Abstract
Overexploitation and subsequent collapse of major worldwide fisheries has made it clear that marine stocks are not inexhaustible. Unfortunately, the perception remains that marine fishes are resilient to large population reductions, as even a commercially ‘collapsed’ stock will still consist of millions of individuals. Coupled with this notion is the idea that fisheries can, therefore, have little effect on the genetic diversity of stocks. We used DNA from archived otoliths collected between 1924 and 1972 together with 2002 juvenile's tissue to estimate effective population size (Ne) in plaice (Pleuronectes platessa). Ne was estimated at 20 000 in the North Sea and 2000 in Iceland. These values are five orders of magnitude smaller than the estimated census size for the two locations. Populations examined between 1924 and 1960 were in Hardy–Weinberg equilibrium, whereas populations examined after approximately 1970 were not. Extensive testing was performed to rule out genotyping artefacts and Wahlund effects. The significant heterozygote deficiencies found from 1970 onward were attributed to inbreeding. The emergence of inbreeding between 1950 and 1970 coincides with the increase in fishing mortality after World War II. Although the biological mechanisms remain speculative, our demonstration of inbreeding signals the need for understanding the social and mating behaviour in commercially important fishes.
Keywords: plaice, Pleuronectes platessa, inbreeding, Ne, microsatellite, fisheries management
1. Introduction
The collapse of most major fisheries in the world emphasizes the fact that marine stocks are not inexhaustible. In 2002, 75% of the world's marine stocks were fully exploited or overexploited (FAO, www.fao.org). Even after the collapse of the cod and herring fisheries (Hutchings 2000), the perception has remained that marine stocks are fundamentally resilient because a ‘collapsed’ stock may still consist of millions of individuals. The problem is that the census size of a population (N) can be vastly different from its effective population size (Ne), that is, the number of individuals contributing to the next generation (Frankham 1995). In marine fishes, Ne can be several orders of magnitude smaller than N, producing Ne : N ratios varying from 10−5 to 10−3 (Hauser et al. 2002; Turner et al. 2002; Hutchinson et al. 2003). Species with such small ratios can suffer loss of genetic diversity under fishing pressure as has been shown for the orange roughy (Smith et al. 1991), the red drum (Turner et al. 2002), the New Zealand snapper (Hauser et al. 2002) and North Sea cod (Hutchinson et al. 2003).
Plaice is a common flatfish species inhabiting Northern European coastal waters and a main target of the North Sea beam-trawl fisheries (Rijnsdorp & Millner 1996). North Sea catches last century increased from a stable 55 000 t in the first half of the century to a record 170 000 t in 1989, after which fisheries collapsed to 82 000 t in 2001 (Rijnsdorp & Millner 1996; ICES 2003). In Iceland, a 50% decline of the stock has been observed these last 10 years (Anonymous 2003).
Two recent genetic surveys of plaice populations in the Northern East Atlantic, based on microsatellite loci (Hoarau et al. 2002b) and mitochondrial DNA (Hoarau et al. 2004), revealed weak but significant genetic differentiation consistent with large-scale homogeneity and recent recolonization of the North Atlantic over the past 15 000 years. Significant heterozygote deficiencies were also found in these surveys. Although heterozygote deficiencies have been commonly observed in marine bivalves (Zouros & Foltz 1984) and fishes (Waldman & McKinnon 1993), most explanations have been attributed to genotyping or sampling artefacts (Wahlund effect). If such artefacts can be ruled out, then biological explanations such as selection or inbreeding must be considered.
Thus the objectives of the present study were to: (i) estimate effective population size (Ne) in plaice; and (ii) analyse the causes for heterozygote deficiencies including the possibility of inbreeding. In order to accomplish these objectives, we used DNA extracted from archived otolith samples collected between 1924 and 1970, and from a time-series of juvenile cohorts collected in 2002. Further, we rigorously evaluated microsatellite data-quality with respect to technical artefacts associated with genotyping and sampling.
2. Materials and methods
(a) Sampling
Juvenile cohorts of young-of-the-year plaice were collected from two nursery grounds at Balgzand in the Dutch Wadden Sea (52°53′–52°59′ N, 4°48′–4°55′ E) and at Alftanes on the west coast of Iceland (64°06′ N, 22°02′ W). These two populations are genetically isolated (Hoarau et al. 2002b, 2004) and thus represent two independent samples. Biweekly sampling was performed in both areas during the settlement season of juvenile plaice using the same type of beam trawl (5 mm mesh). Sampling (referred to as Week 1) began on 26 February 2002 at Balgzand and 22 June 2002 at Alftanes. Samples collected from weeks 1, 3, 7 and 9 in the North Sea and weeks 1, 5, 7 and 11 off Iceland were designated as cohorts (see Electronic Appendix, table S1). Sampling from week 9 in the North Sea was divided into two sub-samples (designated cohorts 4 and 5) based on individual size differences (see Electronic Appendix, table S1). A total of 477 individuals were immediately preserved in 70% ethanol and total length measured to the nearest millimetre. Fertilized eggs (n=64), designated cohort 0, were collected in January 2002 in the southern North Sea using a single haul of high-speed plankton net called a gulf torpedo (Zijlstra 1970).
