Abstract
PERIOD (PER) proteins are central components within the mammalian circadian oscillator, and are believed to form a negative feedback complex that inhibits their own transcription at a particular circadian phase. Phosphorylation of PER proteins regulates their stability as well as their subcellular localization. In a systematic screen, we have identified 21 phosphorylated residues of mPER2 including Ser 659, which is mutated in patients suffering from familial advanced sleep phase syndrome (FASPS). When expressing FASPS-mutated mPER2 in oscillating fibroblasts, we can phenocopy the short period and advanced phase of FASPS patients’ behavior. We show that phosphorylation at Ser 659 results in nuclear retention and stabilization of mPER2, whereas phosphorylation at other sites leads to mPER2 degradation. To conceptualize our findings, we use mathematical modeling and predict that differential PER phosphorylation events can result in opposite period phenotypes. Indeed, interference with specific aspects of mPER2 phosphorylation leads to either short or long periods in oscillating fibroblasts. This concept explains not only the FASPS phenotype, but also the effect of the tau mutation in hamster as well as the doubletime mutants (dbtS and dbtL) in Drosophila.
Keywords: Circadian, phosphorylation, PER, FASPS
Circadian clocks are endogenous oscillators that regulate the temporal organization of physiology, metabolism, and behavior. They provide organisms ranging from unicellular algae to humans with an internal representation of local time, thus allowing them to anticipate their environment. Circadian clocks oscillate in a self-sustained manner with an endogenous (free-running) period close to 24 h. In a natural environment, these free-running rhythms are synchronized (entrained) to external time cues (Zeitgebers), such as 24-h light–dark and ambient temperature cycles (for review, see Stanewsky 2003; Gachon et al. 2004; Merrow et al. 2005; Brunner and Schafmeier 2006).
These cell-autonomous oscillations are thought to be established by feedback loops involving transcription of clock genes and their subsequent autoregulatory transcriptional repression. In mammals, the transcription factor heterodimer CLOCK–BMAL1 activates the expression of Period (Per1, Per2, and Per3) and Cryptochrome (Cry1 and Cry2) genes via E-box enhancer elements in their promoters. PER and CRY proteins are believed to form complexes that translocate in the nucleus to inhibit their own transcription by directly interacting with the CLOCK–BMAL1 complex.
Critical to the properties of this oscillator is the delay between the production of PER and CRY proteins and their autorepression. Post-translational events such as complex formation among clock proteins, nuclear import and export, regulated degradation, modulation of transcriptional activity, and chromatin modification have all been implicated in the generation of this delay (for a review, see Harms et al. 2004). In many cases, phosphorylation of clock proteins is the key step that both initiates these events and regulates their correct timing. In cyanobacteria, even the core of the circadian oscillator seems to be based on rhythmic phosphorylation and dephosphorylation of clock proteins rather than on a transcriptional–translational feedback loop (Nakajima et al. 2005; Tomita et al. 2005). In Drosophila and mammals, circadian oscillations continue even when some clock proteins normally transcribed in circadian fashion are instead expressed from a constitutive promoter (Yang and Sehgal 2001; Fujimoto et al. 2006; Nishii et al. 2006).
In mammals, many core clock proteins are phospho-proteins in vivo or in vitro (Lee et al. 2001; Eide et al. 2002). For example, PER proteins undergo rhythmic phosphorylation that peaks at times of nuclear accumulation when transcriptional repression is maximal. Substantial evidence has accumulated for the importance of these events to clock properties and function. Genetic studies have identified casein kinase Iε (CKIε) and its Drosophila homolog DOUBLETIME (DBT) as kinases regulating the circadian clock. Null mutations in dbt cause arrhythmia and hypophosphorylation of dPER. In addition, other alleles of doubletime have been shown to affect circadian period in behavior in both directions: dbtS causes a short period, whereas dbtL causes a long period in flies (Kloss et al. 1998; Price et al. 1998). CKIε and its homolog CKIδ have been shown to bind and phosphorylate mammalian PER proteins, thereby regulating their stability and subcellular localization (Yagita et al. 2002; Eide et al. 2005; Shirogane et al. 2005; for a review, see Harms et al. 2004). A semidominant mutation in CKIε called tau causes a short circadian period in hamster (Lowrey et al. 2000). Paradoxically, although the alleles described above can have opposite effects on circadian period (e.g., the two doubletime alleles), they are all correlated with reduced kinase activity in vitro (Lowrey et al. 2000; Preuss et al. 2004).
In humans, two types of dominant familial advanced sleep phase syndrome (FASPS) have been described that are linked to post-translational modification of PER proteins (Jones et al. 1999; Toh et al. 2001; Xu et al. 2005). In both types, FASPS patients show phases of sleep, core body temperature, and melatonin rhythms that are 4–5h earlier than those of unaffected siblings. On the molecular level, one of these syndromes has been mapped to an amino acid substitution in a phosphorylatable residue of hPer2, and in the other the hCKIδ kinase gene itself is affected.
In FASPS affecting hPER2, Ser 662 located in the CKIε-binding region of hPER2 is changed to glycine, a modification that leads to hypophosphorylation of hPER2 by CKIε in vitro (Toh et al. 2001). Although it is not yet known whether Ser 662 is phosphorylated in vivo, it has been proposed that inadequate phosphorylation of FASPS-hPER2 might attenuate its CKIε-mediated degradation and thus accelerate its autorepression, leading to phase-advanced molecular rhythms (Toh et al. 2001). Alternatively, an altered phosphorylation profile of hPER2 could affect the timing of nuclear accumulation of hPER2 similar to effects described in dbt mutants (Bao et al. 2001) or the tau mutant hamster (Dey et al. 2005).
To better understand the role of PER phosphorylation in clock function and in disease, we performed a systematic screen to identify phosphorylated residues in mPER2 using a novel mass spectrometric approach. We identified 21 positions in mPER2 as being phosphorylated in living cells, including the serine residue corresponding to FASPS. By introducing the FASPS allele into oscillating fibroblasts, we were able to reproduce on a molecular level the early phase and the short period in FASPS patients’ behavior. Surprisingly, though, we find that the FASPS PER2 protein is less stable than wild-type PER2 and more sensitive to CKIε-mediated degradation. Immunocytochemistry studies reveal that the FASPS mutation leads to an accelerated nuclear clearance of PER2. Combining our experiments with mathematical modeling, we show that PER proteins undergo at least two functionally discernible phosphorylation events, one primarily leading to degradation, the other leading to nuclear retention. This model would explain how reduced phosphorylation states of PER proteins could affect circadian period in opposite directions. Furthermore, we show that predictions of this model about the hamster's tau (CKIε) mutation turn out to be correct when tested experimentally.
Results
Phosphorylation site mapping of mPER2
The importance of PER protein phosphorylation for the circadian systems of Drosophila and mammals is widely accepted. However, little is known about the number and location of phosphorylated PER protein residues. Here, we performed a systematic screen to identify phosphorylated sites in mouse PER2 using a novel mass spectrometric technique that we have developed previously (Schlosser et al. 2005). To this end, we generated a HEK293 cell line that stably expresses mouse PER2 protein containing a V5 epitope tag at its C terminus. HEK293 cells are especially suited for this purpose, since they are able to grow in liquid culture, facilitating the generation of sufficient protein for mass spectrometric analysis. Since phosphorylation of overexpressed mPER2 is performed by endogenous kinases in these cells, it is possible that we miss the detection of some phosphorylation sites due to a low kinase-to-substrate ratio. Nevertheless, we thereby decrease the likelihood of detecting false positives in the screen.
Total cell extracts were subjected to immunoprecipitation using anti-V5 antibody, followed by multiprotease digestion, titansphere nanocolumn phosphopeptide enrichment, and tandem mass spectrometry. Analyses of mPER2 digests prior to phosphopeptide enrichment resulted in an overall sequence coverage >90% (cf. 25% with conventional single-protease techniques) (Schlosser et al. 2005), suggesting that we have obtained an almost complete phosphosite map of mPER2.
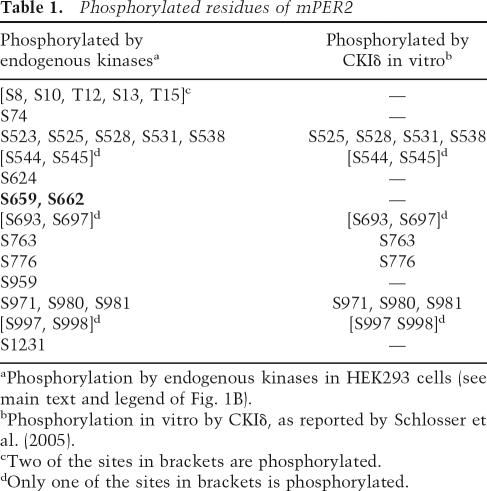
At 21 of 247 serine or threonine residues, we find mPER2 phosphorylated (Table 1; Fig. 8A, below), whereas no phosphorylation at tyrosine residues was detected. Many of the phosphorylation sites are located close to the CKIε-binding domain (amino acids 555–754) (Akashi et al. 2002) or around one of the three nuclear export signals (amino acid 983–990) (Yagita et al. 2002). Whereas some phosphorylation sites were detected close to the N and C termini, there are no phosphorylation sites in the region of the functionally important PAS protein–protein interaction domains.
