Abstract
Comprehensive gene networks in Ciona intestinalis embryos provide a foundation for characterizing complex developmental processes, such as the initial phases of chordate heart development. The basic helix–loop–helix regulatory gene Ci-Mesp is required for activation of cardiac transcription factors. Evidence is presented that Ci-Ets1/2, a transcriptional effector of receptor tyrosine kinase (RTK) signaling, acts downstream from Mesp to establish the heart field. Asymmetric activation of Ets1/2, possibly through localized expression of FGF9, drives heart specification within this field. During gastrulation, Ets1/2 is expressed in a group of four cells descended from two Mesp-expressing founder cells (the B7.5 cells). After gastrulation, these cells divide asymmetrically; the smaller rostral daughters exhibit RTK activation (phosphorylation of ERK) and form the heart lineage while the larger caudal daughters form the anterior tail muscle lineage. Inhibition of RTK signaling prevents heart specification. Targeted inhibition of Ets1/2 activity or FGF receptor function also blocks heart specification. Conversely, application of FGF or targeted expression of constitutively active Ets1/2 (EtsVp16) cause both rostral and caudal B7.5 lineages to form heart cells. This expansion produces an unexpected phenotype: transformation of a single-compartment heart into a functional multicompartment organ. We discuss these results with regard to the development and evolution of the multichambered vertebrate heart.
Keywords: Ets, heart, Mesp, development, evolution
The invertebrate chordate, Ciona intestinalis, represents an ideal system for investigating early steps in heart development (Davidson and Levine 2003; Satou et al. 2004; Davidson et al. 2005; Davidson and Christiaen 2006; Imai et al. 2006). C. intestinalis is a member of the tunicates, a group of basal chordates that diverged prior to genome duplications within the vertebrate lineage. Thus, most Ciona developmental regulatory factors are represented by a single ortholog, while in vertebrates there are often multiple paralogs with partially redundant functions (Dehal et al. 2002). Tunicate embryos are composed of extremely low cell numbers. Gastrulation begins at the 110 cell stage, at which point the heart lineage can be traced to two blastomeres (the B7.5 cells) (Hirano and Nishida 1997; Davidson and Levine 2003). Newly hatched larvae are composed of <2500 cells, including a fully differentiated notochord and dorsal neural tube (two defining chordate traits) (Satoh 2003). The larval heart rudiment consists of only 12–32 undifferentiated cells (B. Davidson, unpubl.). This rudiment differentiates into a single-compartment myocardium surrounded by a pericardial coelom during metamorphosis, and is first evident as a beating organ in juveniles (Robb 1965).
Despite this extraordinary simplicity, Ciona heart development appears to maintain ancestral characters also found in vertebrate cardiogenesis. As in vertebrates, Ciona heart precursor cells form bilateral rudiments that migrate anteriorly and ventrally along the endoderm to fuse at the ventral midline (Davidson et al. 2005). Although Ciona myocytes differ in structure from their vertebrate counterparts (Robb 1965), homology is suggested by the expression of a variety of molecular markers, including a specific splice isoform of Troponin I (MacLean et al. 1997). There is every indication that the earliest phases of cardiac mesoderm specification are conserved in Ciona and vertebrates.
The differentiation of cardiac mesoderm is controlled by a highly conserved cassette of regulatory genes, including Nkx2.5, Hand, and Gata4 (Cripps and Olson 2002). In vertebrates and Ciona (but not Drosophila), Mesp functions upstream of the core cardiac regulatory network during the earliest phases of heart specification (Kitajima et al. 2000; Satou et al. 2004; Davidson et al. 2005). However, the Mesp expression domain extends beyond the limits of the definitive heart progenitors (Saga et al. 1996; Sawada et al. 2000). In vertebrates, initial Mesp expression extends through much of the nascent mesoderm. In Ciona, Mesp is expressed in a single pair of blastomeres (B7.5) whose progeny acquire two distinct fates; rostral descendants migrate to form the bilateral heart rudiments, while caudal descendents remain in the tail, forming the proximal tail muscles.
The decision to form heart or tail muscle within the Mesp lineage is most easily explained by localized induction. Studies in flies and vertebrates have identified a number of signaling factors as candidates for this inductive event. Wnt signaling has been characterized as both an activator and repressor of heart fate (Lockwood and Bodmer 2002; Pandur et al. 2002; Foley and Mercola 2005; Eisenberg and Eisenberg 2006). BMP signaling has a role in the activation or maintenance of Nkx2.5 and Gata4 expression (Halfon et al. 2000; Cripps and Olson 2002; Lien et al. 2002; Lee et al. 2004). FGF is thought to work in conjunction with BMP to drive heart specification (Alsan and Schultheiss 2002; Stathopoulos et al. 2004).
In Ciona embryos, FGF signaling is essential for early neural and mesodermal patterning events (Bertrand et al. 2003; Miya and Nishida 2003; Pasini et al. 2006). FGF9/16/20 (hereafter referred to as FGF9) is expressed in the endoderm during early cleavage stages and has documented roles in the induction of notochord, neural plate, and mesenchyme (a mesodermal lineage that forms rudiments for a number of adult tissues) (Tokuoka et al. 2004). These inductive events require Ets family transcription factors, known mediators of FGF signaling (Miya and Nishida 2003). Ci-Ets1/2 (also referred to as ets/pointed2) is the only Ciona ortholog of the vertebrate Ets1 and Ets2 transcription factors that contains both an Ets DNA-binding domain and a pointed domain. Ci-Ets1/2 also contains a well-conserved ERK docking site and mitogen activated protein kinase (MAPK) phosphorylation site. Recent work on the regulation of Ci-Otx has demonstrated that activation by FGF9 is mediated through Ci-Ets1/2 (Bertrand et al. 2003).
Here we investigate the role of Ets1/2 in the early specification of cardiac mesoderm. Apparently, expression of Ets1/2 downstream from Mesp in the B7.5 lineage establishes the heart field. Ets1/2 is asymmetrically activated in the rostral portion of the heart field through receptor tyrosine kinase (RTK) signaling downstream from FGF. Activated Ets1/2 is required for heart cell migration and differentiation. Ectopic activation of Ets1/2 in the caudal B7.5 lineage is sufficient to transform proximal tail muscles into supernumerary heart cells. In some cases the expanded heart rudiment forms a functional two-compartment heart. We discuss this observation with regard to the evolution of the multichambered vertebrate heart.