Otolith samples from Iceland (1924, 1948 and 1972) and the North Sea (1950 and 1970) were obtained from the HAFRO (Reykjavik, Iceland) and RIVO (IJmuiden, The Netherlands) fisheries institutes, respectively.
(b) DNA extraction
DNA was extracted from juveniles using a CTAB protocol for muscle tissue (Hoarau et al. 2002b) and from eggs using the following Chelex protocol. Eggs were air dried, incubated at 55 °C in 50 μl of a 6% Chelex 100 (BioRad) solution containing 1.2 U proteinase K (Promega) for 2 h, followed by 100 °C for 10 min. Samples were centrifuged (2 min at 20 000g) and the crude DNA supernatant used for PCR.
Historical DNA was extracted from the dry tissues surrounding the otolith (Hutchinson et al. 1999) in a laboratory where no previous DNA-work had ever been conducted. DNA-free pipettes and filter tips were also used to avoid contamination. Negative controls were used at every step from extraction to genotyping. Extracted DNA was of good quality with an overall amplification failure of only 1.4%, a level comparable to DNA extracted from muscle tissue.
(c) Genotyping
Eight microsatellite loci (see Electronic Appendix, table S1) were used (Watts et al. 1999; Hoarau et al. 2002a,b). Details of PCR conditions are reported elsewhere (Hoarau, et al. 2002a,b). Genotyping was performed on an ABI377 (Applied Biosystems), using internal lane standards and the software Genescan to determine allele sizes.
The possibility of genotyping artefacts was examined in several ways. First, a set of homozygous individuals was re-amplified for each of the eight loci under relaxed annealing temperatures (4 °C lower) to verify that additional alleles had not been missed. Second, the expected frequency of null alleles (r; Brookfield 1996, eq. 4) was used to calculate the expected frequency of null homozygotes (r2). Expected counts were then compared with observed counts in the dataset. If the observed were less than the expected, it was concluded that null alleles were not a factor. Third, we used Microchecker (Van Oosterhout et al. 2004) to test for stuttering and large allele drop-out. These tests are based on the size distribution of the genotypes. Fourth, we redesigned new primers for loci, PL142 and PL167 (PL142newF: 5′-GCCTCATTTTCACACTGTTACC-3′, PL142newR: 5′-GGGCAATTACTTGAGATGAAAAAG-3′, PL167newF: 5′-GGGAATACACCAGACAAAATG-3′, PL167newR: 5′-GCACATGTCAAGCTGCAGTCCC-3′). These two loci were chosen because of their higher level of stuttering. All homozygotes were re-scored using these new primer pairs and compared against results from the original primers as a further test.
(d) Genetic diversity and departures from Hardy–Weinberg equilibrium
Observed and unbiased expected heterozygosity (He; Nei 1978) were computed for each locus individually and as a multilocus estimate for each of the cohorts. Single and multilocus FIS were estimated using Weir and Cockerham's f (Weir & Cockerham 1984) and significance was tested using 2000 permutations. Heterogeneity of heterozygote deficiency among loci was tested according to Gafney et al. (1990). All of the computations were performed using Genetix 4.04 (Belkhir et al. 2003). Sequential Bonferroni corrections (Rice 1989) for multiple comparisons were applied where necessary.
(e) Estimating Ne
Variance effective population sizes (Ne) were estimated using a likelihood-based temporal method developed by Wang (2001). The variance effective population size is the size of an ideal population (i.e. one with no selection, random mating and a Poisson-distributed reproductive success) that has the same properties with respect to allele frequency variance (genetic drift) as the actual population. A maximum-likelihood approach is superior to those based on F-statistics because it can use more of the information in the data, that is, the presence of many low-frequencies alleles, which are typical for microsatellite loci. The accuracy of the Ne estimate increases with the number of alleles, the sample size and the interval of sampling (Wang 2001). We used the software Mne 1.0 (Wang 2001) with four points in time for the Iceland samples (i.e. 1924, 1948, 1972 and 2002) and three points in time for the North Sea samples (i.e. 1950, 1970 and 2002). We used a generation time of 7.7 years (Rijnsdorp 1993) and we used 828 individuals, 6 loci and 353 alleles.
(f) Cohort differentiation
Population differentiation among juvenile cohorts was analysed with Wright's FST (Wright 1969) using Weir and Cockerham's estimator, θ (Weir & Cockerham 1984). Global FST was used to detect differentiation among cohorts within each nursery. Significance of all θ estimates was tested using 2000 permutations.
