfunction, the telomere attrition associated with long-term liver damage could be a main factor in HCC.
Material and methods
Mice and induction of Cre
TRF2F/− mice (Celli and de Lange 2005) were crossed with Mx-Cre transgenic mice (Jackson Laboratories; Kuhn et al. 1995) to obtain compound Mx-Cre TRF2F/− mice. Mice were maintained in a mixed 129/Sv C57/Bl6 genetic background. For the induction of Cre, 8-to 10-wk-old mice were injected intraperitoneally with pI-pC (250 mg, Pharmacia) for a total of three times with 48-h intervals.
Genotyping and immunoblotting
For DNA isolation, tissues were incubated overnight in 100 mM NaCl, 10 mM Tris (pH 8.0), 1 mM EDTA, 1% SDS, and 1 mg/mL proteinase K. DNA was sequentially used to detect the TRF2 locus by PCR and genomic blotting as previously described (Celli and de Lange 2005). Protein was isolated from frozen tissues by homogenization in extraction buffer (50 mM Tris-HCl at pH 7.4, 1% Triton X-100, 0.1% SDS, 400 mM NaCl, 1 mM EDTA, 1 mM DTT, supplemented with protease inhibitors), protein concentrations were determined based on Bradford assay, and 20 μg of protein lysate was separated on a 8% SDS-PAGE gel. After immunoblotting, membranes were blocked in PBS with 0.1% Tween and 5% nonfat dry milk and incubated overnight at 4°C with a affinity-purified rabbit antibodies raised against GST-mTRF2 (1254) and GST-mRAP1 (1252). Blots were developed with enhanced chemiluscence (Amersham).
IF, FISH, and IF-FISH
Tissues were frozen in dry ice and embedded in OCT tissue TEC (Sakura). For IF, 4-μm cryosections were fixed for 20 min in 4% paraformaldehyde, blocked for 30 min in blocking solution (1 mg/mL BSA, 3% goat serum, 0.1% Triton X-100, 1 mM EDTA), and incubated for 2 h at room temperature with the following primary antibodies diluted in blocking solution: Rap1, 1252; γ-H2AX (Upstate Biotechnology); phospho-Histone H3 Ser10 (Cell Signaling); pericentrin (rabbit polyclonal a gift from A. Levine). Secondary antibodies mouse and rabbit IgG were labeled with Rhodamine Red-X (RRX, Jackson Laboratories). For IF-FISH (Herbig et al. 2004), cryosections were fixed in 4% paraformaldehyde, incubated for 20 min in 100% methanol at −20°C, and blocked in blocking solution (see above) after rehydration. After incubation with anti-γ-H2AX antibody and secondary antibody, slides were fixed in 2% paraformaldehyde, dehydrated through serial incubation in 70%, 95%, and 100% ethanol, and dried completely. At this point, slides were incubated with a FITC-TelC PNA probe (FITCH-OO-[CCCTAA]3) diluted in hybridization solution (70% formaldhyde, 0.5% blocking reagent [Roche], 10 mM Tris-HCl at pH 7.2). After 2 min of denaturation at 80°C, slides were incubated for 2 h at room temperatute in the dark. Finally, slides were washed twice in 70% formaldehyde, 10 mM Tris-HCl (pH 7.2), and three times in PBS. Y chromosome FISH hybridization was performed on frozen sections using a STAR*FISH whole Y chromosome probe (Open Biosystem) according to the manufacturers’ instructions.
IHC
Tissues were fixed in 4% paraformaldehyde overnight at 4°C, dehydrated, and embedded in paraffin. Five-micrometer sections were deparaffinized in xylene, rehydrated in serial ethanol rinses (100%, 70%, 50%), and rinsed in H20. To unmask epitopes, slides were boiled for 15 min in 10 mM Na Citrate (pH 6.0), incubated in 0.5% H2O2 to quench endogenous peroxidases, and blocked for 30 min in blocking solution (see above). Slides were incubated in the following primary antibodies diluted in blocking solution: γ-H2AX (rabbit polyclonal, Upstate Biotecnology); phospso Histone H3 Ser10 (mouse monoclonal); anti-phosho p53 (Ser15) (Cell Signaling); cleaved Caspase 3 (Asp175) (Cell Signaling); BrdU (BD Biosciences). For detection of mouse antibodies, we used the ABC and DAB kit (Vector Laboratories), while for rabbit antibodies we used the EnVision+ kit (Dako Cytomation). Sections were counterstained with hematoxylin (Sigma), rinsed in xylene, and mounted with VactaMount (Vector Laboratories).