Table 1.
Phosphorylated residues of mPER2
Figure 8.
Schematic representation of PER2 phosphorylation sites and their functional impact on the circadian oscillator. (A) Linear map of the PER2 protein with positions of phosphorylated residues identified by mass spectrometry and additional functionally important domains (see also Table 1). (Blue lines) Sites phosphorylated by endogenous kinases in cells; (red asterisks) sites also phosphorylated in vitro by CKIδ. Functional domains: (PAS) PER-ARNT-SIM domain; (NES) nuclear export sequence; (NLS) nuclear localization signal; (CKIε) CKIε-binding domain; (CRY) CRY1/2-binding domain. (B) Model for the differential effects of PER2 phosphorylation on circadian oscillations. PER2 contains at least two functionally different sets of phosphorylation sites—one primarily mediating proteasomal degradation (green), the other nuclear retention (purple). In FASPS-PER2 (right side of the panels), the latter cannot be phosphorylated because Ser 659 is mutated to glycine (Toh et al. 2001). At the beginning of the circadian cycle (morning/ midday), newly synthesized PER2 protein shuttles between nucleus and cytoplasm, where it is phosphorylated by kinases such as CKIε/δ at sites that target it for rapid proteasomal degradation in the cytoplasm. Later (afternoon/early night), complex formation with CRY proteins enhances the nuclear localization of the PER2–CRY complex and likely activates or recruits additional kinases to the PER2–CRY complex. These yet-unknown kinases phosphorylate PER2 at the FASPS site, which serves as a priming site for CKIε/δ phosphorylation at downstream residues. Together, this leads to nuclear accumulation of the PER2–CRY complex and thereby to transcriptional repression of CLOCK–BMAL1 transactivation. At the end of the circadian cycle (late night), the PER2–CRY repression is released because the PER2–CRY complex is degraded in the cytoplasm after nuclear export. In FASPS-PER2, however, the region responsible for nuclear retention cannot be phosphorylated (red crosses), leading to premature nuclear export of the PER2–CRY complex, and thus to an earlier cytosolic degradation and to a faster circadian cycle.
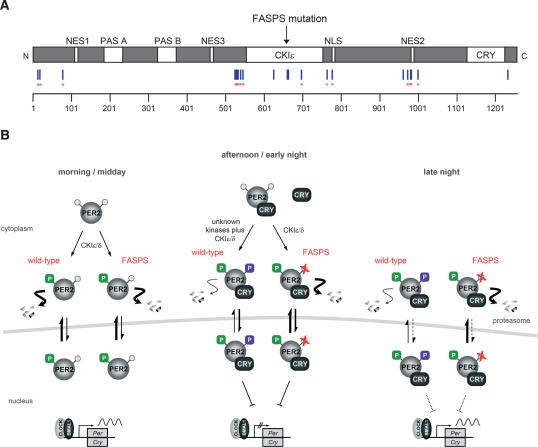
Two of the identified phosphorylation sites, serine residues 659 and 662 (see Supplementary Fig. S1), corresponding to serine residues 662 and 665 in human PER2 (Fig. 1A,B), are particularly well conserved among vertebrates and among PER protein paralogs. In affected members of a family of patients with FASPS, Ser 662 is mutated to glycine in one hPer2 allele (Toh et al. 2001), so it has been speculated that the FASPS phenotype is caused by a phosphorylation defect at this and possibly downstream positions. Our assay represents the first direct evidence that this phosphorylation occurs in living cells.
Figure 1.
Mouse PER2 protein is phosphorylated at a serine residue, which causes FASPS in humans. (A) The alignment of the amino acid sequence of human and mouse PER2 shows a highly conserved cluster of serine and threonine residues in the region where the FASPS mutation has been identified. In patients suffering from FASPS, one copy of hPER2 is mutated at position 662 (serine to glycine). Those phosphorylatable residues—which are identical in PER1, PER2, and PER3—are bold and underlined. (B) The FASPS site of mPER2 is phosphorylated in tissue culture cells. Whole-cell lysate from HEK293 cells stably expressing V5-epitope-tagged mPER2 was used to immunoprecipitate mPER2. After separation by gel electrophoresis, mPER2 protein was digested with proteases, and the resulting phosphopeptides were enriched using titansphere chromatography and subsequently identified using nano-liquid chromatography followed by tandem mass spectrometry (nanoLC-MS/ MS), as described (Schlosser et al. 2005). (C) PCR-generated mutations were made in the conserved serine–threonine cluster of mPER2 for subsequent comparison with wild type in various assays.
Cell culture model for FASPS
To investigate the consequences of this phosphorylation on a biochemical as well as cell biological level, we took a reductionist approach and developed a cell culture model for FASPS in oscillating fibroblasts. Fibroblast cell lines such as rat-1 and NIH3T3 have been reported to show self-sustained circadian oscillations (Balsalobre et al. 1998; Nagoshi et al. 2004) with properties very similar to the oscillator in the SCN (Yagita et al. 2001). Indeed, fibroblasts containing mutations at circadian loci qualitatively recapitulate the impact of clock gene mutation on period length displayed by the mice from which they were taken (Yagita et al. 2001; Pando et al. 2002; Brown et al. 2005).
Therefore, we generated NIH3T3 cell lines that express several mutant forms of mPER2-V5 (Fig. 1C). We exchanged Ser 659 for glycine as in FASPS PER2 and also for aspartic acid to potentially mimic the negative charge of the phosphorylated serine (S659D). In addition, we mutated downstream serine and threonine residues to alanine residues (mut-7) since they comprise a canonical CKIε/δ substrate motif (Flotow et al. 1990), which may be phosphorylated after priming phosphorylation at the FASPS site.
To allow the expression of different PER2 variants from the same chromosomal location of the fibroblast genome, we generated a NIH3T3 host cell line harboring a Flp Recombinase Target (FRT) site (O'Gorman et al. 1991). By recombining the expression constructs to this cassette, it is ensured that potential differences in the properties of the cell lines and protein variants are not artifacts due to chromosomal position effects or variation of integration events. This host cell line also contains a circadian reporter consisting of luciferase driven by an SV40 promoter and six Per1-derived E-box elements. Importantly, the resulting cell lines still contain endogenous wild-type PER2 in order to reflect the heterozygous nature of all known FASPS-afflicted humans. Quantitative PCR analyses revealed that the expression levels of the Per2 variant transcripts in unsynchronized cells were about fourfold higher compared with endogenous wild-type Per2 and did not significantly differ among cell lines (data not shown).
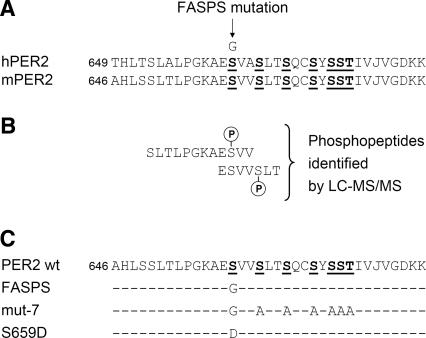
After synchronization with dexamethasone, cell lines expressing PER2 wild-type or an irrelevant control protein show sustained circadian oscillations, whereas the oscillations of FASPS and mut-7-expressing cell lines damp rapidly (Fig. 2A), a phenotype that suggests a dominant effect of these mutations on the circadian oscillator. Although it is difficult to analyze circadian period from these time series, a clear 4-h advance in the phase of the first peak of FASPS and mut-7 cell lines could be seen compared with wild-type cells. This magnitude of phase advance is similar to that of human behavior in FASPS patients (Toh et al. 2001).
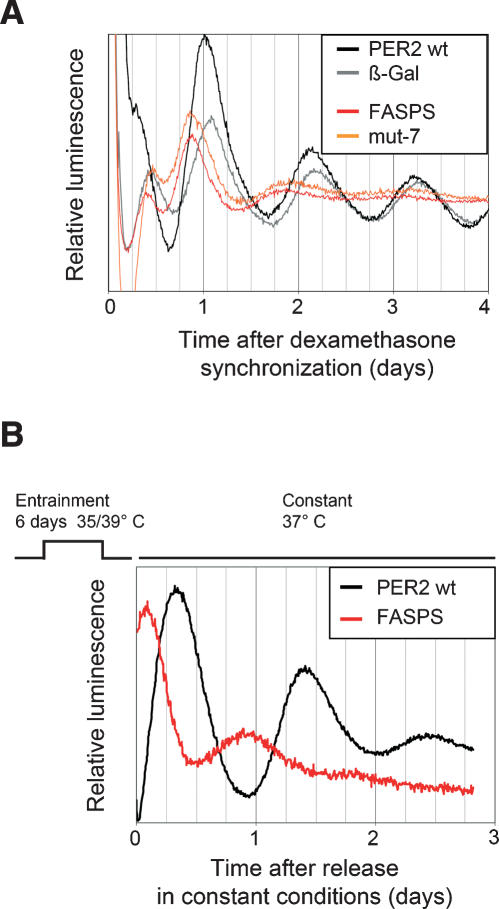
Figure 2.