Results
Asymmetric fates of rostral and caudal B7.5 lineages
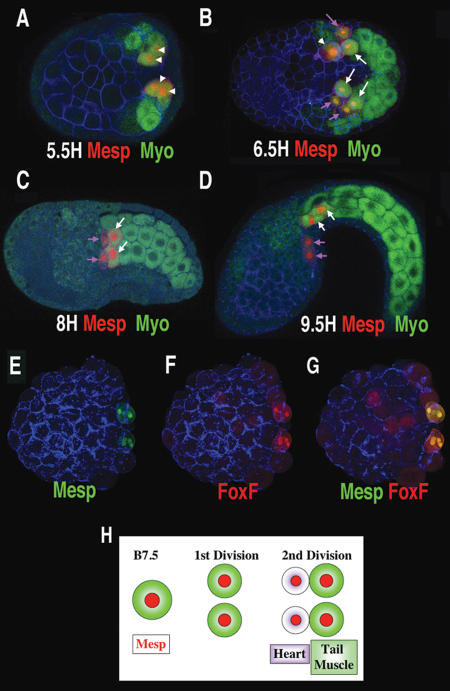
Detailed examination of the B7.5 cell lineage reveals an asymmetric cell division that separates heart and proximal tail muscle lineages (Fig. 1). The first division of the B7.5 cells occurs during gastrulation, 5 h post-fertilization at 19°C (5H), and is symmetric, leading to similarly sized daughter cells and an equal distribution of autofluorescent myoplasm (Fig. 1A, arrowheads). However, during the next division (6.5H) a clear asymmetry is established, whereby myoplasm is differentially segregated into the larger caudal daughter cells (Fig. 1B–D, white arrows). Analysis of confocal sections indicate that this asymmetric division occurs oblique to the anterior–posterior axis, so that the smaller rostral daughter cells are both anterior and ventral to their larger sister cells (data not shown). Typically, this cell division occurs asynchronously on the left and right sides of the embryo. This can be seen in the embryo shown in Figure 1B. On the bottom half (Fig. 1B, right side) of the embryo, both the B7.5 lineage cells have divided asymmetrically, while on the upper half (Fig. 1B, left side), only one of these cells has divided. In the lagging cell (Fig. 1B, arrowhead), it is apparent that myoplasm segregation occurs prior to division. At later stages the smaller rostral daughters migrate to form the heart (Fig. 1B–D, purple arrows), while the larger, myoplasm-containing caudal daughters differentiate as tail muscles (Fig. 1D, white arrows).
Figure 1.
An asymmetric cell division separates heart and proximal tail muscle lineages. (A–D) Confocal projections of transgenic Mesp-RFP Ciona embryos (red), autofluorescent myoplasm (green), and phalloidin (blue). Time of fixation is indicated in hours post-fertilization (H) at 19°C. A and B are ventral views, and C and D are viewed laterally. Arrowheads indicate B7.5 lineage cells prior to the asymmetric cell division, white arrows indicate myoplasm containing tail muscle lineage cells, and purple arrows indicate heart precursor cells. (E–G) Dorsal confocal projections of a transgenic embryo (Mesp-GFP/ FoxF-RFP) arrested at the 110-cell stage (4H) through treatment with cytochalasin, fixed at 10H. The FoxF enhancer drives some reporter expression in the epidermis (faint red staining). Anterior is to the left in all panels. (H) Summary illustrating cell division within the emerging heart lineage. It appears that these cell divisions occur earlier in Ciona savignyi, as eight Mesp-expressing cells are detected by the late gastrula stage (Satou et al. 2004).
Inhibition of cell division was employed to determine whether asymmetric distribution of myoplasm is essential for heart specification. Classical studies on the asymmetric distribution of localized determinants involved the use of drugs that inhibit cell division, such as cytochalasin. An enhancer from the FoxF gene drives reporter gene expression in the heart lineage and ventral head ectoderm, but not in the proximal tail muscle lineage (see Fig. 3 [below]; J. Beh, unpubl.). Treatment of 4H embryos with cytochalasin causes a block in the division of the B7.5 blastomeres at the 110-cell stage. Despite this block, FoxF reporter gene expression is observed in the B7.5 cells “on schedule,” ~10 h after fertilization at the equivalent of the tailbud stage (Fig. 1E–G). FoxF activation is obtained even though these cells retain autofluorescent myoplasm, due to the loss of the asymmetric division that segregates myoplasm away from the future heart cells.
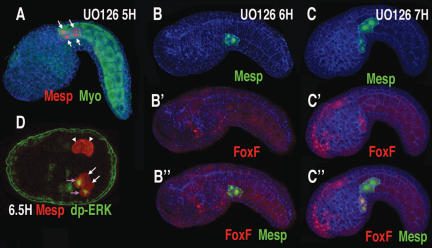
Figure 3.
RTK signaling is required for heart induction. Transgenic embryos (Mesp-RFP or Mesp-GFP/FoxF-RFP), phalloidin (blue) shown laterally, anterior to the left, except for D, which is a ventral view. (A) Tailbud embryo treated at midgastrula stage (5H) with U0126. (B,B′,B″) Tailbud embryo coelectroporated with Mesp-GFP (green) and FoxF-RFP (red) fusion genes, treated at late gastrula stage (6H) with U0126. (B) Green channel. (B′) Red channel. Note FoxF reporter expression in the epidermis (FoxF is expressed in both heart precursors and epidermis; see http://ghost.zool.kyoto-u.ac.jp/tagtscriptg. html). (B″) Red and green channels. (C,C′,C″) Same as B, B′, and B″, except that U0126 was applied 1 h later, during neurulation (7H). Similar results were obtained on application of DMSO alone or when U0126 was applied at 8H or 9H. (D) Late gastrula stage Mesp-GFP transgenic embryo (6.5H) stained with rabbit anti-GFP (red) and mouse anti-dpERK (green); purple arrows indicate heart precursor cells showing nuclear staining for dpERK, white arrows indicate tail muscle precursors, and arrowheads indicate B7.5 lineage cells just prior to the asymmetric division.
Expression of Ets1/2
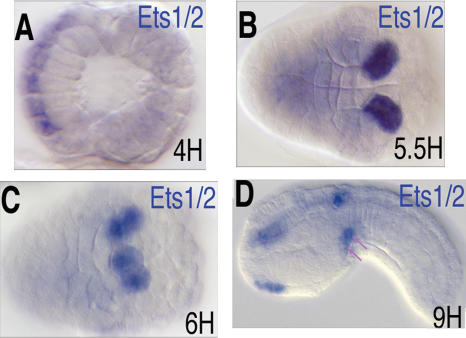
The Ets family transcription factor, Hr-Ets, appears to be expressed in the developing heart lineage of the distantly related tunicate, Halocynthia roretzi (Miya and Nishida 2003). The orthologous gene, Ci-Ets1/2, was examined in C. intestinalis embryos (Fig. 2). The gene first exhibits clear zygotic expression in the A-line CNS at the onset of gastrulation (Fig. 2A). As gastrulation proceeds, Ets1/2 is expressed in both daughters of the B7.5 cells (Fig. 2B). The Ets1/2 expression pattern mirrors the earlier expression of Mesp (Satou et al. 2004), and preliminary data suggests direct regulation by Mesp (B. Davidson, unpubl.). Ets1/2 expression in this lineage remains strong throughout gastrulation and neurulation (Fig. 2C). By the early tail-bud stage, Ets1/2 expression in the B7.5 lineage becomes restricted to migrating cardioblasts (Fig. 2D, arrows).
Figure 2.
Ets1/2 is expressed in the B7.5 lineage cells. (A–D) Ciona embryos hybridized with antisense probes for Ets1/2. Developmental time is indicated in lower right corner; anterior is to the left. (A) Early gastrula, dorsal view. (B) Late gastrula, ventral view. Note the similar position of B7.5 lineage cells in Figure 1A. (C) Early neurula, ventra–lateral view, just prior to asymmetric division. Note the similar position of B7.5 lineage cells in Figure 1B. (D) Early tailbud, lateral view; migrating heart precursor cells are indicated by purple arrows.
