Abstract
The quorum-sensing bacterium Vibrio harveyi produces and responds to three autoinducers (AIs), and this sensory information converges to control the expression of bioluminescence, biofilm formation, type III secretion (TTS), and protease production. The AIs are detected by cognate sensor histidine kinases that all relay phosphate to the shared response regulator LuxO. LuxO indirectly represses the master regulator of quorum sensing, LuxR, through the activation of multiple genes encoding small regulatory RNAs (called qrr genes for Quorum Regulatory RNA). Here we use differential fluorescence induction to identify 50 quorum-sensing-controlled promoters. Some promoters only showed significant responses in the simultaneous presence of all three AIs, while others displayed substantial responses to the individual AIs. A differential response to each AI input state was also observed for qrr and luxR expression and LuxR protein production. Individual cell analyses revealed that, in each case, all the bacteria in the population respond in unison to the various AI inputs. We propose that the V. harveyi quorum-sensing transition is not switch-like but rather operates in a graded manner, and that this signaling arrangement, which uses shared regulatory proteins, nonetheless provides V. harveyi a mechanism to respond uniquely to different AI input states.
Keywords: Quorum sensing, autoinducer, sRNA, DFI
In a process known as quorum sensing, bacteria communicate using secreted signal molecules called autoinducers (AIs) (Waters and Bassler 2005). Quorum-sensing bacteria alter gene expression in response to the accumulation of AIs, which reflects an increase in cell population density. This process is believed to provide bacteria a means to coordinately control the gene expression of the group, giving them multicellular characteristics. Often, bacteria make and respond to multiple AIs. Vibrio harveyi, a free-living marine bacterium, produces at least three distinct AIs to control bioluminescence, biofilm formation, Type III Secretion (TTS), and protease production (Hammer and Bassler 2003; Mok et al. 2003; Henke and Bassler 2004a).
The three V. harveyi AIs are HAI-1, an acyl homoserine lactone; AI-2, a furanosyl-borate-diester; and CAI-1, of unknown structure (Cao and Meighen 1989; Bassler et al. 1994a; Chen et al. 2002; Henke and Bassler 2004b).
Other bacterial species release molecules with HAI-1, AI-2, or CAI-1 activity as judged by a set of V. harveyi reporter strains engineered to respond exclusively to only one AI signal. Interestingly, the different groups of bacteria that produce these three autoinducer activities have varying degrees of relatedness to V. harveyi. To date, HAI-1 has only been shown to be produced by V. harveyi and its most closely known relative, Vibrio parahaemolyticus, suggesting that HAI-1 is an intraspecies signal (Bassler et al. 1997). CAI-1 activity is produced predominantly by vibrios, suggesting it could be an intragenera signal (Henke and Bassler 2004b). Finally, AI-2 production is widely distributed in the bacterial world, suggesting this is a relatively nonspecific, interspecies bacterial communication molecule (Xavier and Bassler 2003). Consistent with this idea, structural studies show that different chemical moieties are used to convey AI-2 information to different bacteria, but the various AI-2s interconvert and are all derived from a common precursor molecule (Chen et al. 2002; Miller et al. 2004).
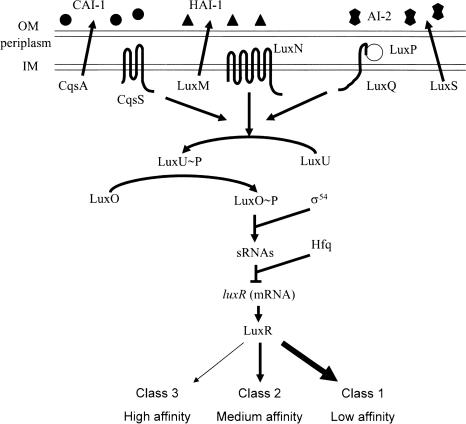
V. harveyi responds to the AIs via a two-component phospho-relay cascade that ultimately controls production of the master regulator LuxR (Fig. 1). HAI-1, CAI-1, and AI-2 are detected by the membrane-bound, two-component sensors LuxN, CqsS, and LuxQ, respectively (Bassler et al. 1993, 1994a; Freeman et al. 2000; Henke and Bassler 2004b). LuxQ requires the periplasmic binding protein LuxP to respond to AI-2 (Bassler et al. 1994a). In the absence of the AIs, the sensors are kinases that send phosphate to the histidine phospho-transfer protein LuxU, which in turn, passes the phosphate to the response regulator LuxO (Bassler et al. 1994b; Freeman and Bassler 1999a,b; Freeman et al. 2000). Phospho-LuxO, together with σ54, activates the expression of the genes encoding five regulatory small RNAs (sRNAs) called Qrr1–5 (Quorum Regulatory RNA) (Lilley and Bassler 2000; Lenz et al. 2004). In conjunction with the RNA chaperone Hfq, the sRNAs destabilize the mRNA encoding the master quorum-sensing regulator LuxR. The presence of the AIs converts the sensors from kinases to phosphatases (Fig. 1), resulting in dephosphorylation and inactivation of LuxO, no qrr sRNA expression, and stabilization of the luxR mRNA, leading to LuxR production. LuxR, directly or indirectly, controls the expression of numerous genes encoding functions involved in processes such as bioluminescence, TTS, biofilm production, and protease expression (Swartzman et al. 1992; Hammer and Bassler 2003; Mok et al. 2003; Henke and Bassler 2004a).
FIGURE 1.
Model of the V. harveyi quorum-sensing system. Three sensory systems converge to control the levels of the master regulator, LuxR. See text for details. Circles, triangles, and double pentagons represent CAI-1, HAI-1, and AI-2, respectively. Arrows denote the direction of phosphate flow in the low-cell-density state.
Preliminary analyses of the above quorum-sensing-controlled genes demonstrated significant regulation in the simultaneous presence of all the AIs, but not under any other AI input state, suggesting that the V. harveyi circuit may function as a coincidence detector. This requirement for all three AIs was hypothesized to be a mechanism to filter out noise generated by other similar molecules in the environment (Mok et al. 2003; Henke and Bassler 2004a). However, the previous screens for quorum-sensing-controlled genes were not saturated and were performed under conditions that would make it difficult to identify genes controlled exclusively by one particular AI. Thus, it remained possible that V. harveyi possesses genes that are controlled by different combinations of AIs. If so, V. harveyi could use the three AIs to extract information about the species composition in the vicinity, and transduce this information into discrete patterns of gene expression.
Here we have identified 50 AI-regulated promoters controlling at least 71 genes from V. harveyi and examined their responses to different combinations of AIs. We find genes that respond to each single AI input as well as genes that respond only when all the signals are present simultaneously. We show that the differential responses are a consequence of the different binding affinities of the various quorum-sensing-regulated promoters for LuxR. Differential AI responses were also observed for qrr expression and LuxR protein production. Analysis of gene expression in response to AIs in individual cells showed that the quorum-sensing response of V. harveyi is graded and not switch-like. These results suggest that V. harveyi discriminates between the different AIs and couples the presence or absence of particular signals into unique patterns of gene expression.
Results
A screen to identify autoinducer-regulated genes
Previous analyses of autoinducer (AI)-regulated genes in V. harveyi indicated that the V. harveyi quorum-sensing circuit differentiates between the simultaneous presence of all autoinducers (HAI-1 + AI-2 + CAI-1) and all other possible AI input states (Mok et al. 2003; Henke and Bassler 2004a). These screens were conducted in V. harveyi strains lacking one AI, and differential gene expression was monitored in the presence and absence of that AI. Because two other AIs were always present, the screens enabled the identification of genes differentially controlled in the presence of two versus three AIs. This experimental design, while having the potential to identify genes regulated exclusively by only one AI, likely enriched for genes that are coordinately controlled by all the AIs as they function synergistically.
To extend the above analysis, differential fluorescence induction (Valdivia and Falkow 1996; Rediers et al. 2005) and fluorescence activated cell sorting (FACS) were used to screen a library of random V. harveyi genomic DNA fragments from the wild-type strain BB120 fused to a promoterless green fluorescent protein (gfp) in the presence and absence of exogenously supplied AIs. Because the V. harveyi genome is not sequenced, more convenient procedures such as microarrays could not be used. The screen was carried out in the V. harveyi luxM, luxS (HAI-1−, AI-2−) double-mutant strain KM413. A triple AI mutant, while available, was not used because the third AI, CAI-1, has only a minor influence on quorum-sensing-controlled gene expression (see below) in typical laboratory conditions (Henke and Bassler 2004b), the CAI-1 molecule has not yet been purified, and the triple HAI-1−, AI-2−, CAI-1− mutant has a modest growth defect, while the double HAI-1−, AI-2− mutant does not.
