Abstract
Damage to the mitotic spindle and centrosome dysfunction can lead to cancer. To prevent this, cells trigger a succession of checkpoint responses, where an initial mitotic delay is followed by slippage without cytokinesis, spawning tetraploid G1 cells that undergo a p53-dependent G1/S arrest. We describe the importance of Lats2 (Large Tumor Suppressor 2) in this checkpoint response. Lats2 binds Mdm2, inhibits its E3 ligase activity, and activates p53. Nocodazole, a microtubule poison that provokes centrosome/mitotic apparatus dysfunction, induces Lats2 translocation from centrosomes to the nucleus and p53 accumulation. In turn, p53 rapidly and selectively up-regulates Lats2 expression in G2/M cells, thereby defining a positive feedback loop. Abrogation of Lats2 promotes accumulation of polyploid cells upon exposure to nocodazole, which can be prevented by direct activation of p53. The Lats2–Mdm2–p53 axis thus constitutes a novel checkpoint pathway critical for the maintenance of proper chromosome number.
Keywords: Lats2, Mdm2, p53, tetraploidy checkpoint
The p53 tumor suppressor gene is subject to frequent mutational alterations in human cancer. The p53 protein is a sequence-specific transcription factor, present in very low amounts in normal cells. In response to various types of stress, p53 is hyperactivated to modulate the expression of numerous target genes (Vousden and Lu 2002; Oren 2003; Harris and Levine 2005; Vousden and Prives 2005). p53 activation can lead to a variety of changes in cell fate, most notably induction of cell cycle arrest and apoptosis. In nonstressed cells, inappropriate p53 activation is prevented through a network of tight controls. Key to this network is Mdm2, which participates in a negative feedback loop with p53, fueled by the transcriptional activation of the mdm2 gene by p53 (Michael and Oren 2003; Bond et al. 2005). Mdm2 is an E3 ubiquitin ligase, which ubiquitinates p53 and promotes its rapid proteasomal degradation. Furthermore, ubiquitination of p53 by Mdm2 expels p53 from the nucleus, keeping it away from its transcriptional targets. The Mdm2–p53 loop is subjected to multiple checkpoint regulations. Checkpoints exist at various stages of the cell cycle to prevent propagation of damaged cells and thereby preserve genomic integrity. For instance, genotoxic agents trigger a DNA damage checkpoint, leading to p53 stabilization and activation. p53 has also been implicated in a checkpoint triggered in tetraploid cells ensuing from defective mitosis. This checkpoint functions to prevent the proliferation of cells with polyploid genomes, and is particularly important in cancer, where polyploidy often precedes aneuploidy, associated with high malignancy and poor prognosis (Rajagopalan and Lengauer 2004 and references therein).
Hyperploidy can arise in mammalian cells as a result of centrosomal perturbations, errors in mitosis, or treatment with anticancer drugs, including inhibitors of microtubule organization (Lanni and Jacks 1998; Nigg 2002; Andreassen et al. 2003; Ditchfield et al. 2003; Barr et al. 2004; Rajagopalan and Lengauer 2004; Giet et al. 2005). Prolonged treatment of cells with spindle-damaging agents such as nocodazole leads to a transient preana-phase arrest due to failure to satisfy the spindle assembly checkpoint (Barr et al. 2004; Morrison and Rieder 2004; Rieder and Maiato 2004). Irrespective of p53 status, these cells usually exit from mitosis in an aberrant fashion, without sister chromatid segregation or cytokinesis, a process known as “mitotic slippage.” Cells possessing functional wild-type p53 typically undergo a p53-dependent growth arrest at the subsequent G1/S boundary. Since this prevents replication of cells with tetraploid genomes, it is sometimes referred to as the “G1 tetraploidy checkpoint” (Margolis et al. 2003). Direct damage to the centrosome, as well as induction of centrosomal stress by other means such as disruption of the microtubule network, also triggers p53 activation and culminates in a p53-dependent G1/S arrest (Doxsey et al. 2005). In contrast, cells lacking functional p53 progress into S phase to acquire polyploid genomes, and often eventually undergo apoptosis. In this way, p53 acts in a post-mitotic checkpoint that protects cells from potentially hazardous polyploidization (Meek 2000; Castedo et al. 2004; Rajagopalan and Lengauer 2004; Duensing and Duensing 2005). The molecular mechanisms driving this checkpoint are still poorly understood. However, the triggering event is probably not simply the presence of tetraploid genomes, but rather a stress signal emanating from dysfunction of the mitotic machinery, which activates p53 in due course to prevent G1/S transition (Stukenberg 2004; Uetake and Sluder 2004; Wong and Stearns 2005).
Lats (Large Tumor Suppressor) (WARTS; Wts) was first found as a tumor suppressor gene in Drosophila (Justice et al. 1995; Xu et al. 1995). Two mammalian homologs, Lats1 and Lats2, have been identified. Lats1-deficient mice develop tumors (St John et al. 1999). The gene encoding Lats2, also known as Kpm (Hori et al. 2000; Yabuta et al. 2000), resides in a chromosomal region that exhibits frequent loss of heterozygosity (LOH) in human cancers (Yabuta et al. 2000), consistent with the idea that it, too, is a tumor suppressor. Human Lats2 is a centrosomal protein (Toji et al. 2004), and recent work suggests that it plays a role in regulating the formation of the mitotic spindle apparatus (Abe et al. 2006). Clues from overexpression experiments suggest that excess Lats2 can cause G2/M arrest (Kamikubo et al. 2003), G1/S arrest (Li et al. 2003), or apoptosis (Ke et al. 2004). Extensive Lats2 overexpression in human lung cancer cells induces apoptosis via down-regulation of the anti-apoptotic proteins Bcl-2 and Bcl-xL (Ke et al. 2004). Lats2 knockout is embryonic lethal in mice; cells from such mice display defects in contact inhibition and cytokinesis, coupled with centrosome amplification, multipolar mitotic spindles, and genomic instability (McPherson et al. 2004). Lats2 is a serine/threonine kinase, and its kinase activity has been implicated in negative regulation of CyclinE/CDK2 and in tumor suppression (Li et al. 2003).
We report that Lats2 interacts physically with Mdm2 to inhibit p53 ubiquitination and promote p53 activation. This interaction is enhanced in cells treated with nocodazole, which causes microtubule and mitotic spindle damage. Nocodazole causes Lats2 translocation from the centrosome into the nucleus, followed by p53 activation. A moderate excess of wild-type but not kinase-dead Lats2 potentiates the p53-dependent G1 arrest of nocodazole-treated cells, thereby preventing tetraploidization; whereas, down-regulation of Lats2 compromises the p53-mediated G1 tetraploidy checkpoint. Furthermore, the Lats2 gene is itself a target for positive transcriptional regulation by p53. Nocodazole, but not DNA damage, rapidly and selectively induces p53-dependent Lats2 transcription. Thus, the Lats2–Mdm2–p53 axis constitutes a novel positive feedback checkpoint pathway, critical for the maintenance of proper chromosome number following mitotic apparatus dysfunction.
Results
Lats2 interacts with Mdm2
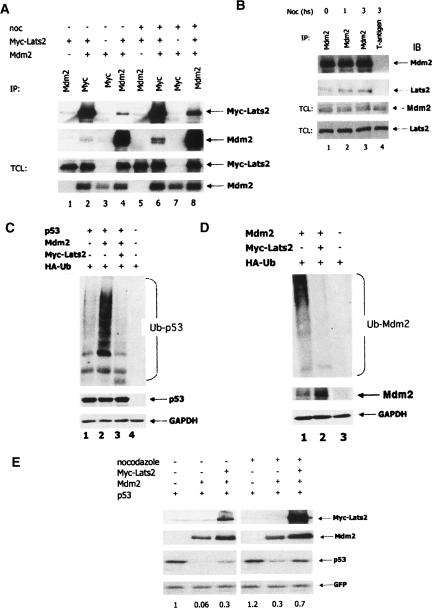
A fragment of human Mdm2 comprising the central acidic domain was used in a Ras recruitment yeast two-hybrid screen (Aronheim 2001). Several novel and established Mdm2 interactors were isolated (data not shown). One interacting clone corresponded to a fragment of Lats2. To determine whether Lats2 could interact with Mdm2 within mammalian cells, HEK293 cells were cotransfected with expression plasmids encoding Mdm2 and Myc-epitope-tagged human Lats2 (Myc-Lats2). Cell lysates were subjected to immunoprecipitation with antibodies directed against either Mdm2 or the Myc epitope, followed by Western blot analysis. As seen in Figure 1A (upper two panels), Mdm2 and Myc-Lats2 coprecipitated specifically (for Myc-Lats2 cf. lanes 1,4, and for Mdm2, cf. lanes 2,3). This interaction was enhanced in cells treated with the microtubule poison nocodazole (noc) (for Myc-Lats2, see Fig. 1A, cf. lanes 5,8, and for Mdm2, cf. lanes 6,7). Importantly, an interaction was also found between endogenous Lats2 and Mdm2 (Fig. 1B); Lats2 coprecipitated with antibodies directed against Mdm2 (Fig. 1B, lanes 1–3), but not against the negative control, SV40 large T antigen (Fig. 1B, lane 4). Of note, nocodazole also promoted the endogenous Mdm2–Lats2 interaction (Fig. 1B, cf. lanes 2,3 and 1).
Figure 1.
