Abstract
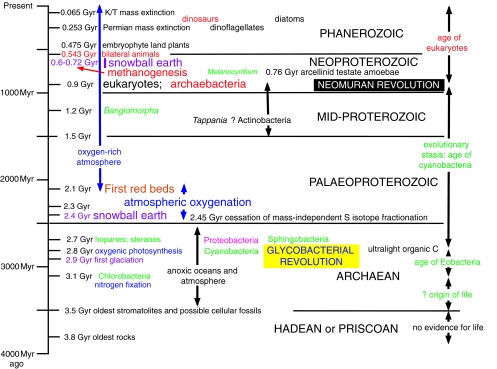
This synthesis has three main parts. The first discusses the overall tree of life and nature of the last common ancestor (cenancestor). I emphasize key steps in cellular evolution important for ordering and timing the major evolutionary innovations in the history of the biosphere, explaining especially the origins of the eukaryote cell and of bacterial flagella and cell envelope novelties. Second, I map the tree onto the fossil record and discuss dates of key events and their biogeochemical impact. Finally, I present a broad synthesis, discussing evidence for a three-phase history of life. The first phase began perhaps ca 3.5 Gyr ago, when the origin of cells and anoxic photosynthesis generated the arguably most primitive prokaryote phylum, Chlorobacteria (=Chloroflexi), the first negibacteria with cells bounded by two acyl ester phospholipid membranes. After this ‘chlorobacterial age’ of benthic anaerobic evolution protected from UV radiation by mineral grains, two momentous quantum evolutionary episodes of cellular innovation and microbial radiation dramatically transformed the Earth's surface: the glycobacterial revolution initiated an oxygenic ‘age of cyanobacteria’ and, as the ozone layer grew, the rise of plankton; immensely later, probably as recently as ca 0.9 Gyr ago, the neomuran revolution ushered in the ‘age of eukaryotes’, Archaebacteria (arguably the youngest bacterial phylum), and morphological complexity. Diversification of glycobacteria ca 2.8 Gyr ago, predominantly inhabiting stratified benthic mats, I suggest caused serial depletion of 13C by ribulose 1,5-bis-phosphate caboxylase/oxygenase (Rubisco) to yield ultralight late Archaean organic carbon formerly attributed to methanogenesis plus methanotrophy. The late origin of archaebacterial methanogenesis ca 720 Myr ago perhaps triggered snowball Earth episodes by slight global warming increasing weathering and reducing CO2 levels, to yield runaway cooling; the origin of anaerobic methane oxidation ca 570 Myr ago reduced methane flux at source, stabilizing Phanerozoic climates. I argue that the major cellular innovations exhibit a pattern of quantum evolution followed by very rapid radiation and then substantial stasis, as described by Simpson. They yielded organisms that are a mosaic of extremely conservative and radically novel features, as characterized by De Beer's phrase ‘mosaic evolution’. Evolution is not evenly paced and there are no real molecular clocks.
Keywords: neomura, snowball Earth, archaebacteria, eobacteria, eukaryote origin, glycobacteria
1. Introduction
Living organisms can be studied in immensely more detail than can fossils. Even without fossils, comparative phylogenetic studies now provide remarkably detailed and accurate reconstructions of evolutionary relationships across the whole tree of life and rigorously inferred ancestral states for most of it. However, reconstructing the history of life requires integrating insights from palaeontology and neontology: comparative study of extant organisms. Without palaeontological evidence the tree lacks a time-scale; inferences would lack the important control of the temporally layered evidence in the rocks; and we would lack direct knowledge of entirely extinct groups. Temporal evidence from palaeontology also helps place the root on the universal tree, and valuably checks inferences about the temporal direction of changes in many parts of it. Moreover, because we all sometimes make interpretative mistakes, each discipline offers the other a valuable external perspective to help correct them.
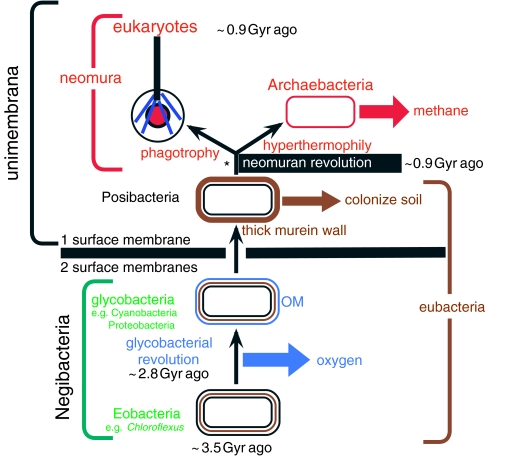
Molecular and cell biological evidence on the tree of life is much more congruent with the fossil record than often supposed; most apparent conflict arises unnecessarily from interpretative errors by neontologists and palaeontologists (Cavalier-Smith 2002a,b). In particular, using steranes as a biomarker for eukaryotes is problematic, as several phylogenetically diverse bacteria can make sterols, including some actinobacteria, which I consider ancestral to both eukaryotes and archaebacteria (i.e. to all neomura); I recently argued that no pre-Neoproterozoic fossils are confidently assignable to any eukaryote phylum or even indubitably eukaryotic (Cavalier-Smith 2002a). Understanding early eukaryote evolution has greatly improved since then, especially evidence for the root of the tree (Stechmann & Cavalier-Smith 2002, 2003; Richards & Cavalier-Smith 2005) and for the new supergroup Rhizaria, which includes abundant fossil foraminifera and radiolaria (Nikolaev et al. 2004), while molecular cell biology has advanced so greatly that the origin of eukaryotes is more specifically explicable than ever before. Cavalier-Smith (2006) placed the topology of the bacterial part of the universal tree of life on firmer grounds and explained how several transitions within it can be confidently polarized by strong evolutionary arguments. I concluded that the root of the tree of life is within negibacteria: Gram-negative eubacteria possessing two envelope membranes. On that interpretation the single surface membrane in posibacteria, archaebacteria and eukaryotes is a derived evolutionary condition that arose by losing the OM (figure 1). That the first cells were double-enveloped negibacteria and that their OM was lost only once in the history of life are not new ideas (Blobel 1980; Cavalier-Smith 1987a,b, 1991a, 2001, 2002a, 2004a), but is increasingly favoured by these new polarizations of major evolutionary transitions and new evidence for monophyly of Posibacteria.