Asymmetric activation of RTK signaling
Ets1/2 is known to mediate FGF signaling during early specification events in Ciona and other ascidians (Bertrand et al. 2003; Miya and Nishida 2003). We therefore tested whether RTK signaling downstream from FGF was involved in Ciona heart cell induction. When gastrulating embryos (4–6H) were treated with the MEK inhibitor, U0126, the rostral cells do not migrate out of the tail (cf. arrows in Figs. 3A and 1D) and fail to activate the FoxF enhancer (Fig. 3, cf. B,B′,B″ and C,C′,C″). Additionally, these cells lack expression of heart marker genes such as Hand-like (Satou et al. 2004; data not shown), but express tail muscle markers (Supplementary Fig. S1). Similar results were obtained through targeted expression of a dominant-negative form of Ras (data not shown).
To determine whether RTK signaling was specifically required at the time when distinct lineages are formed, U0126 was applied sequentially at varying time points. Although U0126 blocked heart induction up to the late gastrula stage (6H) (Fig. 3A–B; Supplementary Fig. S1), application at later time points (7–9H) failed to block induction (Fig. 3C,C′,C″). Thus, the requirement for RTK induction persists until the early neurula stage (6–7H), the stage when the asymmetric division between the heart and nonheart lineages occurs (Fig. 1B).
An anti-dpERK antibody was used to pinpoint the timing of RTK signaling during heart specification (Fig. 3D). Analysis of staged embryos reveals specific staining in the newly formed heart progenitors immediately following the asymmetric division of the B7.5 lineage (Fig. 3D, purple arrows, lower half of embryo). No staining is observed in the precursor cells immediately prior to the asymmetric division (Fig. 3D, arrowheads). These data suggest that the RTK signaling pathway is activated precisely at the time when the lineages are distinguished, and activation is restricted to the rostral heart precursors.
The timing of heart cell induction was further confirmed through disassociation experiments. When transgenic Mesp-GFP/FoxF-RFP embryos were disassociated at the gastrula stage (5H), most B7.5 lineage cells (marked by expression of GFP) fail to coexpress the FoxF reporter gene at 12H, indicating that cell–cell-mediated induction has not occurred in gastrula stage embryos (Fig. 4A,D). When embryos were disassociated at 6–7H, ~50% of B7.5 lineage cells express the FoxF reporter gene, confirming asymmetric induction of the rostral lineage at this time (Fig. 4D).
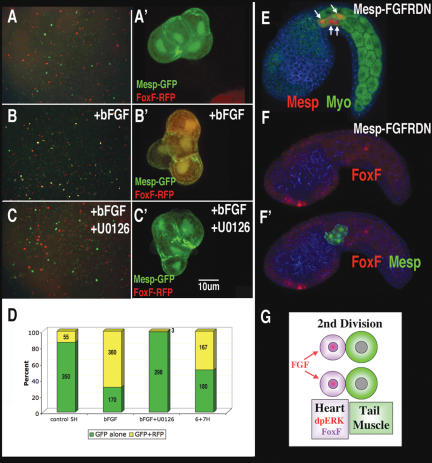
Figure 4.
FGF is both necessary and sufficient for heart specification. (A–C) Low-magnification view (A,B,C) and high-magnification view (A′,B′,C′) of cell clones from embryos electroporated with Mesp-GFP/FoxF-RFP, disassociated at 5H, and fixed at 12H. (B,B′) Immediately after disassociation, cells were placed in seawater with bFGF (100 ng/mL). (C,C′) Immediately after disassociation, cells were placed in sea-water with both bFGF and U0126. (D) Percentage of Mesp-GFP-expressing cells either with (yellow) or without (green) coexpression of FoxF-RFP. Within each colomn is the combined number of cells counted during two trials. (E,F,F′) Same as Figure 3, A, B′, and B″, except that the embryos were coelectroporated with Mesp–FGFRDN and not treated with U0126. (G) Summary diagram of proposed role for FGF in asymmetric cell division.
FGF acts upstream of the RTK signaling pathway to mediate heart specification
To test whether FGF acts upstream of RTK-mediated heart induction, bFGF was applied to isolated cells from Mesp-GFP/FoxF-RFP transgenic embryos disassociated at 5H (prior to endogenous induction). This treatment greatly increased the proportion of B7.5 lineage cells expressing the FoxF reporter gene (Fig. 4B,D). The ability of bFGF to induce FoxF reporter expression was completely blocked by inhibition of the RTK signaling pathway (Fig. 4C,D). Application of bFGF to whole embryos had no effect (data not shown). At the time of induction, the B7.5 lineage cells lie beneath a layer of epidermis, which may prevent them from responding to external application of FGF.
To determine whether FGF signaling is necessary for heart induction, we constructed a dominant-negative form of the sole Ciona FGF receptor (FGFRDN). Targeted expression of FGFRDN in the B7.5 lineage blocks migration of the anterior heart precursors (cf. arrows in Figs. 4E and 1D) and prevents activation of the FoxF enhancer in these cells (cf. Figs. 4F,F′ and 3C″). As with the previous manipulations, all B7.5 lineage cells express tail muscle markers and fail to express the heart lineage marker, Hand-like (Supplementary Fig. S1). Thus, it appears that RTK signaling downstream from FGF is both necessary and sufficient for heart induction in Ciona.
Ets1/2 activation functions downstream from FGF during heart specification
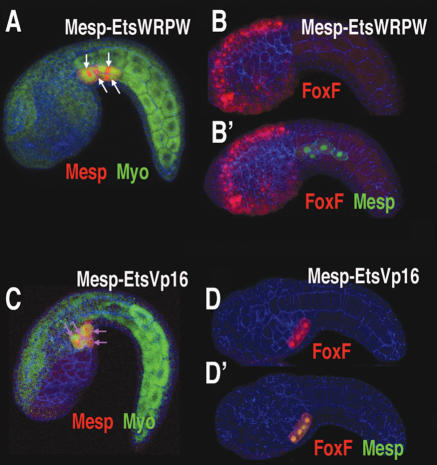
To test the role of Ets1/2 in heart formation, constitutive activator and repressor forms of the protein were selectively expressed in the B7.5 lineage using the Mesp enhancer. The repressor form, EtsWRPW, causes a block in heart migration, so that both the anterior and posterior B7.5 lineages remain in the tail (cf. arrows in Figs. 5A and 1D). The anterior lineage fails to activate the FoxF enhancer (cf. Figs. 5B′ and 3C″). Additionally, most Mesp–EtsWRPW transgenic juveniles do not form a heart after metamorphosis (Supplementary Table S1; Supplementary Movie S1). Conversely, a constitutively activated form of Ets1/2, EtsVp16, causes both lineages to migrate into the head (Fig. 5C, arrows), activate the FoxF enhancer (Fig. 5D,D′) and express the heart lineage marker Hand-like (Supplementary Fig. S1).
Figure 5.