The FASPS mutation leads to an advanced phase and a shorter period in oscillating fibroblasts. (A) Real-time circadian oscillations of luciferase reporter activity in dexamethasone-synchronized NIH3T3 fibroblasts, which stably express either PER2 wild-type or the PER2 mutants FASPS and mut-7 as well as β-galactosidase as a control from the same chromosomal locus (see main text and Materials and Methods). Shown are representative detrended time series from at least five independent experiments. (B) PER2 wild-type and FASPS-expressing NIH3T3 cell lines were entrained with temperature cycles consisting of 12 h at 35°C, and 12 h at 39°C for 6 d, as indicated. After transfer to constant 37°C, real-time circadian oscillations of luciferase reporter activity were recorded. Shown are representative detrended time series from at least three independent experiments.
This correlation prompted us to test whether we could phenocopy the advanced phase of FASPS patients in an entrainment regime. To this end, we subjected the cells to temperature cycles, since such a Zeitgeber cycle has been shown to entrain both SCN slices (Herzog and Huckfeldt 2003) and fibroblast cells (Brown et al. 2002) to a period corresponding to that of the temperature regime.
We entrained FASPS and PER2 wild-type expressing cells for 6 d with a temperature cycle (12 h at 39°C, 12 h at 35°C) and subsequently transferred them to constant temperature (37°C). To bypass temperature-dependent recording fluctuations (caused by changes in the efficiency of the luciferase reaction used to produce light and concomitant variations in sensitivity and background of the photomultiplier tubes used to measure it), the first peak of circadian luminescence in constant temperature was used to infer the phase of entrainment. PER2 FASPS-expressing fibroblasts had a circadian phase that was 6 h earlier than that of PER2 wild-type expressing cells (Fig. 2B). In addition, the peak-to-peak distance of the first two peaks in constant temperature (as a rough estimate for period) was ~6 h smaller for FASPS cells compared with wild type. These data imply that the FASPS phenotype is likely to be caused by a phosphorylation defect in hPER2, and is based on altered fundamental properties of the core molecular oscillator rather than on changes in the light signaling or output pathways of the human circadian system.
Phosphorylation at the FASPS site stabilizes PER2
To investigate the molecular basis for this phenotype, we first tested whether the stability of FASPS, mut-7, S659D, and wild-type mPER2 might be different. For that purpose, NIH3T3 cell lines expressing the different mPER2-V5 variants were treated with or without the translational inhibitor cycloheximide (CHX), and the turnover rates of mPER2 variants were determined over a time course of 8 h by anti-V5 Western blots (Fig. 3A).
Figure 3.
The FASPS mutation leads to a destabilization of the PER2 protein. (A) NIH3T3 cells stably expressing either PER2 wild-type or the indicated PER2 mutant proteins from the same chromosomal locus were treated with the protein translation inhibitor CHX. Cells were harvested at the indicated times after CHX treatment, and the amount of PER2 protein was determined by Western blotting. Treatment of the cells with solvent did not lead to a decrease of protein amount (not shown). (B) Quantification of the PER2 protein from the experiments described above. Error bars represent the range from two independent experiments.
In contrast to the hypothesis by Toh et al. (2001) and others (Piggins 2002; Cermakian and Boivin 2003; Gachon et al. 2004) that FASPS hPER2 is hypophosphorylated in vivo and thereby less prone to proteasomal degradation, we find that FASPS mPER2 and—even more pronounced—mut-7 mPER2 show a substantially reduced stability toward proteasomal degradation. In this assay, the half-lives of FASPS and mut-7 PER2 are decreased to 0.5–1 h compared with 2.5–3 h for wild-type mPER2 and S659D (Fig. 3B). Exchanging serine 659 to aspartic acid to mimic phosphorylation (S659D) almost completely rescues the destabilizing effect of the FASPS mutation.
Next, we tested whether the FASPS mutation has an effect on the ability of PER2 to inhibit CLOCK–BMAL1 transactivation activity. Wild-type and FASPS PER2 inhibited CLOCK–BMAL1-mediated transcriptional activation to the same extent in HEK293 cells (to ~50%) (Supplementary Fig. S2). In addition, the specific CKI-inhibitor CKI-7 had no effect on the inhibitory activity of PER2 in this assay (data not shown). Together, these data render it unlikely that S659 phosphorylation directly modulates repression activity of PER2, although such modulation at nonhomologous sites has been described for Drosophila PER (Nawathean and Rosbash 2004).
PER2 in FASPS is more sensitive toward CKIε-mediated degradation
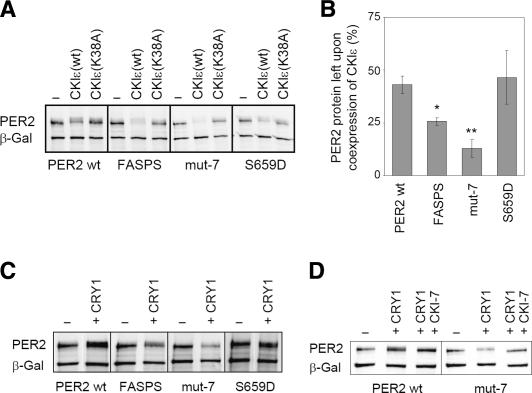
The reduced stability of FASPS and mut-7 PER2 may be caused by an increased sensitivity toward CKIε-mediated degradation. To test this hypothesis, the different PER2 variants were transiently expressed in HEK293 cells with or without wild-type CKIε or a kinase-dead form of CKIε(K38A) (Akashi et al. 2002). The steady-state protein abundance levels were then compared by using anti-V5 Western blots (Fig. 4A). Upon coexpression of CKIε(wt) but not the dominant-negative kinase-dead form CKIε(K38A), wild-type PER2 shows a decreased abundance and an increase in phosphorylation state evident by a reduced electrophoretic mobility. This destabilization is significantly more pronounced for FASPS and mut-7 PER2 (Fig. 4B), again in cells expressing CKIε(wt) but not the dominant-negative CKIε(K38A) protein. This effect is rescued when blocking the proteasome by the specific inhibitor MG-132 (Supplementary Fig. S3). Together, our data indicate that the reduced stability of FASPS and mut-7 PER2 is indeed caused by their higher sensitivity toward CKIε/δ phosphorylation and subsequent proteasomal degradation.
Figure 4.
The FASPS mutation leads to an increased sensitivity of PER2 toward CKIε-mediated degradation. (A) HEK293 cells coexpressing the indicated PER2 variants and either CKIε(wt) or the kinase-dead variant CKIε(K38A) were analyzed for PER2 protein abundance by Western blotting. Plasmid expressing β-galactosidase was cotransfected for normalization. (B) Quantification of the experiments described above with β-galactosidase band intensity used for normalization. The ratio between PER2 protein abundance when coexpressed with CKIε(wt) and PER2 protein abundance without coexpression of the kinase (average ± SEM, n = 2–5) is plotted. For FASPS and mut-7, the coexpression of CKIε leads to a significantly larger decrease in protein abundance than for PER2 wild type ([*] p < 0.05; [**] p < 0.01; unpaired homoscedastic t-test). (C) HEK293 cells expressing the indicated PER2 variants with or without CRY1 were analyzed for PER2 protein abundance by Western blotting. (D) HEK293 cells coexpressing CRY1 and either PER2 wild type or mut-7 were treated with the specific casein kinase I inhibitor CKI-7 or solvent. The effect on PER2 protein abundance was analyzed by Western blotting.
Complex formation of PER proteins with CRYs has been reported both to stabilize the PER proteins and to target them to the nucleus (Kume et al. 1999; Shearman et al. 2000; Yagita et al. 2002). To evaluate whether CRY proteins would equally stabilize FASPS and mut-7 PER2, we transiently expressed the different PER2 variants with or without CRY1 and analyzed steady-state PER2 protein abundance by Western blot. Surprisingly, whereas wild-type and S659D PER2 are stabilized by CRY1, FASPS and mut-7 PER2 are rather destabilized (Fig. 4C). This destabilization is likely due to an increased sensitivity toward phosphorylation by endogenous CKIε/δ, because upon CRY1 expression, an electrophoretic mobility shift in PER2 is visible. This shift can be reversed by -phosphatase treatment (data not shown). In addition, the CRY1-mediated destabilization of FASPS and mut-7 PER2 can be rescued by treatment of the cells with the specific CKI-inhibitor CKI-7 (Fig. 4D).
Phosphorylation at the FASPS position leads to an increased nuclear retention of PER2
Thus our data indicate that phosphorylation of PER2 has at least two functionally different effects. Phosphorylation of PER2 at the FASPS position and immediate downstream positions causes PER2 protein stabilization, whereas phosphorylation by CKIε/δ at other sites leads to PER2 protein degradation. To investigate the basis for the stabilizing effect, we tested whether the FASPS mutation influences the subcellular localization of PER2. For that purpose, we used our cell culture model for FASPS and first asked whether the nuclear import might be regulated by phosphorylation at this position.