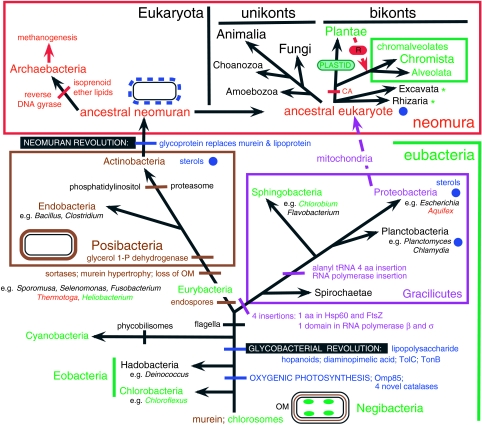
Figure 1.
The tree of life emphasizing major revolutions in cell structure. The most basic distinction in cell biology is between negibacteria, with a cell envelope of two distinct lipid bilayer membranes, and unimembrana, with but one surface membrane. Contrary to long-standing assumptions that cells first had only one membrane, phylogenetic analysis and palaeontology jointly show that the last common ancestor of all life was a negibacterium with two surface membranes (Cavalier-Smith 1987b, 2002a, 2006a), and that unimembrana evolved from them by losing the outer membrane (OM). Within negibacteria and unimembrana two revolutions in cell-surface chemistry created radically novel bacteria whose diversification transformed the biosphere and biogeochemical cycles. The glycobacterial revolution, soon after a photosystem duplication enabling oxygenic photosynthesis, complexified the OM by adding lipopolysaccharide and novel transport machinery. Nearly 2 Gyr later, the neomuran revolution made cell surfaces potentially more flexible by replacing the rigid eubacterial corset of cross-linked murein peptidoglycan by separate glycoproteins. The neomuran revolution was closely followed by the origin of the eukaryote cell; this entailed the origin of phagotrophy, the endomembrane system and endoskeleton and enslavement of a negibacterium (specifically an α-proteobacterium) to create mitochondria (not shown: see figures 5 and 6; Cavalier-Smith 1987b, 2002c, in press). At about the same time, life belatedly colonized hot, acid environments by evolving the ancestrally hyperthermophilic archaebacteria, sisters—not ancestors—of eukaryotes; distinctly later, one archaebacterial lineage evolved biological methanogenesis. The asterisk shows a widely assumed, incorrect position for the root of the tree that was based on a few paralogue trees for a subset of proteins that underwent an episode of such extensive quantum evolution during the neomuran revolution as to fabricate that misleading position of the root (between neomura and eubacteria) by a long-branch phylogenetic reconstruction artefact (Cavalier-Smith 2002a, 2006a). Metabolic enzyme paralogue trees typically place the root within Negibacteria, but in inconsistent places (Peretó et al. 2004).
In this paper I attempt to map the tree of life onto the fossil record, arguing that they are broadly congruent, and sketch a synthesis. I discuss evidence for a three-phase history of life.
Before discussing the tree, I outline three major dichotomies in cell organization among bacteria, each almost as important as the distinction between bacteria and eukaryotes for understanding the major steps in cell evolution, but less generally appreciated. I use ‘bacteria’ as in classical terminology for all prokaryotes; it is misleading and confusing to restrict ‘bacteria’ to eubacteria alone, excluding archaebacteria from the concept.
I shall argue that endogenous biological limitations to megaevolutionary innovations in cell structure probably dominated timing of the most fundamentally innovative changes in the biosphere. The immense difficulty of the neomuran revolution and eukaryogenesis accounts for the late origin of plants (by cyanobacterial enslavement), animals (by evolving epithelia and connective tissue), fungi (by evolving tip-growing chitin walls) and chromalveolates (by secondary enslavement of a red alga), and soon thereafter their Cambrian explosive radiations. The relatively early origin of cyanobacteria and proteobacteria, and far later origin of methanogenic archaebacteria, was probably responsible, respectively, for the Palaeoproterozoic and Neoproterozoic snowball Earth episodes by altering greenhouse gas balances.
2. Fundamental cell diversity
(a) The contrast between neomura and eubacteria
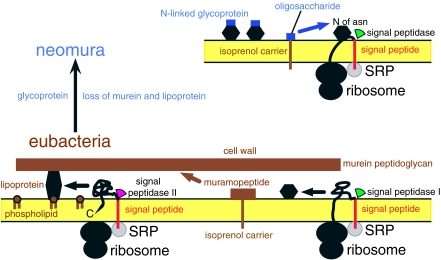
The largest single phenotypic and genetic dichotomy in the living world is between prokaryotes and eukaryotes, but an earlier, more basic dichotomy is between eubacteria (i.e. negibacteria plus posibacteria) and neomura (archaebacteria and eukaryotes; Cavalier-Smith 1987b). The transition between them involved major changes in the cell surface and types of conjugated proteins synthesized cotranslationally, i.e. during protein synthesis itself (figure 2). In all cells, membrane proteins and some or all secreted proteins are made by ribosomes attached to a membrane by a ribonucleoprotein particle—the signal recognition particle (SRP). The SRP possibly evolved even before cells (Cavalier-Smith 2001) and is conserved throughout life, but more complex in neomura than eubacteria, one of many reasons for considering neomura more advanced and to have evolved from eubacteria, not the reverse (Cavalier-Smith 2002a). Neomura and eubacteria also differ greatly in their DNA-handling enzymes—those mediating DNA replication and repair, or transcribing genes. Differences in replication enzymes are sometimes used to suggest that DNA replication involved independently in eubacteria and neomura. This exaggerates their significance. Much more likely, a bout of quantum evolution (Simpson 1944) rapidly modified pre-existing enzymes in the cenancestor of neomura; I suggested this was connected with an inferred origin of core histones then (Cavalier-Smith 2002a). Discovery of crenarchaeote core histone genes (Cubonova et al. 2005) strengthens that inference. Of 19 derived neomuran characters, 11 are attributable to their common ancestor having been thermophilic (Cavalier-Smith 2002a).
Figure 2.