Ets1/2 activity drives heart specification. (A,B,B′) Tailbud embryos (lateral view) that were electroporated with a Mesp–EtsWRPW fusion gene and Mesp-RFP (red) (A) or Mesp-GFP (green) and FoxF-RFP (red) (B,B′). (B) Red channel. (B′) Red and green channels. (C,D,D′) Same as A, B, and B′ except embryos were electroporated with a Mesp–EtsVp16 fusion gene. (D) Note that FoxF-RFP reporter expression in the B7.5 lineage is greatly increased relative to expression in the epidermis, to the extent that epidermal staining can no longer be visualized simultaneously. Anterior is to the left in all panels.
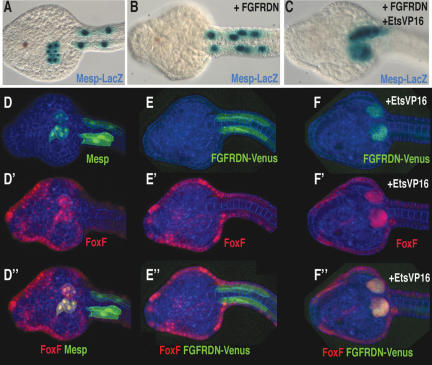
Epistasis assays were conducted to determine whether Ets1/2 acts downstream from FGF signaling (and the FGF receptor) to control heart formation. As seen previously, a dominant-negative form of the FGF receptor (FGFRDN) causes a block in heart cell migration and differentiation when expressed throughout the heart field using the Mesp B7.5 enhancer (Fig. 6, cf. B,E″ and A,D″). Migration and FoxF reporter gene activity were restored when both EtsVp16 and FGFRDN were simultaneously expressed in the B7.5 lineage (Fig. 6, cf. C,F″ and B,E″). Thus, EtsVp16 effectively bypasses the block in FGF signaling, suggesting that Ets1/2 activation is central to heart cell specification.
Figure 6.
Ets1/2 activation bypasses the requirement for FGF signaling in heart specification. (A–C) Transgenic Mesp-LacZ embryos. (B) Embryo coelectroporated with the Mesp–FGFRDN construct. (C) Embryo coelectroporated with Mesp–FGFRDN and Mesp–EtsVp16 constructs. (D,D′,D″) Transgenic Mesp-GFP, FoxFRFP embryo. (D) Green channel. (D′) Red channel. (D″) Green and red channels. (E,E′,E″) Transgenic Mesp–FGFRDN–Venus, FoxF-RFP embryo. (E) Green channel. (E′) Red channel. (E″) Green and red channels. (F) Transgenic Mesp–FGFRDN–Venus, Mesp–EtsVp16, FoxF-RFP embryo. (F) Green channel. (F′) Red channel. (F″) Green and red channels. Note that membrane localization of FGFRDN–Venus is lost in migrating cells, compared with E. All embryos were fixed at the prehatching larval stage (12H); ventral views, anterior to the left in all panels.
Symmetric activation of Ets1/2 drives compartmentalization of the heart
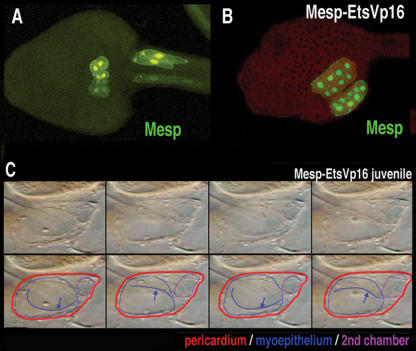
Symmetric activation of Ets1/2 in the B7.5 lineage doubles the number of heart progenitor cells by transforming the proximal tail muscle lineage into supernumerary heart cells (e.g., Fig. 5D′). To determine the effect of this transformation on subsequent development we traced heart lineage cell behavior and heart morphology in larval and juvenile stages, respectively. In wild-type larvae, heart precursor cells migrate to the ventral mid-line and undergo a characteristic asymmetric cell division (Fig. 7A). Typical Mesp–EtsVp16 transgenic tad-poles display some disruption of normal cardiac cell behavior (e.g., see Figs. 5C, 6F). However, supernumerary heart cells are capable of completing characteristic migration and cell division (Fig. 7, cf. B and A). The newly recruited caudal heart lineage remains tightly associated with their rostral sisters. During metamorphosis, the heart rudiment differentiates into a single pulsatile myocardial compartment enclosed within a pericardial sheath (Supplementary Movie S2). Juveniles derived from tadpoles expressing Mesp–EtsVp16 often exhibit disorganized hearts and contain excess cardiac tissue (Supplementary Table S1). Some of the juveniles derived from tadpoles expressing the Mesp–EtsVp16 transgene display a far more dramatic phenotype. Nine percent (46/ 542) contain beating hearts with two distinct myocardial compartments within a single pericardium (Fig. 6C; Supplementary Fig. S2; Supplementary Movies S3–S6). The two compartments can function in synchrony to drive blood flow efficiently through the juvenile body cavity. The clear functional connection between these compartments is indicated by the movement of individual blood cells from one compartment into the other before exiting into the general circulation (Supplementary Movies S3–S5). The independence of the two compartments is made evident by periodic bouts of asynchronous beating (Supplementary Movies S3–S6).
Figure 7.
Supplemental heart progenitor cells generate a second myocardial compartment. (A) Transgenic Mesp-GFP tadpole, ventral view. (B) Transgenic Mesp-GFP, Mesp–EtsVp16 tadpole, ventra–lateral view. (C) Sequential frames from a movie of a Mesp–EtsVp16 transgenic juvenile heart (Supplementary Movie S3). In the bottom row, the pericardium is outlined in red and the myoepithelium is outlined in blue. The blue line indicates a peristaltic contractile wave visualized as it meets the plane of focus. The second chamber is outlined in purple. (Second and fourth panels) Note how rhythmic expansion of the small upper chamber is synchronous with progression of the peristaltic wave within the larger lower compartment (blue arrows). See Supplementary Movies for dynamic visualization of the distinct heart phenotypes; independent contraction of the two compartments is particularly evident in Supplementary Movies S4–S6.
Discussion
Gene network for Ciona heart specification
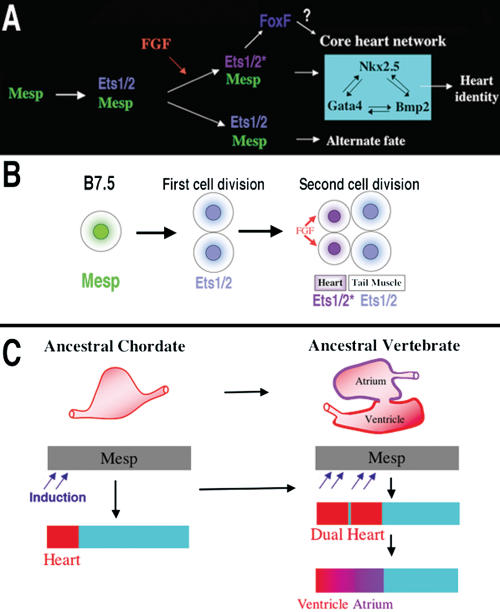
We propose a provisional gene network governing the earliest phases of heart specification (summarized in Fig. 8A,B). Mesp acts through Ets1/2 to create a field of potential heart cells. FGF signaling initiates cardiac specification within a subset of cells in this field through the localized induction of Ets1/2 activity. Activated Ets1/2 then either directly or indirectly regulates cardiac target genes such as Ci-FoxF, Ci-Nkx, and Ci-GataA (Ciona orthologs of vertebrate FoxF, Nkx2.5, and Gata4).