Lats2 binds Mdm2, inhibits its E3 ligase activity, and stabilizes p53. (A) Transfected Mdm2 and Lats2 interact in cells. HEK293 cells were transiently transfected with the indicated combinations of expression plasmids encoding Myc epitope-tagged human Lats2 (Myc-Lats2) and Mdm2. Twenty-four hours later, cells were treated with 0.2 μg/mL nocodazole for an additional 24 h, or mock-treated. Cell lysates were immunoprecipitated either with a mix of 4B2 and 4B11 anti-Mdm2 monoclonal antibodies (MoAbs) or with 9E10 anti-Myc MoAb. Immunoprecipitated proteins were resolved by SDS-PAGE and subjected to Western blot analysis, along with 2.5% of each total cell lysate (TCL). Mdm2 was detected with a mix of the anti-Mdm2 MoAbs 4B2 and 4B11; Myc-Lats2 was detected using the 9E10 MoAb. (B) Endogenous Mdm2 and Lats2 interact within cells. Endogenous Mdm2 was precipitated from U2OS cell extracts using a mix of 4B2, 4B11, and 2A9 MoAbs. Immunoprecipitation with MoAb against T-antigen (PAb419) was used as a negative control. Immunoprecipitated proteins and 3% of each TCL were detected using antibodies against Mdm2 (4B2, 4B11, and 2A9) or Lats2 (LA-2). (C) Lats2 inhibits p53 ubiquitination. p53/mdm2 double-null MEFs were transiently transfected with the indicated combinations of plasmids encoding p53, Mdm2, Myc-Lats2, and HA-tagged ubiquitin (400 ng, 200 ng, 8 μg, and 1.5 μg, respectively). Forty-eight hours later, cells were harvested. Extracts were denatured and immunoprecipitated with antibodies against p53 (mix of DO1 and PAb1801). Ubiquitinated forms of precipitated p53 were visualized using an anti-HA polyclonal Ab. Western blot of TCL was carried out as described in B. (D) Lats2 inhibits Mdm2 ubiquitination. p53/mdm2 double-null MEFs were transiently transfected with the indicated combinations of plasmids encoding Mdm2, MycLats2, and HA-tagged ubiquitin (400 ng, 8 μg, and 1.5 μg, respectively). Forty-eight hours later, cells were harvested, and extracts were denatured and immunoprecipitated with antibodies against Mdm2 (mix of 4B2, 4B11, and 2A9). Ubiquitinated forms of precipitated Mdm2 were visualized using an anti-HA polyclonal Ab. Western blot of TCL was carried out as described in B. (E) Lats2 overexpression increases p53 levels. p53/mdm2 double-null MEFs were transiently transfected with GFP (100 ng) expression plasmid together with the indicated combinations of p53, Mdm2, and Myc-Lats2 expression plasmids (40 ng, 40 ng, and 5 μg, respectively). Twenty-four hours later, cells were treated with nocodazole (0.2 μg/mL) or mock-treated for 20 h, harvested, and subjected to Western blot analysis. Quantification of relative p53 levels is shown below each lane.
Lats2 inhibits the E3 ligase activity of Mdm2 and promotes p53 stabilization
Mdm2 is an E3 ubiquitin ligase. In this capacity, it ubiquitinates several cellular proteins, most notably p53, and also undergoes autoubiquitination. As expected, overexpression of Mdm2 promoted p53 ubiquitination in transfected cells (Fig. 1C, lane 2). However, coexpression of Lats2 virtually prevented this from occurring (Fig. 1C, lane 3). Similarly, Lats2 overexpression led to a pronounced decrease in polyubiquitinated Mdm2 (Fig. 1D), most probably due to aborted Mdm2 auto-ubiquitination. Hence, Lats2 is a potent inhibitor of the E3 activity of Mdm2.
The experiment in Figure 1C was performed in the presence of the proteasome inhibitor MG-132; when the proteasome was not inhibited, coexpression of Lats2 resulted in significant protection of p53 against destabilizing effects of Mdm2 (Fig. 1E). In line with its ability to block Mdm2 autoubiquitination, excess Lats2 also caused an increase in Mdm2 stability and steady-state levels (data not shown). All together, these data demonstrate that Lats2 can stabilize p53 by alleviating Mdm2-mediated p53 ubiquitination and degradation.
Nocodazole causes nuclear translocation of Lats2, increased Lats2–Mdm2 interaction, and p53 stabilization
Lats2 is localized preferentially to the centrosomes throughout the cell cycle (McPherson et al. 2004; Toji et al. 2004), whereas Mdm2 is predominantly nuclear. We reasoned that Lats2–Mdm2 interaction might occur specifically when centrosome function is compromised. The microtubule poison, nocodazole, affects adversely microtubules, spindles, and centrosome homeostasis (Scolnick and Halazonetis 2000; Dammermann and Merdes 2002; Uzbekov et al. 2002; Burakov et al. 2003). As shown in Figure 1, A and B, nocodazole treatment markedly increased the Mdm2–Lats2 interaction. Of note, augmented interaction between the two endogenous proteins was observed as early as 1 h after nocodazole treatment (Fig. 1B, lane 2).
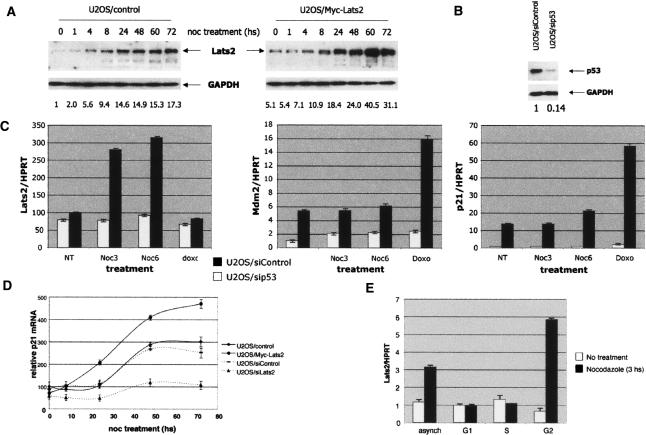
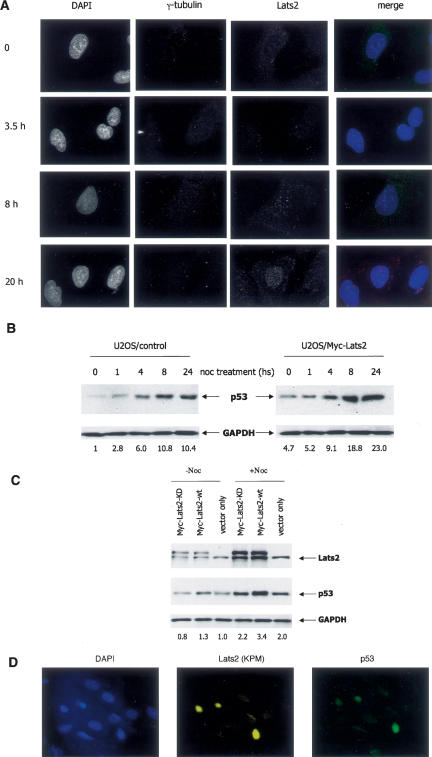
To further explore the impact of nocodazole, we stably transfected U2OS osteosarcoma cells with an expression plasmid encoding Myc-tagged Lats2 (Supplementary Fig. 1). The levels of exogenous Lats2 in these cells were comparable with those of the endogenous protein (see Fig. 3A, below). The Myc-Lats2 protein localized to centrosomes, marked by staining for γ-tubulin (Fig. 2A, top row; Supplementary Fig. 2 shows a higher-resolution picture). Centrosomal localization of Lats2 was visualized with two different Lats2-specific antibodies (data not shown). Faint staining was also observed throughout the cytoplasm, consistent with earlier findings (Li et al. 2003). Of note, the nucleus was devoid of any significant staining. By 3.5 h of nocodazole treatment, some centrosomes were no longer positive for Lats2 (Fig. 2A, second row, arrow indicates a centrosome staining positive for γ-tubulin but negative for Lats2). In parallel, a small amount of Lats2 became discernable in the nucleoplasm, in some cases exhibiting a fine punctate pattern. By 8 h, many cells displayed an easily visible punctate nuclear pattern of Lats2 staining (Fig. 2A, third row). After 20 h, many cells exhibited a morphology indicative of a mitotic arrest (Supplementary Fig. 3A), but nonmitotic cells displayed intense nuclear Lats2 staining (Fig. 2A, fourth row). Within this timeframe, Mdm2 staining was augmented and remained nuclear (data not shown). Of note, in untreated mitotic cells, Lats2 was retained at the centrosome (Supplementary Fig. 3B). In contrast to nocodazole, the DNA damaging agent doxorubicin did not induce Lats2 nuclear translocation (Supplementary Fig. 3C). Notably, unlike doxorubicin, nocodazole did not cause any detectable DNA damage, as assessed by nuclear γH2AX foci (Supplementary Fig. 3D). These observations suggest that Lats2 responds distinctly to microtubule damage; neither mitosis per se nor DNA damage leads to a similar response. As early as 1 h after nocodazole addition, an increase in steady-state p53 levels already could be discerned (Fig. 2B, left panel). This increase became more pronounced at 4 h, and continued to escalate thereafter. Hence p53 accumulation, presumably reflecting elevated protein stability, is initiated at a time when Lats2 is just beginning to appear in the nucleus and intensifies gradually in parallel with the increased presence of nuclear Lats2. A similar picture was observed in the stable Myc-Lats2 transfectants, except that both basal and induced absolute levels of p53 were higher (Fig. 2B, right panel). Moreover, the kinase activity of Lats2 was essential for optimal nocodazole-induced p53 accumulation: A kinase-dead Lats2 mutant was deficient in p53 induction (Fig. 2C).
Figure 3.