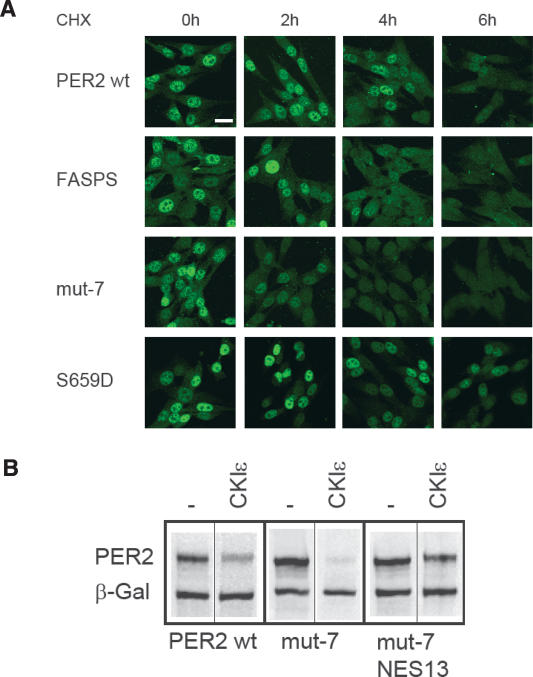
Our PER2 variant expressing NIH3T3 cell lines were treated with CHX for 14 h to clear PER2 protein from the cells. Subsequently, CHX was removed to reinduce PER2 protein synthesis and simultaneously synchronize the cells (Yamaguchi et al. 2003). PER2 protein is rapidly formed, and appreciable phosphorylation was detected only ~4 h after induction (Supplementary Fig. S4A). In immunocytochemistry experiments, we detected nuclear PER2 as early as 1.5 h after induction of protein synthesis, when the proteins were still hypophosphorylated. No gross difference was detected when comparing FASPS and mut-7 PER2 with wild-type and S659D PER2 (Supplementary Fig. S4B). These data suggest that non- or hypophosphorylated PER2 can enter the nucleus immediately after its synthesis and that phosphorylation at the FASPS position does not seem to influence nuclear import.
In contrast, when we investigated nuclear clearance rates, we see a substantially faster nuclear clearance of FASPS and mut-7 PER2 compared with wild-type and S659D PER2 (Fig. 5A). To detect this, we again treated our cell lines with CHX over a course of 6 h and determined the time course of average nuclear PER2 staining during this period. Whereas FASPS and mut-7 PER2 levels in the nucleus are substantially reduced after 4 h and barely detectable after 6 h, wild type and S659D are present in the nucleus much longer, with appreciable nuclear staining even after 6 h of CHX treatment. This result is reminiscent of findings of Dey et al. (2005), who also report an earlier nuclear clearance of PER2 in the tau mutant hamster.
Figure 5.
The FASPS mutation leads to a premature nuclear clearance. (A) PER2 variant-expressing NIH3T3 cell lines were treated with the protein translation inhibitor CHX. Cells were fixed at the indicated time points after treatment and immunostained with anti-V5 and fluorescently labeled secondary antibody. Shown are representative fields of two experiments. Bar, 20 μm. (B) HEK293 cells were transiently transfected with either PER2 wild type, mut-7, or a mut-7 variant, whose nuclear localization signals 1 and 3 (mut-7 NES13) were mutated. CKIε was coexpressed where indicated, and PER2 protein abundance was detected by Western blotting.
The effects that we observe for PER2-FASPS phosphorylation site mutations on protein stability and nuclear export might be either independent or related phenomena. To address these alternatives explicitly, we next asked whether constitutive nuclear localization might rescue PER2-FASPS from CKIε/δ-mediated degradation. We mutated the nuclear export sequences (NES) 1 and 3 (Vielhaber et al. 2001; Yagita et al. 2002) of the PER2 mut-7 variant in order to target it constitutively to the nucleus. Whereas PER2 mut-7 was almost completely degraded upon coexpression with CKIε in HEK293 cells, the nuclear-export-deficient PER2 mut-7 variant was substantially stabilized (Fig. 5B). These results suggest that phosphorylation at the FASPS and downstream positions stabilizes PER2 protein by preventing premature nuclear clearance and thereby cytosolic degradation.
Testing a mathematical model of multisite PER2 phosphorylation
Taken together, our results are consistent with a model containing at least two functionally different phosphorylation events in PER2. To better understand the implications of such a scenario in both a conceptual and a quantitative manner, we constructed a mathematical model of PER2 phosphorylation in the circadian system based on our data (Fig. 6A).
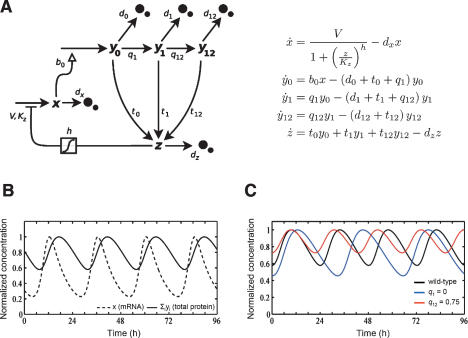
Figure 6.
Mathematical model of PER2 phosphorylation. (A) Scheme and differential equations of our conceptual model of the circadian oscillator with emphasis on phosphorylation events. The variables y 0, y 1, and y 12 represent the different phosphorylated protein species with y 0 being nonphosphorylated protein, y 1 phosphorylated at “one” site, and y 12 phosphorylated at “two” sites. The variables x and z describe mRNA and nuclear protein, respectively. (Black arrows) Net fluxes of the different species; (open arrow) translation; (chopped arrow with box) transcriptional inhibition using a Hill term; (black dots) degradation. The differential equations are chosen as simply as possible and are essentially an extension of the Goodwin oscillator model (Goodwin 1965). For a full reasoning of the model design and parameter choice, see main text and Supplemental Material. (B) With the parameters chosen, the model shows sustained 24-h oscillations with the mRNA peak preceding the peak of total protein by ~6 h, thus reproducing experimental results. (C) Interference with the phosphorylation on the “second” site, as is the case in FASPS, is modeled by reducing the respective phosphorylation rate constant parameter q 12, leading to a shorter circadian period (red) compared with a wild-type situation (black). In contrast, if phosphorylation of “both” sites is blocked (modeled by reducing q 1 to 0), it is predicted that the circadian period lengthens.
The model is aimed to conceptualize our and others’ key findings and does not want to describe the transcriptional/translational feedback loops of the circadian oscillator in all its complexity. Therefore, we deliberately designed the model to be as simple as possible by not modeling parameters irrelevant to our findings, and starting from the simple oscillator described 40 yr ago by Goodwin (1965). We used mostly linear kinetics (with a Hill function for transcriptional repression) and only five variables, which represent mRNA (x), nonphosphorylated protein (y 0), protein phosphorylated at “one” site (y 1), protein phosphorylated at “two” sites (y 12), and nuclear protein (z). Since this is a conceptual model, the “protein” variables may represent any of the various components of the negative feedback loop including CRY, PER, and CKIε/δ proteins and complexes thereof.
Our experimental findings were then translated to constrain the parameters of the model. For example, the degradation rate constant d 1 is bigger than d 12, describing the fact that FASPS PER2 is more rapidly degraded than wild-type PER2. Similarly, the rate constant of the net flux into the nucleus t (import minus export) is higher for the fully phosphorylated protein y 12 than for y 1. This describes the accelerated nuclear clearance of PER2 in our FASPS cell line and in tau mutant hamsters’ SCN neurons (Dey et al. 2005). For a description of all parameter choices, please see our Supplemental Material.
In the “wild-type” situation, our model shows robust, self-sustained oscillations with a 24-h period and an ~6-h advanced oscillation in mRNA compared with total protein, a scenario that nicely reproduces experimental findings (Fig. 6B). For the FASPS phenotype, we assume that the phosphorylation specifically of the “second” site (y 12) is severely attenuated. This describes the fact that the putative priming phosphorylation site (Ser 662 in humans) is missing, but PER2 is still phosphorylated at other sites (Fig. 4A). To achieve this reduction, we substantially decreased the phosphorylation rate constant q 12 for the “second” phosphorylation. This perturbation leads to a period shortening (Fig. 6C; Supplementary Fig. S5) as observed in FASPS patients. In contrast, reducing the phosphorylation rate constants q 1 and q 12 simultaneously, which corresponds to kinase inactivation, leads to a lengthening of the oscillation period (Fig. 6C; Supplementary Fig. S5). Thus, the concept of “two” functionally different phosphorylation sites in PER proteins can explain the experimental observation of opposite period effects in different kinase mutants (e.g., dbtS and dbtL in Drosophila).