Fundamental contrasts and continuities between eubacteria and neomura in surface chemistry and protein secretion. The older eubacteria (below) typically have walls of murein peptidoglycan and lipoprotein. Neomura have a surface coat (eukaryotes) or wall (archaebacteria) of N-linked glycoprotein (oligosaccharide covalently attached to protein via asparagine: Asn). Simple surface proteins in eubacteria (lower right hexagon) and protein parts of the conjugated glycoproteins and lipoproteins have hydrophobic N-terminal signal peptides recognized by homologous signal recognition particles (SRP: grey) that then link them to SRP-receptor proteins in the membrane and transfer the signal peptide into cross-membrane protein translocation channels (not shown). Ribosomes making surface proteins temporarily stick to the membrane at ribosome receptors, forcing the growing polypeptide chain through the channel. After synthesis, signal peptidases remove signal peptides of secreted proteins: signal peptidase I, used for simple eubacterial proteins, is homologous and ancestral to the neomuran signal peptidase. The very different signal peptidase II, specific for eubacterial lipoproteins, was lost during the neomuran revolution, as were lipoproteins and murein. Hydrophilic muramopeptide precursors of eubacteria and core oligosaccharides of neomura are moved across the membrane by long-chain amphipathic isoprenol carriers (dolichol in neomura, undecaprenol in eubacteria) to which homologous enzymes temporarily attach them via N-acetylglucosamine (GlcNAc; Cavalier-Smith 2006a) before being incorporated covalently into the wall. SRPs are more complex in neomura, with a novel protein, 19P, binding to an extra helix 6 in SRP RNA that is absent from the simpler, more primitive eubacterial SRP; this extra complexity stops proteins growing further until the ribosome/SRP complex correctly docks onto the membrane, an improvement to cotranslational protein secretion associated with novel cotranslational glycoprotein synthesis in the ancestral neomuran (Cavalier-Smith 2002a). For eubacteria, the condition in Posibacteria only is shown: the more complex negibacterial envelope has an OM outside the murein (figure 3). Neomura and eubacteria also differ substantially in DNA-supercoiling machinery: in both, DNA is similarly negatively supercoiled (i.e. underwound to help strands separate in transcription and replication), but eubacteria achieve this actively by the enzyme DNA gyrase, hydrolysing ATP, whereas neomura wrap DNA passively around core histones as nucleosome particles resembling beads on a string. Replacing DNA gyrase by histones had marked coevolutionary repercussions on DNA-handling enzymes, which were substantially modified during the neomuran revolution; archaebacteria alone invented reverse gyrase by fusing two eubacterial genes, a hyperthermophilic adaptation, showing they and hyperthermophily must be more recent (Cavalier-Smith 2002a).
Despite the notable differences between eubacteria and neomura, most cell properties are not nearly as sharply distinct (Cavalier-Smith 2002a); both groups share over a thousand fundamentally similar characters, ruling out separate origin, sometimes postulated (Koga et al. 1998; Woese 1998; Martin & Russell 2003)—their common ancestor was a complex, highly developed cell (Cavalier-Smith 2002b; Peretó et al. 2004), definitely a eubacterium (Cavalier-Smith 2006a). Two key cellular innovations only made the ancestral archaebacterium: novel acid-stable flagella and novel acid- and heat-stable isoprenoid ether lipids, both secondary adaptations to hyperthermophily (Cavalier-Smith 2002a). Their unique flagella evolved from type IV secretion proteins previously used to make eubacterial pili (Cavalier-Smith 2006a). As eubacteria make isoprenoids, the archaebacterial novelty was to add them to glycerol phosphate with new stereochemistry to make isoprenoid-ether glycerophosphate lipids; the enzyme doing this is related to one found in posibacteria (only), and thus involved no major innovation (Peretó et al. 2004); some archaebacteria even retained the key enzyme making acyl ester lipids. Neither change involved really novel proteins.
Classification of proteins into fold superfamilies reflects their fundamental distinctiveness. Only one novel protein fold superfamily is universal among archaebacteria: that of the enzyme making their unique tRNA base, archaeosine; even this is related to five other fold superfamilies (Yang et al. 2005). By contrast 21 novel fold superfamilies arose during the neomuran revolution (two universal in neomura, i.e. never secondarily lost), whereas eukaryotes have 153 fold superfamilies (14 universal) never in bacteria (Yang et al. 2005). The degree of novel protein evolution during eukaryote origins (eukaryogenesis) was far greater than for archaebacterial origins. Treating archaebacteria as a separate kingdom or superkingdom is unjustified. Their differences from other bacteria are greatly exaggerated by quantitative differences in rRNA, ribosomal proteins and DNA-handling enzymes. The perception of archaebacteria as a third form of life, based on rRNA quantitative differences, has been profoundly misleading; in most characters they are no more distinctive than any other eubacterial phylum and they are younger, despite their name (Cavalier-Smith 2002a). Their two subphyla (Euryarchaeota, Crenarchaeota) were both probably ancestrally anaerobic respirers energetically dependent on sulphur compounds, a much more ancient physiology in eubacteria. By combining this with hyperthermophily, archaebacteria expanded the biosphere in a relatively minor way into previously inaccessible hot/acid environments. No unique protein fold superfamilies are found in all eubacteria but absent from neomura (Yang et al. 2005), as expected if eubacteria are paraphyletic (i.e. ancestral to at least one other group) and the only primary domain of life, and neomura derived. The major richness of the 1244 protein fold superfamilies probably arose in stem bacteria before the cenancestor; the greatest bout of protein innovation since then was during eukaryogenesis, reinforcing the classical distinction between eukaryotes and prokaryotes.
(b) The contrast between negibacteria and posibacteria
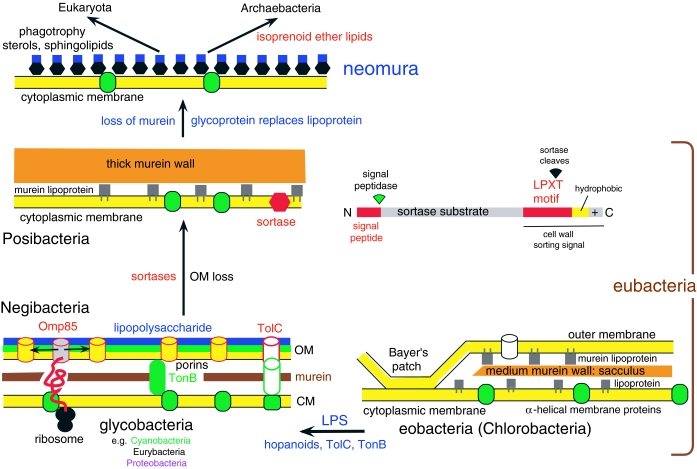
The most fundamental distinction within eubacteria is between Posibacteria, invariably bounded by one membrane (e.g. Bacillus, Streptomyces), and Negibacteria, always having two distinct membranes (e.g. Escherichia, Salmonella, cyanobacteria, spirochaetes; figure 3). The negibacterial inner membrane is homologous to the single cytoplasmic membrane (CM) of posibacteria and archaebacteria (collectively called unibacteria; Cavalier-Smith 2002a), whereas their OM is unique and homologous to the OM of mitochondria and chloroplasts, which evolved from enslaved negibacteria (Cavalier-Smith 2002a, 2006b, in press). All OM lipids are made in the CM and exported to the OM, probably through specific contact sites, Bayer's patches. OM proteins are made by ribosomes on the CM cytosolic face and transported across the periplasmic space (that between the CM and OM) helped by specific chaperones. These lipid- and protein-export mechanisms may be homologous for all negibacteria, but are well studied only in proteobacteria. In negibacteria, the murein peptidoglycan wall, when present—as it was ancestrally (lost only within planctobacteria and by mitochondria and most chloroplasts)—is much thinner than in posibacteria and lies between the CM and OM, to which it is attached by lipoproteins.