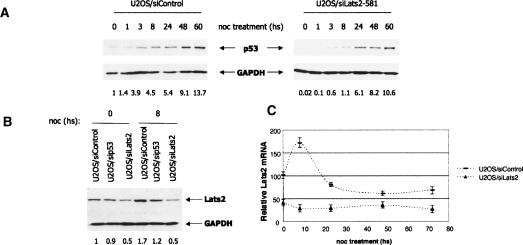
Rapid transcriptional activation of Lats2 is p53 dependent, specific for nocodazole, and occurs preferentially at the G2/M phase of the cell cycle. (A) Kinetics of Lats2 induction by nocodazole. U2OS/Myc-Lats2 cells and control cells were treated and analyzed as in Figure 2B. Lats2 (endogenous as well as stably transfected Myc-Lats2) was detected with LA-2 anti-Lats2 antibodies; endogenous GAPDH was used as a loading control. Quantification of relative Lats2 levels is shown below each lane. (B) U2OS cells were infected with a recombinant retrovirus encoding p53-specific shRNA (U2OS/sip53) or shRNA against lacZ (U2OS/siControl). From 24 h after infection, cells were maintained under puromycin selection. Endogenous p53 levels were monitored by Western blot analysis using DO1 + PAb1801; endogenous GAPDH was used as a loading control. Quantification of relative p53 levels is shown below each lane. (C) lats2 mRNA levels increase in a p53-dependent manner upon nocodazole treatment. U2OS cells with shRNA against lacZ (U2OS/siControl) and their p53-depleted derivatives (U2OS/sip53) were treated with 0.2 μg/mL nocodazole (noc) and harvested after 3 h (Noc3) or 6 h (Noc6) of treatment or exposed to 1 μM doxorubicin (Doxo) for 6 h. RNA was extracted and subjected to quantitative real-time RT–PCR, using primers specific for lats2, mdm2, and p21 mRNA. All values were normalized for the amount of HPRT RNA in the same sample. (D) Basal Lats2 levels modulate the extent of p21 mRNA induction by nocodazole. Stably transfected U2OS/control (transfected with pcDNA3), U2OS/Myc-Lats2, U2OS/siControl (shRNA against mouse PUMA, nonreactive with human PUMA mRNA), or U2OS/siLats2 (shRNA against lats2) cells were treated with 0.2 μg/mL nocodazole and harvested at the indicated times. RNA was extracted from each sample and subjected to quantitative real-time RT–PCR analysis, using primers specific for p21 mRNA. Values were normalized for the amount of HPRT RNA in the same sample. “relative p21 mRNA” represents normalized RNA levels compared with t = 0 levels in U2OS/control cells. (E) Lats2 transcription is induced by nocodazole solely in cells positioned in the G2/M phases of the cell cycle. U2OS cells were fractionated by centrifugal elutriation (see Supplementary Fig. 6). Mock-elutriated asynchronous cells (asynch), or G1-, S-, or G2/M-enriched populations were harvested after 3 h of treatment with 0.2 μg/mL nocodazole. RNA was extracted from each sample and subjected to quantitative real-time RT–PCR analysis, using primers specific for lats2 mRNA. Values were normalized for the amount of HPRT RNA in the same sample.
Figure 2.
Nocodazole induces Lats2 nuclear translocation and accumulation of Lats2 and p53. (A) Nocodazole induces Lats2 nuclear translocation. U2OS cells stably transfected with Myc-tagged Lats2 (U2OS/Myc-Lats2) were seeded on cover-slips. Twenty-four hours later, nocodazole (0.2 μg/mL) was added, and cells were fixed at the indicated times thereafter and subjected to immunofluorescence staining. Centrosomes were visualized with anti-γ-tubulin antibodies; Myc-Lats2 was visualized with anti-KPM (Santa Cruz Bio-technology) antibodies; nuclei were visualized with DAPI. Representative fields are shown (magnification 100×). The arrow indicates a centrosome negative for Lats2 staining. (B) Kinetics of p53 induction by nocodazole. U2OS/Myc-Lats2 cells and control cells (U2OS stably transfected with pcDNA3 empty vector) were treated with 0.2 μg/mL nocodazole (noc) and harvested at the indicated times. Endogenous p53 was detected with a mix of DO1 and PAb1801 MoAbs; endogenous GAPDH was used as a loading control (Chemicon). Quantification of relative p53 levels is shown below each lane. (C) Kinase activity of Lats2 is essential for p53 induction by nocodazole. U2OS cells were transiently transfected with wild-type (wt) or kinase-dead (KD) Lats2 or vector-only control. Twenty-four hours after transfection, transfected cells were selected for 3 d with 1 μg/mL puromycin. On the second day of puromycin treatment, nocodazole (0.2 μg/ mL) was added for 24 h. Endogenous p53 was visualized with DO1 and PAb1801 MoAbs; endogenous GAPDH was used as a loading control. Quantification of relative p53 levels is shown below each lane. (D) Nuclear accumulation of Lats2 correlates with p53 induction. U2OS cells were exposed to nocodazole, fixed after 8 h (magnification 40×), and analyzed as in A. Endogenous p53 and Lats2 were visualized with DO1 + PAb1801 or anti-KPM antibodies, respectively. A representative field is shown.
The observations described above are consistent with a scenario wherein nocodazole-induced stress causes Lats2 to depart from the centrosome and translocate to the nucleus, where it encounters Mdm2, inactivates it, and promotes p53 stabilization. In agreement with this scenario, staining intensities of nuclear endogenous Lats2 and p53 staining coincided in nocodazole-treated cells (Fig. 2D).
Lats2 expression is induced by nocodazole as part of a p53-mediated positive feedback loop
Extended treatment of U2OS cells with nocodazole resulted in a pronounced increase in Lats2 protein levels (Fig. 3A, left panel). A mild increase was already observed after 4 h of treatment. Thus, the onset of Lats2 increase follows the accumulation of p53 (cf. Fig. 2B). The pattern was similar in the Myc-Lats2 transfectants, except that overall Lats2 levels (endogenous + transfected) were higher, as expected (FIG. 3A, right panel).
Transcription of the lats2 gene has been reported to be up-regulated by p53 (Kostic and Shaw 2000). We therefore generated derivatives of U2OS cells in which endogenous p53 was stably knocked down by an integrated plasmid expressing p53-specific short hairpin RNA (shRNA) (Fig. 3B; Brummelkamp et al. 2002). Control and knockdown cells were treated with nocodazole, and the RNA was subjected to real-time quantitative RT–PCR analysis. Basal lats2 mRNA levels were not affected by p53 status (Fig. 3C, left panel). lats2 mRNA was induced rapidly by nocodazole, but only in cells carrying wild-type p53, where substantial lats2 mRNA induction was seen within 3 h. In contrast, lats2 expression was only marginally increased in p53-null cells. Thus, nocodazole leads to a rapid p53-dependent increase in lats2 mRNA. This pattern of induction was unique to nocodazole; exposure to doxorubicin (doxo, 1 μM) did not promote lats2 mRNA induction. Conversely, p21 and mdm2, two classical p53 response genes, exhibited p53-dependent basal transcript levels, and a robust p53-dependent induction in response to doxorubicin treatment. Neither p21 nor mdm2 was up-regulated within 3 h of nocodazole treatment (Fig. 3C), although a gradual p53-dependent induction of both transcripts occurred later (Fig. 3C,D; data not shown). Importantly, optimal induction of p21 at late time points was dependent on both Lats2 and p53, since p21 mRNA levels were markedly attenuated in U2OS cells stably knocked down for either gene (Fig. 3C,D; data not shown). It is noteworthy that the increase in Lats2 protein upon nocodazole treatment was more pronounced than the increase in lats2 mRNA (Fig. 3, cf. A and C), arguing that post-translational mechanisms also contribute to Lats2 elevation.
Essentially identical results were obtained with p53-positive HCT116 colorectal cancer cells and their derivatives in which the p53 gene had been somatically knocked out (Bunz et al. 1999). Here, too, prominent p53-dependent induction of lats2 mRNA was observed within 3 h of nocodazole treatment (Supplementary Fig. 4), whereas mdm2 and p21 mRNA were only minimally induced within this timeframe. Transcript levels of Lats1, the other member of the mammalian Lats family, were unaffected within the first 8 h of treatment.
In conclusion, nocodazole orchestrates a p53-dependent transcription program that is very distinct from that elicited by DNA damage; whereas the former entails a rapid up-regulation of lats2 but not p21 or mdm2 mRNA, the latter induces transcription of p21 and mdm2 but not lats2.
Nocodazole causes depolymerization of microtubules, which can result in the disorganization of major cellular constituents including the Golgi and endoplasmic reticulum. However, in cells approaching mitosis, a major consequence of nocodazole treatment is disruption of the mitotic spindle, which prevents the cell from satisfying the spindle assembly checkpoint. Theoretically, the effect of nocodazole on lats2 mRNA induction could be uniform throughout the cell cycle, implying a general nonspecific stress response, or it could be associated with a particular phase of the cell cycle. To distinguish between these possibilities, populations of cells enriched at different stages of the cell cycle were separated by centrifugal elutriation (Supplementary Fig. 5). The different cell populations were treated with nocodazole for 3 h and analyzed by quantitative real-time RT–PCR (Fig. 3E). In untreated cells, basal Lats2 mRNA levels were unaffected by cell cycle stage. Hence, elevated Lats2 gene expression is not merely a consequence of altered cell cycle distribution of nocodazole-treated cells. The rapid elevation of Lats2 mRNA upon nocodazole treatment was clearly dependent on cell cycle position at the time of drug addition; only the G2/M-enriched population (G2 in Fig. 3E, corresponding to FR 120 in Supplementary Fig. 5) mounted such a response. This is consistent with a rapid and specific induction of Lats2 only in cells approaching mitosis with spindle damage.