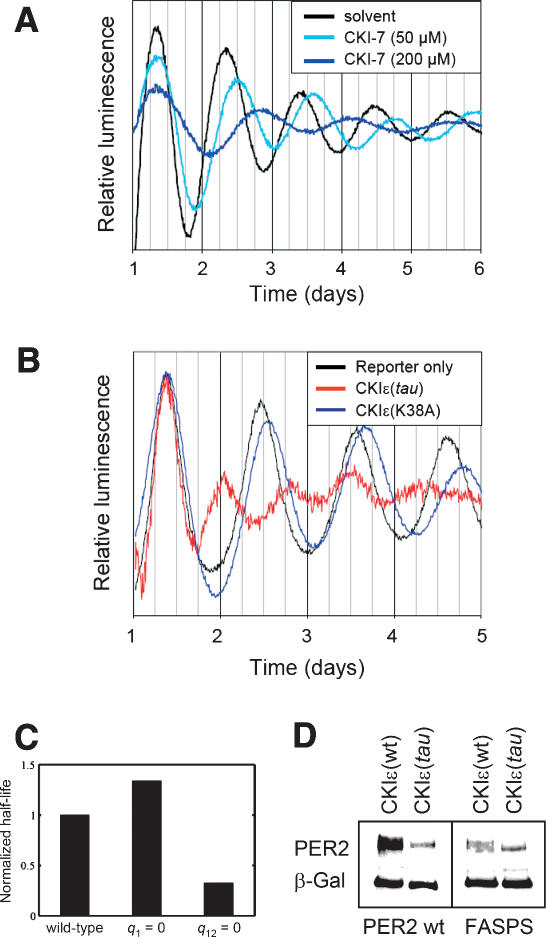
We tested the validity of this concept by examining the effects on circadian period length of pharmacological inactivation of CKIε in our fibroblast system. We treated NIH3T3 reporter cells with the specific CKI-inhibitor CKI-7 and analyzed the effect on circadian period. As predicted, a dose-dependent period lengthening of up to ~34 h for the highest dose (200 μM) was observed (Fig. 7A). Two other highly specific CKI-inhibitors (D4476 and IC261) also resulted in substantial period lengthening (Eide et al. 2005; data not shown). In addition, we see several hours’ delay in phase when we subject these cells to a temperature entrainment paradigm (data not shown).
Figure 7.
Experimental test of the model's predictions. (A) Dexamethasone-synchronized NIH3T3 reporter cells were treated with the specific CKI-inhibitor CKI-7 in different concentrations or with solvent (DMSO) only. Shown are representative detrended time series from at least five independent experiments, which were aligned to the first peak for period comparison. (B) NIH3T3 cells were stably transfected with circadian luciferase reporter and either CKIε(tau) or the kinase-dead version CKIε(K38A) and synchronized with dexamethasone. Shown are representative detrended and normalized time series from two independent experiments, which were aligned to the first peak for period comparison. (C) Prediction of the model for the effect of CKIε(tau) on PER2 protein half-life. The reduced kinase activity of CKIε(tau) in vitro is modeled by either reducing the “first” phosphorylation (q 1 = 0), the “second” phosphorylation (q 12 = 0), or “both” (q 1 = 0). Since only the reduction of the “second” phosphorylation event (q 12 = 0) results in a short circadian period as seen in hamsters, the model predicts a destabilization of PER2 by CKIε(tau) in comparison with CKIε(wt). For the calculation of half-lives, see Supplemental Material. (D) Experiments verify the model's predictions: CKIε(wt) or CKIε(tau) was coexpressed with wild-type or FASPS PER2, and steady-state protein abundance levels were analyzed in Western blot experiments. CKIε(tau) destabilizes mPER2 to a much higher extent than CKIε(wt).
Next, we wanted to know whether this model can also describe the tau hamster's phenotype, which has a short circadian period in vivo, while having a reduced CKIε activity in vitro (Lowrey et al. 2000). Three alternative hypotheses were considered: Either the reduced kinase activity of CKIε(tau) affects “both” phosphorylation events, only one of them (“first” or “second”), or only the other. Again, this assumption is modeled by decreasing the phosphorylation rate constants q 1 and q 12 either simultaneously or individually. Only the decrease of q 12 alone is able to model the short circadian period in tau mutant hamsters (Supplementary Fig. S5). Experimentally, these consequences were verified by expressing either CKIε(tau) or the kinase-inactive protein CKIε(K38A) in NIH3T3 reporter cells. For both forms of the kinase, dominant behavior is expected due to an unaltered ability to bind to PER proteins, thereby probably preventing wild-type kinases from phosphorylation. While the addition of the kinase-inactive form CKIε(K38A) evokes an ~1.5-h period lengthening, CKIε(tau) leads to a drastic period shortening to ~17 h and a substantially reduced amplitude in this experiment (Fig. 7B).
The biochemical prediction from this model is that in these cells CKIε(tau) efficiently phosphorylates the “first” phosphorylation site of PER2 but is debilitated in its ability to phosphorylate the “second” site—i.e., presumably including the residues downstream from Ser 659—leading to a more cytoplasmic localization of the protein. This prediction can be tested experimentally, since it has implications on protein stability: Expression of CKIε(tau) should destabilize PER2 much more than expression of CKIε(wt) (Fig. 7C). Concomitantly, destabilizing effects of CKIε(tau) and CKIε(wt) on FASPS PER2 stability should not be different from each other. This model prediction turns out to be correct. When we coexpressed PER2 wild-type or FASPS PER2 with CKIε(wt) or CKIε(tau) in cultured cells, the steady-state levels of PER2 coexpressed with CKIε(tau) is much lower than that coexpressed with CKIε(wt), a result recently also reported by others (Gallego et al. 2006). Steady-state levels of FASPS PER2, however, are equally affected by both kinases (Fig. 7D).
Taken together, the data presented here demonstrate that mPER2 phosphorylation has at least two discernible roles in the circadian oscillator, which are differentially affected in PER2 and CKIε/δ (or dbt) mutants. One is CKIε-mediated and targets the protein to proteasomal degradation, and the other is leading to nuclear retention and is associated with the phosphorylation of Ser 659 (FASPS site) and downstream residues. Interference with these phosphorylation events can result in opposite circadian period phenotypes.
Discussion
Post-translational control of clock proteins is a crucial step in the molecular function of circadian oscillators. In cyanobacteria, rhythmic phosphorylation and dephosphorylation of clock proteins seem to be sufficient to produce circadian oscillations, even in vitro (Nakajima et al. 2005; Tomita et al. 2005). Similarly to our results, in Neurospora, the differential regulation of the phosphorylation state of the core clock protein Frequency (FRQ) plays a pivotal role in circadian rhythmicity. Phosphorylation of FRQ by casein kinase 1a (a Neurospora homolog of CKIε) at one site (in the PEST-1 region) targets the protein to proteasomal degradation (Görl et al. 2001), whereas phosphorylation at other sites (in the PEST-2 region) is required for a distinct subcellular localization of FRQ; i.e., FRQ matures from a nuclear repressor toward a cytosolic activator (Schafmeier et al. 2006).
In Drosophila and mammals, rhythmic phosphorylation of PER proteins has been shown to be essential for rhythm generation itself as well as for correct circadian period and phase. By beginning with a general screen for PER2 phosphorylation sites and then focusing on a detailed molecular analysis of mutations in a few biologically important residues, we suggest that different phosphorylation events having distinct effects on PER protein dynamics may be necessary for the correct regulation of circadian rhythms in vivo.
PER2 is phosphorylated at many positions including the FASPS site
So far, there has been no systematic effort to map the exact residues in mammalian PER proteins that are phosphorylated during the circadian cycle. To accomplish this task, we developed a novel mass spectrometric approach for the comprehensive mapping of phosphorylation sites of clock proteins (Schlosser et al. 2005). This screen identified 21 residues of mPER2 phosphorylated by endogenous kinases in cells including the position that is mutated in humans suffering from FASPS (Fig. 8A). Interestingly, the FASPS position may not be phosphorylated by CKIε/δ in vivo, since CKIδ could not phosphorylate the FASPS position (and seven other sites) in an in vitro phosphorylation experiment (see Table 1 and our previous publication Schlosser et al. 2005). Moreover, the FASPS position does not comprise a canonical substrate recognition site (Flotow et al. 1990; Flotow and Roach 1991). Once phosphorylated, however, Ser 659 is likely to serve as a priming site for a cascade of subsequent downstream phosphorylation by CKIε/δ (Fig. 1A), since the motif pS-X-X-S is a known CKIε/δ substrate recognition site (pS, phosphorylated serine; X, any amino acid). This is in agreement with the results of Toh et al. (2001), who have shown that FASPS mutant hPER2 is less efficiently phosphorylated by CKIε in vitro. It should be noted, however, that the same mutation when introduced in PER1 causes hyperphosphorylation of mPER1 in an in vitro CKIε phosphorylation study (Takano and Nagai 2006). The discrepancy of these two results may be due either to substrate specificity problems with in vitro phosphorylation studies or to different properties of hPER2 and mPER1.
How does the phosphorylation of PER2 influence the circadian oscillator mechanism?
Our studies of the effects of the S659G (FASPS) phosphorylation site defect suggest that this mutation has two distinct effects: It leads to a premature PER2 nuclear clearance, and it destabilizes the PER2 protein. These two findings are likely to be interdependent. Proteasomal degradation of PER2 happens primarily in the cytoplasm, since inhibiting the nuclear export of mut-7 PER2 rescues its destabilization. Thus, the destabilization of FASPS PER2 is more a result of PER2 protein localization than of an altered degradation mechanism. The same could also be true for the unexpectedly strong destabilizing effect of CKIε(tau) on PER2 protein, an effect predicted by our mathematical model and confirmed by experiments. In this scenario, localization of PER2 in tau mutant hamsters is likely to be altered, with PER2 being more in the cytoplasm and thereby susceptible to increased degradation simply due to its increased concentration there. In addition, the in vivo kinase activity of CKIε(tau) might itself be increased, as suggested by Gallego et al. (2006). Both of these possibilities are discussed further below.