Figure 3.
Membrane evolution and the fundamental contrast between negibacteria and posibacteria. Earliest cells were negibacteria bounded by two lipoprotein membranes: an inner cytoplasmic membrane (CM) impermeable to small hydrophilic molecules (with alpha-helical membrane proteins: green) and an OM, more permeable because of large proteinaceous pores, made by porin proteins. Between the two negibacterial membranes is the murein sacculus or cell wall: a gigantic hollow bag-shaped peptidoglycan molecule covalently cross-linked in three dimensions, rigidly enough to resist high internal osmotic pressures of more than 20 atm. The simplest negibacterial envelope (right) is in Chlorobacteria, which lack Omp85 that inserts OM β-barrel proteins in glycobacteria (left) and Hadobacteria (not shown: intermediate in complexity between chlorobacteria and glycobacteria), in both of which all known OM proteins are β-barrels. In eobacteria, the CM and OM both consist of acyl ester phospholipids plus embedded proteins. Glycobacteria retained these, but also have hopanoids that make membranes more rigid, and replaced phospholipids in the OM outer leaflet by the immensely more complex lipopolysaccharide (LPS), which so dramatically reduces OM permeability that they simultaneously evolved novel protein export machinery (using the β-barrel TolC protein) and the TonB importer of small molecules (Cavalier-Smith 2006a). For clarity, lipoproteins and Bayer's patches are not shown for glycobacteria. Posibacteria lack an OM, but have a much thicker wall of murein peptidoglycan supplemented by other polymers (teichoic acids in Endobacteria; a disparate array in Actinobacteria). Murein walls can grow and divide only by cleavage of covalent bonds by murein hydrolases. The transition from negibacteria to posibacteria involved loss of the cell wall and the origin of novel families of sortase enzymes that recognize a C-terminal LPXT motif on numerous extracellular proteins which they cleave and use for attaching them covalently to the cell wall, preventing their escape as would otherwise be likely without an OM. Posibacterial wall-attached proteins include murein lipoproteins with hydrophobic lipid tails embedded in the outer leaflet of the cytoplasmic membrane, not in the OM inner leaflet as in negibacteria.
The lipid tails of posibacterial lipoproteins are in the CM outer leaflet; their protein part is covalently attached to the thick murein wall by sortase enzymes that recognize cell wall sorting signals near the C-terminus of its precursor, after its export to the periplasmic space by SRP (Comfort & Clubb 2004). Prior to this, sortases cleave the hydrophobic C-terminal sequence that temporarily anchors it to the CM. Hundreds of cell wall proteins, mostly not lipoproteins, are thus anchored to the thick murein of posibacteria before the N-terminal signal sequence is cleaved. Most posibacteria have sortases of 2–4 subfamilies with different substrate specificities (Comfort & Clubb 2004). Sortases are a synapomorphy for posibacteria (comprising subphyla Endobacteria and Actinobacteria), strongly supporting their monophyly (Cavalier-Smith 2006a). They probably evolved to attach the wall firmly to the CM and prevent soluble periplasmic proteins escaping when their cenancestor arose by losing the negibacterial OM.
Endobacteria include ancestrally endospore-forming Gram-positive bacteria, e.g. Bacillus, Clostridium, Staphylococcus, and the parasitic Mollicutes (mycoplasmas, spiroplasmas) that secondarily lost murein, lipoproteins and the sortase machinery, and miniaturized their genomes. Endobacteria also exclude the ancestrally endospore-producing Eurybacteria (e.g. Heliobacteria, Sporomusa), which group with them on most trees (Woese et al. 1985), and so are often also misleadingly called low-GC Gram-positives. Eurybacteria are neither Gram-positive nor posibacteria, but negibacteria with OM and endospores; as discussed below, they are probably ancestral to posibacteria (Cavalier-Smith 2006a). Thermotogales are also Eurybacteria, despite frequent, probably artefactual, grouping on sequence trees with neomura and/or Aquificales (Cavalier-Smith 2006a).
(c) The contrast between Eobacteria and glycobacteria
Typical negibacteria (e.g. Proteobacteria and cyanobacteria) rigidify their OM by inserting very complex LPS molecules into the outer leaflet of its lipid bilayer instead of simple phospholipids. I call the six negibacterial phyla ancestrally possessing LPS, invented by their a common ancestor but secondarily lost within Sphingobacteria and Spirochaetae, glycobacteria (Cavalier-Smith 2002a, 2006a). Glycobacteria also use hopanoids to rigidify their CM. Two negibacterial phyla, Chlorobacteria and Hadobacteria, which uniformly lack LPS and hopanoids are grouped as Eobacteria (Cavalier-Smith 2002a, 2006a). Their simpler cell envelopes lack TolC, a unique tubular channel that mediates type I protein-secretion across the OM and TonB—involved in molecular import across the OM, coupling function of OM and CM proteins (figure 3). Cavalier-Smith (2006a) argued that absence of these four complex characters from Eobacteria is the primitive state for bacteria and that the root of the universal tree is within Eobacteria, specifically between Hadobacteria and Chlorobacteria. (Chlorobacteria are widely called Chloroflexi, a name published much later and validated slightly later than Chlorobacteria, thus not used here.)
3. The tree of life and its root
(a) Problems with sequence trees
Sequence trees for single molecules, e.g. rRNA, invaluably cluster together relatively closely related organisms, but suffer from three fundamental problems. First, they have insufficient information to resolve the order of closely spaced branches, e.g. those diverging in rapid radiations—the norm for major classes within a phylum or major phyla within a kingdom. Thus, the branching order of eubacterial phyla is unresolved on single-gene trees, as are most basal branches among eukaryotes; the archaebacterial tree is better resolved (Gribaldo & Brochier 2006), probably because it has only two basal branches—interchanging them makes no difference. Second, some genes can occasionally be transferred laterally among unrelated organisms, so their tree partially reflects horizontal transfer of individual genes, not vertical phylogeny of the organisms containing them (Philippe & Douady 2003; Lerat et al. 2005). Third, systematic biases in evolutionary rates and modes of virtually all genes in at least parts of the tree can make the branching order partly incorrect (Gribaldo & Philippe 2002; Lopez et al. 2002). Problems of random error through insufficient sequence sampling and misleading lateral transfers can be largely solved by making trees from several or many different genes. Multigene trees may also reduce systematic biases by cancelling them out, when they vary randomly among genes and taxa. But multigene trees can be positively misleading and mathematically certain to give the wrong topology, given sufficiently great genome-wide systematic biases in sequence evolutionary rates or modes among taxa or biases shared by many genes (Gribaldo & Philippe 2002; Lopez et al. 2002). Such problems are more widespread than assumed in the early days of tree reconstruction. They are exacerbated in trees with relatively few taxa or by phylogenetic algorithms with assumptions that poorly model the actual processes of sequence divergence (and convergence—a serious problem with long branches). Sequence trees also lack inherent evidence of evolutionary direction or the position of the root.