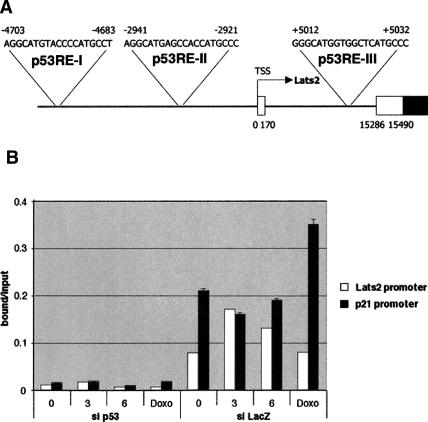
To further explore the up-regulation of Lats2 expression, we investigated the effect of nocodazole on recruitment of p53 to the Lats2 promoter, as assayed by chromatin immunoprecipitation (ChIP). The Lats2 gene harbors three putative p53-responsive elements (p53REs) (El-Deiry et al. 1992), two in the promoter region (p53REI/−4703 to −4683 and p53RE-II/−2941to −2921) and one within the first intron (p53RE-III/+5012 to +5032) (Fig. 4A). p53 was found to associate selectively with the DNA region comprising p53RE-I, even without nocodazole treatment (Fig. 4B). Moreover, 3 h of nocodazole treatment increased this association. In contrast, no increase in occupancy of the p21 promoter by p53 was seen even at 6 h. Conversely, doxorubicin (Doxo) led to an increase in p53 binding to the p21 promoter, but not the Lats2 promoter. p53RE-II also selectively associated with p53, but was largely unaffected by nocodazole treatment, whereas p53RE-III showed no detectable p53 binding (data not shown). Thus, p53 is selectively recruited to specific regions of the Lats2 promoter early after exposure to nocodazole, presumably giving rise to rapid selective triggering of Lats2 transcription.
Figure 4.
p53 associates with the Lats2 promoter in vivo. (A) Location and sequence of predicted p53REs in the Lats2 gene. Numbers represent nucleotide positions relative to the accepted 5′ end of the Lats2 mRNA transcription start site (TSS). Open boxes represent untranslated regions, and dark boxes represent coding regions. (B) ChIP analysis of DNA derived from stably transfected U2OS/sip53 or U2OS/siControl (siLacZ) cells. Cells were treated with 0.2 μg/mL nocodazole (noc) for 3 or 6 h or with 1 μM doxorubicin (Doxo) for 6 h. p53-bound chromatin was precipitated using CM1 polyclonal antibody (Novocastra). Chromatin-derived DNA was subjected to real-time PCR analysis with primer pairs bracketing the 5′ p53RE of the p21 gene and the Lats2 p53RE-I (see A). Values represent the ratio of the value obtained for bound DNA over that obtained for total input (chromatin before immunoprecipitation) DNA for each PCR primer pair (bound/input).
The contribution of Lats2 to induction of p53 by nocodazole treatment was further assessed through the use of U2OS cells stably expressing Lats2 shRNA (siLats2-581). Although Lats2 could be efficiently knocked down transiently (Supplementary Fig. 6B), stable transfectants exhibited only approximately a twofold reduction. As seen in Figure 5A, even this modest decrease in Lats2 was sufficient to substantially delay the early nocodazole-induced p53 induction. Similar results were obtained with a second Lats2 shRNA (siLats2-1438) (Supplementary Fig. 7A). Importantly, Lats2 knockdown compromised p53 induction in response to nocodazole also in nontransformed human diploid WI-38 fibroblasts (Supplementary Fig. 7B).
Figure 5.
Lats2 is required for optimal induction of p53, p21, and Lats2 itself in response to nocodazole. (A) Optimal induction of p53 protein by nocodazole is compromised by Lats2 knockdown. Stably transfected U2OS/siControl and U2OS/siLats2-581 cells were treated with 0.2 μg/mL nocodazole (noc) and harvested at the indicated times. Endogenous p53 was detected using DO1 + PAb18O1 antibodies, and GAPDH was used as a loading control. Quantification of relative p53 levels is shown below each lane. (B) Full induction of Lats2 by nocodazole is dependent on p53 and Lats2 itself. Stably transfected U2OS cells subjected to the indicated shRNA knockdown were treated with 0.2 μg/mL nocodazole (noc) and harvested after 8 h. Endogenous Lats2 was visualized using LA-2 antibodies, and GAPDH was used as a loading control. (C) Basal Lats2 is required for efficient induction of subsequent Lats2 mRNA by nocodazole (positive feedback). Stably transfected U2OS/siControl and U2OS/siLats2 cells were treated with 0.2 μg/mL nocodazole (noc) and harvested at the indicated times. RNA was analyzed by quantitative RT–PCR, as in Figure 3D. “Relative Lats2 mRNA” represents HPRT-normalized RNA levels compared with t = 0 levels in siControl cells.
The involvement of Lats2 in p53 induction, on the one hand (Fig. 5A), and the role of p53 in up-regulating Lats2 expression (Fig. 3C), on the other hand, suggest a positive feedback loop. To further explore this notion, U2OS/ siLats2, U2OS/sip53, and control U2OS cells were subjected to nocodazole treatment. As expected, maximal up-regulation of Lats2 was dependent on p53 status (Fig. 5B). Quantitative real-time RT–PCR analysis revealed that Lats2 mRNA expression peaked rapidly, followed by a return to basal levels by 24 h (Figs. 3C, 5C); the persistent increase in Lats2 protein levels (Fig. 3A) may be due to increased translation and/or protein stability. In contrast, nocodazole-treated U2OS/siLats2 cells failed to exhibit significant increases in Lats2 protein (Fig. 5B) or RNA (Fig. 5C). This is not likely to be due to complete ablation of newly synthesized Lats2 transcripts by the resident siRNA, since this siRNA was relatively inefficient in reducing basal Lats2 mRNA levels in untreated cells (Fig. 5C, time = 0). Hence, even a moderate reduction of basal Lats2 protein is enough to render the cells incapable of up-regulating Lats2 mRNA upon nocodazole treatment. This implies that the positive p53-mediated feedback loop governing Lats2 transcription in response to microtubule damage is very finely tuned; Lats2 protein below a certain level might not suffice to fuel this loop.
Lats2 is required to prevent polyploidization in response to microtubule damage
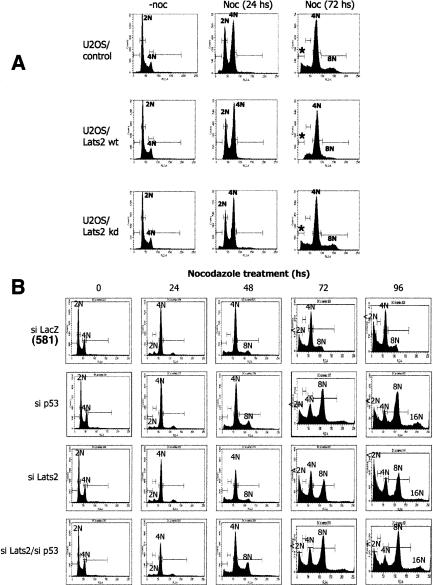
Dysfunction of the mitotic apparatus, which can occur as a result of microtubule damage, prevents cells from satisfying the spindle checkpoint and leads to a p53-independent delay of the cell in mitosis. Cells ultimately exit mitosis (“mitotic slippage”) and reenter G1 without cytokinesis. This triggers a p53-mediated arrest at the subsequent G1/S boundary (“G1 tetraploidy checkpoint”) (Dash and El-Deiry 2004; Rieder and Maiato 2004; Stukenberg 2004), which acts to prevent further DNA replication and emergence of polyploid genomes. To assess the functional outcome of the Lats2–p53 loop, flow cytometry was used to compare cells with different genotypes after nocodazole treatment. Untreated cells expressing wild-type Myc-Lats2 (Lats2-wt) or kinase-dead Myc-Lats2 (Lats2-kd) exhibited a similar cell cycle distribution as control cells (−noc in Fig. 6A). Within 24 h of nocodazole treatment, a prominent fraction of cells with 4N DNA content became apparent in all cases. By 72 h, the majority of cells of all three genotypes were arrested with a 4N DNA content, consistent with activation of the tetraploidy checkpoint. However, control U2OS displayed also a significant subpopulation with 8N DNA content, implying that these cells had undergone an additional cycle of DNA replication despite cytokinesis failure. Hence, U2OS cells have a “leaky” tetraploidy checkpoint response. In contrast, only a minute fraction of cells supplemented with Myc-Lats2 became polyploid, arguing that the increased Lats2 levels equip “leaky” cancer cells with a more effective checkpoint. Unlike wild-type Lats2, Lats2-kd failed to rescue the checkpoint defect, indicating that the kinase activity of Lats2 is required for efficient checkpoint function. Interestingly, at this late time, control cultures contained a noticeable fraction of cells with sub-G1 DNA content (asterisk), indicative of apoptosis, whereas very few of the Lats2-wt cells were apoptotic. Lats2-kd failed to rescue this apoptotic phenotype.
Figure 6.
Lats2 modulates polyploidy and apoptosis upon prolonged treatment with nocodazole. (A) Lats2 overexpression prevents polyploidy and apoptosis in U2OS cells in response to nocodazole treatment. U2OS cells stably transfected with vector only (control), wild-type Myc-Lats2 (Lats2-wt), or kinase-dead Myc-Lats2 (Lats2-kd) were treated with 0.2 μg/mL nocodazole (noc) and harvested at the indicated times. Fixed cells were stained with propidium iodide (PI) to measure DNA content and subjected to FACS analysis. Horizontal bars mark areas quantified to determine the proportion of cells with the indicated DNA content. Asterisks indicate apoptotic cells with sub-G1 DNA content. (B) Cells lacking Lats2 reduplicate their DNA similarly to p53-null cells. Stably transfected U2OS/siControl (with shRNA against LacZ), U2OS/sip53, U2OS/siLats2 (using siLats2-581), and U2OS/siLats2/sip53 cells were treated with 0.2 μg/mL nocodazole (noc) and harvested at the indicated times. Fixed cells were stained with propidium iodide (PI) to measure DNA content and subjected to FACS analysis.