H&E staining and determination of nuclear surface area
Paraffin-embedded slides were deparaffinizzed and dehydrated as described above. Slides were rinsed in water, stained for 30 sec in Mayer's Hematoxylin (Sigma), and extensively washed in water, and then incubated for 15 sec in 70% ethanol. Slides were stained for 5 sec in Eosin Y (Richard-Allan Scientific), washed twice in 95% ethanol and absolute ethanol, rinsed in xylene, and mounted with VactaMount (Vector Laboratories). Nuclear size was calculated from digital images using the Openlab (Improvision) advanced measure module.
DNA (Fuelgen) content measurements
Paraffin-embedded sections were deparafinnized in xylene and rehydrated in serial ethanol rinse (100%, 90%, 70%). After 1 h of fixation in 4% buffered formaline and washing in PBS, sections were incubated for 60 min in 5 N HCl. After three washes in PBS, sections were incubated in Schiff's reagent (Sigma) for 20 min, washed three times for 5 min in 0.5% sodium bisulfite and dehydrated in ethanol (70%, 90%, 100%). After a short incubation in xylene, slides were mounted with Vacta-Mount (Vector Laboratories). Digital images were acquired as described above. Analysis was carried out using the advanced measurement module for Openlab (Improvision). For each sample, at least 300 nuclei were measured and DNA content was automatically calculated based on 300 reference kidney nuclei from the same mouse.
CHEF analysis of telomeric DNA
Telomeric DNA was analyzed as described previously (Celli and de Lange 2005).
Assays for liver function
Liver function assays were performed on serum isolated from Retro-orbital-collected blood by the Laboratory of Comparative Pathology of Cornell University.
Partial hepatectomy and administration of BrdU
Mice were anaesthetized with an intraperitoneal (i.p.) injection of 0.015 mL of 2.5% Avertin per gram of body weight, an abdominal transversal incision was performed, and the large left and two median lobes of the liver (collectively corresponding to ~70% of the liver) were ligated and removed. Following partial hepatectomy, BrdU (150 mg/kg) was administrated at 12-h intervals by i.p. injections.
Acknowledgments
We thank Devon White, Alan Li, and Megan Fitzgerald for mouse husbandry and genotyping. Arnie Levine is thanked for a gift of anti-pericentrin antibody, Kristian Helin for a gift of anti-p53 antibody, and Sara Buonomo for providing helpful discussion. G.B.C. was supported by the Leukemia and Lymphoma Society. E.L.D was supported by an Irma T. Hirschl fellowship. This work was supported by grants from the NIH (GM49046 and AG16642).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1453606.
References
- Blasco, M.A., Lee, H.W., Hande, M.P., Samper, E., Lansdorp, P.M., De-Pinho, R.A., Greider, C.W. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- Celli, G., de Lange, T. DNA processing not required for ATM-mediated telomere damage response after TRF2 deletion. Nat. Cell Biol. 2005;7:712–718. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- Celli, G.B., Denchi, E.L., de Lange, T. Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat. Cell Biol. 2006;8:885–890. doi: 10.1038/ncb1444. [DOI] [PubMed] [Google Scholar]
- d'Adda di Fagagna, F., Reaper, P.M., Clay-Farrace, L., Fiegler, H., Carr, P., Von Zglinicki, T., Saretzki, G., Carter, N.P., Jackson, S.P. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- de Lange, T. Shelterin: The protein complex that shapes and safeguards human telomeres. Genes & Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- Farazi, P.A., Glickman, J., Jiang, S., Yu, A., Rudolph, K.L., DePinho, R.A. Differential impact of telomere dysfunction on initiation and progression of hepatocellular carcinoma. Cancer Res. 2003;63:5021–5027. [PubMed] [Google Scholar]