Cladistic analyses of morphological or macromolecular characters sufficiently complex to rule out convergence are often the most reliable way of establishing relationships, though can be confused by secondary evolutionary losses. Cladistic analysis of genetic characters (e.g. shared lateral gene transfers, gene replacements, gene fusions) is also important. Best phylogenetic practice seeks congruence between all types of evidence. One should try to understand reasons for any conflicts and biases and give more weight to the strongest evidence and most reliable methods.
(b) Rooting the universal tree
This is the most difficult of all phylogenetic problems (Cavalier-Smith 2002a, 2006a). Even for subtrees, e.g. eukaryotes alone, placing the root is more difficult than obtaining a reliable unrooted topology. This is particularly so for sequence trees when these conditions apply: outgroups have long branches; some ingroup taxa have very long branches; multiple basal radiations occur in a small fraction of overall evolutionary time. For many genes and most groups, all three complicating conditions are the rule not the exception, making judicious weighing and intellectual synthesis of conflicting evidence mandatory. Gene duplications can, in principle, provide direction because they occurred after the unduplicated state; combining trees from duplicated sisters in one analysis makes each outgroup to the other, and can theoretically place the root. Unfortunately, gene duplication commonly involves marked differentiation in function, so duplicates diverge greatly prior to diversification of their descendants, typically giving such a long stem to each subtree that long-branch tree reconstruction artefacts dominate, giving a false topology and false, often conflicting, rooting (Cavalier-Smith 2002a, 2006a).
Two more reliable approaches can polarize trees, and thereby root them: transition analysis and the fossil record. Transition analysis of sufficiently complex characters can often polarize evolutionary change, as recently explained for the origin of neomura (Cavalier-Smith 2002a) and numerous transitions within bacteria (Cavalier-Smith 2006a). Section 3c uses the origin of posibacteria to illustrate how transition analysis can polarize a major evolutionary step. Transition analysis can establish ancestor–descendant relationships. It gives a relative time-scale to biological evolution, analogous to that in geology yielded by the principle that younger sedimentary strata generally overlie older ones. But it cannot give absolute times to evolution as radiometric clocks can for igneous rocks. Nor does sequence evolution give an absolute time-scale. The idea of a molecular clock for sequences is misleadingly bad terminology. Rates of nucleotide substitution can vary million-fold among molecules when functions change dramatically (as often following gene duplication), and can change thousands-fold for a single molecule with basically unchanged function (Cavalier-Smith 2002b). Sometimes, for short evolutionary periods and restricted lineages, rates of change may be steadier and useful for rough interpolation between dates established by fossils. But fossil dates must be paramount. Extrapolating beyond them is always risky and sometimes grotesquely wrong.
(c) Negibacteria preceded posibacteria
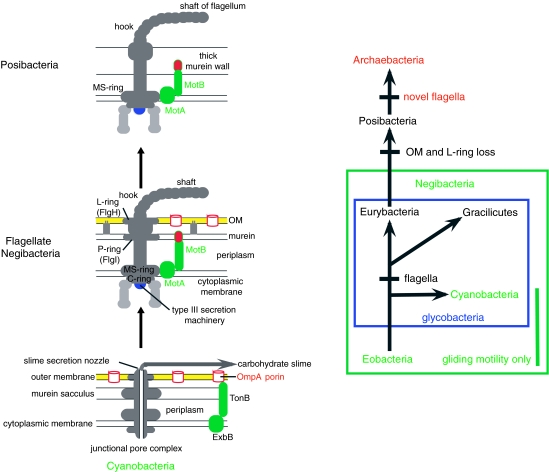
The transition between negibacteria and posibacteria occurred in cells possessing eubacterial flagella (Cavalier-Smith 2006a). Determining whether flagella originally evolved in posibacteria or in negibacteria polarizes the transition. Eubacterial flagella have three parts: a static motor embedded in the CM, typically covalently attached to the murein wall; the cylindrical basal body, rotated by a proton current through the associated motor; the helical shaft outside the CM passively transmits rotation to the environment. As evolutionary precursors of both basal body and motor exist only in negibacteria, never posibacteria, flagella evolved first in negibacteria and were transmitted vertically to the first posibacterium (figure 4). This unambiguously polarizes evolution from negibacteria to posibacteria, not the reverse, so the ancestral posibacterium lost the OM; the simplest mechanistic cause of this loss was sudden murein hypertrophy thickening the wall enough to break contacts between the CM and OM at the Bayer's patches, thus preventing transfer of lipids and LPS to the OM, causing its irreversible loss (Cavalier-Smith 1987b). OM loss had two coevolutionary consequences: loss of flagellar L-rings that in negibacteria except spirochaetes embed the basal body in the OM, and origin of the posibacterial sortase enzymes to attach periplasmic proteins to the thick murein wall, thereby preventing their loss to the environment when the OM disappeared.
Figure 4.
Flagellar origin unambiguously polarizes evolution from negibacteria to posibacteria. The TonB importer and its force-providing proton channel ExbB are homologues of the motor proteins MotB and MotA of eubacterial flagella and probably their evolutionary precursors (all in green); they are found in all glycobacterial phyla, including the totally non-flagellate Cyanobacteria, but never in Posibacteria or Eobacteria. The other key flagellar precursor was probably the hollow cylindrical junctional pore complex of cyanobacteria that carries a slime secretion nozzle used for gliding motility, which arguably became the flagellar basal body (Cavalier-Smith 2006a). Eubacterial flagella probably originated by novel functional association between these previously unrelated precursors: proton inflow through the MotA channel now powers basal body rotation instead of molecular import across the OM like ExcB. Both TonB and MotB are fixed to the rigid murein layer. To enable the cell to swim and not merely rotate relative to its secreted slime, it evolved the hook and shaft, made from a protein family that arose by gene duplication from unknown precursors (slime nozzle constituent?). As motor and basal body precursors are both totally absent from posibacteria, and MotB betrays a chimaeric ancestry from TonB and OM protein OmpA, also never in posibacteria, flagella cannot have arisen in a cell lacking an OM. They must have arisen in a negibacterium and been inherited by posibacteria. Therefore, posibacteria are derived from negibacteria. Evolution occurred in the direction of the arrows not its reverse. The L-ring bushing that lodges the distal end of the basal body in the OM of typical negibacteria was probably lost after the OM, when its function disappeared in the first posibacterium (Cavalier-Smith 2006a). Archaebacterial flagella are unrelated (see text); eubacterial flagella were lost with murein during the neomuran revolution. The phylogeny on the right shows the three taxa with eubacterial flagella in black; Gracilicutes include Proteobacteria and three other phyla (see figure 5). Cyanobacteria are sisters to these flagellate taxa, not their direct ancestors; therefore, the junctional pore complex evolved prior to their common ancestor.