Any discussion of nucleocytoplasmic shuttling of PER proteins would not be complete without mentioning the partners of PERs, the CRY proteins. In mammals, CRY proteins seem to be the primary nuclear repressors of CLOCK–BMAL1 transactivation (Griffin et al. 1999; Kume et al. 1999); however, PERs are required for CRY protein nuclear accumulation (Lee et al. 2001) and vice versa (Shearman et al. 2000), and the level of PER protein abundance is rate limiting for PER–CRY complex formation and subsequent transport to the nucleus (Lee et al. 2001).
Here, we show that complex formation with CRY1 not only stabilizes PER2, but also induces phosphorylation. We rationalize this observation by suggesting that upon complex formation with CRY1, PER2 is phosphorylated at sites that prevent nuclear export, thereby leading to nuclear accumulation and protein stabilization. Since this site is missing in FASPS PER2, the opposite occurs, a result that we also show. In Cry1/Cry2 double-mutant mice, PER2 is not detected in SCN neurons despite high transcript levels (Shearman et al. 2000), a result that indicates that PER2 can be phosphorylated and rapidly degraded even in the absence of CRY proteins. Thus, CRYs are not required for the type of PER2 phosphorylation that is responsible for degradation.
Thus, we propose the following model (Fig. 8B): In the mammalian circadian oscillator, PER proteins (or at least PER2) are primarily responsible for a proper timing of the nuclear appearance and nuclear repression of CRY proteins. The molecular setscrew for this is likely to be the phosphorylation pattern of PER proteins at a given circadian phase. It determines the amount of PERs available for complex formation with CRYs by regulating PER protein degradation. At the beginning of the circadian cycle, PER2 protein shuttles between nucleus and cytoplasm, where it is rapidly phosphorylated by kinases such as CKIε/δ at sites that are required for targeting it to the proteasomal degradation pathway. Upon complex formation with CRYs, we speculate that additional kinases are activated or recruited to the PER2–CRY complex. These phosphorylate PER2 at functionally different sites including the FASPS region. Here, CKIε/δ likely has a second role in phosphorylation of the serine residues downstream from the FASPS site. This prevents a premature nuclear clearance and thereby stabilizes PER2 and possibly the PER2–CRY complex, which ensures the correct timing of transcriptional repression.
It is tempting to speculate that the short- and long-period phenotypes of Cry1 −/− and Cry2 −/− mutant mice may also be due to alterations in the phosphorylation patterns of PER proteins. The lack of CRY1 or CRY2 would differentially influence the phosphorylation state of PER proteins and thus could modulate the circadian period in opposite directions. To test this hypothesis, a dynamic and quantitative phosphorylation site mapping of PER proteins in a Cry1- or a Cry2-deficient background would be needed.
Perturbing the PER phosphorylation pattern can lead to opposite period phenotypes
We used mathematical modeling to conceptualize our experimental findings and to analyze their implications for circadian oscillator dynamics. We reduced the complex molecular network of the circadian oscillator as far as possible to key elements for that purpose. Basically, we assume three different phosphorylation states—i.e., a nonphosphorylated state, a phosphorylation state one, and a phosphorylation state two. These individual states have different molecular consequences, which is modeled as variations in degradation rates and net nuclear fluxes. Those parameters were chosen according to the main findings of our experiments. If we reduce only the phosphorylation that is perturbed in FASPS (the second state y 12), we see a short circadian period. In contrast, reducing PER phosphorylation more globally; i.e., both phosphorylation states (y 1 and y 12), results in long circadian periods. This led to the prediction that expressing a dominant-negative form of CKIε—which can still bind to PER proteins but prevents wild-type kinases from phosphorylation—or using specific inhibitors leads to long periods and delayed phases, which was indeed the case when tested experimentally.
With this model, we can also explain the short-period phenotypes of the tau mutant hamster and our tau-over-expressing cells as well as the second form of FASPS, in which the CKIδ gene is mutated, also leading to a less active kinase in vitro (Xu et al. 2005). Here, we assume that both mutations specifically and semidominantly reduce the phosphorylation of PER proteins at positions responsible for nuclear localization. Molecularly, our simple assumption is supported by data from Dey et al. (2005), who found that nuclear clearance of PER proteins is accelerated in SCN neurons of tau hamsters. Our model predicted that CKIε(tau) should destabilize PER2 much more than wild-type CKIε, a consequence of the fact that proteasomal degradation is a cytoplasmic event. Indeed, Lee et al. (2001) found lower PER2 protein abundance in the tau hamster. When we tested the model's prediction here by coexpressing CKIε(tau) with PER2 in cultured cells, we saw a substantially stronger destabilization of PER2 than with CKIε(wt) coexpression. FASPS PER2, which cannot be phosphorylated at positions responsible for nuclear localization, was not further destabilized by CKIε(tau). Together, these data suggest that CKIε(tau) is debilitated in its ability to phosphorylate the positions immediately downstream from the FASPS position but is still able to phosphorylate positions required for proteasomal degradation. It is conceivable, however, that these mutations have additional effects on not yet identified in vivo substrates of CKIε, one of which may be BMAL1, a substrate of CKIε in vitro (Eide et al. 2002).
A recently published mathematical model by Gallego et al. (2006) predicted that CKIε(tau) has increased kinase activity in vivo, which is claimed to be necessary to explain the short circadian period of the tau mutant hamster. This contrasts with our assumption that the tau mutation selectively decreases the phosphorylation of PER specifically at sites that promote nuclear localization. For the available data about the tau mutation, both of these models are capable of recapitulating the observed phenotype, and, indeed, a combination of both mechanisms is possible. For the FASPS mutation, the localization data that we present strongly favor the localization-dependent model that we postulate.
This mathematical model can be also used for the circadian oscillator in Drosophila. As mentioned in the introduction, CKIε homolog doubletime mutant alleles dbtS and dbtL have strong but opposite effects on circadian period (Kloss et al. 1998; Price et al. 1998), but both lead to an enzyme with reduced kinase activity in vitro (Preuss et al. 2004). In dbtS mutant flies, the nuclear accumulation dynamics of PER is altered, whereas in dbtL mutant flies, phosphorylation-dependent PER turnover is decreased (Price et al. 1998; Rothenfluh et al. 2000; Bao et al. 2001). Doubletime may also be involved in functionally different phosphorylation events that result in opposite effects on the circadian period, one being primarily responsible for degradation, the other for nuclear localization. Thus, the mechanism that we have uncovered may be generally important to circadian regulation.
Materials and methods
Phosphorylation site mapping of mPER2
HEK293 cells were stably transfected with mPer2-pDEST40 (Invitrogen) coding for a V5-epitope-tagged mPER2 protein. Cells (8 × 107) were harvested and lysed in immunoprecipitation buffer (20 mM Tris/HCl at pH 8.0, 140 mM NaCl, 1 mM TCEP, 1.5 mM MgCl2, 1% Triton X-100, 10% glycerine). Immunoprecipitation was carried out with anti-V5 antibody (Invitrogen) and G-protein-coupled agarose beads (Santa Cruz Biotechnology). Phosphorylation site mapping using mass spectrometry was performed as described in Schlosser et al. (2005).
Plasmids
The mPer2 coding sequence was cloned by PCR from a Per2pCMVSport vector kindly provided by Chuck Weitz (Harvard Medical School, Boston, MA). The mCKIε PCR product was derived from cDNA reverse-transcribed from NIH3T3 RNA. Both genes were inserted into pENTR/D-TOPO (Gateway System; Invitrogen) and verified by DNA sequencing. Afterward, the mPer2 and mCKIε cDNAs were recombined into different expression vectors (pcDNA-DEST40; pDEST26, pEFDEST51, pEF5/FRT/V5-DEST; for vector maps, see http://www.invitrogen.com/content.cfm?pageid=94).The luciferase reporter vector (pGL3_E6S) containing six E-boxes of the mPer1 promoter in the pGL3-Promoter vector (Promega; for vector map, see http://www.promega.com/vectors/pgl3prom.txt) was generated as follows. A vector containing the three E-boxes of the mPer1 promoter (pGL3_E3S) was provided by Chuck Weitz. In order to duplicate the E-box-containing fragment, this region was excised using the restriction enzymes SacI and BsgI and introduced at the Ecl136II site of the pGL3_E3S vector. The resulting luciferase reporter vector harboring six E-boxes (pGL3_E6S) was verified by DNA sequencing.
Site-directed mutagenesis
Amino acids were substituted in the mPER2 FASPS region (FASPS S659G; S659D; mut-7 S659G S662A S665A S668A S670A S671A T672A), in the mPER2 nuclear export sequences 1 and 3 (NES1 L113A L116A; NES3 I464A L467A), in the mCKIε catalytic region (K38A), and to generate the mCKIε(tau)(R178C) by site-directed mutagenesis (QuickChange Site-Directed Mutagenesis Kit; Stratagene). The sequences of the mutagenesis primers are given in the Supplemental Material. The presence of the corresponding base-pair substitutions was confirmed by DNA sequencing.