Cells expressing GFP were isolated using repeated rounds of FACS in the absence and presence of saturating levels of both HAI-1 and AI-2 to simulate low and high cell density, respectively. We reasoned that any gene that responds to HAI-1 or AI-2 alone would also respond to both HAI-1 and AI-2 together because the AIs act synergistically. The sorted GFP-producing cells were grown into colonies, individually arrayed in wells of microtiter dishes, and again screened for differential GFP production in the presence and in the absence of both HAI-1 and AI-2. Eight thousand colonies were screened, half from each of the two growth conditions. Sixty-four unique genomic inserts showed greater than twofold regulation by AI. These inserts corresponded to 50 unique promoters that regulate at least 71 genes (including potential operons). Of the 50 promoters isolated, 11 were induced by AI, and 39 were repressed. The genomic inserts were sequenced, and the closest homolog in the database was determined by BLAST (Table 1). Fourteen of the sequences had no homology with any available DNA sequence.
TABLE 1.
AI regulated promoters
The isolation of three of the five qrr sRNA promoters (qrr2, qrr3, and qrr4) validated this strategy as a means to identify quorum-sensing-regulated promoters. Two qrr sRNA promoters (qrr1 and qrr5) were not isolated by this method. Complementary analyses have demonstrated that qrr1 and qrr5 are only weakly expressed and do not drive detectable levels of gfp expression in this plasmid system (C.K. Tu and B.L. Bassler, unpubl.). Promoters to some other known quorum-sensing-regulated genes were also not isolated in this screen, including those for luxR and luxC. We note, however, that the screen has not been saturated.
Promoters to genes encoding a variety of functions were obtained (Table 1), suggesting a role for quorum sensing in the regulation of a wide array of cellular activities in V. harveyi. Consistent with our previous results, genes located in a TTS operon that had been predicted to exist in V. harveyi based on the V. parahaemolyticus genome sequence were identified as repressed by AI (clone 209) (Henke and Bassler 2004a). As in Vibrio fischeri, AI-repressed flagellar biosynthesis genes (clones 349, 353) were identified (Lupp and Ruby 2005). Other identified genes encoded regulatory factors including a σ-cross-reacting protein with homologs in organisms spanning bacteria to humans (clone 222) (Ueshima et al. 1992; Chang and Gilbert 1997; Scott et al. 1997), a putative response regulator (clone 214), and two proteins possessing GGDEF motifs (clones 275, 342). These motifs direct the synthesis of the intracellular second messenger cyclic-di-GMP that regulates colony morphology, promotes biofilm formation, and inhibits motility (Romling et al. 2005). AI addition represses biofilm production in vibrios, and repression of these GGDEF proteins could be associated with this phenotype (Hammer and Bassler 2003; Zhu and Mekalanos 2003). Several promoters for metabolic enzymes were AI-repressed, indicating that information about the surrounding community is used by V. harveyi to adjust its overall metabolic program. Finally, a subset of the promoters mapped to the anti-sense strand of putative ORFs, suggesting that these may be potential sRNA regulators. Similarly, a recent screen for novel regulatory sRNAs in Escherichia coli identified putative cis-encoded antisense sRNAs (Kawano et al. 2005).
LuxR controls all AI-regulated genes
The AI response of every gene previously shown to be a member of the V. harveyi quorum-sensing regulon requires LuxR (Mok et al. 2003; Henke and Bassler 2004a). To test if the promoters identified here also lie downstream from LuxR in the quorum-sensing cascade, we measured the expression of each promoter–gfp fusion in a luxR-null mutant and a luxO-null mutant that constitutively produces LuxR. Almost all of the promoters showed a pattern consistent with regulation by LuxR (Supplementary Fig. 1). Specifically, AI-repressed promoters exhibited higher expression in the luxR mutant than in the luxO mutant, while AI-activated promoters showed the opposite pattern of expression. These results imply that, irrespective of whether a gene in the V. harveyi quorum-sensing cascade is activated or repressed by AI, LuxR is required for its regulation. A small number of AI-repressed promoters showed only minimal expression differences in these two strains. Notably, these were the promoters that we isolated with the weakest regulation by AI (approximately twofold), making reliable determinations of differences in expression difficult.
LuxR directly regulates nine promoters
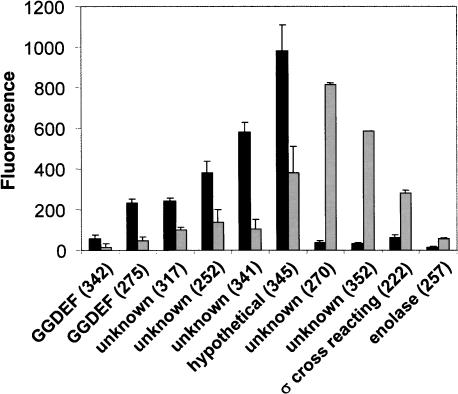
To determine if any of the AI-controlled promoters is directly regulated by LuxR, we measured the expression of each of the promoter–gfp fusions in an E. coli strain harboring a cosmid that constitutively expresses luxR. Ten promoters showed at least 2.5-fold control by LuxR; six were repressed, and four were activated (Fig. 2). LuxR regulation varied from 2.5 -fold repression to >20-fold activation. To verify a direct role for LuxR, the 10 promoter DNA sequences that displayed significant regulation by LuxR in E. coli were assayed for LuxR binding in gel mobility shift assays with purified LuxR protein. LuxR bound the DNA encoding all of the promoters except that from clone 257 (data not shown). We therefore conclude that these nine promoters are directly regulated by LuxR in V. harveyi.
FIGURE 2.
LuxR directly regulates quorum-sensing-controlled genes. Expression of AI-regulated promoters fused to gfp was measured in an E. coli strain harboring a vector carrying luxR (light bars) or a vector control (black bars). Error bars indicate the standard deviations.
Sequence alignments of the cloned V. harveyi DNA regions exhibiting direct regulation by LuxR failed to identify any consistent motif. Consistent with this result, previous studies identifying putative LuxR/HapR (HapR is the Vibrio cholerae LuxR homolog)-binding sites indicate that these LuxR-type proteins have rather promiscuous DNA-binding capabilities (Kovacikova and Skorupski 2002; Lin et al. 2005). LuxR could be directly involved in the regulation of other promoters identified in this study, but this regulation could not be observed in E. coli, possibly due to a requirement for additional V. harveyi-specific factors.
Different AI inputs elicit discrete responses from quorum-sensing-regulated promoters
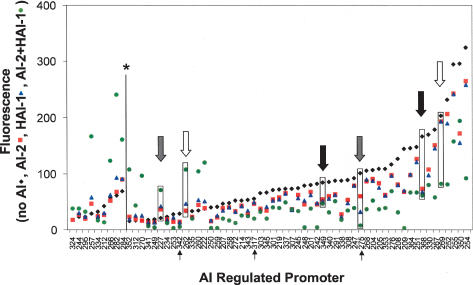
In V. harveyi, all quorum-sensing information is funneled to LuxO and, through the sRNAs, to LuxR, obscuring any obvious mechanism for differential regulation of genes in response to the different AIs (Lenz et al. 2004). To test whether V. harveyi quorum-sensing-controlled genes have distinct responses to the different combinations of AI inputs, we measured the expression of the promoter–gfp fusions isolated in the DFI screen in V. harveyi strain KM413 (HAI-1−, AI-2−) grown to stationary phase under four different AI conditions. The promoter–gfp fusions are arranged along the X-axis of Figure 3 in the order of lowest fluorescence production in the absence of AI on the left to highest fluorescence production on the right. Plotted on the Y-axis of Figure 3 is the fluorescence from each promoter fusion in four test conditions: no addition of AIs (black diamonds), AI-2-only (red squares), HAI-1 only (blue triangles), and HAI-1 and AI-2 together (green circles). Repression by AI results in points below the black triangles, while activation yields points above them.
FIGURE 3.
Differential AI-regulation of quorum-sensing-controlled genes. Fluorescence production from the promoter gfp fusions isolated in this study was measured in the absence of AI (black diamonds), or in the presence of 10 μM AI-2 (red squares), 10 μM HAI-1 (blue triangles), or 10 μM each AI-2 and HAI-1 (green circles). Each AI-regulated promoter is depicted on the X-axis with the values for fluorescence emission plotted on the Y-axis. The fluorescence of the promoters to the left of the line marked with the asterisk (*) were multiplied by a factor of 5 so that they could be visualized on the same axis. White, gray, and black arrows show examples of class 1, 2, and 3 regulation, respectively. The four values corresponding to each arrow are boxed. The black arrows below the X-axis indicate the fusions analyzed in Figure 8.
The promoters displayed three types of responses to the AIs. First, more than half of the promoters exhibited coincidence behavior, which we term the class 1 response: Only when both HAI-1 and AI-2 were added simultaneously did significant activation or repression occur (examples of one activated and one repressed class 1 promoter are indicated by the white arrows in Fig. 3 with the corresponding points highlighted by a box). Second, many promoters showed significant alterations in expression in response to either HAI-1 or AI-2, and the response to both AIs together was more dramatic than to either AI alone (class 2 genes) (Fig. 3, gray arrows). Third, in a few cases, equivalent GFP expression occurred following the addition of either or both AIs (class 3) (Fig. 3, black arrows). Class 3 consisted entirely of repressed genes that showed a modest response (two- to fivefold repression). Additionally, this class of promoters often showed transient AI-regulation that depended on growth phase.