(g) Wahlund effect
Tests for Wahlund effects among juvenile cohorts were performed using PartitionML (Belkhir & Bonhomme 2002). The method searches for the best way to partition the samples into two (or more) panmictic clusters and provides the likelihood for such a partition. Significance was tested using permutations (Castric et al. 2002). In order to test if the partition into two clusters was more likely than no subdivision, we generated the null distribution of the likelihood in 100 panmictic pseudo-samples and compared them with the observed likelihood. The panmictic pseudo-samples were generated by permuting the monolocus genotypes (Belkhir & Bonhomme 2002) using all cohorts within one nursery.
(h) Testing for inbreeding
We tested for inbreeding by comparing the distribution of individual multi-locus heterozygosity (MLH; number of heterozygous loci per individual) with the expected distribution under random mating (Castric et al. 2002). Inbred individuals are expected to be more homozygous over all loci, leading to lower MLH values than expected and a shift in the distribution towards lower MLH values. The expected MLH distribution (MLHexp) was generated by permutation, 1000 pseudo-samples were generated by permuting alleles and the difference between the observed (MLHobs) and expected (MLHexp) frequencies was computed for each MLH class. A homozygote excess was computed: (MLHobs−MLHexp)/MLHexp. Significance was tested by comparing the mean observed MLH with the means of the 1000 pseudo-samples.
We also performed a simulation in order to test what level of inbreeding would be required to obtain the MLHobs distribution, given the allele frequencies. We used the following formula for each locus: H=Σpi2(1−F)+F where H is the proportion of homozygotes, pi the frequency for allele i and F the inbreeding coefficient (Hedrick 2000). In this manner we generated the MLH distribution given a specified F. The simulated distribution was then compared with the MLHobs distribution to determine the best fit of F.
(i) Relatedness
Relatedness was estimated by the pairwise identity coefficient (I; Mathieu et al. 1990) using Identix (Belkhir et al. 2002). Identity was used because it has a smaller variance as compared with other estimators of relatedness (Belkhir et al. 2002). The mean and the variance of I were estimated for each cohort and compared with their expected distribution under the null hypothesis of no relatedness. These expected distributions were generated by permutation of genotypes (n=1000). Sequential Bonferroni corrections (Rice 1989) for multiple comparisons were applied where necessary.
(j) Selection against heterozygotes
Selection against heterozygotes was tested by plotting the multilocus FIS against the mean body-size found in each cohort. If selection was a factor, we predicted an increase in the heterozygote deficiency with size, which can serve as a proxy for age.
3. Results
(a) Potential artefacts
All microsatellite loci were extensively tested for genotyping artefacts (see Electronic Appendix, table S2) that might produce null alleles. An amplification test using relaxed annealing temperatures for all loci did not decrease the number of observed homozygotes. Following Brookfield's eq. 4 (N×r2), the expected number of null homozygotes (Nnull_exp=12) was greater than the actual number observed (Nnull_obs=4), in six of the eight loci (see Electronic Appendix, table S2), allowing us to reject the possibility of nulls. For loci PL06 and PL09, null alleles could not be rejected and these loci were subsequently eliminated from further analyses. No evidence for large allele drop-out was found for any of the loci. All loci displayed clean signals with minimal stuttering (see Electronic Appendix, figure S1). Post-PCR treatment with T4-polymerase (Ginot et al. 1996) of the original primers as well as reamplification with the new primers confirmed earlier results, i.e. no change in the observed number of homozygotes. We therefore concluded that genotyping artefacts could not explain the departure from Hardy–Weinberg equilibrium (HWE) observed in our dataset.
A temporal Wahlund effect could also be rejected for all juvenile cohorts (p≥0.11) as none of the PartitionML log-likelihood values departed from the null distribution of a homogeneous population (data not shown).
(b) Genetic differentiation
No genetic differentiation was detected among 2002 cohorts from the same location (Balgzand: θ=0.0021, p=0.06; Alftanes: θ=0.0018, p=0.14). Between Iceland and the North Sea there was a significant differentiation stable over time (1950s: θ=0.0201, p<0.001; 1970s: θ=0.0233, p<0.001; 2002: θ=0.0207, p<0.001).
(c) Genetic diversity
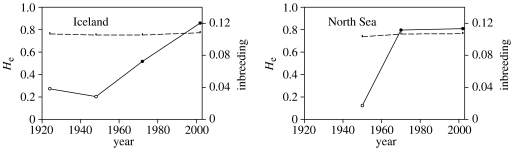
Genetic variation was moderate to high with 5–40 alleles per locus per cohort, and He ranged from 0.3182 to 0.9678 (see Electronic Appendix, table S1). Icelandic samples were less diverse than North Sea samples. There was no evidence for a loss of genetic diversity (He) at either location (figure 1) throughout the twentieth century. Once corrected for sample size, there was also no evidence for a loss of allelic diversity (data not shown).
Figure 1.
Temporal changes in genetic diversity He (dashed line) and inbreeding (solid line). Open circles: n.s., filled circles: significant at p<0.001. Inbreeding is defined as the excess of homozygotes (MLHobs−MLHexp)/MLHexp).