Our findings predict that the effect of Lats2 inactivation on aberrant polyploidization should share common features with the effect of p53 inactivation. To test this prediction, U2OS cells stably knocked down for both Lats2 and p53 together (U2OS/siLats2/sip53) were compared with their single knockdown counterparts. Twenty-four hours of nocodazole treatment caused all cell types to uniformly accumulate with 4N DNA content (Fig. 6B). Even after 96 h, control cells carrying wild-type levels of p53 and Lats2 (U2OS/siLacZ) largely remained arrested with 4N DNA content. In contrast, many of the siLats2 and sip53 cells, as well as double-knockdown cells, reduplicated their DNA and accumulated more than 4N DNA content. U2OS/double-knock-down cells exhibited a similar phenotype to that of each of the single knockdowns, in line with our proposal that Lats2 and p53 act epistatically. Quantification of the relevant effects of Lats2 up-regulation as well as Lats2 and p53 knockdown, as deduced from multiple experiments, is presented in Supplementary Figure 8D. Practically identical results were obtained with a different Lats2 shRNA (Supplementary Fig. 8A). Moreover, Lats2 knockdown and p53 knockout had a similar effect on the tetraploidy checkpoint response also in HCT116 colorectal cancer cells (Supplementary Fig. 8B). Hence, both Lats2 and p53 are essential for preventing polyploidy. Of note, Lats2 was instrumental for preventing polyploidy also in nontransformed WI-38 cells (Supplementary Fig. 8C). Here, practically no 8N DNA population was present after 72 h of nocodazole treatment of control cells. Thus, in contrast to the two cancer cell lines, these non-transformed fibroblasts possess a robust tetraploidy checkpoint. Consistent with this notion, ablation of p53 resulted in only a modest checkpoint failure (9.8% polyploid cells). Importantly, Lats2 ablation had a very similar effect.
Interestingly, many siLats2 cells underwent p53-dependent apoptosis after failure to arrest effectively (Fig. 6B, cf. siLats2 and siLats2/sip53 at 96 h), suggesting that proper levels of Lats2 are necessary for protective cell survival in the face of mitotic apparatus dysfunction. This conclusion was confirmed by direct analysis of cell death (Supplementary Fig. 9).
Further support for a functional link between Lats2 and p53 was gained with the aid of the drug Nutlin-3 (Vassilev et al. 2004; Tovar et al. 2006), which directly activates p53 irrespective of upstream signals. As seen in Supplementary Figure 10, in Lats2-depleted cells but not cells lacking p53, Nutlin almost completely abolished nocodazole-induced polyploidy. The fact that the defect caused by Lats2 depletion could be rescued by direct p53 activation corroborates that these two genes act epistatically and that Lats2 is necessary for p53 activation to prevent polyploidization.
Unlike the response to nocodazole, the cell cycle effects elicited by inhibition of ribonucleotide reductase (hydroxurea, HU), endoplasmic reticulum stress (thapsigargin, TG) or ionizing radiation (IR), all of which trigger G1/S and/or G2/M checkpoints in a manner that is at least partially p53-dependent, were unaffected by Lats2 modulation (Supplementary Fig. 11). Hence, Lats2 is specialized in its ability to promote p53-dependent checkpoint activation selectively in response to stress induced by microtubule damage.
Together, our findings demonstrate the existence of a positive regulatory loop involving Lats2, Mdm2, and p53. Lats2 binds and inhibits Mdm2; this activates p53, which, in turn, increases Lats2 transcription. Moreover, this signaling pathway is activated selectively in response to mitotic apparatus damage. Optimal Lats2 expression is required for efficient orchestration of a checkpoint response that prevents polyploidy and protects cells from p53-dependent death that presumably occurs when such damaged cells attempt to progress further into the next cell cycle.
Discussion
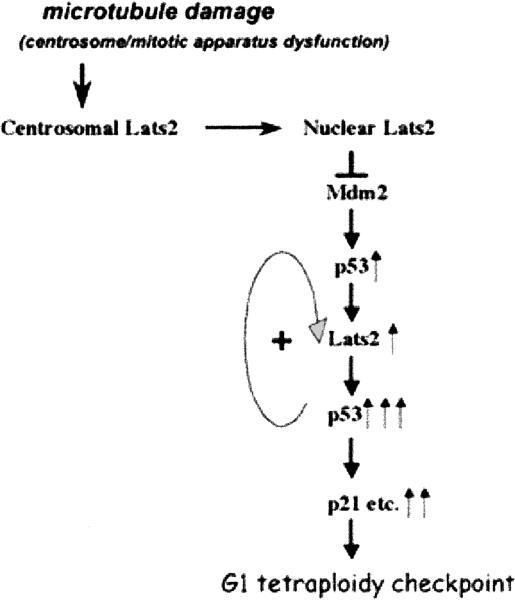
Aneuploidy is a hallmark of many advanced tumors. Establishment of aneuploidy occurs much more readily in cells lacking p53. The ability of p53 to prevent accrual of aneuploidy and polyploidy is, to a great extent, due to its ability to arrest cell cycle progression in the face of mitotic machinery dysfunction, via the so-called G1 tetraploidy checkpoint (Meek 2000; Castedo et al. 2004; Rajagopalan and Lengauer 2004; Duensing and Duensing 2005). The findings described here implicate Lats2 as a key mediator of this response. More specifically we suggest that, in response to stress signals generated by mitotic apparatus dysfunction, Lats2 translocates from centrosomes into the nucleus. There, it binds and inactivates Mdm2 (Fig. 7), leading to rapid initial p53 activation. The activated p53 binds preferentially to the Lats2 promoter, and triggers prompt, selective induction of Lats2 gene expression in cells approaching mitosis (see Fig. 3E). This further elevates nuclear Lats2 protein, further driving p53 activation. Eventually, classical p53 target genes such as p21 are induced, leading to a cell cycle arrest at the G1/S boundary. Our data implicate the kinase activity of Lats2 in p53 induction following mitotic apparatus stress (Fig. 2C) and in enforcement of an effective G1 tetraploidy checkpoint (Fig. 6A). At present, the relevant substrate(s) of that kinase activity remain unknown; neither p53 nor Mdm2 could be phosphorylated in vitro by Lats2 in our hands (data not shown). The notion that p53 and Lats2 are components of a common checkpoint pathway is also supported by the finding that the loss of either protein gives rise to a very similar cellular phenotype, namely, centrosome amplification, multipolar spindles, and genomic instability (Fukasawa et al. 1996; Tarapore et al. 2001; McPherson et al. 2004).
Figure 7.
Model for the proposed roles of Lats2 and p53 in the response to microtubule-damage-induced stress. In response to microtubule damage and ensuing centrosome/mitotic apparatus dysfunction, Lats translocates from the centrosomes to the nucleus, where it binds Mdm2 and inactivates it. This leads to p53 activation, which selectively turns on Lats2 transcription, resulting in general up-regulation of nuclear Lats2 levels and further stabilization and induction of p53. The potent p53 activation leads to transcription of many other p53 target genes, including p21, and consequent arrest of tetraploid cells at the G1/S boundary, practically constituting a “G1 tetraploidy checkpoint.” Upward arrows indicate activation; the number of arrows indicates the relative extent of the activation. The positive feedback loop between p53 and Lats2 is also indicated.
We show that, in cells encountering microtubule damage and mitotic apparatus dysfunction, p53 activation is initiated very rapidly, well before mitotic slippage occurs. Further p53 activation builds up gradually thereafter, through escalation of the output of the Lats2–p53 positive feedback loop. Accordingly, optimal levels of inhibitory p53 activity may be achieved only at a time when cells have already slipped out of mitosis and reentered G1. Furthermore, the arrest may not occur before the G1/S boundary because it is only then that p21 finds and inhibits Cyclin E/Cdk2. Thus, although the checkpoint serves to effectively halt tetraploid cells from continuing into polyploidy, the triggering event occurs long before tetraploidy is actually encountered. Cell fate in response to mitotic spindle/centrosome dysfunction may be dictated by the balance between Lats2 and p53. Cells lacking either Lats2 or p53 do not activate efficiently the tetraploidy checkpoint, and tend to accumulate 8N DNA content. However, failure to activate this checkpoint (as in the case of Lats2 depletion) in cells possessing wild-type p53 results in augmented apoptosis. Hence, p53 might act to shunt “escaper” polyploid cells toward apoptosis. The reverse is also true; although Lats2 helps activate p53 in response to spindle/centrosome stress, mild overexpression of Lats2 actually decreases the portion of dead cells. This suggests that tetraploid cells that undergo a successful G1 arrest can avoid catastrophic cell death.
Overexpression of Lats2 in HeLa cells, which lack functional p53, can cause G2/M arrest and apoptosis (Hori et al. 2000; Kamikubo et al. 2003). In contrast, in transformed NIH-3T3 cells, which presumably possess wild-type p53, excess Lats2 inhibits G1/S transition in conjunction with down-regulation of Cyclin E/Cdk2 kinase activity (Li et al. 2003); it is conceivable that this G1/S arrest is mediated, at least in part, via the p53–p21 axis, as demonstrated in the present study. In concordance with this notion, we observed that elevated levels of Lats2 result in enhanced repression of Cyclin E mRNA levels upon extended nocodazole treatment of U2OS cells, whereas knockdown of Lats2 leads to elevated Cyclin E mRNA (data not shown).