A second argument polarizing the transition from negibacteria to posibacteria is the mechanistic difficulty of adding an OM in one step to a flagellate posibacterium; nobody has explained how this could be done while plausibly allowing OM biogenesis. The difficulty of evolution is that hypothetical direction is compounded by the closest negibacterial relatives of posibacteria being Eurybacteria, which are glycobacteria with a much more complex OM than eobacteria, which are arguably more primitive (Cavalier-Smith 2006a). If evolution were in that direction, the OM must immediately have evolved hugely complex LPS and other complexities shared by glycobacteria, e.g. hopanoids, porins, TolC. If instead the first negibacteria were eobacteria, especially chlorobacteria with much the simplest OM, their origin is much simpler; I explained how the OM could have evolved simply and gradually to generate the first eobacterium (Cavalier-Smith 2001).
(d) Eobacteria preceded glycobacteria
The strongest argument that eobacteria are older than glycobacteria concerns OM protein biogenesis (Cavalier-Smith 2006a). In all glycobacteria and in the eobacterial Hadobacteria (e.g. Deinococcus, Thermus), the diverse OM proteins are all β-barrel proteins; β-barrel membrane proteins do not occur in the CM, being specific for the OM of negibacteria (and the OM of mitochondria and chloroplasts, once enslaved glycobacteria). Negibacteria make them by ribosomes on the CM, but after signal peptide cleavage they traverse the periplasmic space, helped by periplasmic chaperone proteins, and are inserted into the OM by a catalyst, Omp85, itself a β-barrel protein embedded in the OM. In these bacteria, OM biogenesis absolutely requires Omp85; if it is mutationally removed Escherichia coli cells die (Gentle et al. 2005). Homologues of Omp85 (Sam50 in mitochondria, Omp75 in chloroplasts) are essential for biogenesis of the profoundly modified, formerly negibacterial envelopes of these organelles, even though all their β-barrel proteins are now made in the host cytosol; deleting the yeast gene for Sam50 kills them (Gentle et al. 2005). As these organellar envelopes lost so many negibacterial components (e.g. murein, LPS, lipoprotein), but obligately retain Omp85 homologues for inserting β-barrel proteins (e.g. themselves and porins), Omp85 could probably never be lost in evolution—unless the whole OM were lost as when posibacteria originated, unless every OM β-barrel protein was lost first.
I argued that Omp85 was never lost by any organism that retained the OM; its apparent absence from chlorobacteria, alone among negibacteria, means that chlorobacteria are the most primitive negibacteria, originating before the Omp85-mediated mechanism for inserting OM β-barrel proteins (Cavalier-Smith 2006a). Thus, chlorobacteria are the most ancient negibacteria. Combining this argument with the polarization from negibacteria to posibacteria, chlorobacteria must also be the first eubacteria. Chlorobacteria are also simpler than Hadobacteria and glycobacteria in lacking 10 other widespread bacterial features (Cavalier-Smith 2006a).
(e) Archaebacteria are sisters to eukaryotes not their ancestors
Although it is sometimes suggested or assumed that archaebacteria are ancestral to eukaryotes, extensive and varied evidence strongly supports archaebacterial holophyly (being sisters to eukaryotes); none of any consequence supports their paraphyly (being our ancestors).
Ribosomal RNA trees and all well-resolved and taxonomically well-sampled sequence trees for single proteins, and for combined data from many proteins show eukaryotes as sisters to archaebacteria, not nested within archaebacteria (Clarke et al. 2002). Holophyly of archaebacteria was strongly favoured by their unique isoprenoid ether lipids (Cavalier-Smith 1987b). If the common ancestor of archaebacteria and eukaryotes had such lipids, but no acyl ester lipids (present throughout eubacteria), the ancestral eukaryote must have reacquired acyl ester lipids and totally lost isoprenoid ethers, a very non-parsimonious assumption. As the eukaryote cenancestor already had a mitochondrion (Cavalier-Smith 2002b, 2003b, 2004b, 2006b; Stechmann & Cavalier Smith 2003; Embley 2006), a protoeukaryote with only isoprenoid ether lipids could theoretically have replaced them entirely by acyl ester lipids from the enslaved proteobacterium (Martin & Müller 1998), but this is mechanistically and evolutionarily extremely improbable. A further flaw in that improbable hypothesis is that proteobacteria lack phosphatidylinositol, one of the most important eukaryote phospholipids, required for eukaryote-specific cell signalling. The only eubacteria having phosphatidylinositol are Actinobacteria, which are ancestral or sister to neomura according to much other evidence (Cavalier-Smith 2002a, 2006a). Thus, eukaryote membrane lipids probably came vertically from an actinobacterial ancestor, archaebacterial lipids originating in their cenancestor after it diverged from eukaryotes (Cavalier-Smith 1987b). Archaebacteria, eukaryotes and neomura are all holophyletic.