(d) Effective population size
The maximum likelihood estimated Ne for Iceland was 1733 individuals (1063; 3598 at 95% CI) and for the North Sea 19 535 individuals (3435; 70 000 at 95% CI). Adult (>3 years old) census sizes (N) were estimated to be ∼108 for Iceland (J Palsson, Icelandic Fisheries, personal communication) and ∼109 for the North Sea (ICES 2003). The Ne : N ratios for both populations were similar, at approximately 2×10−5.
(e) Temporal departures from HWE and inbreeding
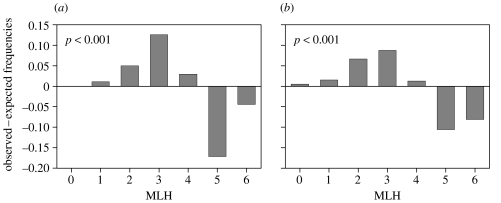
In 2002, the frequency of low MLH classes was higher than expected under random mating at both locations (p<0.001; figure 2). Consequently, individuals were more homozygous over all six loci than expected under random mating, indicative of inbreeding. The same analysis performed on each cohort separately (data not shown) showed a significant shift towards low MLH classes as well (p≤0.004). Inbreeding was also found in North Sea and Icelandic populations from 1970s (p<0.001; figure 1). In sharp contrast, the individual MLH distributions did not differ significantly from those expected under random mating in either the North Sea or Icelandic populations in assays from samples taken ≤1950 (figure 1). Homogeneity of heterozygote deficiency among the six loci could not be rejected (χ2=54.9, d.f.=59), further supporting inbreeding. In other words, there was strong evidence that inbreeding developed between 1950 and 1970. Simulations for Iceland and North Sea 2002 populations yielded an F of, respectively, 0.13 and 0.09. Such F values are on the order of half-sib matings.
Figure 2.
Distribution of (MLHobs−MLHexp) frequencies (loci PL06 and PL09 excluded). (a) all cohorts from Balgzand (Dutch Wadden Sea), (b) all cohorts from Alftanes (Iceland).
(f) Relatedness
Relatedness is a prerequisite for inbreeding. Evidence for relatedness was found in 2 of the 10 juvenile cohorts, i.e. cohort 3 at Balgzand (mean I=0.2040, p<0.05) and cohort 1 at Alftanes (mean I=0.1229, p<0.05). Relatedness was also found in adult samples taken from two spawning grounds, that is, Terschellinger Bank, North Sea (I=0.2102, p<0.05) and central Irish Sea (I=0.1459, p<0.05). The data used for this calculation were taken from Hoarau et al. (2002b).
(g) Selection
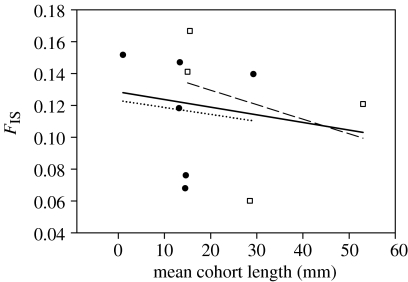
The relationship between multilocus FIS and mean fish size within each juvenile cohort (mm) appears to decrease with age (figure 3). Although suggestive of selection against homozygotes, the trend is not significant.
Figure 3.
Changes in multilocus FIS (loci PL06 and PL09 excluded) with mean fish size within a cohort (mm). Filled circle, Balgzand (Dutch Wadden Sea); open square, Alftanes (Iceland); dotted line, Balgzand; dashed line, Alftanes; solid line, all. None of the trends is significant.
4. Discussion
While it is hard to imagine how inbreeding could be a factor when plaice populations in the North Sea are estimated at 109 adults, we will argue that moderately low Ne in combination with social behaviour and fishing on the spawning grounds during seasonal spawning aggregations may make plaice exceptionally vulnerable to potential inbreeding when exploitation is strong.
(a) Low effective population size
Ne is estimated to be as low as 2000 individuals in Iceland and 20 000 in the North Sea. The Ne : N ratios for both populations are similar at approximately 2×10−5. Although this ratio is very small, it is within the range found for other marine fishes with a similar life history (Hauser et al. 2002; Turner et al. 2002; Hutchinson et al. 2003). Low Ne : N ratios thus appear to be common in marine fishes characterized by type III survivorship curves, i.e. high fecundity and high juvenile mortality. Female fecundity in plaice is estimated at 20 000–600 000 eggs per female (Rijnsdorp 1991), which is offset by high levels of daily mortality for eggs of up to 20% (Rijnsdorp & Jaworski 1990) and of juveniles up to 4% (Van der Veer et al. 1990). Both of these factors may produce high variance in individual reproductive success and, subsequently, to large discrepancies between Ne and N (Hedgecock 1994). Sexual dimorphism in maturation and growth of plaice makes males more vulnerable to fishing mortality, thus producing a skewed sex ratio towards females (Rijnsdorp 1994) that would further reduce Ne.
From a theoretical standpoint, however, Ne estimates of thousands of individuals would still be far too large for the effects of genetic drift, loss of variability and inbreeding to play a significant role under the assumption of random mating. But what if there is cryptic social structure and mating is not random?