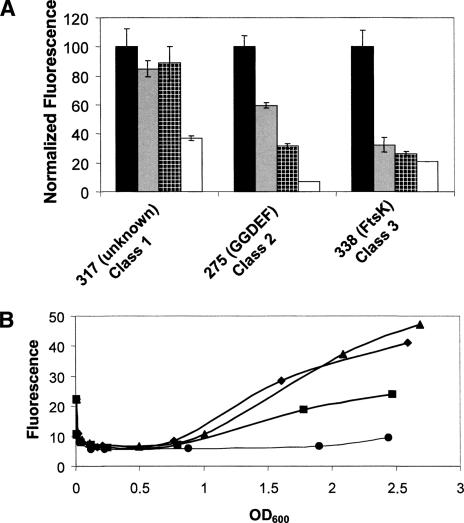
The detailed AI-response profile for one representative promoter from each of the three classes is shown in Figure 4A. The fluorescence values are normalized to the expression level when no AI was added. The fusion designated “unknown” (no significant sequence homology with any known sequence) shows the class 1 response. Addition of only HAI-1 or only AI-2 has no effect, whereas addition of HAI-1 and AI-2 together causes a threefold reduction in GFP production. The fusion to a gene encoding a GGDEF domain-containing protein is shown as a representative class 2 promoter. This fusion is repressed, twofold, threefold, and 14-fold in response to AI-2, HAI-1, and HAI-1 + AI-2, respectively. The promoter for FtsK represents a class 3 promoter. This fusion responds similarly to all three inputs (three-, four-, and fivefold repression in response to the addition of AI-2, HAI-1, and both HAI-1 and AI-2 together, respectively). The AI response profiles for all the promoters identified in this work are shown in Supplementary Figure 2.
FIGURE 4.
Three classes of AI-regulated promoters. (A) Fluorescence production from three promoters, indicated on the X-axis, in response to no AI (black bars), 10 μM AI-2 (gray bars), 10 μM HAI-1 (hatched bars), or 10 μM each AI-2 and HAI-1 (white bars). Fluorescence values are expressed as the percentage measured in the absence of AIs. Error bars indicate the standard deviations. (B) Time course of fluorescence production from clone 275 fused to an unstable GFP (LVA) variant under the four AI conditions listed in A. The OD600 of each culture is plotted on the X-axis. (Triangles) No AI; (diamonds) AI-2; (squares) HAI-1; (circles) AI-2 and HAI-1.
Expression dynamics of AI-regulated genes
In the analyses shown in Figures 3 and 4A, promoter expression was measured at a single time point following addition of AIs (14–16 h). However, gene expression in response to AIs need not be constant throughout growth. To explore this possibility, we measured the response of one promoter (clone 275) to the various AI inputs over time. This particular promoter was chosen because it displays a large dynamic range of expression (14-fold repression), it responds to both the HAI-1 and AI-2 single inputs, and it is directly repressed by LuxR (Fig. 2). For this experiment, the stable GFP variant used for the DFI screen and above expression studies was replaced by an unstable variant of GFP (LVA) that has a shorter half-life (40 min), which is essential for measuring dynamic expression changes (Andersen et al. 1998). The HAI-1−, AI-2− mutant V. harveyi strain containing the plasmid carrying the unstable gfp fusion to clone 275 was grown overnight and subsequently diluted into fresh medium containing no AI or containing saturating levels of AI-2 only, HAI-1 only, or both AI-2 and HAI-1. Figure 4B shows that maximal, steadily increasing expression of the fusion occurs in the absence of any added AIs (triangles), while addition of both HAI-1 and AI-2 together causes full repression of expression at all time points (circles). Minimal repression occurs following AI-2 addition (Fig. 4B, diamonds), while HAI-1 alone causes an intermediate level of repression at each time point (Fig. 4B, squares), consistent with previous findings that indicate HAI-1 is a stronger quorum-sensing signal than is AI-2. Together, the results from Figures 3 and 4A suggest that the strength of signal input into the cell (HAI-1 + AI-2 > HAI-1 > AI-2) is likely translated into the levels of LuxR available for control of target gene expression.
CAI-1 input into the quorum-sensing cascade
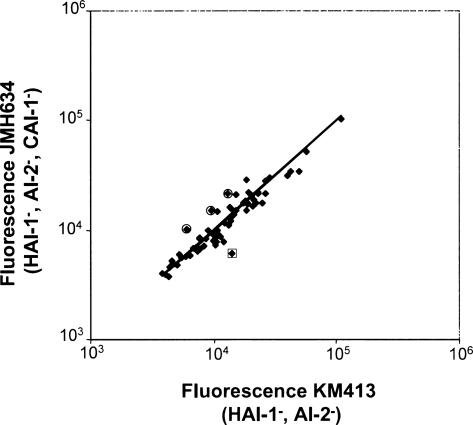
The initial DFI screen was performed in strain KM413 (HAI-1−, AI-2−), which produces the third AI signal, CAI-1. To ensure that the regulation observed in the above experiments is not strongly influenced by this signal, we compared the gfp expression from each promoter obtained in the screen in the triple AI mutant (JMH634: HAI-1−, AI-2−, CAI-1−) to that in the double AI mutant (KM413: HAI-1−, AI-2−) at high cell density. There were less than twofold differences in expression between the two strains in all cases (Fig. 5), verifying that, in our experimental system, CAI-1 plays only a minor role in V. harveyi quorum-sensing gene regulation. Three fusions to the qrr sRNAs consistently showed ∼1.5-fold higher expression in the triple AI synthase mutant JMH634 than in the double AI synthase mutant KM413, as indicated by the circled points in Figure 5. sRNA gene transcription is the immediate output of LuxO regulation (Lenz et al. 2004), and unlike other targets that lie further downstream in the pathway, monitoring qrr expression enables us to observe the most subtle levels of AI regulation (i.e., including regulation by CAI-1). Apparently, targets positioned below LuxR are not as precisely controlled as are the sRNA genes, and so we cannot detect expression differences stemming from CAI-1 input or, alternatively, genes that respond specifically to CAI-1 were not identified in this screen. Lastly, one of the fusions to a TTS promoter consistently showed twofold higher expression in KM413 than in JMH634 (clone 209; note the boxed point in Fig. 5). Because TTS genes are repressed by AIs (Henke and Bassler 2004a), we would expect lower expression in KM413 than in JMH634. Other fusions to TTS genes did not display this behavior. While we do not understand this result, we note that this particular fusion is repressed 50-fold by the addition of both AIs, indicating that the fold difference observed in this experiment is likely not biologically significant.
FIGURE 5.
CAI-1 has no influence on quorum-sensing-regulated targets. Expression of the AI-regulated promoters in the V. harveyi triple synthase mutant JMH634 (Y-axis) and the double AI synthase mutant KM413 (X-axis) is shown. Only one promoter consistently shows twofold or greater regulation (boxed, described in text). The three fusions to qrr promoters are indicated by the circles.
The qrr sRNA genes respond to the individual AI inputs
The qrr genes are, to our knowledge, the only genes directly controlled by LuxO (Lenz et al. 2004). We reasoned that examination of their changes in expression in response to AI should give an indirect readout of the phosphorylation state of LuxO and therefore indicate if different AI inputs translate into discrete levels of phospho-LuxO. The three qrr promoters (qrr2, qrr3, and qrr4) that have detectable expression in our assays displayed a significant response to HAI-1 and to AI-2 when added individually as well as when the AIs were added together (Fig. 6A). A fusion to qrr2 (clone 250) is shown as the representative example in Figure 6, A and B; the other qrr promoters behaved similarly. This result suggests that the immediate mediators of the V. harveyi quorum-sensing response are not controlled by coincident regulation but rather show the class 2 type response. Based on these results, we conclude that significant changes in qrr expression and, in turn, quorum-sensing-controlled gene expression must occur following V. harveyi encountering any combination of AI signals.
FIGURE 6.
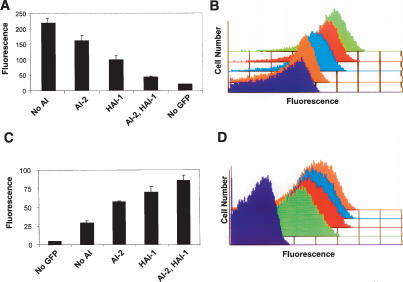
AIs repress qrr expression and activate LuxR production in a graded manner. (A) Expression of a qrr2-gfp promoter (clone 250) in the presence of no AI, 10 μM AI-2, 10 μM HAI-1, or 10 μM each AI-2 and HAI-1. Error bars indicate the standard deviations. (B) Single cell analysis of qrr2-gfp expression measured by flow cytometry. (Purple trace) Background fluorescence; (green trace) no AI; (red trace) AI-2; (blue trace) HAI-1; (orange trace) both AI-2 and HAI-1. (C) Expression of a luxR–gfp translational fusion was assayed in V. harveyi CW2005 (HAI-1−, AI-2−, luxR) following exposure to no AI, 10 μM AI-2, 10 μM HAI-1, or 10 μM each AI-2 and HAI-1. Error bars indicate the standard deviations. (D) Flow cytometry results of individual cells carrying the luxR–gfp translational fusion from C. (Purple trace) Background fluorescence; (green trace) no AI; (red trace) AI-2; (blue trace) HAI-1; (orange trace) both AI-2 and HAI-1.