Figure 8.
Models for the heart specification network and chordate heart evolution. (A) Summary of the gene network controlling heart specification in Ciona. Mesp drives expression of Ets1/2 in all descendants of the B7.5 blastomeres. FGF signaling activates Ets1/2 in the rostral daughters, leading to the expression of FoxF and ultimately to the deployment of the heart differentiation cassette. (B) Summary diagram illustrating heart specification events on the cellular level. (C) Diagram illustrating a model of chordate heart evolution. According to this model, expansion of induction within a broad heart field led to the emergence of a dual heart phenotype (as illustrated experimentally through manipulation of Ets1/2 activation in Ciona embryos). In basal vertebrates, this transitional organ was patterned and modified to form two distinct chambers.
The identity of the FGF ligand involved in Ciona heart specification remains unresolved. However, there is mounting evidence supporting a role for FGF9. There are only six FGF genes in the Ciona genome (Imai et al. 2004). Of these, only FGF9 is expressed in cells neighboring the B7.5 lineage at the time of induction. At the early neurula stage, FGF9 is expressed in a stripe of epidermis just anterior/ventral to the dividing B7.5 lineage cells (K. Imai, pers. comm.). This expression domain is complementary to the plane of division in the neighboring B7.5 lineage cells and would place the FGF9-expressing cells in contact with the rostral heart precursors and not their caudal sister cells. Studies of FGF9 function in Ciona neural induction indicate that the extent of cell–cell contact between the secreting and target cells is a decisive factor (Tassy et al. 2006). Thus, FGF9 may provide the spatial cue for asymmetric activation of Ets1/2 in the heart lineage. Morpholino knockdown of Ci-FGF9 disrupts expression of heart lineage markers, further supporting a role for this factor in heart induction (Imai et al. 2006).
Mesp and Ets1/2 may have a conserved role in establishing the chordate heart field
FGF signaling is essential for early heart specification in vertebrate embryos. Disruption of FGF signaling through drug treatment or dominant-negative receptors prevents expression of heart marker genes (Reifers et al. 2000; Alsan and Schultheiss 2002). However, the factors that mediate the response to FGF during heart specification remain undefined. The present study suggests that Mesp mediates competence to respond to FGF during early heart specification by up-regulating Ets1/2. In particular, the bypass of FGFRDN with constitutively activated Ets1/2 indicates that FGF signaling promotes heart specification through a transcriptional path centered on Ets1/2 activation. Perhaps a similar mechanism applies to early cardiac specification in vertebrates.
In early vertebrate embryos, Mesp is broadly expressed in the nascent mesoderm prior to gastrulation (Saga et al. 1996; Sawada et al. 2000). Previous work on Mesp function in both mice and Ciona demonstrate that Mesp has a conserved role in heart specification (Kitajima et al. 2000; Satou et al. 2004; Davidson et al. 2005). The ease of inducing ectopic heart tissue in vertebrate embryos through disruption of Wnt signaling indicates that much of the naïve mesoderm is competent to form heart tissue (Schneider and Mercola 2001; Eisenberg and Eisenberg 2004). Taken together, these lines of evidence support the hypothesis that Mesp creates a broad potential heart field in the nascent mesoderm through regulation of Ets family transcription factors.
An asymmetric cell division is associated with Ciona heart specification
We have visualized an intriguing asymmetric cell division that distinguishes cardioblasts and tail muscle cells within the B7.5 lineage (e.g., Fig. 1B). This asymmetry appears to require RTK signaling downstream from FGF. Treatment with U0126 at 5H seems to disrupt the asymmetry of this division, as does targeted expression of FGFRDN and EtsVp16 (Figs. 3A, 4F′, 5D). However, treatment with U0126 at 6H, as well as targeted expression of EtsWRPW, disrupts heart specification without any apparent affect on this asymmetric division (Figs. 3B, 5A,B′; Supplementary Fig. S1). A more systematic analysis of how manipulations of FGF/RTK signaling effect this cell division pattern is now underway.
The functional importance of this asymmetry remains unclear. The associated loss of myoplasm from the heart precursors is not required for the initial phases of heart development (Fig. 1G). Interestingly, a similar asymmetry is observed in other blastomeres undergoing FGF9-mediated induction in early tunicate embryos (Nishida 2005). Future work will focus on how this asymmetry contributes to proper heart development, and whether the link between FGF and asymmetric cell division is also present during vertebrate heart development.
Symmetrical Ets1/2 activation can convert the anterior tail muscle lineage into a supplemental myocardial compartment
Targeted manipulations of Ets1/2 demonstrate that cell fate decisions within the B7.5 lineage hinge on Ets1/2 activation. Most dramatically, expression of a constitutively activated form of Ets1/2 in the B7.5 lineage transforms the anterior tail muscle lineage into heart. Doubling of the heart rudiment not only generates a larger heart field but can also lead to the formation of a functional dual-compartment heart. This two-compartment phenotype is clearly distinct from the supplemental heart tissue resulting from manipulations of Mesp activity, whereby migration defects generate a separate, disorganized mass of beating heart tissue in the tail rudiment (Davidson et al. 2005). This phenotype is also distinct from cardia bifida, in that the two compartments are contained in a single pericardium and constitute a single tube. We have defined the novel compartment on a strictly functional level, namely the ability to beat independently of the original compartment. There is no reason to expect that symmetrical activation of Ets1/2 would generate novel structural complexity, such as valves, or genetic complexity, such as chamber-specific gene expression. The mechanisms by which the expanded heart progenitor field forms two compartments remain ambiguous. The newly recruited posterior cardioblasts may fail to fuse completely with their anterior sisters, or there may be intrinsic mechanisms of cell counting, so that the expanded progenitor cells separate into two groups. A clear understanding of two-compartment heart formation requires an in-depth analysis of heart morphogenesis in wild-type Ciona juveniles.