(b) Social structure and mating behaviour
Previous generalizations about the apparent lack of social structure in type III commercial species have been challenged by a study which showed direct evidence for such structure in two other temperate species of Pleuronectes, P. yokohamae and P. herzensteini (Gomelyuk & Shchetkov 1999). On the spawning grounds, aggregations of plaice males are visited by gravid females (Rijnsdorp 1989). Courting behaviour of plaice (Forster 1953) and flounder (Stoner et al. 1999) involves circling and ventral pairing. Recent observations of non-commercial flatfish species also indicate strong territoriality and complex mating behaviours including the possibility of female choice (Stoner et al. 1999; Carvalho et al. 2003). For plaice, sex-specific behaviour has been hypothesized to explain the greater catchability of spawning males (Solmundsson et al. 2003)—another possible factor that could reduce Ne. If it turns out that female choice is an important factor in plaice mating, then anything that disrupts this process could have strong effects on fertilization success and population fitness for the local aggregation.
Indirect evidence for group living in plaice is at least suggested from our finding of kin structure (relatedness) among juvenile plaice in 2 of the 10 cohorts sampled, as well as among adults sampled on feeding and spawning grounds. The results from our simulations imply mating between related individuals, at the half-sib level (F∼1/8). Although direct observational evidence in plaice may be technically difficult to get, there is already enough evidence available for flatfish to conclude that some form of subtle social structure is maintained. Kin structure has also been found in several other fishes including perch (Gerlach et al. 2001), rainbowfish (Arnold 2000) and tilapia (Pouyaud et al. 1999); the latter also revealed inbreeding.
Fidelity of plaice to their spawning grounds is well-documented (De Veen 1978; Hunter et al. 2003), but how do plaice know how to find their spawning ground given that spawning larvae are quickly advected from their offshore natal site? One possibility would be through larval attachment (natal philopatry), which would promote strong population differentiation. This can be rejected, however, because recent genetic studies on plaice indicate that spawning populations are only weakly differentiated and strongly connected by gene flow (Hoarau et al. 2002b, 2004). A second possibility suggested by Arnold & Metcalfe (1995) is that immature fish accompany mature fish to the spawning ground in order to learn the location, and to associate some feature of the mature spawning fish (e.g. pheromone) with physical features of the seabed. Learning behaviour of this type is considered to be the most plausible explanation in herring (reviewed in Corten 2002), a species that shares many life history traits with plaice. We hypothesize that if groups of related plaice stay together from the juvenile stage, then individuals will learn the same migrating route and share the same spawning ground, ultimately leading to an extended family structure.
The apparent contradiction of simultaneously finding small-scale genetic heterogeneity and large-scale genetic homogeneity (Hoarau et al. 2002b, 2004) can, on the one hand, be explained by plaice homing behaviour. In Iceland, site fidelity has been shown to be high, with between 72 and 94% of the females returning repeatedly to the same spawning sites (Solmundsson et al. in preparation). On the other hand, an ‘unfaithfulness’ of 6–28% of the individuals is also strong enough to homogenize allelic frequencies among spawning grounds. The two observations are, therefore, not mutually exclusive.
(c) Effects of heavy fishing pressure
The emergence of inbreeding in the North Sea coincides with the introduction of the beam trawl in the beginning of the 1960s. Although plaice has been heavily exploited for a century, the past fishing activities targeted juveniles and young adults. Expansions of the fishery after World War II and the introduction of more powerful vessels led to a greater focus on fishing mature individuals on the spawning grounds in winter. If, as we hypothesize, kinship-based social structure is associated with the spawning grounds, then fishing-mediated reductions in spawning aggregations in combination with fishing-mediated disruptions of behaviour during the mating season may force non-random mating to occur.
The effect of increased fishing could be direct, by reducing the size of the spawning aggregations, and/or indirectly, by disrupting courtship behaviours that are necessary for successful mating. The effects of noise and vibration from fishing gear have been shown to affect mating in several fish species, including cod, which use squeaks and grunts to locate, warn and attract one another (reviewed in Rowe & Hutchings 2003). It is estimated that the entire North Sea bottom is trawled at least four times per year (Rijnsdorp et al. 1998). During the spawning season such a stress could readily disturb social behaviour.
Since fisheries activities account for 80% of plaice mortality (Rijnsdorp & Millner 1996), the rise of inbreeding after the 1970s is most probably related to the threefold increase in landings of plaice over the last 50 years. The potential for inbreeding may be inherent in the life history of plaice, but is only likely to become an issue under severe exploitation.