Although not proving so formally, our data suggest that the effects of inactivation of either Lats2 or p53 on the biological response to nocodazole are partially overlapping, consistent with the two proteins being components of the same checkpoint pathway. However, additional proteins beyond Lats2 might relay microtubule damage and centrosome/mitotic apparatus dysfunction stress signals to p53, and some of the effects of Lats2 may be exerted in a p53-independent manner. Of particular interest is Lats1, which shares 52% overall sequence identity with Lats2, with 85% sequence identity over the kinase domain (Yabuta et al. 2000). Lats1 also resides on interphase centrosomes (Nishiyama et al. 1999; Hirota et al. 2000). Indeed, overexpression of kinase-dead Lats1, presumably acting in a dominant-negative fashion, causes prolonged mitotic delay and impairment of the G1 tetraploidy checkpoint as a result of inefficient p53 induction (Iida et al. 2004). Hence, Lats1 may perform functions similar to those described here for Lats2. Yet, it remains possible that these effects of the kinase-dead Lats1 might also be due to dominant-negative inhibition of endogenous Lats2, for example, by depletion of upstream activators or coactivators common to Lats1 and Lats2, such as Mats, a direct interactor of Wts (the fly ortholog of Lats1/Lats2), which is rate limiting for Wts biochemical activation (Lai et al. 2005). Numerous observations indicate that Lats1 and Lats2 have nonidentical biological functions. For instance, the consequences of their overexpression appear to be different (Xia et al. 2002; Li et al. 2003). In particular, whereas modest Lats2 overexpression in U2OS cells delays mitotic exit in nocodazole-treated cells (data not shown), similar Lats1 overexpression facilitates mitotic exit (Bothos et al. 2005). Moreover, while Lats2 overexpression protects nocodazole-treated cells from death (Fig. 6), Lats1 actually has the opposite effect (Iida et al. 2004). The precise interplay between Lats1 and Lats2 and its impact on regulation of p53 activity remain to be explored.
One open question is how Lats2 is alerted to the stress elicited by microtubule damage. Lats2 is subject to regulation by numerous proteins. In addition to Aurora A, which phosphorylates Lats2 directly (Toji et al. 2004), Lats2 may also be targeted by kinases such as MST2, which phosphorylates the fly Lats2 ortholog Wts (O'Neill et al. 2005), and interacting proteins such as Mats/Mob1A, required for the activation of Wts as well as of Lats1 (Bothos et al. 2005; Lai et al. 2005; Hergovich et al. 2006). It remains to be determined whether any of these proteins play a role in the nuclear translocation of Lats2.
Recently, Lats2 has also been implicated in the induction of p53-dependent senescence in response to Ras activation (Voorhoeve et al. 2006). In that context, Lats2 was found to act downstream from p53, somehow enabling the inactivation of cyclin-dependent kinases by p21. Although in our experimental model, driven by microtubule dysfunction, Lats2 is required for efficient induction of p21 expression (Fig. 3D), it is quite likely that an effect on p21 or cyclin E/Cdk2 function might also contribute to its important role in the tetraploidy checkpoint. Lats2 may thus operate at multiple levels downstream from and upstream of p53, further underscoring the tight functional link between these two tumor suppressors.
Normal cells likely contain optimal levels of Lats2, sufficient for potent activation of p53 in response to centrosome/mitotic apparatus dysfunction and effective prevention of polyploidization. However in many tumor cells, which often harbor inherent genomic instability, Lats2 expression may become suboptimal and rate limiting. This Lats2 shortage may be due to LOH, promoter hypermethylation, or other mechanisms that suppress Lats2 transcription or otherwise render the protein less functional (Jimenez-Velasco et al. 2005; Takahashi et al. 2005). In fact, our data suggest that U2OS cells may also possess suboptimal Lats2 levels, therefore slipping into polyploidy (Fig. 6A) despite their having wild-type p53. Hence, tumors may conceivably bypass optimal G1 tetraploidy checkpoint control and accrue aneuploidy by disabling either p53 or Lats2. Consequently, compromised Lats2 functionality might reduce the selective pressure for p53 inactivation during tumor progression, as demonstrated recently (Voorhoeve et al. 2006).
Materials and methods
Plasmids
The plasmid for expression of 6xMyc-Lats2-wt was described before (Toji et al. 2004). The expression plasmid for human Mdm2 (Hdm2) was kindly provided by B. Vogelstein, and expression plasmids for Mdm2 were kindly provided by B. Wasylyk. Loading and transfection controls, GFP and Flag-H2B, were described previously (Minsky and Oren 2004). Plasmids expressing Lats2 shRNA were generated as described in Supplementary Figure 2A. For infections, inserts were transferred to an EcoRI– HindIII-digested pRetroSuper vector (Brummelkamp et al. 2002).
Cell culture, transfections, and viral infections
All cells were maintained at 37°C in a 5% CO2 humidified incubator. HEK293, U2OS, and MCF7 cells and mouse embryonic fibroblast (MEF) cells were grown in Dulbecco's modified Eagle's medium (DMEM), and HCT116 cells were grown in Mc-Coy's medium supplemented with 2 mM L-glutamine (Biological Industries) and 10% heat-inactivated fetal bovine serum (HIFBS; Sigma) and antibiotics. HEK293 were transfected by the calcium phosphate coprecipitation method; MEFs, HCT116, and MCF7 were transfected using JetPEI (PolyPlus Transfection); U2OS were transfected using either JetPEI or FuGene (Roche), according to the manufacturers’ instructions. To generate stable clones, transfected cells were diluted and selected with the appropriate drug. Single colonies were isolated and validated for proper expression patterns. Alternatively, U2OS cells stably expressing ectropic receptor were infected with retrovirus and drug-selected to generate siLats, siLacZ, sip53, and siPUMAmouse (siControl) cell pools. hTERT-immortalized WI-38 human embryonic fibroblasts (gift of V. Rotter) were infected with pRS-Lats2#2 (Voorhoeve et al. 2006), pRS-p53, or with a retrovirus expressing mouse PUMA shRNA.
Nocodazole (Sigma) was used at 0.2 μg/mL; treatment was generally performed 24 h after cell seeding. The proteasome inhibitor MG-132 (Calbiochem) was administered either for 20 h (10 μM final) or for 4 h (25 μM final).
Isolation of total RNA and quantitative real-time PCR
Total RNA was isolated using a NucleoSpin RNA II kit (Macherey-Nagel). A 2-μg aliquot of the total RNA was reverse-transcribed using Moloney murine leukemia virus reverse transcriptase (Promega) and random hexamer primers (Amersham). Real-time PCR was done using SYBR Green Master Mix (Applied Biosystems) on an ABI 7000 instrument (Applied Biosystems). The values for specific genes were normalized to either GAPDH or HPRT housekeeping controls. Primer sequences were as follows: (1) GAPDH forward, 5′-ACCCACTCCTCCACCTTTGA-3′; reverse, 5′-ACGAATTTGGCTACAGCAACAG-3′; (2) HPRT forward, 5′-TGACACTGGCAAA ACAATGCA-3′; reverse, 5′-GGTCCTTTTCACCAGCAAGCT-3′; (3) Mdm2 forward, 5′-AGGCAAATGTGCAATACCAACA-3′; reverse, 5′-GGTTA CAGCACCATCAGTAGGTACAG-3′; (4) p21 forward, 5′-GGC AGACCAGCATGACAGATT-3′; reverse, 5′-GCGGATTAG GGCTTCCTCTT-3′; (5) Lats2 forward, 5′-GCAGATTGTGC GGGTCATTA-3′; reverse, 5′-GGCATGAGCCCCTTTCCT-3′; (6) Lats1 forward, 5′-TCATCAGCAGCGTCTACATCG-3′; reverse, 5′-TCCAACCCGCATCATTTCAT-3′.
Yeast two-hybrid screen
To identify proteins that interact with the human Mdm2 acidic domain in Saccharomyces cerevisiae, we used the cytoplasmic yeast two-hybrid system designated the Ras recruitment system (RRS) (Aronheim 2001). The bait was made as a hybrid protein that included the entire acidic domain with a few flanking amino acids (amino acids 222–304), fused C-terminally to the activated Ras protein (L61) lacking the CAAX box. Yeast cdc25-2 cells were cotransformed with this plasmid and with a human testis cDNA library encoding protein fragments fused to the myristylation signal (Stratagene). We identified several peptides capable of recruiting the acidic domain–Ras fusion protein to the membrane; one of those was a fragment of 200 amino acids derived from the N-terminal nonkinase domain of Lats2.
Coimmunoprecipitation analysis
The monolayer was gently washed twice with ice-cold PBS and lysed on ice for 30 min with NP-40 lysis buffer (50 mM Tris-HCl at pH 8.0, 150 mM NaCl, 1.0% NP-40, and protease inhibitor mix [Sigma]). For denatured immunoprecipitation, cell pellets were lysed in 1% SDS, 10 mM Tris-HCl (pH 7.5), 150 mM NaCl, and protease inhibitor mix (Sigma) by consecutive vigorous vortex and boiling. Protein A Sepharose beads (Repligen), pre-blocked with BSA, were incubated with either anti-Mdm2 antibodies (mix of 4B2, 2A9, and 4B11) (Chen et al. 1993), DO1 and PAb1801 anti-p53 antibodies, anti-Myc antibodies (9E10), or anti-T-antigen antibodies as a negative control. Lysates were incubated with bound antibodies for 2 h at 4°C, washed with NP-40 lysis buffer, released from beads by boiling, and resolved by SDS-PAGE.
Fluorescence-activated cell sorting (FACS) analysis
For cell cycle analysis, cells were detached with trypsin, collected with supernatant, and washed once with ice-cold PBS. Cells were fixed in cold methanol for no less than 30 min, washed, rehydrated, and then resuspended in PBS containing 50 μg/mL propidium iodide (PI) and 50 μg/mL RNase A. Samples were analyzed by flow cytometry using a FACS sorter (Becton Dickinson).