Evolutionary origins of the multichambered vertebrate heart
Our findings support the hypothesis that a key transition in the emergence of dual-chambered hearts in the ancestral vertebrate involved recruitment of additional heart precursor cells (Fig. 8C). All extant vertebrate species have hearts with at least two chambers. In basal vertebrates (lamprey and teleosts), the heart already contains both ventricular and atrial chambers. Developmental studies indicate that the left ventricle represents the ancestral chordate heart compartment (Christoffels et al. 2004; Buckingham et al. 2005; Simoes-Costa et al. 2005). Progenitor cells of the atrium lie posterior to the ventricular field and will revert to a ventricular fate in the absence of retinoic acid signals or atrial-specific gene expression (Hochgreb et al. 2003). Modularity in the cis-regulatory elements of vertebrate Nkx2.5 genes suggests that new compartments arose in a “progressive” manner (Schwartz and Olson 1999). There are no species, in the extant or fossil fauna, representative of the transitional stage between the dual chambered heart of basal vertebrates and single-compartment hearts of invertebrate chordates, such as Ciona. Our study demonstrates that subtle changes in inductive signaling are sufficient to increase cardiac recruitment within a broad heart field (delineated by Mesp expression). Furthermore, this recruitment can potentiate the formation of new compartments through an intrinsic mechanism. This primitive multicompartment organ would then be gradually modified to exploit the selective advantage of independent inflow and outflow compartments (Moorman and Christoffels 2003; Simoes-Costa et al. 2005), leading to the formation of an ancestral dual-chambered vertebrate heart. Recent work indicates that the subsequent evolution of the right ventricle and outflow tract may also depend on the recruitment of a “secondary” progenitor population, neighboring the ancestral ventricular/atrial field (Christoffels et al. 2004).
Implications of the dual heart phenotype for mechanisms of evolutionary change
Compartmentalization of the Ciona heart in transgenic EtsVp16 juveniles provides a dramatic demonstration of how subtle changes in embryonic gene activity can potentiate the formation of novel adaptive traits. The evolutionary diversification of external appendages, including beak morphology in Darwin's finches, have also been mimicked experimentally through perturbing gene activity within embryonic progenitor fields (Sanz-Ezquerro and Tickle 2003; Abzhanov et al. 2004; Harris et al. 2005; Kassai et al. 2005). These cases illustrate how shifts in proliferation or recruitment patterns within embryonic progenitor fields can generate novel structural complexity. Our study differs from these previous examples in that it involves an internal organ and relies primarily on shifts in patterns of recruitment rather than growth. Increased proliferation of primordia is likely to be highly constrained within the more rigid confines surrounding internal organs. Therefore, altering the distribution of progenitor cells represents a more suitable mechanism for potentiating diversification of internal morphology. We propose that variation in patterns of progenitor cell recruitment may have a general role in the evolution of novel internal structures, particularly those arising from interconnected fields, such as the pancreas, liver, and lung (Deutsch et al. 2001; Serls et al. 2005; Tremblay and Zaret 2005).
Materials and methods
Ascidians: collection, handling, and experimental techniques
Collection, rearing, fertilization, dechorionation, in situ hybridization, electroporation, and lacZ staining were conducted as described previously (Corbo et al. 1998; Davidson et al. 2005). Treatments with U0126 and cytochalasin also followed published protocols (Kim and Nishida 2001). For disassociation, embryos were allowed to settle at the bottom of a 2-mL tube (USA Scientific #1480-2700) and rinsed in calcium/magnesium-free seawater (CMF; 449 mM NaCl, 9 mM KCl, 33 mM Na2SO4, 2.15 mM NaHCO3, 10 mM Tris at pH 8, 2.5 mM EGTA) three times. The embryos were then disassociated by repeated passage through a Pasteur Pipette for 3 min. Isolated cells were then spun down at 1000 rpm for 2 min in a tabletop centrifuge and resuspended in CMF. Cells were then passed through a 35-μm mesh (the cap from Falcon tube #352235) directly into 3-mL dishes of seawater with 0.1% BSA, BSA + 100 ng/mL bFGF (Sigma, F0291), or 100 ng/mL U0126 (EMD Biosciences, 662,005). Rearing of transgenic juveniles involved transferring dechorionated embryos at the time when twitching of the tail is first observed to uncoated petri dishes filled with antibiotic-filtered seawater (50 μg/mL ampicillin and kanamycin). Water was changed after fixation and rotation of juvenile organ rudiments (48 h after transfer) and subsequently every 24–48 h.
Immunohistochemistry
Ciona embryos were fixed in 4% paraformaldehyde, 0.5 M NaCl, 0.1 M MOPS (pH 7.5), 2 mM MgSO4, and 1 mM EGTA at 4°C overnight. Staining of dpErk was carried out sequentially with mouse anti-dpErk (1:1000, Sigma, M9692) using TSA-fluorescein amplification and rabbit anti-GFP (1:200, Molecular Probes).
Construction of vectors for transgenesis
The RFP used in this study (optimized for use in Ciona) was obtained from the Zeller laboratory (Zeller et al. 2006). Vectors and techniques used to subclone new constructs are as described previously (Davidson et al. 2005); further details are provided below.
Recombinant Ets1/2, DN–Ras, and FGFRDN constructs
The Ets1/2 DNA-binding domain was amplified from an Ets1/2 EST clone (R1CiGC01h16) obtained from the Ciona intestinalis Gene Collection Release 1 (Satou et al. 2005) using the following primers (lower case indicates restriction site and flanking base pairs): EtsDBDf, aaagctagcGCCTATACTGACACCTTT GAAGCC; EtsDBDr, aaaactagtATCATCACTTAGTCGATCT TCAGGC. It was then fused in-frame into modified pCES vectors containing the full-length Mesp enhancer and 3′ activator (Vp16) or repressor (WRPW) domains (modified from Mesp–MespVp16) (Davidson et al. 2005). The WRPW domain consists of the following sequence replacing Vp16 by ligation at the Nhe1 and Spe1 sites in Mesp–MespVp16: CAGATCAAGGAG GAGGAGCAGCCCTGGCGGCCCTGGTAATAA.
The dominant-negative form of Ci-FGFR (R1CiGC32j14) lacking the intracellular domain was generated using the following primer pair: 5′-aatggcggccgcaATGATACAACTACA AAATACGTTT-3′ and 3′-ggaattcTTAGTTCCCGAACAACA TCACC-5′.
The DN–Ras construct contained the dominant-negative mutant form (N17) of human c-Ha-Ras1. Both FGFRDN and DN–Ras were fused in-frame downstream from the Mesp enhancer, replacing the lacZ coding region of Mesp-lacZ. The FGFRDN Venus construct was constructed by fusing Venus YFP (Nagai et al. 2002) to the 3′ end of the Mesp–FGFRDN construct employing EcoR1 and Blp1 sites.
Confocal microscopy
Transgenic embryos with GFP/RFP-expressing cells were fixed for1hin 0.3% formaldehyde seawater, stained with Alexa Fluor 635 phalloidin (per manufacturer's protocol; Invitrogen), mounted in Vectashield mounting medium (Vector Laboratories), and stored at −20°C. Confocal images were obtained on a Leica TCS SL1 laser scanning confocal microscope. Images were processed using the BitPlane Imaris 3.3 software package.
Acknowledgments
We thank Yutaka Satou, Karou Imai, Eric Olson, Richard Losick, and Nipam Patel for their comments and suggestions, as well as Bob Zeller for providing the optimized RFP base construct. This study was funded by grants from the NIH and NSF to M.L. and a NIH post-doctoral fellowship to B.D.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1467706.