(d) Genetic diversity through the twentieth century
No decline in genetic diversity (He) was detected in either the Icelandic or North Sea plaice populations (figure 1). Loss of genetic diversity has, however, been recorded for New Zealand snapper and North Sea cod populations (Hauser et al. 2002; Hutchinson et al. 2003), where effective populations sizes are one to two orders of magnitude smaller than plaice. All three species have been commercially important from the onset of the industrialization of the fisheries in the mid-1800s, with cod having suffered the highest fishing mortality. Theoretically, a Ne of 50 is all that is required to maintain short term genetic diversity, and a Ne of 500 individuals to maintain longer-term stability (Van Dyke 2003). For plaice, Ne is still sufficiently large to maintain diversity. However, this conclusion should be treated with caution as a very recent bottleneck would not have yet led to a detectable reduction of diversity.
(e) Potential consequences of inbreeding
The consequences of inbreeding are potentially severe, leading to population decline and possible extinction (Saccheri et al. 1998). As inbreeding in plaice is less than five generations old, inbreeding depression or reduced fitness of inbred individuals could be a result (Charlesworth & Charlesworth 1987). The effects of inbreeding depression are expected to be most severe just after the onset of inbreeding notably as deleterious recessive alleles are not yet purged by selection. The negative, albeit non-significant, trend line between FIS and size (figure 1) suggests that inbreeding depression may be occurring in juvenile plaice.
5. Conclusion
Effective population sizes in plaice are small and have provided evidence for the emergence of inbreeding between 1950 and 1970. Although the biological mechanism underlying the observed results remains speculative, the emergence of inbreeding coincides with the development of industrial scale fishing on the spawning grounds in the 1960s. As long as spawning aggregations remain large, inbreeding will remain unlikely; but if effective population sizes are also reduced, directly and/or as a consequence of behavioural disruptions, then inbreeding may develop. Although we cannot prove that industrial-scale fishing has caused the observed inbreeding, under the precautionary principle, we recommend that fishing on the spawning grounds be restricted.
Acknowledgments
We thank F. Weissing for help with the simulation. We also thank N. Bierne, F. Bonhomme, M. Chevolot, J. Coyer, F. Volckaert for their many useful discussions and comments on earlier versions of this manuscript. This research was supported by NWO-Prioriteit Programma ‘Sustainable Use of Marine Natural Resources’ Project Nr. 885-10.101-P.
Footnotes
As this paper exceeds the maximum length normally permitted, the authors have agreed to contribute to production costs.
Supplementary Material
References
- Anonymous, 2003 Nytjastofnar sjávar 2002/2003. Aflahorfur fiskveiðiárið 2003/2004. State of marine stocks in icelandic waters 2002/2003. Prospects for the quota year 2003/2004. Hafrannsóknastofnunin, Fjölrit nr. 97, p. 173 [English summary].
- Arnold K.E. Kin recognition in rainbow fish (Melanotaenia eachamensis): sex, sibs and shoalings. Behav. Ecol. Sociobiol. 2000;48:385–391. [Google Scholar]
- Arnold G.P, Metcalfe J.D. Seasonal migrations of plaice (Pleuronectes platessa) through Dover Strait. Mar. Biol. 1995;127:151–160. [Google Scholar]
- Belkhir K, Bonhomme F. Université de Montpellier. See; Montpellier, France: 2002. PartitionML: a maximum likelihood estimation of the best partition of a sample into panmictic units. http://www.univ-montp2.fr/~genetix/partitionml.htm. [Google Scholar]
- Belkhir K, Castric V, Bonhomme F. Identix, a software to test for relatedness in a population using permutation methods. Mol. Ecol. Notes. 2002;2:611. [Google Scholar]
- Belkhir, K., Borsa, P., Chikhi, L., Raufaste, N. & Bonhomme F. 2003 Genetix v. 4.04, Logiciel sous WindowsTM pour la Génétique des Populations. Laboratoire Génome et Population, Université Montpellier 2, Montpellier. See http://www.univ-montp2.fr/~genetix/genetix/genetix.htm
- Brookfield J.F.Y. A simple method for estimating null allele frequency from heterozygote deficiency. Mol. Ecol. 1996;5:453–455. doi: 10.1111/j.1365-294x.1996.tb00336.x. [DOI] [PubMed] [Google Scholar]
- Carvalho N, Afonso P, Santos R.S.A. The haremic mating system and mate choice in the wide-eyed flounder, Bothus podas. Environ. Biol. Fish. 2003;66:249–258. [Google Scholar]