Three proteins found only in archaebacteria, plus archaebacterial flagella, are synapomorphies for archaebacteria and evidence for their holophyly (Cavalier-Smith 2002a). Splitting the genes for RNA polymerase A (=β′ in eubacteria) and glutamate synthetase (GS) to code two and three proteins, respectively, is also a shared derived character for archaebacteria (Nesbo et al. 2001; Cavalier-Smith 2002a; two halophilic archaebacteria, Natronomonas and Haloarcula reacquired an unsplit GS gene by lateral transfer from a eubacterium; their GS is far more similar to that of the halophilic Salinibacter ruber (57% identity) than to archaebacterial ones—my Blast results). My database Blast analysis reveals no exceptions yet to the rule that neomuran polymerase A is split into separate A′ and A″ proteins in archaebacteria (Bartlett et al. 2004); this would have to have been reversed by gene fusion if, contrary to my arguments, archaebacteria were ancestral to eukaryotes, as no eubacterium could have provided this characteristically neomuran molecule to the cenancestral eukaryote by lateral gene transfer (LGT; nor could the mitochondrial ancestor). Another argument for archaebacterial holophyly comes jointly from MreB and the largest subunit of RNA polymerase. MreB, the probable eubacterial ancestor of actin and actin-related proteins (Cavalier-Smith 2002a), is found in most eubacteria (all phyla) but is rare in archaebacteria, found only in Methanopyrus and Methanothermobacter; probably other archaebacteria lost it—four times to generate the major archaebacterial clades, none of which could have been ancestral to eukaryotes as they lack MreB, which initiated eukaryogenesis by becoming actin for phagotrophy. RNA polymerase B is split in most archaebacteria including these two; such archaebacteria could not be ancestral to eukaryotes unless the split were reversed. The polymerase B split and MreB losses together exclude all archaebacterial branches as eukaryotes ancestors; the only remaining possibility (a stem lineage near the crenarchaeote/euryarchaeote divergence and first divergence within euryarchaeotes) would be disallowed if MreB entered Methanopyrus/Methanothermobacter by lateral transfer, not vertical inheritance. Scores of universal proteins and 310 protein domain superfamilies shared by eukaryotes and eubacteria are never found in archaebacteria (Cavalier-Smith 2002a; Yang et al. 2005); none could have entered eukaryotes vertically if archaebacteria were our ancestors. Probably most were vertically inherited by eukaryotes from a eubacterial ancestor prior to separation of pre-eukaryote and pre-archaebacterial lineages.
(f) Neomura evolved from actinobacteria
Eubacteria are monophyletic with a common ancestor having walls of peptidoglycan murein and lipoprotein. Neomura are also monophyletic with a common ancestor with N-linked glycoprotein, no murein or lipoprotein, and almost certainly acyl ester but no isoprenoid ether membrane lipids. This ancestor had only one membrane, like Posibacteria. It is far more parsimonious that neomura evolved from a posibacterium than that their unimembranous state arose independently (by OM loss or independent origin of the cell envelope). The possibility of independent membrane origins in eubacteria and neomura (Koga et al. 1998; Martin & Russell 2003) is refuted by numerous integral membrane proteins associated with the most basic functions of lipid membranes being homologous and shared between eubacteria and archaebacteria, e.g. SRP receptors, ribosome receptors/protein translocation channels, cytochrome b, and by cytoskeletal/division proteins (e.g. FtsZ) that act on lipid membranes. A specific relationship between neomura and the posibacterial Actinobacteria was suggested by shared protein characters (Cavalier-Smith 1987b, 2002a).
Of key importance is the proteasome, a tiny cylindrical protein-digestion chamber shared by all neomura and many actinobacteria only. Other eubacteria have no homologue or else a simpler one, HslV. HslV protease is a squat hollow cylinder of 12 identical proteins comprising two laterally attached hexameric rings. The proteasome core has two different proteins, related to HslV and each other, arranged as four heptameric rings making a longer and wider digestion chamber than HslV (Gille et al. 2003). I argued that core proteasomes evolved by duplicating HslV and its divergence into functionally different subunits (α, β) in the common ancestor of neomura and actinobacteria with proteasomes (Cavalier-Smith 2006a). The inner rings (β-subunits) retained protease activity but lost the ATPase-binding activity of HslV, whereas α-subunits lost protease activity and became able to bind a novel regulatory ATPase (Cavalier-Smith 2006a). Reverse evolution from this complex differentiated proteasome core to HslV by losing α-subunits and de novo acquisition by β-subunits of ATPase-binding capacity is mechanistically and selectively most unlikely; hypothetical reversal also leaves unexplained the origin of the proteasome. Thus, neomura plus actinobacteria are a clade; the root of the tree of life is therefore among other eubacteria (figure 5; Cavalier-Smith 2006a).
Figure 5.
The tree of life. All 10 bacterial phyla that I currently recognize are shown, with Posibacteria split into subphyla Endobacteria and Actinobacteria (for more detailed classification see Cavalier-Smith 2002a, 2006a). It is uncertain whether Actinobacteria are paraphyletic ancestors of neomura, as shown, or their holophyletic sisters; evidence for this otherwise fully resolved bacterial phylogeny is detailed in Cavalier-Smith (2006a). Bars mark major innovations. Cortical alveoli (CA) unite Plantae and chromalveolates as corticates (Cavalier-Smith 2003b); the bikont basal trifurcation remains unresolved. Ancestrally photosynthetic taxa are in green or purple (it is likely, but not certain, that Chlorobacteria were ancestrally photosynthetic; claims for lateral transfer of photosynthesis among bacteria are unsound; see Cavalier-Smith 2006a). Chlorarachnea within Rhizaria and many euglenoids within Excavata have chloroplasts acquired by green algal enslavement; whether this happened independently (asterisks) or, as I think more likely, in a putative common ancestor of Rhizaria and Excavata (Cavalier-Smith 1999) is uncertain. For clarity, the line connecting cyanobacteria to the ancestral plastid is omitted; dashed lines show the implantation of a red algal slave (R) into the ancestral chromalveolate and mitochondria into the protoeukaryote. Secondary losses of the murein wall within Endobacteria (generating Mollicutes) and Planctobacteria are not shown: nor are more numerous losses of flagella and photosynthesis within phyla. Two hyperthermophilic eubacterial groups (Thermotogales, Aquificales) often misplaced on sequence trees, probably by long-branch attraction to the ancestrally hyperthermophilic archaebacteria, are in red. In addition to characters discussed by Cavalier-Smith (2006a), gene arrangements support the grouping Gracilicutes (including Aquifex) and exclusion of Thermotoga (Kunisawa 2006). Blue blobs mark the four taxa that include species able to make sterols. Phylogenetic arrows are not to scale—either of time or magnitude of change; only topology and the position of the root are significant. Contrary to their misleading name, Archaebacteria is the youngest bacterial phylum by many hundreds of millions of years; methanogenesis arose even later. Glycerol 1-P dehydrogenase probably evolved slightly earlier than indicated as it is also present in the eurybacterium Thermotoga.
(g) Chlorobacterial root of the universal tree
Combining the above arguments establishes the root of the tree of the life either between Chlorobacteria and all other organisms or within Chlorobacteria. In either case, the last common ancestor of life was a negibacterium and eobacterium with OM of acyl ester phospholipids but no Omp85 mechanism, a murein peptidoglycan and lipoprotein wall, but no flagella. The root is probably beside chlorobacteria not within them, but further evidence is needed to prove this (Cavalier-Smith 2006a). If this is true, the last common ancestor of life performed anoxygenic photosynthesis with one photosystem, as do chlorobacteria, and trapped light by chlorosomes. If the root were within chlorobacteria, it could either have been photosynthetic (most likely, I think) or heterotrophic, but even then the first form of photosynthesis would have been chlorobacterial.