References
- Abzhanov, A., Protas, M., Grant, B.R., Grant, P.R., Tabin, C.J. Bmp4 and morphological variation of beaks in Darwin's finches. Science. 2004;305:1462–1465. doi: 10.1126/science.1098095. [DOI] [PubMed] [Google Scholar]
- Alsan, B.H., Schultheiss, T.M. Regulation of avian cardiogenesis by Fgf8 signaling. Development. 2002;129:1935–1943. doi: 10.1242/dev.129.8.1935. [DOI] [PubMed] [Google Scholar]
- Bertrand, V., Hudson, C., Caillol, D., Popovici, C., Lemaire, P. Neural tissue in ascidian embryos is induced by FGF9/16/20, acting via a combination of maternal GATA and Ets transcription factors. Cell. 2003;115:615–627. doi: 10.1016/s0092-8674(03)00928-0. [DOI] [PubMed] [Google Scholar]
- Buckingham, M., Meilhac, S., Zaffran, S. Building the mammalian heart from two sources of myocardial cells. Nat. Rev. Genet. 2005;6:826–835. doi: 10.1038/nrg1710. [DOI] [PubMed] [Google Scholar]
- Christoffels, V.M., Burch, J.B., Moorman, A.F. Architectural plan for the heart: Early patterning and delineation of the chambers and the nodes. Trends Cardiovasc. Med. 2004;14:301–307. doi: 10.1016/j.tcm.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Corbo, J.C., Fujiwara, S., Levine, M., Di Gregorio, A. Suppressor of hairless activates Brachyury expression in the Ciona embryo. Dev. Biol. 1998;203:358–368. doi: 10.1006/dbio.1998.9067. [DOI] [PubMed] [Google Scholar]
- Cripps, R.M., Olson, E.N. Control of cardiac development by an evolutionarily conserved transcriptional network. Dev. Biol. 2002;246:14–28. doi: 10.1006/dbio.2002.0666. [DOI] [PubMed] [Google Scholar]
- Davidson, B., Christiaen, L. Linking chordate gene networks to cellular behavior in ascidians. Cell. 2006;124:247–250. doi: 10.1016/j.cell.2006.01.013. [DOI] [PubMed] [Google Scholar]
- Davidson, B., Levine, M. Evolutionary origins of the vertebrate heart: Specification of the cardiac lineage in Ciona intestinalis . Proc. Natl. Acad. Sci. 2003;100:11469–11473. doi: 10.1073/pnas.1634991100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson, B., Shi, W., Levine, M. Uncoupling heart cell specification and migration in the simple chordate Ciona intestinalis . Development. 2005;132:4811–4818. doi: 10.1242/dev.02051. [DOI] [PubMed] [Google Scholar]
- Dehal, P., Satou, Y., Campbell, R.K., Chapman, J., Degnan, B., De Tomaso, A., Davidson, B., Di Gregorio, A., Gelpke, M., Goodstein, D.M., et al. The draft genome of Ciona intestinalis: Insights into chordate and vertebrate origins. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- Deutsch, G., Jung, J., Zheng, M., Lora, J., Zaret, K.S. A bipotential precursor population for pancreas and liver within the embryonic endoderm. Development. 2001;128:871–881. doi: 10.1242/dev.128.6.871. [DOI] [PubMed] [Google Scholar]
- Eisenberg, L.M., Eisenberg, C.A. An in vitro analysis of myocardial potential indicates that phenotypic plasticity is an innate property of early embryonic tissue. Stem Cells Dev. 2004;13:614–624. doi: 10.1089/scd.2004.13.614. [DOI] [PubMed] [Google Scholar]
- Wnt signal transduction and the formation of the myocardium. Dev. Biol. 2006;293:305–315. doi: 10.1016/j.ydbio.2006.02.014. ———. [DOI] [PubMed] [Google Scholar]
- Foley, A.C., Mercola, M. Heart induction by Wnt antagonists depends on the homeodomain transcription factor Hex. Genes & Dev. 2005;19:387–396. doi: 10.1101/gad.1279405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfon, M.S., Carmena, A., Gisselbrecht, S., Sackerson, C.M., Jimenez, F., Baylies, M.K., Michelson, A.M. Ras pathway specificity is determined by the integration of multiple signal-activated and tissue-restricted transcription factors. Cell. 2000;103:63–74. doi: 10.1016/s0092-8674(00)00105-7. [DOI] [PubMed] [Google Scholar]
- Harris, M.P., Williamson, S., Fallon, J.F., Meinhardt, H., Prum, R.O. Molecular evidence for an activator-inhibitor mechanism in development of embryonic feather branching. Proc. Natl. Acad. Sci. 2005;102:11734–11739. doi: 10.1073/pnas.0500781102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano, T., Nishida, H. Developmental fates of larval tissues after metamorphosis in ascidian Halocynthia roretzi. I Origin of mesodermal tissues of the juvenile. Dev. Biol. 1997;192:199–210. doi: 10.1006/dbio.1997.8772. [DOI] [PubMed] [Google Scholar]
- Hochgreb, T., Linhares, V.L., Menezes, D.C., Sampaio, A.C., Yan, C.Y., Cardoso, W.V., Rosenthal, N., Xavier-Neto, J. A caudorostral wave of RALDH2 conveys anteroposterior information to the cardiac field. Development. 2003;130:5363–5374. doi: 10.1242/dev.00750. [DOI] [PubMed] [Google Scholar]
- Imai, K.S., Hino, K., Yagi, K., Satoh, N., Satou, Y. Gene expression profiles of transcription factors and signaling molecules in the ascidian embryo: Towards a comprehensive understanding of gene networks. Development. 2004;131:4047–4058. doi: 10.1242/dev.01270. [DOI] [PubMed] [Google Scholar]
- Imai, K.S., Levine, M., Satoh, N., Satou, Y. Regulatory blueprint for a chordate embryo. Science. 2006;312:1183–1187. doi: 10.1126/science.1123404. [DOI] [PubMed] [Google Scholar]
- Kassai, Y., Munne, P., Hotta, Y., Penttila, E., Kavanagh, K., Ohbayashi, N., Takada, S., Thesleff, I., Jernvall, J., Itoh, N. Regulation of mammalian tooth cusp patterning by ectodin. Science. 2005;309:2067–2070. doi: 10.1126/science.1116848. [DOI] [PubMed] [Google Scholar]
- Kim, G.J., Nishida, H. Role of the FGF and MEK signaling pathway in the ascidian embryo. Dev. Growth Differ. 2001;43:521–533. doi: 10.1046/j.1440-169x.2001.00594.x. [DOI] [PubMed] [Google Scholar]
- Kitajima, S., Takagi, A., Inoue, T., Saga, Y. MesP1 and MesP2 are essential for the development of cardiac mesoderm. Development. 2000;127:3215–3226. doi: 10.1242/dev.127.15.3215. [DOI] [PubMed] [Google Scholar]
- Lee, K.H., Evans, S., Ruan, T.Y., Lassar, A.B. SMAD-mediated modulation of YY1 activity regulates the BMP response and cardiac-specific expression of a GATA4/5/6-dependent chick Nkx2.5 enhancer. Development. 2004;131:4709–4723. doi: 10.1242/dev.01344. [DOI] [PubMed] [Google Scholar]