Cell culture and transfection
NIH3T3 and HEK293 cells were maintained in DMEM supplemented with 10% fetal bovine serum and antibiotics (100 μg/ mL penicillin and streptomycin). Stable and transient transfections were carried out using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. For transient transfections, HEK293 cells were seeded in 24-well plates in medium without antibiotics. Transfection was performed after reaching 80%–90% confluence. The total amount of transfected DNA was 1.2 μg (composed of 0.6 μg of empty vector and 0.6 μg of proteinogenic vector with 0.2 μg of mPer2 and 0.05 μg of CKIε). Cells were harvested 24–48 h after transfection. For generation of stable CKIε(K38A)- and CKIε(tau)-expressing NIH3T3 cell lines, NIH3T3 cells were cotransfected with CKIε(K38A)_pDEST51 and pGL3_E6S or CKIε(tau)_pDEST51 and pGL3_E6S, respectively. Stable transfectants were obtained by blasticidin selection.
Flp-In host and expression cell lines
The generation of Flp-In host and expression cell lines was done in accordance to the manufacturerer's protocol (Flp-In System; Invitrogen). Briefly, NIH3T3 cells were stably cotransfected with the Flp recombination target site vector pFRT/NeoR and the luciferase reporter vector pGL3_E6S, and a single clone was chosen as the host cell line. For the generation of expression cell lines by homologous recombinations at the FRT sites, plasmids containing PER2 variants (pEF5/mPER2variant/FRT-V5-DEST) were cotransfected with pOG44 (Flp-recombinase expression vector) to derive the expression cell lines for PER2, FASPS, mut-7, and S659D. Selection was performed with hygromycin.
Synchronization and measurement of circadian rhythms
Cells were either synchronized by a single pulse of dexamethasone (Sigma; 1 μM, 2 h) or entrained to temperature cycles (6 d: 12 h 35°C/12 h 39°C; constant conditions 37°C). E-box luciferase reporter activity was measured in the Lumicycle (Actimetrics), while maintaining the cells in phenol red-free culture medium supplemented with 0.1 mM luciferin (P.J.K.) and 1% DMSO. To determine the effects of CKIε/δ inhibition on the circadian oscillator, CKI-inhibitor CKI-7 (USBioUnited States Biological) was added to the culture medium in different concentrations (50 μM, 200 μM). Raw luminescence data were detrended by subtracting the 24-h running average from each value of the time series. Normalization of the data was performed when indicated by matching the first maxima of the time series to be compared.
CHX assays
For measurement of PER2 half-lives and for the PER2 nuclear clearance assay, PER2 expression cell lines were treated with CHX (Sigma; 710 μM, 355 μM, respectively) for up to 8 h. The PER2 protein amount was quantified after SDS-PAGE and Western blot with the Lumi-Imager (Roche). Nuclear clearance was investigated by immunocytochemistry.
Immunocytochemistry
Cells were seeded on eight chamber slides (Lab-Tek II Chamber Slides; Nunc). After reaching 80%–90% confluence, CHX treatment was performed. Cells were fixed for 15 min with 3% paraformaldehyde in PBS and permeabilized with 0.25% Triton X-100 in PBS for 10 min. Blocking was performed by incubating the cells with 5% goat serum in TBS for 1 h. To detect PER2 protein, cells were incubated at room temperature for up to 2 h with mouse anti-V5 monoclonal antibody (1:200; Invitrogen) and Alexa Fluor-488 goat anti-mouse antibody (1:200; Molecular Probes) as primary and secondary antibody, respectively. DAPI (1 μg/mL; Sigma) was used for nuclear staining. Cells were mounted with Fluoromount-G (Southern Biotech). Images were captured using a confocal fluorescence microscope (Olympus FV-1000, software Olympus Fluoview).
Western blotting
Cells were harvested in RIPA buffer (1% Igepal CA-630, 0.5% Sodium-Deoxycholat, 0.1% SDS in 1× PBS) supplemented 1:100 with protease and phosphatase inhibitor cocktails (Sigma). After centrifugation (12,000 rpm, 4°C, 30 min), equal amounts of protein were separated by SDS-PAGE and transferred to nitrocellulose membranes. Detection was performed using mouse anti-V5 antibody (1:5000; Invitrogen) and goat anti-mouse IgG-HRP antibody (1:1000; Santa Cruz Biotechnology). Protein abundance was detected with the Lumi-Imager (Roche) and quantified using the LumiAnalyst software (Roche).
Numerical calculations
All numerical solutions to the mathematical model were calculated using MATLAB (The Mathworks, Inc.).
Acknowledgments
We thank Jürgen Ripperger, Ueli Schibler, and Chuck Weitz for the generous gift of materials; Nadine Thierfelder for help with the kinase inhibitor assays; Astrid Grudziecki for excellent technical assistance; and Steve Brown for helpful discussions and critical reading of the manuscript. The Deutsche Forschungsgemeinschaft is acknowledged for financial support (Emmy-Noether program for A.K., grants SFB 618/A4 for A.K. and H.H., and SCHL 613/1-1 for A.S.). Our work is supported by the 6th EU framework program EUCLOCK.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.397006.
References
- Akashi, M., Tsuchiya, Y., Yoshino, T., Nishida, E. Control of intracellular dynamics of mammalian period proteins by casein kinase I ε (CKIε) and CKIδ in cultured cells. Mol. Cell. Biol. 2002;22:1693–1703. doi: 10.1128/MCB.22.6.1693-1703.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsalobre, A., Damiola, F., Schibler, U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- Bao, S., Rihel, J., Bjes, E., Fan, J.Y., Price, J.L. The Drosophila double-timeS mutation delays the nuclear accumulation of period protein and affects the feedback regulation of period mRNA. J. Neurosci. 2001;21:7117–7126. doi: 10.1523/JNEUROSCI.21-18-07117.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, S.A., Zumbrunn, G., Fleury-Olela, F., Preitner, N., Schibler, U. Rhythms of mammalian body temperature can sustain peripheral circadian clocks. Curr. Biol. 2002;12:1574–1583. doi: 10.1016/s0960-9822(02)01145-4. [DOI] [PubMed] [Google Scholar]
- Brown, S.A., Fleury-Olela, F., Nagoshi, E., Hauser, C., Juge, C., Meier, C.A., Chicheportiche, R., Dayer, J.M., Albrecht, U., Schibler, U. The period length of fibroblast circadian gene expression varies widely among human individuals. PLoS Biol. 2005;3 doi: 10.1371/journal.pbio.0030338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner, M., Schafmeier, T. Transcriptional and post-transcriptional regulation of the circadian clock of cyanobacteria and Neurospora . Genes & Dev. 2006;20:1061–1074. doi: 10.1101/gad.1410406. [DOI] [PubMed] [Google Scholar]
- Cermakian, N., Boivin, D.B. A molecular perspective of human circadian rhythm disorders. Brain Res. Brain Res. Rev. 2003;42:204–220. doi: 10.1016/s0165-0173(03)00171-1. [DOI] [PubMed] [Google Scholar]
- Dey, J., Carr, A.J., Cagampang, F.R., Semikhodskii, A.S., Loudon, A.S., Hastings, M.H., Maywood, E.S. The tau mutation in the Syrian hamster differentially reprograms the circadian clock in the SCN and peripheral tissues. J. Biol. Rhythms. 2005;20:99–110. doi: 10.1177/0748730404274264. [DOI] [PubMed] [Google Scholar]
- Eide, E.J., Vielhaber, E.L., Hinz, W.A., Virshup, D.M. The circadian regulatory proteins BMAL1 and cryptochromes are substrates of casein kinase Iε. J. Biol. Chem. 2002;277:17248–17254. doi: 10.1074/jbc.M111466200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide, E.J., Woolf, M.F., Kang, H., Woolf, P., Hurst, W., Camacho, F., Vielhaber, E.L., Giovanni, A., Virshup, D.M. Control of mammalian circadian rhythm by CKIε-regulated proteasome-mediated PER2 degradation. Mol. Cell. Biol. 2005;25:2795–2807. doi: 10.1128/MCB.25.7.2795-2807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flotow, H., Roach, P.J. Role of acidic residues as substrate determinants for casein kinase I. J. Biol. Chem. 1991;266:3724–3727. [PubMed] [Google Scholar]
- Flotow, H., Graves, P.R., Wang, A.Q., Fiol, C.J., Roeske, R.W., Roach, P.J. Phosphate groups as substrate determinants for casein kinase I action. J. Biol. Chem. 1990;265:14264–14269. [PubMed] [Google Scholar]
- Fujimoto, Y., Yagita, K., Okamura, H. Does mPER2 protein oscillate without its coding mRNA cycling?: Post-transcriptional regulation by cell clock. Genes Cells. 2006;11:525–530. doi: 10.1111/j.1365-2443.2006.00960.x. [DOI] [PubMed] [Google Scholar]
- Gachon, F., Nagoshi, E., Brown, S.A., Ripperger, J., Schibler, U. The mammalian circadian timing system: From gene expression to physiology. Chromosoma. 2004;113:103–112. doi: 10.1007/s00412-004-0296-2. [DOI] [PubMed] [Google Scholar]