Immunofluorescence
Cells growing on 12-mm coverslips were fixed with ice-cold methanol for 20 min. After gradual rehydration with PBS, cells were permeabilized with 0.5% Triton X-100 in PBS, washed with PBS, and then blocked with 3% BSA in PBST (phosphate-buffered saline + 0.05% Tween 20). Samples were then incubated for 1 h with primary antibody, washed, and then incubated with secondary antibody and DAPI (2.5 μg/mL final) for 40 min in the dark. To detect Lats2, anti-KPM/Lats2 (Santa Cruz Biotechnology) or LA-2 antibodies were used. Affinity-purified anti-Lats2 polyclonal antibody (LA-2) (N. Yabuta and H. Nojima, unpubl.) was raised in a rabbit against the GST-fused N-terminal portion of human Lats2 (amino acids 79–257). To detect p53, a mix of antibodies DO1 + PAb1801 was used; an antibody against γ-tubulin (Sigma) was used to locate centrosomes.
ChIP
ChIP analysis was performed as described previously (Minsky and Oren 2004). The following PCR primers were used to detect promoter sequences from immunoprecipitated chromatin: Lats2 promoter (p53RE1) forward, 5′-TGCAACCTCCACTT CCTGGGCTC-3′; reverse, 5′-CCAACGTAGCGGAACCCTTT CTC-3′; (2) p21 promoter forward, 5′-GCACTCTTGTCCC CCAG-3′; reverse, 5′-TCTATGCCAGAGCTCAACAT-3′.
Centrifugal elutriation
Centrifugal elutriation of 5 × 105 U2OS cells was performed using a J-6 MI centrifuge and JE-5 rotor. Cells were loaded at a flow rate of 20 mL/min. After equilibration at 40 mL/min for 15 min, 150-mL samples were taken at 10 mL/min increments. Asynchronous control cells were handled in parallel but did not undergo centrifugation. Immediately after elutriation, cells were counted, and then replated with or without nocodazole (0.2 μg/ mL), or taken for FACS analysis.
Signal quantification
Band intensity was quantified using the ImageJ program. Values were standardized to GFP or GAPDH loading controls and normalized to the control, untreated lane of each blot.
Acknowledgments
We thank S. Wilder, L. Sananes, and R. Shalgi for helpful contributions to the experimental work and generation of valuable reagents; N. Raver-Shapira for ChIP advice; A.J. Levine for antibodies; R. Agami for cell lines and for pRS-Lats2#2; V. Rotter for hTERT-immortalized WI-38 cells; and B. Vogelstein, B. Wasylyk, and R. Agami for the gift of plasmids. This work was supported in part by grant R37 CA40099 from the National Cancer Institute, EC FP6 grant LSHC-CT-2004-503576, and grants from the Center for Excellence Program of the Israel Science Foundation, the Robert Bosch Foundation (Germany), and the Yad Abraham Center for Cancer Diagnosis and Therapy. N.Y. and H.N. were supported by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Culture of Japan. The EC is not liable for any use that may be made of the information contained herein.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1447006.
References
- Abe, Y., Ohsugi, M., Haraguchi, K., Fujimoto, J., Yamamoto, T. LATS2–Ajuba complex regulates γ-tubulin recruitment to centrosomes and spindle organization during mitosis. FEBS Lett. 2006;580:782–788. doi: 10.1016/j.febslet.2005.12.096. [DOI] [PubMed] [Google Scholar]
- Andreassen, P.R., Lohez, O.D., Margolis, R.L. G2 and spindle assembly checkpoint adaptation, and tetraploidy arrest: Implications for intrinsic and chemically induced genomic instability. Mutat. Res. 2003;532:245–253. doi: 10.1016/j.mrfmmm.2003.08.020. [DOI] [PubMed] [Google Scholar]
- Aronheim, A. Membrane recruitment systems for analysis of protein–protein interactions. Methods Mol. Biol. 2001;177:319–328. doi: 10.1385/1-59259-210-4:319. [DOI] [PubMed] [Google Scholar]
- Barr, F.A., Sillje, H.H., Nigg, E.A. Polo-like kinases and the orchestration of cell division. Nat. Rev. Mol. Cell Biol. 2004;5:429–440. doi: 10.1038/nrm1401. [DOI] [PubMed] [Google Scholar]
- Bond, G.L., Hu, W., Levine, A.J. MDM2 is a central node in the p53 pathway: 12 years and counting. Curr. Cancer Drug Targets. 2005;5:3–8. doi: 10.2174/1568009053332627. [DOI] [PubMed] [Google Scholar]
- Bothos, J., Tuttle, R.L., Ottei, M., Luca, F.C., Halazonetis, T.D. Human Lats1 is a mitotic exit network kinase. Cancer Res. 2005;65:6568–6575. doi: 10.1158/0008-5472.CAN-05-0862. [DOI] [PubMed] [Google Scholar]
- Brummelkamp, T.R., Bernards, R., Agami, R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Bunz, F., Hwang, P.M., Torrance, C., Waldman, T., Zhang, Y., Dillehay, L., Williams, J., Lengauer, C., Kinzler, K.W., Vogelstein, B. Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J. Clin. Invest. 1999;104:263–269. doi: 10.1172/JCI6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burakov, A., Nadezhdina, E., Slepchenko, B., Rodionov, V. Centrosome positioning in interphase cells. J. Cell Biol. 2003;162:963–969. doi: 10.1083/jcb.200305082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castedo, M., Perfettini, J.L., Roumier, T., Andreau, K., Medema, R., Kroemer, G. Cell death by mitotic catastrophe: A molecular definition. Oncogene. 2004;23:2825–2837. doi: 10.1038/sj.onc.1207528. [DOI] [PubMed] [Google Scholar]
- Chen, J.D., Marechal, V., Levine, A.J. Mapping of the p53 and mdm-2 interaction domains. Mol. Cell. Biol. 1993;13:4107–4114. doi: 10.1128/mcb.13.7.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammermann, A., Merdes, A. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J. Cell Biol. 2002;159:255–266. doi: 10.1083/jcb.200204023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash, B.C., El-Deiry, W.S. Cell cycle checkpoint control mechanisms that can be disrupted in cancer. Methods Mol. Biol. 2004;280:99–161. doi: 10.1385/1-59259-788-2:099. [DOI] [PubMed] [Google Scholar]
- Ditchfield, C., Johnson, V.L., Tighe, A., Ellston, R., Haworth, C., Johnson, T., Mortlock, A., Keen, N., Taylor, S.S. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 2003;161:267–280. doi: 10.1083/jcb.200208091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxsey, S., Zimmerman, W., Mikule, K. Centrosome control of the cell cycle. Duensing, A. and Duensing, S. 2005. Guilt by association? p53 and the development of aneuploidy in cancer. Trends Cell Biol. Biochem. Biophys. Res. Commun. 2005;15331:303–311. 694–700. doi: 10.1016/j.tcb.2005.04.008. [DOI] [PubMed] [Google Scholar]
- El-Deiry, W.S., Kern, S.E., Pietenpol, J.A., Kinzler, K.W., Vogelstein, B. Definition of a consensus binding site for p53. Nat. Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- Fukasawa, K., Choi, T., Kuriyama, R., Rulong, S., Vande Woude, G.F. Abnormal centrosome amplification in the absence of p53. Science. 1996;271:1744–1747. doi: 10.1126/science.271.5256.1744. [DOI] [PubMed] [Google Scholar]
- Giet, R., Petretti, C., Prigent, C. Aurora kinases, aneuploidy and cancer, a coincidence or a real link? Trends Cell Biol. 2005;15:241–250. doi: 10.1016/j.tcb.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Harris, S.L., Levine, A.J. The p53 pathway: Positive and negative feedback loops. Oncogene. 2005;24:2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- Hergovich, A., Schmitz, D., Hemmings, B.A. The human tumour suppressor LATS1 is activated by human MOB1 at the membrane. Biochem. Biophys. Res. Commun. 2006;345:50–58. doi: 10.1016/j.bbrc.2006.03.244. [DOI] [PubMed] [Google Scholar]
- Hirota, T., Morisaki, T., Nishiyama, Y., Marumoto, T., Tada, K., Hara, T., Masuko, N., Inagaki, M., Hatakeyama, K., Saya, H. Zyxin, a regulator of actin filament assembly, targets the mitotic apparatus by interacting with h-warts/ LATS1 tumor suppressor. J. Cell Biol. 2000;149:1073–1086. doi: 10.1083/jcb.149.5.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori, T., Takaori-Kondo, A., Kamikubo, Y., Uchiyama, T. Molecular cloning of a novel human protein kinase, kpm, that is homologous to warts/lats, a Drosophila tumor suppressor. Oncogene. 2000;19:3101–3109. doi: 10.1038/sj.onc.1203659. [DOI] [PubMed] [Google Scholar]
- Iida, S., Hirota, T., Morisaki, T., Marumoto, T., Hara, T., Kuninaka, S., Honda, S., Kosai, K., Kawasuji, M., Pallas, D.C., et al. Tumor suppressor WARTS ensures genomic integrity by regulating both mitotic progression and G1 tetraploidy checkpoint function. Oncogene. 2004;23:5266–5274. doi: 10.1038/sj.onc.1207623. [DOI] [PubMed] [Google Scholar]
- Jimenez-Velasco, A., Roman-Gomez, J., Agirre, X., Barrios, M., Navarro, G., Vazquez, I., Prosper, F., Torres, A., Heiniger, A. Downregulation of the large tumor suppressor 2 (LATS2/KPM) gene is associated with poor prognosis in acute lymphoblastic leukemia. Leukemia. 2005;19:2347–2350. doi: 10.1038/sj.leu.2403974. [DOI] [PubMed] [Google Scholar]