All of the above experiments rely on measurements of the average behavior of the population. While these analyses help us understand how groups of V. harveyi switch between low- and high-cell-density states, they do not address whether the cells behave homogeneously or whether a large distribution of behaviors gives rise to an average population behavior. To address these possibilities, we used flow cytometry to measure qrr expression in individual cells in response to the AI input signals. Figure 6B shows the result for the qrr2-gfp promoter in KM413 (clone 250) grown to stationary phase. Background autofluorescence is indicated by the purple trace. Maximal expression occurs in the absence of AI (Fig. 6B, green trace), and consistent with Figure 6A, addition of either AI-2 (Fig. 6B, red trace) or HAI-1 (Fig. 6B, blue trace) shifts the qrr2-gfp expression to a lower, intermediate level. Finally, addition of both HAI-1 and AI-2 together maximally represses expression (Fig. 6B, orange trace). Importantly, addition of each AI represses qrr2 expression in the entire population of cells to the same extent. However, full repression only occurs in the presence of all the signals. Therefore, the response to AI, at least in terms of qrr expression at high cell density, is not switch-like (on–off). Rather, V. harveyi cells can express a range of levels of the qrr genes, suggesting that their responses to the AIs are graded.
The different AI inputs translate into distinct levels of LuxR
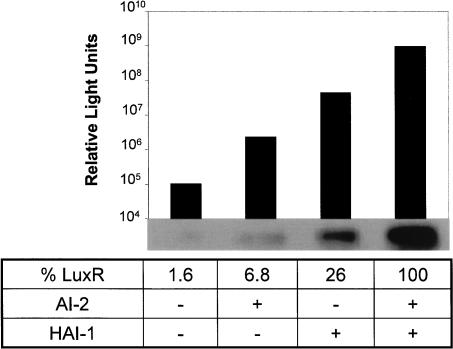
Since the qrr sRNAs control luxR mRNA stability, and because each AI-input combination produces a distinct level of qrr expression (Fig. 6A,B), it follows that the different AI inputs should translate into distinct levels of LuxR production. To test this hypothesis, we measured bioluminescence expression (as a readout of LuxR activity) and LuxR protein production in V. harveyi KM413 in the absence of added AI or supplemented with saturating levels of AI-2, HAI-1, or both AI-2 and HAI-1. Only background levels of light were produced in the absence of added AI, and the addition of both AIs together resulted in a 10,000-fold increase in light emission (Fig. 7). As we have shown previously (Mok et al. 2003), addition of only AI-2 or only HAI-1 cause 50- and 250-fold increases in light emission over background, respectively. Analogous to qrr expression and bioluminescence, each AI combination resulted in production of a distinct level of LuxR protein as determined by Western blotting (Fig. 7). These LuxR levels follow the expected pattern for what is known about the relative strength of the AI signals. In the absence of exogenous AI, KM413 makes 1.6% as much LuxR protein as is made when both HAI-1 and AI-2 are present. Addition of AI-2 alone leads to 6.8%, and HAI-1 alone leads to 26% of the amount of LuxR present when both AIs are present simultaneously. Thus, increased AI signaling strength is represented internally in increasing LuxR production. LuxR levels fluctuate over a 100-fold range, whereas a 10,000-fold change in light production occurs in response to AIs, implying that a large amplification step exists between LuxR and lux expression and/or luciferase activity.
FIGURE 7.
Each AI induces a distinct level of bioluminescence and LuxR production. Strain KM413 (HAI-1−, AI-2−) was exposed to no AI, 10 μM AI-2, 10 μM HAI-1, or 10 μM each AI-2 and HAI-1, and subsequently bioluminescence expression (Relative Light Units: counts per minute per milliliter at OD600nm) and LuxR protein levels were determined. Protein levels from the Western blot were quantified and are expressed as a percentage of the LuxR produced in the presence of both HAI-1 and AI-2.
We considered two potential molecular mechanisms that could lead to the observed levels of LuxR under different signaling conditions. First, if the V. harveyi circuit has switch-like properties, the intermediate levels of LuxR observed in the single AI input states could result from some percentage of the cells in the population switching from the uninduced to the fully induced state. Alternatively, if the circuit functions by a graded mechanism, when only one AI is present, the entire population could shift to production of an intermediate level of LuxR. This latter possibility follows from what was observed for qrr expression (Fig. 6B). To distinguish between these two possibilities, we constructed a LuxR–GFP protein fusion. The fusion is driven from the native luxR promoter and ribosome-binding site and has the first 10 codons of luxR fused directly to the initiation codon of gfp. In a control experiment, we verified that LuxR–GFP production is repressed by the qrr sRNAs by assaying GFP production in E. coli expressing the cloned qrr1 gene (data not shown). LuxR autorepresses its own expression (Chatterjee et al. 1996; Lin et al. 2005). To eliminate this mode of regulation and thereby exclusively measure repression of luxR by the Qrr sRNAs, luxR was deleted from strain KM413 to create strain CW2005. The luxR–gfp translational fusion in CW2005 was exposed to the four AI test conditions, and fluorescence was measured by flow cytometry (Fig. 6C [populations], D [individual cells]). In the absence of added AI (Fig. 6D, green trace), expression from this construct was low but higher than in control cells without GFP (Fig. 6D, purple trace). This result shows that even in the absence of AI-2 and HAI-1, the Qrr sRNAs do not fully repress luxR expression. Addition of AI-2 (Fig. 6D, red trace) and HAI-1 (Fig. 6D, blue trace) resulted in increasing amounts of fluorescence, and, as expected, addition of both AI-2 and HAI-1 resulted in the maximal fluorescence observed (Fig. 6D, orange trace). The flow cytometric single-cell analysis convincingly demonstrates that, in each case, exposure to AI resulted in the entire population shifting to an intermediate level of GFP production as opposed to a fraction of the cells remaining uninduced and another fraction becoming fully induced. This result is similar to the single-cell analysis of the qrr promoters (Fig. 6A,B), leading us to conclude that, at least under the conditions of our experiments, the V. harveyi AI response is graded rather than switch-like.
Promoter affinities determine AI responses
Together, the results show that the extracellular AI inputs are translated internally into different amounts of LuxR protein. This finding suggests a possible molecular mechanism for the discrete responses of quorum-sensing-regulated genes to the various AI inputs. Specifically, class 3 genes, which display maximal responses to each AI individually, must be responsive to lower levels of LuxR than are class 2 genes, which respond additively to the individual AIs. Similarly, class 2 genes must respond to lower levels of LuxR than do class 1 genes, which only respond when both AIs are present simultaneously. Therefore, we predict that class 3 promoters have the highest affinity for LuxR binding, class 2 genes have intermediate affinities, and class 1 genes have the lowest affinities (Fig. 1). This principle can also apply to promoters not directly regulated by LuxR based on the response of their upstream regulator to LuxR. To test this prediction, we examined LuxR binding at three AI-repressed promoters, one class 1 gene (317) (Fig. 4A) and two class 2 genes (275 and 342) (Figs. 3, 4A; Supplementary Fig. 2). No class 3 gene was identified that is directly repressed by LuxR, so class 3 promoters could not be tested for LuxR affinity. We could not carry out a similar analysis for LuxR-activated promoters because every directly activated promoter we identified showed the class 1 response.