- Gallego, M., Eide, E.J., Woolf, M.F., Virshup, D.M., Forger, D.B. An opposite role for tau in circadian rhythms revealed by mathematical modeling. Proc. Natl. Acad. Sci. 2006;103:10618–10623. doi: 10.1073/pnas.0604511103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin, B.C. Oscillatory behavior in enzymatic control processes. Adv. Enzyme Regul. 1965;3:425–438. doi: 10.1016/0065-2571(65)90067-1. [DOI] [PubMed] [Google Scholar]
- Görl, M., Merrow, M., Huttner, B., Johnson, J., Roenneberg, T., Brunner, M. A PEST-like element in FREQUENCY determines the length of the circadian period in Neurospora crassa . EMBO J. 2001;20:7074–7084. doi: 10.1093/emboj/20.24.7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin, E.A., Jr., Staknis, D., Weitz, C.J. Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science. 1999;286:768–771. doi: 10.1126/science.286.5440.768. [DOI] [PubMed] [Google Scholar]
- Harms, E., Kivimae, S., Young, M.W., Saez, L. Post-transcriptional and posttranslational regulation of clock genes. J. Biol. Rhythms. 2004;19:361–373. doi: 10.1177/0748730404268111. [DOI] [PubMed] [Google Scholar]
- Herzog, E.D., Huckfeldt, R.M. Circadian entrainment to temperature, but not light, in the isolated suprachiasmatic nucleus. J. Neurophysiol. 2003;90:763–770. doi: 10.1152/jn.00129.2003. [DOI] [PubMed] [Google Scholar]
- Jones, C.R., Campbell, S.S., Zone, S.E., Cooper, F., DeSano, A., Murphy, P.J., Jones, B., Czajkowski, L., Ptacek, L.J. Familial advanced sleep-phase syndrome: A short-period circadian rhythm variant in humans. Nat. Med. 1999;5:1062–1065. doi: 10.1038/12502. [DOI] [PubMed] [Google Scholar]
- Kloss, B., Price, J.L., Saez, L., Blau, J., Rothenfluh, A., Wesley, C.S., Young, M.W. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iε. Cell. 1998;94:97–107. doi: 10.1016/s0092-8674(00)81225-8. [DOI] [PubMed] [Google Scholar]
- Kume, K., Zylka, M.J., Sriram, S., Shearman, L.P., Weaver, D.R., Jin, X., Maywood, E.S., Hastings, M.H., Reppert, S.M. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- Lee, C., Etchegaray, J.P., Cagampang, F.R., Loudon, A.S., Reppert, S.M. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- Lowrey, P.L., Shimomura, K., Antoch, M.P., Yamazaki, S., Zemenides, P.D., Ralph, M.R., Menaker, M., Takahashi, J.S. Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science. 2000;288:483–492. doi: 10.1126/science.288.5465.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrow, M., Spoelstra, K., Roenneberg, T. The circadian cycle: Daily rhythms from behaviour to genes. EMBO Rep. 2005;6:930–935. doi: 10.1038/sj.embor.7400541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagoshi, E., Saini, C., Bauer, C., Laroche, T., Naef, F., Schibler, U. Circadian gene expression in individual fibroblasts: Cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119:693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- Nakajima, M., Imai, K., Ito, H., Nishiwaki, T., Murayama, Y., Iwasaki, H., Oyama, T., Kondo, T. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- Nawathean, P., Rosbash, M. The doubletime and CKII kinases collaborate to potentiate Drosophila PER transcriptional repressor activity. Mol. Cell. 2004;13:213–223. doi: 10.1016/s1097-2765(03)00503-3. [DOI] [PubMed] [Google Scholar]
- Nishii, K., Yamanaka, I., Yasuda, M., Kiyohara, Y.B., Kitayama, Y., Kondo, T., Yagita, K. Rhythmic post-transcriptional regulation of the circadian clock protein mPER2 in mammalian cells: A real-time analysis. Neurosci. Lett. 2006;401:44–48. doi: 10.1016/j.neulet.2006.03.022. [DOI] [PubMed] [Google Scholar]
- O'Gorman, S., Fox, D.T., Wahl, G.M. Recombinase-mediated gene activation and site-specific integration in mammalian cells. Science. 1991;251:1351–1355. doi: 10.1126/science.1900642. [DOI] [PubMed] [Google Scholar]
- Pando, M.P., Morse, D., Cermakian, N., Sassone-Corsi, P. Phenotypic rescue of a peripheral clock genetic defect via SCN hierarchical dominance. Cell. 2002;110:107–117. doi: 10.1016/s0092-8674(02)00803-6. [DOI] [PubMed] [Google Scholar]
- Piggins, H.D. Human clock genes. Ann. Med. 2002;34:394–400. doi: 10.1080/078538902320772142. [DOI] [PubMed] [Google Scholar]
- Preuss, F., Fan, J.Y., Kalive, M., Bao, S., Schuenemann, E., Bjes, E.S., Price, J.L. Drosophila doubletime mutations which either shorten or lengthen the period of circadian rhythms decrease the protein kinase activity of casein kinase I. Mol. Cell. Biol. 2004;24:886–898. doi: 10.1128/MCB.24.2.886-898.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, J.L., Blau, J., Rothenfluh, A., Abodeely, M., Kloss, B., Young, M.W. double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell. 1998;94:83–95. doi: 10.1016/s0092-8674(00)81224-6. [DOI] [PubMed] [Google Scholar]
- Rothenfluh, A., Abodeely, M., Young, M.W. Short-period mutations of per affect a double-time-dependent step in the Drosophila circadian clock. Curr. Biol. 2000;10:1399–1402. doi: 10.1016/s0960-9822(00)00786-7. [DOI] [PubMed] [Google Scholar]
- Schafmeier, T., Kaldi, K., Diernfellner, A., Mohr, C., Brunner, M. Phosphorylation-dependent maturation of Neurospora circadian clock protein from a nuclear repressor toward a cytoplasmic activator. Genes & Dev. 2006;20:297–306. doi: 10.1101/gad.360906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlosser, A., Vanselow, J.T., Kramer, A. Mapping of phosphorylation sites by a multi-protease approach with specific phosphopeptide enrichment and NanoLC-MS/MS analysis. Anal. Chem. 2005;77:5243–5250. doi: 10.1021/ac050232m. [DOI] [PubMed] [Google Scholar]
- Shearman, L.P., Sriram, S., Weaver, D.R., Maywood, E.S., Chaves, I., Zheng, B., Kume, K., Lee, C.C., van der Horst, G.T., Hastings, M.H., et al. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288:1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- Shirogane, T., Jin, J., Ang, X.L., Harper, J.W. SCFβ–TRCP controls clock-dependent transcription via casein kinase 1-dependent degradation of the mammalian period-1 (Per1) protein. J. Biol. Chem. 2005;280:26863–26872. doi: 10.1074/jbc.M502862200. [DOI] [PubMed] [Google Scholar]
- Stanewsky, R. Genetic analysis of the circadian system in Drosophila melanogaster and mammals. J. Neurobiol. 2003;54:111–147. doi: 10.1002/neu.10164. [DOI] [PubMed] [Google Scholar]
- Takano, A., Nagai, K. Serine 714 might be implicated in the regulation of the phosphorylation in other areas of mPer1 protein. Biochem. Biophys. Res. Commun. 2006;346:95–101. doi: 10.1016/j.bbrc.2006.05.082. [DOI] [PubMed] [Google Scholar]
- Toh, K.L., Jones, C.R., He, Y., Eide, E.J., Hinz, W.A., Virshup, D.M., Ptacek, L.J., Fu, Y.H. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291:1040–1043. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- Tomita, J., Nakajima, M., Kondo, T., Iwasaki, H. No transcription–translation feedback in circadian rhythm of KaiC phosphorylation. Science. 2005;307:251–254. doi: 10.1126/science.1102540. [DOI] [PubMed] [Google Scholar]
- Vielhaber, E.L., Duricka, D., Ullman, K.S., Virshup, D.M. Nuclear export of mammalian PERIOD proteins. J. Biol. Chem. 2001;276:45921–45927. doi: 10.1074/jbc.M107726200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y., Padiath, Q.S., Shapiro, R.E., Jones, C.R., Wu, S.C., Saigoh, N., Saigoh, K., Ptacek, L.J., Fu, Y.H. Functional consequences of a CKIδ mutation causing familial advanced sleep phase syndrome. Nature. 2005;434:640–644. doi: 10.1038/nature03453. [DOI] [PubMed] [Google Scholar]
- Yagita, K., Tamanini, F., van der Horst, G.T., Okamura, H. Molecular mechanisms of the biological clock in cultured fibroblasts. Science. 2001;292:278–281. doi: 10.1126/science.1059542. [DOI] [PubMed] [Google Scholar]
- Yagita, K., Tamanini, F., Yasuda, M., Hoeijmakers, J.H., van der Horst, G.T., Okamura, H. Nucleocytoplasmic shuttling and mCRY-dependent inhibition of ubiquitylation of the mPER2 clock protein. EMBO J. 2002;21:1301–1314. doi: 10.1093/emboj/21.6.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, S., Isejima, H., Matsuo, T., Okura, R., Yagita, K., Kobayashi, M., Okamura, H. Synchronization of cellular clocks in the suprachiasmatic nucleus. Science. 2003;302:1408–1412. doi: 10.1126/science.1089287. [DOI] [PubMed] [Google Scholar]
- Yang, Z., Sehgal, A. Role of molecular oscillations in generating behavioral rhythms in Drosophila . Neuron. 2001;29:453–467. doi: 10.1016/s0896-6273(01)00218-5. [DOI] [PubMed] [Google Scholar]