- Hemann, M.T., Rudolph, K.L., Strong, M.A., DePinho, R.A., Chin, L., Greider, C.W. Telomere dysfunction triggers developmentally regulated germ cell apoptosis. Mol. Biol. Cell. 2001;12:2023–2030. doi: 10.1091/mbc.12.7.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbig, U., Jobling, W.A., Chen, B.P., Chen, D.J., Sedivy, J.M. Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21(CIP1), but not p16(INK4a) Mol. Cell. 2004;14:501–513. doi: 10.1016/s1097-2765(04)00256-4. [DOI] [PubMed] [Google Scholar]
- Karlseder, J., Broccoli, D., Dai, Y., Hardy, S., de Lange, T. p53-and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science. 1999;283:1321–1325. doi: 10.1126/science.283.5406.1321. [DOI] [PubMed] [Google Scholar]
- Kuhn, R., Schwenk, F., Aguet, M., Rajewsky, K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- Latella, L., Lukas, J., Simone, C., Puri, P.L., Bartek, J. Differentiation-induced radioresistance in muscle cells. Mol. Cell. Biol. 2004;24:6350–6361. doi: 10.1128/MCB.24.14.6350-6361.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechel, A., Satyanarayana, A., Ju, Z., Plentz, R.R., Schaetzlein, S., Rudolph, C., Wilkens, L., Wiemann, S.U., Saretzki, G., Malek, N.P., et al. The cellular level of telomere dysfunction determines induction of senescence or apoptosis in vivo. EMBO Rep. 2005;6:275–281. doi: 10.1038/sj.embor.7400352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H.W., Blasco, M.A., Gottlieb, G.J., Horner, J.W., II, Greider, C.W., DePinho, R.A. Essential role of mouse telomerase in highly proliferative organs. Nature. 1998;392:569–574. doi: 10.1038/33345. [DOI] [PubMed] [Google Scholar]
- Margolis, R.L., Lohez, O.D., Andreassen, P.R. G1 tetraploidy checkpoint and the suppression of tumorigenesis. J. Cell. Biochem. 2003;88:673–683. doi: 10.1002/jcb.10411. [DOI] [PubMed] [Google Scholar]
- Maser, R.S., DePinho, R.A. Connecting chromosomes, crisis, and cancer. Science. 2002;297:565–569. doi: 10.1126/science.297.5581.565. [DOI] [PubMed] [Google Scholar]
- Plentz, R.R., Caselitz, M., Bleck, J.S., Gebel, M., Flemming, P., Kubicka, S., Manns, M.P., Rudolph, K.L. Hepatocellular telomere shortening correlates with chromosomal instability and the development of human hepatoma. Hepatology. 2004;40:80–86. doi: 10.1002/hep.20271. [DOI] [PubMed] [Google Scholar]
- Plentz, R.R., Schlegelberger, B., Flemming, P., Gebel, M., Kreipe, H., Manns, M.P., Rudolph, K.L., Wilkens, L. Telomere shortening correlates with increasing aneuploidy of chromosome 8 in human hepatocellular carcinoma. Hepatology. 2005;42:522–526. doi: 10.1002/hep.20847. [DOI] [PubMed] [Google Scholar]
- Rudolph, K.L., Chang, S., Millard, M., Schreiber-Agus, N., DePinho, R.A. Inhibition of experimental liver cirrhosis in mice by telomerase gene delivery. Science. 2000;287:1253–1258. doi: 10.1126/science.287.5456.1253. [DOI] [PubMed] [Google Scholar]
- Smogorzewska, A., Karlseder, J., Holtgreve-Grez, H., Jauch, A., de Lange, T. DNA ligase IV-dependent NHEJ of deprotected mammalian telomeres in G1 and G2. Curr. Biol. 2002;12 doi: 10.1016/s0960-9822(02)01179-x. [DOI] [PubMed] [Google Scholar]
- Takai, H., Smogorzewska, A., de Lange, T. DNA damage foci at dysfunctional telomeres. Curr. Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- van Steensel, B., Smogorzewska, A., de Lange, T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- Wirth, K.G., Wutz, G., Kudo, N.R., Desdouets, C., Zetterberg, A., Taghybeeglu, S., Seznec, J., Ducos, G.M., Ricci, R., Firnberg, N., et al. Separase: A universal trigger for sister chromatid disjunction but not chromosome cycle progression. J. Cell Biol. 2006;172:847–860. doi: 10.1083/jcb.200506119. [DOI] [PMC free article] [PubMed] [Google Scholar]