- Castric V, Bernatchez L, Belkhir K, Bonhomme F. Heterozygote deficiencies in small lacustrine populations of brook charr Salvelinus fontinalis Mitchill (Pisces, Salmonidae): a test of alternative hypothesis. Heredity. 2002;89:27–35. doi: 10.1038/sj.hdy.6800089. [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B. Inbreeding depression and its evolutionary consequences. Annu. Rev. Ecol. Syst. 1987;18:237–268. [Google Scholar]
- Corten A. The role of “conservatism” in herring migrations. Rev. Fish Biol. Fish. 2002;11:339–361. [Google Scholar]
- De Veen J.F. On selective tidal transport in the migration of North Sea plaice (Pleuronectes platessa) and other flatfish species. Neth. J. Sea Res. 1978;12:115–147. [Google Scholar]
- Forster G.R. The spawning behaviour of plaice. J. Mar. Biol. Assoc. UK. 1953;31:319. [Google Scholar]
- Frankham R. Effective population size/adult population size ratios in wildlife: a review. Genet. Res. 1995;66:95–107. doi: 10.1017/S0016672308009695. [DOI] [PubMed] [Google Scholar]
- Gaffney P.M, Scott T.M, Koehn R.K, Diehl W.J. Interrelationships of heterozygosity, growth rate and heterozygote deficiencies in the coot clam, Mulinia lateralis. Genetics. 1990;124:687–699. doi: 10.1093/genetics/124.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach G, Schardt U, Eckmann R, Meyer A. Kin-structured subpopulations in Eurasian perch (Perca fluviatilis L.) Heredity. 2001;86:213–221. doi: 10.1046/j.1365-2540.2001.00825.x. [DOI] [PubMed] [Google Scholar]
- Ginot F, Bordelais I, Nguyen S, Gyapay G. Correction of some genotyping errors in automated fluorescent microsatellite analysis by enzymatic removal of one base overhangs. Nucleic Acids Res. 1996;24:540–541. doi: 10.1093/nar/24.3.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomelyuk V.E, Shchetkov S.Y. Small-scale spatial structure of two flatfish species, in Peter the Great Bay, Sea of Japan. J. Mar. Biol. Assoc. UK. 1999;79:509–520. [Google Scholar]
- Hauser L, Adcock G.J, Smith P.J, Ramirez J.H.B, Carvalho G.R. Loss of microsatellite diversity and low effective population size in an overexploited population of New Zealand snapper (Pagrus auratus) Proc. Natl Acad. Sci. USA. 2002;99:11742–11747. doi: 10.1073/pnas.172242899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgecock D. Does variance in reproductive success limit effective population sizes of marine organisms? In: Beaumont A.R, editor. Genetics and evolution of aquatic organisms. Chapman & Hall; London: 1994. pp. 122–134. [Google Scholar]
- Hedrick P.W. Jones and Bartlett Publishers; Sudbury: 2000. Genetics of populations. [Google Scholar]
- Hoarau G, Cook C, Stam W.T, Olsen J.L. New microsatellite primers for plaice, Pleuronectes platessa L. (Teleostei: Pleuronectidae) Mol. Ecol. Notes. 2002;2:60–61. [Google Scholar]
- Hoarau G, Rijnsdorp A.D, Van der Veer H.W, Stam W.T, Olsen J.L. Population structure of plaice (Pleuronectes platessa L.) in Northern Europe: microsatellites revealed large scale spatial and temporal homogeneity. Mol. Ecol. 2002;11:1165–1176. doi: 10.1046/j.1365-294x.2002.01515.x. [DOI] [PubMed] [Google Scholar]
- Hoarau G, Piquet A.M.-T, Rijnsdorp A.D, Van Der Veer H.W, Stam W.T, Olsen J.L. Population structure of plaice (Pleuronectes platessa) in northern Europe: a comparison of resolving power between microsatellite and mtDNA data. J. Sea Res. 2004;51:183–190. [Google Scholar]
- Hunter E, Metcalfe J.D, Reynolds J.D. Migration route and spawning area fidelity by North Sea plaice. Proc. R. Soc. B. 2003;270:2097–2103. doi: 10.1098/rspb.2003.2473. 10.1098/rspb.2003.2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchings J.A. Collapse and recovery of marine fishes. Nature. 2000;406:882–885. doi: 10.1038/35022565. [DOI] [PubMed] [Google Scholar]
- Hutchinson W.F, Carvalho G.R, Rogers S.I. A non destructive technique for the recovery of DNA from dried fish otoliths for subsequent molecular genetic analysis. Mol. Ecol. 1999;8:893–894. [Google Scholar]
- Hutchinson W.F, van Oosterhout C, Rogers S.I, Carvalho G.R. Temporal analysis of archived samples indicates marked genetic changes in declining North Sea cod (Gadus morhua) Proc. R. Soc. B. 2003;270:2125–2132. doi: 10.1098/rspb.2003.2493. 10.1098/rspb.2003.2493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ICES Working Group on the Assessment of Demersal Stocks in the North Sea and Skagerrak. ICES CM 2003/ACFM:2.