- Justice, R.W., Zilian, O., Woods, D.F., Noll, M., Bryant, P.J. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes & Dev. 1995;9:534–546. doi: 10.1101/gad.9.5.534. [DOI] [PubMed] [Google Scholar]
- Kamikubo, Y., Takaori-Kondo, A., Uchiyama, T., Hori, T. Inhibition of cell growth by conditional expression of kpm, a human homologue of Drosophila warts/lats tumor suppressor. J. Biol. Chem. 2003;278:17609–17614. doi: 10.1074/jbc.M211974200. [DOI] [PubMed] [Google Scholar]
- Ke, H., Pei, J., Ni, Z., Xia, H., Qi, H., Woods, T., Kelekar, A., Tao, W. Putative tumor suppressor Lats2 induces apoptosis through downregulation of Bcl-2 and Bcl-x(L) Exp. Cell Res. 2004;298:329–338. doi: 10.1016/j.yexcr.2004.04.031. [DOI] [PubMed] [Google Scholar]
- Kostic, C., Shaw, P.H. Isolation and characterization of sixteen novel p53 response genes. Oncogene. 2000;19:3978–3987. doi: 10.1038/sj.onc.1203747. [DOI] [PubMed] [Google Scholar]
- Lai, Z.C., Wei, X., Shimizu, T., Ramos, E., Rohrbaugh, M., Nikolaidis, N., Ho, L.L., Li, Y. Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell. 2005;120:675–685. doi: 10.1016/j.cell.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Lanni, J.S., Jacks, T. Characterization of the p53-dependent postmitotic checkpoint following spindle disruption. Mol. Cell. Biol. 1998;18:1055–1064. doi: 10.1128/mcb.18.2.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Pei, J., Xia, H., Ke, H., Wang, H., Tao, W. Lats2, a putative tumor suppressor, inhibits G1/S transition. Oncogene. 2003;22:4398–4405. doi: 10.1038/sj.onc.1206603. [DOI] [PubMed] [Google Scholar]
- Margolis, R.L., Lohez, O.D., Andreassen, P.R. G1 tetraploidy checkpoint and the suppression of tumorigenesis. J. Cell. Biochem. 2003;88:673–683. doi: 10.1002/jcb.10411. [DOI] [PubMed] [Google Scholar]
- McPherson, J.P., Tamblyn, L., Elia, A., Migon, E., Shehabeldin, A., Matysiak-Zablocki, E., Lemmers, B., Salmena, L., Hakem, A., Fish, J., et al. Lats2/Kpm is required for embryonic development, proliferation control and genomic integrity. EMBO J. 2004;23:3677–3688. doi: 10.1038/sj.emboj.7600371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek, D.W. The role of p53 in the response to mitotic spindle damage. Pathol. Biol. (Paris) 2000;48:246–254. [PubMed] [Google Scholar]
- Michael, D., Oren, M. The p53–Mdm2 module and the ubiquitin system. Semin. Cancer Biol. 2003;13:49–58. doi: 10.1016/s1044-579x(02)00099-8. [DOI] [PubMed] [Google Scholar]
- Minsky, N., Oren, M. The RING domain of Mdm2 mediates histone ubiquitylation and transcriptional repression. Mol. Cell. 2004;16:631–639. doi: 10.1016/j.molcel.2004.10.016. [DOI] [PubMed] [Google Scholar]
- Morrison, C., Rieder, C.L. Chromosome damage and progression into and through mitosis in vertebrates. DNA Repair (Amst.) 2004;3:1133–1139. doi: 10.1016/j.dnarep.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Nigg, E.A. Centrosome aberrations: Cause or consequence of cancer progression? Nat. Rev. Cancer. 2002;2:815–825. doi: 10.1038/nrc924. [DOI] [PubMed] [Google Scholar]
- Nishiyama, Y., Hirota, T., Morisaki, T., Hara, T., Marumoto, T., Iida, S., Makino, K., Yamamoto, H., Hiraoka, T., Kitamura, N., et al. A human homolog of Drosophila warts tumor suppressor, h-warts, localized to mitotic apparatus and specifically phosphorylated during mitosis. FEBS Lett. 1999;459:159–165. doi: 10.1016/s0014-5793(99)01224-7. [DOI] [PubMed] [Google Scholar]
- O'Neill, E.E., Matallanas, D., Kolch, W. Mammalian sterile 20-like kinases in tumor suppression: An emerging pathway. Cancer Res. 2005;65:5485–5487. doi: 10.1158/0008-5472.CAN-05-1453. [DOI] [PubMed] [Google Scholar]
- Oren, M. Decision making by p53: Life, death and cancer. Cell Death Differ. 2003;10:431–442. doi: 10.1038/sj.cdd.4401183. [DOI] [PubMed] [Google Scholar]
- Rajagopalan, H., Lengauer, C. Aneuploidy and cancer. Nature. 2004;432:338–341. doi: 10.1038/nature03099. [DOI] [PubMed] [Google Scholar]
- Rieder, C.L., Maiato, H. Stuck in division or passing through: What happens when cells cannot satisfy the spindle assembly checkpoint. Dev. Cell. 2004;7:637–651. doi: 10.1016/j.devcel.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Scolnick, D.M., Halazonetis, T.D. Chfr defines a mitotic stress checkpoint that delays entry into metaphase. Nature. 2000;406:430–435. doi: 10.1038/35019108. [DOI] [PubMed] [Google Scholar]
- St John, M.A., Tao, W., Fei, X., Fukumoto, R., Carcangiu, M.L., Brownstein, D.G., Parlow, A.F., McGrath, J., Xu, T. Mice deficient of Lats1 develop soft-tissue sarcomas, ovarian tumours and pituitary dysfunction. Nat. Genet. 1999;21:182–186. doi: 10.1038/5965. [DOI] [PubMed] [Google Scholar]
- Stukenberg, P.T. Triggering p53 after cytokinesis failure. J. Cell Biol. 2004;165:607–608. doi: 10.1083/jcb.200405089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, Y., Miyoshi, Y., Takahata, C., Irahara, N., Taguchi, T., Tamaki, Y., Noguchi, S. Down-regulation of LATS1 and LATS2 mRNA expression by promoter hypermethylation and its association with biologically aggressive phenotype in human breast cancers. Clin. Cancer Res. 2005;11:1380–1385. doi: 10.1158/1078-0432.CCR-04-1773. [DOI] [PubMed] [Google Scholar]
- Tarapore, P., Tokuyama, Y., Horn, H.F., Fukasawa, K. Difference in the centrosome duplication regulatory activity among p53 ‘hot spot’ mutants: Potential role of Ser 315 phosphorylation-dependent centrosome binding of p53. Oncogene. 2001;20:6851–6863. doi: 10.1038/sj.onc.1204848. [DOI] [PubMed] [Google Scholar]
- Toji, S., Yabuta, N., Hosomi, T., Nishihara, S., Kobayashi, T., Suzuki, S., Tamai, K., Nojima, H. The centrosomal protein Lats2 is a phosphorylation target of Aurora-A kinase. Genes Cells. 2004;9:383–397. doi: 10.1111/j.1356-9597.2004.00732.x. [DOI] [PubMed] [Google Scholar]
- Tovar, C., Rosinski, J., Filipovic, Z., Higgins, B., Kolinsky, K., Hilton, H., Zhao, X., Vu, B.T., Qing, W., Packman, K., et al. Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: Implications for therapy. Proc. Natl. Acad. Sci. 2006;103:1888–1893. doi: 10.1073/pnas.0507493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetake, Y., Sluder, G. Cell cycle progression after cleavage failure: Mammalian somatic cells do not possess a ‘tetraploidy checkpoint.’. J. Cell Biol. 2004;165:609–615. doi: 10.1083/jcb.200403014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzbekov, R., Kireyev, I., Prigent, C. Centrosome separation: Respective role of microtubules and actin filaments. Biol. Cell. 2002;94:275–288. doi: 10.1016/s0248-4900(02)01202-9. [DOI] [PubMed] [Google Scholar]
- Vassilev, L.T., Vu, B.T., Graves, B., Carvajal, D., Podlaski, F., Filipovic, Z., Kong, N., Kammlott, U., Lukacs, C., Klein, C., et al. In vivo activation of the p53 pathway by smallmolecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- Voorhoeve, P.M., le Sage, C., Schrier, M., Gillis, A.J., Stoop, H., Nagel, R., Liu, Y.P., van Duijse, J., Drost, J., Griekspoor, A., et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169–1181. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- Vousden, K.H., Lu, X. Live or let die: The cell's response to p53. Nat. Rev. Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- Vousden, K.H., Prives, C. P53 and prognosis: New insights and further complexity. Cell. 2005;120:7–10. doi: 10.1016/j.cell.2004.12.027. [DOI] [PubMed] [Google Scholar]
- Wong, C., Stearns, T. Mammalian cells lack checkpoints for tetraploidy, aberrant centrosome number, and cytokinesis failure. BMC Cell Biol. 2005;6 doi: 10.1186/1471-2121-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, H., Qi, H., Li, Y., Pei, J., Barton, J., Blackstad, M., Xu, T., Tao, W. LATS1 tumor suppressor regulates G2/M transition and apoptosis. Oncogene. 2002;21:1233–1241. doi: 10.1038/sj.onc.1205174. [DOI] [PubMed] [Google Scholar]
- Xu, T., Wang, W., Zhang, S., Stewart, R.A., Yu, W. Identifying tumor suppressors in genetic mosaics: The Drosophila lats gene encodes a putative protein kinase. Development. 1995;121:1053–1063. doi: 10.1242/dev.121.4.1053. [DOI] [PubMed] [Google Scholar]
- Yabuta, N., Fujii, T., Copeland, N.G., Gilbert, D.J., Jenkins, N.A., Nishiguchi, H., Endo, Y., Toji, S., Tanaka, H., Nishimune, Y., et al. Structure, expression, and chromosome mapping of LATS2, a mammalian homologue of the Drosophila tumor suppressor gene lats/warts. Genomics. 2000;63:263–270. doi: 10.1006/geno.1999.6065. [DOI] [PubMed] [Google Scholar]