- Lien, C.L., McAnally, J., Richardson, J.A., Olson, E.N. Cardiac-specific activity of an Nkx2-5 enhancer requires an evolutionarily conserved Smad binding site. Dev. Biol. 2002;244:257–266. doi: 10.1006/dbio.2002.0603. [DOI] [PubMed] [Google Scholar]
- Lockwood, W.K., Bodmer, R. The patterns of wingless, decapentaplegic, and tinman position the Drosophila heart. Mech. Dev. 2002;114:13–26. doi: 10.1016/s0925-4773(02)00044-8. [DOI] [PubMed] [Google Scholar]
- MacLean, D.W., Meedel, T.H., Hastings, K.E. Tissue-specific alternative splicing of ascidian Troponin I isoforms. Redesign of a protein isoform-generating mechanism during chordate evolution. J. Biol. Chem. 1997;272:32115–32120. doi: 10.1074/jbc.272.51.32115. [DOI] [PubMed] [Google Scholar]
- Miya, T., Nishida, H. An Ets transcription factor, HrEts is target of FGF signaling and involved in induction of notochord, mesenchyme, and brain in ascidian embryos. Dev. Biol. 2003;261:25–38. doi: 10.1016/s0012-1606(03)00246-x. [DOI] [PubMed] [Google Scholar]
- Moorman, A.F., Christoffels, V.M. Cardiac chamber formation: Development, genes, and evolution. Physiol. Rev. 2003;83:1223–1267. doi: 10.1152/physrev.00006.2003. [DOI] [PubMed] [Google Scholar]
- Nagai, T., Ibata, K., Park, E.S., Kubota, M., Mikoshiba, K., Miyawaki, A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- Nishida, H. Specification of embryonic axis and mosaic development in ascidians. Dev. Dyn. 2005;233:1177–1193. doi: 10.1002/dvdy.20469. [DOI] [PubMed] [Google Scholar]
- Pandur, P., Lasche, M., Eisenberg, L.M., Kuhl, M. Wnt-11 activation of a non-canonical Wnt signalling pathway is required for cardiogenesis. Nature. 2002;418:636–641. doi: 10.1038/nature00921. [DOI] [PubMed] [Google Scholar]
- Pasini, A., Amiel, A., Rothbacher, U., Roure, A., Lemaire, P., Darras, S. Formation of the ascidian epidermal sensory neurons: Insights into the origin of the chordate peripheral nervous system. PLoS Biol. 2006;4 doi: 10.1371/journal.pbio.0040225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reifers, F., Walsh, E.C., Leger, S., Stainier, D.Y., Brand, M. Induction and differentiation of the zebrafish heart requires fibroblast growth factor 8 (fgf8/acerebellar) Development. 2000;127:225–235. doi: 10.1242/dev.127.2.225. [DOI] [PubMed] [Google Scholar]
- Robb, J.S. Comparative basic cardiology. Grune & Stratton; New York: 1965. [Google Scholar]
- Saga, Y., Hata, N., Kobayashi, S., Magnuson, T., Seldin, M.F., Taketo, M.M. MesP1: A novel basic helix–loop–helix protein expressed in the nascent mesodermal cells during mouse gastrulation. Development. 1996;122:2769–2778. doi: 10.1242/dev.122.9.2769. [DOI] [PubMed] [Google Scholar]
- Sanz-Ezquerro, J.J., Tickle, C. FGF signaling controls the number of phalanges and tip formation in developing digits. Curr. Biol. 2003;13:1830–1836. doi: 10.1016/j.cub.2003.09.040. [DOI] [PubMed] [Google Scholar]
- Satoh, N. The ascidian tadpole larva: Comparative molecular development and genomics. Nat. Rev. Genet. 2003;4:285–295. doi: 10.1038/nrg1042. [DOI] [PubMed] [Google Scholar]
- Satou, Y., Imai, K.S., Satoh, N. The ascidian Mesp gene specifies heart precursor cells. Development. 2004;131:2533–2541. doi: 10.1242/dev.01145. [DOI] [PubMed] [Google Scholar]
- Satou, Y., Kawashima, T., Shoguchi, E., Nakayama, A., Satoh, N. An integrated database of the ascidian, Ciona intestinalis: Towards functional genomics. Zoolog. Sci. 2005;22:837–843. doi: 10.2108/zsj.22.837. [DOI] [PubMed] [Google Scholar]
- Sawada, A., Fritz, A., Jiang, Y.J., Yamamoto, A., Yamasu, K., Kuroiwa, A., Saga, Y., Takeda, H. Zebrafish Mesp family genes, Mesp-a and Mesp-b are segmentally expressed in the presomitic mesoderm, and Mesp-b confers the anterior identity to the developing somites. Development. 2000;127:1691–1702. doi: 10.1242/dev.127.8.1691. [DOI] [PubMed] [Google Scholar]
- Schneider, V.A., Mercola, M. Wnt antagonism initiates cardiogenesis in Xenopus laevis . Genes & Dev. 2001;15:304–315. doi: 10.1101/gad.855601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, R.J., Olson, E.N. Building the heart piece by piece: Modularity of cis-elements regulating Nkx2-5 transcription. Development. 1999;126:4187–4192. doi: 10.1242/dev.126.19.4187. [DOI] [PubMed] [Google Scholar]
- Serls, A.E., Doherty, S., Parvatiyar, P., Wells, J.M., Deutsch, G.H. Different thresholds of fibroblast growth factors pattern the ventral foregut into liver and lung. Development. 2005;132:35–47. doi: 10.1242/dev.01570. [DOI] [PubMed] [Google Scholar]
- Simoes-Costa, M.S., Vasconcelos, M., Sampaio, A.C., Cravo, R.M., Linhares, V.L., Hochgreb, T., Yan, C.Y., Davidson, B., Xavier-Neto, J. The evolutionary origin of cardiac chambers. Dev. Biol. 2005;277:1–15. doi: 10.1016/j.ydbio.2004.09.026. [DOI] [PubMed] [Google Scholar]
- Stathopoulos, A., Tam, B., Ronshaugen, M., Frasch, M., Levine, M. pyramus and thisbe: FGF genes that pattern the mesoderm of Drosophila embryos. Genes & Dev. 2004;18:687–699. doi: 10.1101/gad.1166404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassy, O., Daian, F., Hudson, C., Bertrand, V., Lemaire, P. A quantitative approach to the study of cell shapes and interactions during early chordate embryogenesis. Curr. Biol. 2006;16:345–358. doi: 10.1016/j.cub.2005.12.044. [DOI] [PubMed] [Google Scholar]
- Tokuoka, M., Imai, K.S., Satou, Y., Satoh, N. Three distinct lineages of mesenchymal cells in Ciona intestinalis embryos demonstrated by specific gene expression. Dev. Biol. 2004;274:211–224. doi: 10.1016/j.ydbio.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Tremblay, K.D., Zaret, K.S. Distinct populations of endoderm cells converge to generate the embryonic liver bud and ventral foregut tissues. Dev. Biol. 2005;280:87–99. doi: 10.1016/j.ydbio.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Zeller, R.W., Weldon, D.S., Pellatiro, M.A., Cone, A.C. Optimized green fluorescent protein variants provide improved single cell resolution of transgene expression in ascidian embryos. Dev. Dyn. 2006;235:456–467. doi: 10.1002/dvdy.20644. [DOI] [PubMed] [Google Scholar]