- Mathieu E, Autem M, Roux M, Bonhomme F. Epreuve de validation dans l'analyse de structures génétiques multivariées: comment tester l'équilibre panmictique? Rev. Stat. Appl. 1990;38:47–66. [Google Scholar]
- Nei M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics. 1978;89:583–590. doi: 10.1093/genetics/89.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouyaud L, Desmarais E, Chenuil A, Agnese T.F, Bonhomme F. Kin cohesiveness and possible inbreeding in the mouthbrooding tilapia Sarotherodon melanotheron (Pisces Cichlidae) Mol. Ecol. 1999;8:803–812. [Google Scholar]
- Rice W.R. Analyzing tables of statistical tests. Evolution. 1989;43:223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x. [DOI] [PubMed] [Google Scholar]
- Rijnsdorp A.D. Maturation of male and female North Sea plaice (Pleuronectes platessa L.) J. Cons. Int. Explor. Mer. 1989;46:35–52. [Google Scholar]
- Rijnsdorp A.D. Changes in fecundity of female North Sea plaice (Pleuronectes platessa L.) between three periods since 1900. ICES J. Mar. Sci. 1991;48:253–280. [Google Scholar]
- Rijnsdorp A.D. Fisheries as a large-scale experiment on life-history evolution—disentangling phenotypic and genetic-effects in changes in maturation and reproduction of north-sea plaice, Pleuronectes platessa L. Oecologia. 1993;96:391–401. doi: 10.1007/BF00317510. [DOI] [PubMed] [Google Scholar]
- Rijnsdorp A.D. Population regulating processes during the adult phase in flatfish. Neth. J. Sea Res. 1994;32:207–223. [Google Scholar]
- Rijnsdorp A.D, Jaworski A. Size-selective mortality in plaice and cod eggs—a new method in the study of egg mortality. J. Cons. Int. Explor. Mer. 1990;47:256–263. [Google Scholar]
- Rijnsdorp A.D, Millner R.S. Trends in population dynamics and exploitation of North Sea plaice (Pleuronectes platessa L.) since the late 1800s. ICES J. Mar. Sci. 1996;53:1170–1184. [Google Scholar]
- Rijnsdorp A.D, Buys A.M, Storbeck F, Visser E.G. Micro-scale distribution of beam trawl effort in the southern North Sea between 1993 and 1996 in relation to the trawling frequency of the sea bed and the impact on benthic organisms. ICES J. Mar. Sci. 1998;55:403–419. [Google Scholar]
- Rowe S, Hutchings J.A. Mating systems and the conservation of commercially exploited marine fish. Trends Ecol. Evol. 2003;18:567–572. [Google Scholar]
- Saccheri I, Kuussaari M, Kankare M, Vikman P, Fortelius W, Hanski I. Inbreeding and extinction in a butterfly metapopulation. Nature. 1998;392:491–493. [Google Scholar]
- Smith P.J, Francis R.I.C.C, McVeagh M. Loss of genetic diversity due to fishing pressure. Fish. Res. 1991;10:309–316. [Google Scholar]
- Solmundsson J, Karlsson H, Palsson J. Sexual differences in spawning behaviour and catchability of plaice (Pleuronectes platessa) west of Iceland. Fish. Res. 2003;61:57–71. [Google Scholar]
- Stoner A.W, Bedja A.J, Manderson J.P, Phelan B.A, Stehlik L.L, Pessutti J.P. Behaviour of winter flounder, Pseudopleuronectes americanus, during the reproductive season: laboratory and filed observation on spawning, feeding, and locomotion. Fish. Bull. 1999;97:999–1016. [Google Scholar]
- Turner T.F, Wares J.P, Gold J.R. Genetic effective size is three order of magnitude smaller than adults census size in an abundant, estuarine-dependant marine fish (Scianops ocellatus) Genetics. 2002;162:1329–1339. doi: 10.1093/genetics/162.3.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Veer H.W, Pihl L, Bergman M.J.N. Recruitment mechanisms in North Sea plaice Pleuronectes platessa. Mar. Ecol. Prog. Ser. 1990;64:1–12. [Google Scholar]
- Van Dyke F. McGraw-Hill; New York: 2003. Conservation biology. [Google Scholar]
- Van Oosterhout C, Hutchinson B, Wills D, Shipley P. Microchecker: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes. 2004;4:535–538. [Google Scholar]
- Waldman B, McKinnon J.S. Inbreeding and outbreeding in fishes, amphibians and reptiles. In: Thornbel N.W, editor. The natural history of inbreeding and outbreeding. University of Chicago Press; 1993. [Google Scholar]
- Wang J. A pseudo-likelihood method for estimating effective population size from temporally spaced samples. Genet. Res. 2001;78:243–257. doi: 10.1017/s0016672301005286. [DOI] [PubMed] [Google Scholar]
- Watts P.C, Nash R.D.M, George S.G, Kemp S.J. Isolation and characterization of microsatellite loci in the European plaice, Pleuronectes platessa L. (Teleostei: Pleuronectidae) Mol. Ecol. 1999;8:2151–2152. doi: 10.1046/j.1365-294x.1999.00802-6.x. [DOI] [PubMed] [Google Scholar]
- Weir B.S, Cockerham C.C. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Wright S. The theory of gene frequencies. vol. 2. University of Chicago Press; 1969. Evolution and the genetics of populations. [Google Scholar]
- Zijlstra J.J. Herring larvae in the central North Sea. Ber. Dt. Wiss. Komm. Meeresforsh. 1970;21:92–115. [Google Scholar]
- Zouros E, Foltz D.W. Possible explanation of heterozygote deficiency in bivalve molluscs. Malacologia. 1984;25:583–591. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.