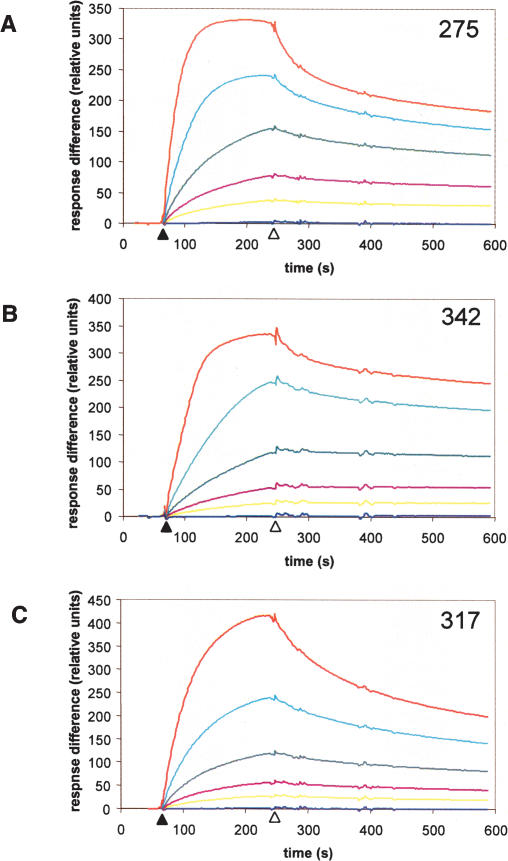
Surface plasmid resonance was used to determine the affinity of LuxR for promoters from clones 317, 275, and 342. Biotin-labeled DNA fragments containing the promoters were attached to a BIAcore streptavidin chip, and LuxR protein from 11.9 to 190 nM was passed over the DNA. The kinetics of binding of LuxR to each promoter are shown in the time interval between the solid and open arrows in Figure 8. The open arrows show when addition of LuxR protein was terminated. Dissociation of LuxR from the promoters was monitored thereafter. Analysis of the dissociation rates reveals that the class 2 promoters 275 and 342 both had lower dissociation constants (7.0 and 9.2 nM, respectively) than the class 1 promoter 317 (38 nM) (Fig. 8). This result preliminarily confirms that class 2 promoters have higher affinities for LuxR than do class 1 promoters. We predict that class 3 promoters would have even higher LuxR-binding affinities.
FIGURE 8.
Class 2 promoters have a higher affinity for LuxR than do Class 1 promoters. Surface plasmon resonance was used to examine the interaction of LuxR with two class 2 promoters (275 and 342, A,B), and one class 1 promoter (317, C). 5′-Biotin end-labeled DNA probes were amplified and attached to a BIAcore streptavidin chip. Purified LuxR at concentrations of 0 nM (dark blue), 11.9 nM (yellow), 23.75 nM (maroon), 47.5 nM (green), 95 nM (light blue), and 190 nM (red) was passed over the chip beginning at the time indicated by the solid arrow on the X-axis. At the time indicated by the open arrow, LuxR flow was terminated, and dissociation of LuxR from the DNA was determined.
Discussion
V. harveyi integrates multiple AI signals, each of different strength and encoding unique information about the neighboring species content, into one phospho-relay signaling system. To understand how V. harveyi integrates this sensory information and translates it into gene expression changes, we identified 50 promoters encoding at least 71 genes that are controlled by quorum sensing (Table 1) and studied their responses to different AI combinations. Because the screen used here is not saturated, additional genes are likely to be controlled by quorum sensing in V. harveyi. The regulon includes genes specifying structural proteins, metabolic processes, and putative regulators (Table 1). The genes in the quorum-sensing regulon, including the qrr and luxR genes, display discrete responses to different AI inputs (Figs. 4, 6–8). Based on these observations, we propose that V. harveyi extracts maximal information from the various combinations of signals and converts that information into unique patterns of gene expression. Importantly, in response to each AI test combination, the entire population of cells shifts in unison to a new discrete level of qrr expression and LuxR production (Fig. 6). We interpret this result to mean that the V. harveyi quorum-sensing transition is graded and not an on-off switch. For an onoff switch mechanism, we would expect subsets of cells to fully switch patterns of gene regulation in response to the different AI conditions.
The majority of the genes identified in this study, while controlled by LuxR, are regulated only indirectly, suggesting that a hierarchy of gene expression exists downstream from LuxR (Fig. 2). One potential mediator in this pathway is AphA, which is directly regulated by the LuxR homolog HapR in V. cholerae (Kovacikova and Skorupski 2002) and is therefore likely positioned immediately downstream from LuxR in V. harveyi (Mok et al. 2003). A σ-cross-reacting protein and two GGDEF proteins identified here also lie directly downstream from LuxR, and like AphA, could be second-tier regulators in the V. harveyi quorum-sensing hierarchy.
To account for our findings that all quorum-sensing information impinges on LuxR, yet quorum-sensing-controlled genes are differentially regulated by the various AI input states, we propose a model in which differential gene expression arises from limiting amounts of LuxR and its particular affinities for target promoters (Fig. 1, bottom arrows). Class 3 genes that respond fully to either AI-2 or HAI-1 are predicted to have promoters with high affinities for LuxR, because this is the condition in which the least LuxR is present (Fig. 7). Class 2 genes show an additive response to autoinducers. We predict that these genes have an intermediate affinity for LuxR because the maximum amount of LuxR obtained by the addition of both AI-2 and HAI-1 is required for a full response, but a significant response is observed in the presence of only AI-2 or HAI-1. Finally, class 1 genes that only respond to the coincident presence of HAI-1 and AI-2 have the lowest affinity for LuxR because they require the highest concentrations of LuxR for any significant response. As an initial test of the model, we showed that representative class 2 promoters have higher affinities for LuxR than does a representative class 1 promoter. We are presently performing genetic screens to identify additional promoters that are directly regulated by LuxR. We are especially interested in identifying a class 3 promoter that is a direct target of LuxR, as we predict this type of promoter would have a higher LuxR-binding affinity than the class 2 and class 1 promoters examined here.
The principles described above can be extended to the CAI-1 system. We know that CAI-1 is the weakest of the three signals, so we predict that genes regulated by LuxR in the exclusive presence of CAI-1 should possess the highest affinity promoters. In this promoter affinity model, genes that are indirectly controlled by LuxR can also show a differential response to the various AI inputs based on the type of AI control of their upstream regulators. The LuxR-affinity model further predicts that all genes responsive to AI-2 must also be responsive to HAI-1 (Fig. 1). Indeed, this prediction is borne out by our finding that no promoter that is responsive to AI-2 fails to respond to HAI-1. By analogy, we expect that genes responsive to CAI-1 must also respond to AI-2 and HAI-1. Genes that respond exclusively to only one AI could exist if unidentified feedback loops are operating in the network. Such genes could not be identified in the present screen because the initial FACS isolation was not performed on cells exposed to only AI-2 or only HAI-1.
Similar observations were reported in Bacillus subtilis for Spo0A, the regulator that controls entry into the sporulation state. Genes directly regulated by Spo0A were classified as either low threshold or high threshold (Fujita et al. 2005). Low-threshold genes showed a significant response to low levels of Spo0A, whereas high-threshold genes required higher concentrations of Spo0A for regulation. Electrophoretic mobility shift assays confirmed that, in most cases, low-threshold genes had a higher affinity for Spo0A than high-threshold genes, similar to the model we propose for LuxR (Fujita et al. 2005). It was later hypothesized that the design of the Spo0A system is critical for precisely ordering gene expression to properly control sporulation (Fujita and Losick 2005).
Sporulation in B. subtilis is ultimately a bi-stable response; cells either do or do not sporulate; there are no intermediate states (Veening et al. 2005). Likewise, ComK-governed competence in B. subtilis is bi-stable (Maamar and Dubnau 2005). In this respect, these processes differ from quorum sensing in V. harveyi, which shows intermediate states in the presence of differing autoinducer signals. The difference between these systems is likely that Spo0A and ComK each initiates a positive feedback loop (Maamar and Dubnau 2005; Veening et al. 2005), which is one known mechanism that generates bi-stability. In contrast, LuxR in V. harveyi is autorepressive (Chatterjee et al. 1996; Lin et al. 2005), a regulation strategy that does not promote bi-stability. However, it should be noted that all of our analyses were performed in stationary phase (following 14–16 h of growth). We are presently performing single-cell analyses of the expression of qrr, luxR, luxC (luciferase), and other quorum-sensing-regulated genes at multiple time points during growth to determine whether the low-to-high-cell-density transition of V. harveyi is always graded.
AI detection in V. harveyi requires the sensors LuxN, LuxQ, and CqsS, which switch from net kinase activity to net phosphatase activity following binding of AI (Fig. 1). Thus, in the absence of a particular AI, the combined phosphatase activity of the two occupied receptors is at least partially offset by the kinase activity of the unoccupied receptor. This signaling arrangement results in a specific level of phosphate flow through the circuit that is less than the maximum possible, which presumably occurs when all three AIs are absent. Because the signals differ in strength, the combination of signals present dictates the discrete level of phosphate flow through the circuit. In each case, however, net phospho-flow through the circuit is represented in the concentration of phospho-LuxO.
Here (Fig. 7) and in a previous study (Mok et al. 2003), light production was analyzed as a canonical readout of phospho-flow through the circuit under differing AI input conditions. In both studies, four discrete levels of light were produced in response to the four AI input conditions. Each AI input condition produced roughly a 100-fold change in light emission. Thus, when all the AIs were present simultaneously, 100-fold more light is produced than when even the weakest signal is removed. At that time, it was not clear if these intermediate levels of light represented biologically significant states or leakage of a noisy system. One possibility was that the V. harveyi circuit functions as a coincidence detector, and the three different AIs buffer the system against molecules in the environment that are similar to one of the bone fide signals (Mok et al. 2003). An alternative hypothesis was that V. harveyi could interpret the blend of chemical signals in its environment as a gauge of genetic relatedness of bacteria in its vicinity. If so, the expectation was that V. harveyi differentially regulates gene expression based on particular combinations of AIs present in any consortium. The results presented here showing that each AI input leads to a distinct level of qrr expression, a discrete level of LuxR production, and a unique pattern of downstream target gene expression lend support to this latter idea. We suggest that the different combinations of signals are, indeed, biologically relevant because they translate into different patterns of gene expression, and furthermore, that these chemical combinations are indicators of the members of the vicinal consortium.
One can envision environments in which V. harveyi would encounter different combinations of AI signals. For example, when V. harveyi initially associates with a mixed species biofilm, AI-2 may be the predominant signal. A mixed vibrio consortium would likely contain significant AI-2 and CAI-1 but an insignificant amount of HAI-1. V. harveyi inhabits the gastrointestinal tracts of marine animals. E. coli, a member of the normal flora of terrestrial organisms, has an avid AI-2 import and degradation system (Xavier and Bassler 2005b), and AI-2 consumption dramatically alters quorum-sensing gene regulation in V. harveyi in mixed V. harveyi–E. coli cultures (Xavier and Bassler 2005a). Although E. coli is not present in the ocean, other bacteria with AI-2 consumption systems could coexist with V. harveyi in the gastrointestinal tracts of marine animals. In such cases, the lack of AI-2 combined with the presence of CAI-1 and HAI-1 could be an indicator to V. harveyi that it is in the gastrointestinal niche. Additionally, accumulation of the autoinducers could also direct the timing of gene expression in mixed- or mono-species environments. Depending on the order in which each AI reaches the concentration required for detection by V. harveyi, distinct subsets of genes should be activated or repressed in a temporal sequence based on the affinities of their promoters for LuxR and/or how many layers of regulators exist between LuxR and a downstream target gene.
We do not expect these complex AI signal integration capabilities or the ability to initiate a precisely ordered cascade of gene expression in response to AIs to be restricted to V. harveyi. Any bacterium that integrates information from multiple AIs could have similar capabilities. In the case of the vibrios, all of the species sequenced to date possess multiple AIs, a LuxR homolog, and many or all of the other V. harveyi-like quorum-sensing regulatory components (Miyamoto et al. 2003; Henke and Bassler 2004a; Lenz et al. 2004). Importantly, every sequenced vibrio species possesses one or more of the qrr sRNA genes, indicating that the focal point of all of these cascades is the regulation of translation of the luxR homologs. Beyond the vibrios, other studied quorum-sensing bacteria make and respond to multiple AIs. For example, the Pseudomonas aeruginosa quorum-sensing system uses three distinct AIs, which together are hypothesized to regulate genes in a specific temporal order (Schuster et al. 2003). While the molecular mechanisms underpinning these other signal relays might not resemble that of V. harveyi, it is likely that these integrated circuits have nonetheless evolved to provide precisely ordered patterns of gene expression.
Materials and methods
Bacterial strains and culture conditions
The V. harveyi strains used in this study are derived from BB120 (Bassler et al. 1997) and include KM413 (ΔluxM, ΔluxS) (Henke and Bassler 2004b), JMH634 (ΔluxM, ΔluxS, ΔcqsA) (Henke and Bassler 2004a), KM816 (ΔluxS) (this study), and JMH363 (ΔluxM) (this study). CW2000 is a ΔluxM, ΔluxS, ΔluxCDA derivative constructed in this study by integrating an unmarked deletion of luxCDA into the chromosome of KM413. Strain CW2005 was derived from KM413 by inserting the cat gene at luxR. V. harveyi strains were grown in Luria-Marine (LM) medium at 30°C with aeration except when selection with tetracycline was used. In those cases, growth was in a 1:1 mixture of LM medium and Autoinducer Bioassay (AB) medium. E. coli S17-1λpir (de Lorenzo and Timmis 1994) was grown at 37°C with aeration, and used for general DNA manipulation and the experiments performed in Figures 2 and 8. Antibiotics were obtained from Sigma and used at the following concentrations: 100 μg/mL kanamycin in E. coli, 250 μg/mL kanamycin in V. harveyi, 10 μg/mL tetracycline, 50 μg/mL polymyxin B, and 10 μg/mL chloramphenicol.
DNA manipulations
All DNA manipulation was performed using standard procedures (Sambrook et al. 1989). T4 DNA ligase and restriction enzymes were obtained from New England Biolabs. E. coli was transformed in 0.2-cm electroporation cuvettes (USA Scientific) using a Bio-Rad Micro Pulser. PCR was performed with Phusion DNA polymerase (New England Biolabs). The vector used for construction of the promoter–gfp library (pCMW1) was constructed from pEVS141 (gift of Eric Stabb, University of Georgia, Athens, GA) (Dunn et al. 2006). The cat gene was removed by AvrII and AflII digestion, the ends were filled in with Klenow DNA polymerase (Roche) in the presence of 12.5 mM dNTPs at room temperature for 30 min, and subsequently, the plasmid was religated. In the absence of insert DNA, pCMW1 shows no detectable gfp expression. The luxR–gfp fusion used in Figure 8 was constructed by amplifying the luxR promoter region from −302 to +30 relative to the translation start site from V. harveyi BB120 genomic DNA. This fragment was inserted into the vector pCMW1 between the SphI and NheI restriction sites, generating an in-frame fusion of the first 10 codons of luxR to gfp. A cat-resistance cassette amplified from plasmid pKD3 using the priming site 1 and 2 sequences (Datsenko and Wanner 2000) was inserted downstream from gfp at the BamHI site. The region from −302 to downstream from the cat gene was then amplified and recombined with the λ red technique into the cosmid pKM699 containing the wild-type luxR gene (Datsenko and Wanner 2000). Plasmids and cosmids were introduced into V. harveyi by conjugation, and exconjugants were selected using the antibiotic resistances carried on the plasmids together with polymyxin B. pCMW275-LVA was constructed by PCR amplification of the gfp-LVA gene from plasmid pRCV-3 (Basu et al. 2005) and insertion into the NheI, BamHI sites of pCMW275.
Construction of promoter trap libraries
Two gfp promoter trap libraries were constructed following published methods (Bartilson et al. 2001). Briefly, genomic DNA from V. harveyi BB120 was digested with DNase I (Sigma) in the presence of 10 mM MnCl2. Overhanging ends were filled in using Klenow DNA polymerase (Roche). Fragments from 100 to 1000 base pairs (bp) were isolated by agarose gel purification using the QiaEx II Gel Extraction Kit (Qiagen), and ligated into StuI-digested pCMW1. This strategy maintains the endogenous gfp ribosome-binding site. Ligation mixtures were transformed into E. coli S17-1λpir. Transformants were harvested from Petri plates, and conjugated into V. harveyi CW2000. Each library had approximately fivefold coverage of the V. harveyi genome. Fusions designated pCMW201–pCMW245 were isolated from one library, while pCMW247–pCMW369 came from the second library.
Differential fluorescence induction
CW2000 carrying the gfp promoter libraries was grown 14–16 h prior to cell sorting in the absence of added AI (to simulate the low-cell-density state) or in the presence of 10 μM each purified AI-2 and HAI-1 (to simulate high cell density). These concentrations of AIs were chosen because they induce maximal light production. Before the initial sort, the percentage of GFP-positive cells varied from 1% to 2%. Generally, three rounds of FACS were performed on a Becton Dickinson FACS Vantage cell sorter to yield a population close to 100% GFP-positive cells. The bacteria were grown 14–16 h between each sort. Following the third sort, the bacteria were plated, and individual colonies were arrayed into Costar 3595 96-well culture plates in 200 μL of LM medium with kanamycin. After growth for 14–16 h, each plate was rearrayed into two new plates, one with 10% cell-free culture fluids lacking AIs and the other with 10% cell-free culture fluids prepared from BB120 grown in LM medium for 14–16 h from a frozen stock. GFP production from the 96-well plates was measured on a 1420 Victor2 multilabel counter (Wallac). Approximately 4000 clones from each condition were individually screened. Clones exhibiting differential gene expression were streaked on fresh medium, and reanalyzed by flow cytometry on a FACScan (Becton Dickinson) flow cytometer a minimum of three times. Vectors carrying candidate AI-regulated promoters were isolated from CW2000 using a QIA-prep spin Miniprep kit and transformed into E. coli S17-1λpir for propagation and sequencing. Gene identities were determined using the BLASTX program from the Vector NTI suite 9.0 software package (InforMax Invitrogen Software Package). All candidate clones were backcrossed into V. harveyi KM413 to confirm the regulation phenotype and for further analyses.
Response to autoinducer and direct LuxR regulation
AI-regulated clones were grown for 14–16 h in 1 mL of LM medium containing kanamycin. Triplicate subcultures were prepared in 1 mL of fresh medium containing no AI, 10 μM synthetically prepared AI-2 (Schauder et al. 2001), 10 μM synthetically prepared HAI-1 (Cao and Meighen 1989), or 10 μM each AI-2 and HAI-1. Following 14–16 h of growth, GFP levels were determined by flow cytometry. For the experiment shown in Figure 5, the AI-regulated promoters were grown in 96-well plates for 14–16 h, and fluorescence was measured with the 1420 Victor2 multilabel counter (Wallac). Representative examples from several identical experiments are shown in Figure 5. In the time course in Figure 4B, a 1:1000-fold back dilution of an overnight culture of KM413 with the plasmid pCMW275-LVA was made in LM medium with kanamycin ± 10 μM AIs. GFP production, measured by flow cytometry, and the absorbance at OD600 were determined every hour. For tests of LuxR regulation, AI-regulated promoters were conjugated into E. coli S17-1λpir containing either pLAFR-2 (Friedman et al. 1982) or pKM699, which is pLAFR-2 carrying a 2.3-kB HindIII fragment containing the luxR gene. Initial analyses were performed in 96-well plates using the 1420 Victor2 (Wallac). Clones that showed differential regulation were further characterized by flow cytometry. LuxR was purified using the IMPACT-CN system (New England Biolabs) as previously described for HapR (Lin et al. 2005). The promoters in clones showing >2.5-fold regulation by LuxR were PCR-amplified with primers directed to the regions adjacent to the StuI site of pCMW1 in the presence of 5 μCi of [α-32P]dATP. Two microliters of these PCR reactions (∼20 ng) was incubated with 50 ng of purified LuxR in the presence of 1 μg of the nonspecific competitor, poly(dI–dC) in a 20-μL volume at 30°C for 15 min. Binding specificity was determined by amplifying a region of the V. harveyi aphA promoter that has previously been shown in V. cholerae to contain a HapR-binding site (from −200 to +115 relative to the transcription start site), and a region that does not contain a HapR-binding site (from −458 to −100). Only the fragment with the binding site was shifted by LuxR. The program YMF 3.0 (Sinha and Tompa 2002) was used in an attempt to identify statistically overrepresented motifs in the promoters directly regulated by LuxR.
Determination of LuxR levels under different AI input conditions and LuxR–GFP fusion analysis
V. harveyi KM413 was grown for 14–16 h at 30°C with aeration in the absence and presence of AIs. Light production was measured using a Wallac 1490 Liquid Scintillation Counter and is defined as counts per minute per milliliter at OD600nm. One milliliter of these cultures was harvested, washed with 1 mL of Dulbecco's PBS (GIBCO), and resuspended in 100 μL of water to lyse V. harveyi. Lysates (5 μL) were analyzed by SDS-PAGE and Western blot as previously described (Henke and Bassler 2004a). Polyclonal antisera to LuxR were adsorbed and used as described (Lenz et al. 2004). LuxR protein was quantified on an Alpha Innotech Fluor Chem SP chemiluminescence image analysis system, ensuring that the images were not saturated. Qrr1 regulation of the luxR–gfp fusion was performed with the qrr1 overexpression construct pDL1605 (Lenz et al. 2004) and the vector control pKK177-3R1 (gift of S. Gottesman, National Institutes of Health, Bethesda, MD). Analysis of the luxR–gfp fusion in CW2005 was performed after 14–16 h of growth.
Surface plasmon resonance
Approximately 500-bp DNA fragments containing the promoters 275, 317, and 342 were amplified with the following 5′-biotin-tagged primers: 275, 5′-CGTTTAACAAGTTCCAA ACT-3′ and 5′-TGACAACTCAGCTGATGAAG-3′; 317, 5′-AGATTCTTTAAATTCGTCTGCG-3′ and 5′-TGACAAAGT TCAACAACGAA-3′; and 342, 5′-AAATACGGCATGAACTA ATACG-3′ and 5′-GCGATACTGTATTTGACGAC-3′. The region of the aphA promoter from −458 to −100, which lacks the LuxR-binding site, was also amplified with biotin-tagged primers and used as a control for nonspecific binding. The DNA was passed over a BIAcore strepavidin chip (Sensor chip SA) in 0.5 M NaCl and 10 mM Tris (pH 8.0) at a rate of 10 μL/min for 10 min in a BIAcore 3000 instrument (Biacore Lifesciences). Each probe showed a 2000–2500 increase in RUs. Purified LuxR was dialyzed against 150 mM NaCl, 10 mM HEPES (pH 7.4), and subsequently washed over the DNA bound to the chips at a flow rate of 10 μL/min for 2 min. Dissociation was measured for 5 min. LuxR binding/dissociation was analyzed at concentrations from 11.9 to 190 nM. The chip was regenerated with 1 M NaCl between the different LuxR concentrations. The background changes in RUs resulting from subtle alterations of the buffer upon protein addition and nonspecific interaction of LuxR with the chip were subtracted from each curve. The binding curves were analyzed with the BIAevaluation software (version 4.1) and fit to a 1:1 (Langmuir) binding model.
Acknowledgments
We thank Ned Wingreen and members of the Bassler laboratory for helpful discussions and suggestions, Kenny Mok and Jen Henke for strain constructions, Christina DeCoste for flow cytometry assistance, Robert Caroll and Andrew Beacham for technical assistance, and Eric Stabb and Susan Gottesman for plasmids. This work was funded by HHMI, and NIH grant R01 GM 065859 to B.L.B. and NIH fellowship F32-AI0586492 to C.M.W.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1466506.
Supplemental material is available at http://www.genesdev.org.
References
- Andersen, J.B., Sternberg, C., Poulsen, L.K., Bjorn, S.P., Givskov, M., Molin, S. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol. 1998;64:2240–2246. doi: 10.1128/aem.64.6.2240-2246.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartilson, M., Marra, A., Christine, J., Asundi, J.S., Schneider, W.P., Hromockyj, A.E. Differential fluorescence induction reveals Streptococcus pneumoniae loci regulated by competence stimulatory peptide. Mol. Microbiol. 2001;39:126–135. doi: 10.1046/j.1365-2958.2001.02218.x. [DOI] [PubMed] [Google Scholar]
- Bassler, B.L., Wright, M., Showalter, R.E., Silverman, M.R. Intercellular signalling in Vibrio harveyiSequence and function of genes regulating expression of luminescence. Mol. Microbiol. 1993;9:773–786. doi: 10.1111/j.1365-2958.1993.tb01737.x. [DOI] [PubMed] [Google Scholar]
- Bassler, B.L., Wright, M., Silverman, M.R. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi Sequence and function of genes encoding a second sensory pathway. Mol. Microbiol. 1994a;13:273–286. doi: 10.1111/j.1365-2958.1994.tb00422.x. [DOI] [PubMed] [Google Scholar]
- Sequence and function of LuxO, a negative regulator of luminescence in Vibrio harveyi . Mol. Microbiol. 1994b;12:403–412. doi: 10.1111/j.1365-2958.1994.tb01029.x. ———. [DOI] [PubMed] [Google Scholar]
- Bassler, B.L., Greenberg, E.P., Stevens, A.M. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi . J. Bacteriol. 1997;179:4043–4045. doi: 10.1128/jb.179.12.4043-4045.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu, S., Gerchman, Y., Collins, C.H., Arnold, F.H., Weiss, R. A synthetic multicellular system for programmed pattern formation. Nature. 2005;434:1130–1134. doi: 10.1038/nature03461. [DOI] [PubMed] [Google Scholar]
- Cao, J.G., Meighen, E.A. Purification and structural identification of an autoinducer for the luminescence system of Vibrio harveyi . J. Biol. Chem. 1989;264:21670–21676. [PubMed] [Google Scholar]
- Chang, H., Gilbert, W. A novel zebrafish gene expressed specifically in the photoreceptor cells of the retina. Biochem. Biophys. Res. Commun. 1997;237:84–89. doi: 10.1006/bbrc.1997.7081. [DOI] [PubMed] [Google Scholar]
- Chatterjee, J., Miyamoto, C.M., Meighen, E.A. Auto-regulation of. The Vibrio harveyilux-operon activator functions as a repressor. Mol. Microbiol. . luxR. 1996;20:415–425. doi: 10.1111/j.1365-2958.1996.tb02628.x. [DOI] [PubMed] [Google Scholar]
- Chen, X., Schauder, S., Potier, N., Van Dorsselaer, A., Pelczer, I., Bassler, B.L., Hughson, F.M. Structural identification of a bacterial quorum-sensing signal containing boron. Nature. 2002;415:545–549. doi: 10.1038/415545a. [DOI] [PubMed] [Google Scholar]
- Datsenko, K.A., Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lorenzo, V., Timmis, K.N. Analysis and construction of stable phenotypes in Gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 1994;235:386–405. doi: 10.1016/0076-6879(94)35157-0. [DOI] [PubMed] [Google Scholar]
- Dunn, A.K., Millikan, D.S., Adin, D.M., Bose, J.L., Stabb, E.V. New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl. Environ. Microbiol. 2006;72:802–810. doi: 10.1128/AEM.72.1.802-810.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman, J.A., Bassler, B.L. A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibrio harveyi . Mol. Microbiol. 1999a;31:665–677. doi: 10.1046/j.1365-2958.1999.01208.x. [DOI] [PubMed] [Google Scholar]
- Sequence and function of LuxU: A two-component phosphorelay protein that regulates quorum sensing in Vibrio harveyi . Mol. Microbiol. 1999b;31:665–677. doi: 10.1128/jb.181.3.899-906.1999. ———. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman, J.A., Lilley, B.N., Bassler, B.L. A genetic analysis of the functions of LuxN: A two-component hybrid sensor kinase that regulates quorum sensing in Vibrio harveyi . Mol. Microbiol. 2000;35:139–149. doi: 10.1046/j.1365-2958.2000.01684.x. [DOI] [PubMed] [Google Scholar]
- Friedman, A.M., Long, S.R., Brown, S.E., Buikema, W.J., Ausubel, F.M. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene. 1982;18:289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- Fujita, M., Losick, R. Evidence that entry into sporulation in Bacillus subtilis is governed by a gradual increase in the level and activity of the master regulator Spo0A. Genes & Dev. 2005;19:2236–2244. doi: 10.1101/gad.1335705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita, M., Gonzalez-Pastor, J.E., Losick, R. High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis . J. Bacteriol. 2005;187:1357–1368. doi: 10.1128/JB.187.4.1357-1368.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer, B.K., Bassler, B.L. Quorum sensing controls biofilm formation in Vibrio cholerae . Mol. Microbiol. 2003;50:101–104. doi: 10.1046/j.1365-2958.2003.03688.x. [DOI] [PubMed] [Google Scholar]
- Henke, J.M., Bassler, B.L. Quorum sensing regulates type III secretion in Vibrio harveyi and Vibrio parahaemolyticus . J. Bacteriol. 2004a;186:3794–3805. doi: 10.1128/JB.186.12.3794-3805.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Three parallel quorum-sensing systems regulate gene expression in Vibrio harveyi . J. Bacteriol. 2004b;186:6902–6914. doi: 10.1128/JB.186.20.6902-6914.2004. ———. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano, M., Reynolds, A.A., Miranda-Rios, J., Storz, G. Detection of 5′- and 3′-UTR-derived small RNAs and cis-encoded antisense RNAs in Escherichia coli . Nucleic Acids Res. 2005;33:1040–1050. doi: 10.1093/nar/gki256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacikova, G., Skorupski, K. Regulation of virulence gene expression in Vibrio cholerae by quorum sensing: HapR functions at the aphA promoter. Mol. Microbiol. 2002;46:1135–1147. doi: 10.1046/j.1365-2958.2002.03229.x. [DOI] [PubMed] [Google Scholar]
- Lenz, D.H., Mok, K.C., Lilley, B.N., Kulkarni, R.V., Wingreen, N.S., Bassler, B.L. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae . Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Lilley, B.N., Bassler, B.L. Regulation of quorum sensing in Vibrio harveyi by LuxO and σ-54. Mol. Microbiol. 2000;36:940–954. doi: 10.1046/j.1365-2958.2000.01913.x. [DOI] [PubMed] [Google Scholar]
- Lin, W., Kovacikova, G., Skorupski, K. Requirements for Vibrio cholerae HapR binding and transcriptional repression at the hapR promoter are distinct from those at the aphA promoter. J. Bacteriol. 2005;187:3013–3019. doi: 10.1128/JB.187.9.3013-3019.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupp, C., Ruby, E.G. Vibrio fischeri uses two quorum-sensing systems for the regulation of early and late colonization factors. J. Bacteriol. 2005;187:3620–3629. doi: 10.1128/JB.187.11.3620-3629.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maamar, H., Dubnau, D. Bistability in the Bacillus subtilis K-state (competence) system requires a positive feedback loop. Mol. Microbiol. 2005;56:615–624. doi: 10.1111/j.1365-2958.2005.04592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, S.T., Xavier, K.B., Campagna, S.R., Taga, M.E., Semmel-hack, M.F., Bassler, B.L., Hughson, F.M. A novel form of the bacterial quorum sensing signal AI-2 recognized by the Salmonella typhimurium receptor LsrB. Mol. Cell. 2004;15:677–687. doi: 10.1016/j.molcel.2004.07.020. [DOI] [PubMed] [Google Scholar]
- Miyamoto, C.M., Dunlap, P.V., Ruby, E.G., Meighen, E.A. LuxO controls luxR expression in. Evidence for a common regulatory mechanism in Vibrio. Mol. Microbiol. . Vibrio harveyi. 2003;48:537–548. doi: 10.1046/j.1365-2958.2003.03453.x. [DOI] [PubMed] [Google Scholar]
- Mok, K.C., Wingreen, N.S., Bassler, B.L. Vibrio harveyi quorum sensing: A coincidence detector for two auto-inducers controls gene expression. EMBO J. 2003;22:870–881. doi: 10.1093/emboj/cdg085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rediers, H., Rainey, P.B., Vanderleyden, J., De Mot, R. Unraveling the secret lives of bacteria: Use of in vivo expression technology and differential fluorescence induction promoter traps as tools for exploring niche-specific gene expression. Microbiol. Mol. Biol. Rev. 2005;69:217–261. doi: 10.1128/MMBR.69.2.217-261.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romling, U., Gomelsky, M., Galperin, M.Y. C-di-GMP: The dawning of a novel bacterial signalling system. Mol. Microbiol. 2005;57:629–639. doi: 10.1111/j.1365-2958.2005.04697.x. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., Maniatis, T. Cold Spring Harbor Laboratory Press, Cold Spring Harbor; NY: 1989. Molecular cloning: A laboratory manual. [Google Scholar]
- Schauder, S., Shokat, K., Surette, M.G., Bassler, B.L. The LuxS family of bacterial autoinducers: Biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 2001;41:463–476. doi: 10.1046/j.1365-2958.2001.02532.x. [DOI] [PubMed] [Google Scholar]
- Schuster, M., Lostroh, C.P., Ogi, T., Greenberg, E.P. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: A transcriptome analysis. J. Bacteriol. 2003;185:2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, H.S., Chen, H., Rossier, C., Lalioti, M.D., Antonarakis, S.E. Isolation of a human gene (HES1) with homology to an Escherichia coli and a zebrafish protein that maps to chromosome 21q22.3. Hum. Genet. 1997;99:616–623. doi: 10.1007/s004390050416. [DOI] [PubMed] [Google Scholar]
- Sinha, S., Tompa, M. Discovery of novel transcription factor binding sites by statistical overrepresentation. Nucleic Acids Res. 2002;30:5549–5560. doi: 10.1093/nar/gkf669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartzman, E., Silverman, M., Meighen, E.A. The luxR gene product of Vibrio harveyi is a transcriptional activator of the lux promoter. J. Bacteriol. 1992;174:7490–7493. doi: 10.1128/jb.174.22.7490-7493.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueshima, R., Fujita, N., Ishihama, A. Identification of Escherichia coli proteins cross-reacting with antibodies against region 2.2 peptide of RNA polymerase σ subunit. Biochem. Biophys. Res. Commun. 1992;184:634–639. doi: 10.1016/0006-291x(92)90636-y. [DOI] [PubMed] [Google Scholar]
- Valdivia, R.H., Falkow, S. Bacterial genetics by flow cytometry: Rapid isolation of Salmonella typhimurium acid-inducible promoters by differential fluorescence induction. Mol. Microbiol. 1996;22:367–378. doi: 10.1046/j.1365-2958.1996.00120.x. [DOI] [PubMed] [Google Scholar]
- Veening, J.W., Hamoen, L.W., Kuipers, O.P. Phosphatases modulate the bistable sporulation gene expression pattern in Bacillus subtilis . Mol. Microbiol. 2005;56:1481–1494. doi: 10.1111/j.1365-2958.2005.04659.x. [DOI] [PubMed] [Google Scholar]
- Waters, C.M., Bassler, B.L. QUORUM SENSING: Cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- Xavier, K.B., Bassler, B.L. LuxS quorum sensing: More than just a numbers game. Curr. Opin. Microbiol. 2003;6:191–197. doi: 10.1016/s1369-5274(03)00028-6. [DOI] [PubMed] [Google Scholar]
- Interference with AI-2-mediated bacterial cell–cell communication. Nature. 2005a;437:750–753. doi: 10.1038/nature03960. ———. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regulation of uptake and processing of the quorum-sensing autoinducer AI-2 in Escherichia coli . J. Bacteriol. 2005b;187:238–248. doi: 10.1128/JB.187.1.238-248.2005. ———. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J., Mekalanos, J.J. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae . Dev. Cell. 2003;5:647–656. doi: 10.1016/s1534-5807(03)00295-8. [DOI] [PubMed] [Google Scholar]