(h) The origin of eukaryotes and root of the eukaryote tree
The origin of eukaryotes involved the most radical changes in cell structure (figure 6a) and division mechanism (figure 6b,c) in the history of life. It is a prime example of quantum evolution (Simpson 1944), with episodically dramatically accelerated evolutionary rates for many genes and, more importantly, massive novel gene creation by duplication, divergence and protein domain shuffling. So many hundreds of gene duplications and functional divergences occurred (e.g. in myosins (Richards & Cavalier-Smith 2005), tubulins, coated vesicle proteins) that it is impossible for evolution to have occurred instead from eukaryotes to prokaryotes, reversing the complex cytological differentiation shown in figure 6. Spliceosomal introns inserted into thousands of genes in the eukaryote cenancestor (Cavalier-Smith 1991b, 2002b) never totally reversed despite reductions to three in trypanosomes; the former belief by some eminent biologists that the prokaryotic absence of spliceosomal introns is a secondarily derived state, despite entering influential textbooks, is not now accepted.
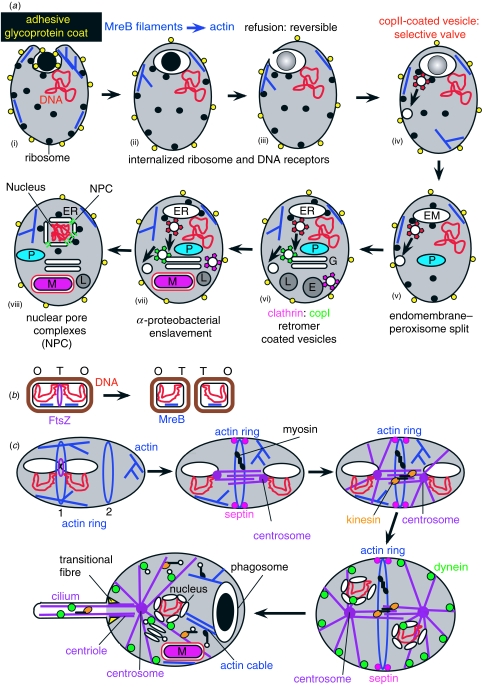
Figure 6.
The phagotrophic origin of eukaryotes. (a) Transformation of growing trophic cells by phagotrophy and consequential endomembrane evolution. (i) A potentially flexible surface coat of N-linked glycoproteins facilitated the origin of phagocytosis by their adhesion to prey and modifying the MreB cortical skeleton (Gitai et al. 2004) into an actin skeleton able to soften locally by filament severing and form engulfing pseudopodia by localized polymerization nucleated by Arp2/3 (actin-related proteins that arose with actin by gene triplication of MreB in the pre-eukaryote; Bretschneider et al. 2004). (ii) Inevitably, internalized food vacuoles would bear attached ribosomes and sometimes also chromosomal DNA. (iii) After digestion, internalized membrane recycled by refusion with the cell surface, so initial internalization of DNA and ribosomes was reversible and impermanent. (iv) A permanent endomembrane system formed by the origin of coated vesicle budding from the internalized membrane, plus membrane return to the surface by fusion with it of transport vesicles produced by their uncoating, instead of reverse fusion of the whole food vacuole (Cavalier-Smith 2002b). Coated vesicle budding, selective for which membrane proteins are included in the budded vesicle, was a selective valve that indirectly caused fundamental differentiation between protoendomembranes and cell surface: continued phagocytosis inevitably rapidly removed all ribosome receptors from the surface, so after non-specific refusion of food vacuoles ceased it could never regain ribosomes or DNA. (v) As bacterial cell-surface derivatives, the now-permanent internal membranes had two protein-insertion mechanisms: SRP/ribosome receptors (figure 2) for cotranslationally inserting unfolded nascent proteins and the twin-arginine translocase (TAT) system for post-synthesis export of folded mature proteins. By chance these segregated into different vesicles; those with SRP receptors became protoendomembranes; those with TAT machinery became peroxisomes (P), whose membrane-embedded pex proteins import native proteins tagged by a C-terminal sequence like that recognized by TAT. (vi) Gene duplications multiplied vesicle coat types, and also the SNARE proteins whose complementary interactions ensure docking specificity onto target membranes, differentiating endomembranes into topologically, chemically and functionally distinct compartments: copI and retromers for retrograde membrane recycling from protoGolgi and endosomes; clathrin for generating endosomes from the cell surface and lysosome precursors from the trans-Golgi. (vii) An ingested α-proteobacterium (M), probably photosynthetic, escaped from its food vacuole by accidental breakage of its membrane, and multiplied in the cytoplasm. The host enslaved it by inserting inner membrane carriers that tapped its photosynthesate for host use and evolving a novel protein-targeting system; carriers probably originated from the peroxisome ATP-importer, implying that mitochondrial enslavement postdated the phagotrophy-dependent autogenous origin of peroxisomes; all theories for the host being a prokaryote are unsound. Enslavement could have started as early as shown, but the major gene transfers into host chromosomes probably postdated the nuclear envelope (Cavalier-Smith in press). (viii) Endoplasmic reticulum (ER) cisternae attached to heterochromatin via the nuclear lamina to protect DNA from shearing damage by new cytoplasmic motors (Cavalier-Smith 2005); evolution of nuclear pore complexes from the same novel eukaryotic gene family as vesicle coats (Devos et al. 2004) prevented lethal complete fusion; plugging their lumen and novel nucleocytoplasmic transport proteins excluded ribosomes, allowing mitochondrial group II introns transferred to host DNA to become spliceosomal introns only afterwards (Cavalier-Smith 1991b)—not before as Martin & Koonin (2006) mistakenly suggest. (b) Logic of the bacterial cell cycle involves DNA attachment to the cell surface by proteins and membrane division between attachment sites (Cavalier-Smith 1981, 1987b). Replication origins (O) separate by moving along a linear MreB track (Gitai et al. 2005a,b). Division of the cell membrane by a GTPase FtsZ ring is precisely between the membrane attachment points of DNA replication termini (T). (c) Origin of eukaryotic cell division, cytoskeleton and cilia. (i) Conversion of linear MreB filaments to Arp2/3-nucleated branched actin filaments and phagotrophic internalization of membranes bearing DNA negated MreB-based chromosome segregation and the FtsZ ring division mechanism, causing mis-segregation and daughters with several or (worse) no chromosomes. A new division mechanism by a ring of overlapping actin filaments nucleated by formins (Ingouff et al. 2005) evolved, but also needed positioning between the chromosomes (1) not to one side (2) to avoid wasteful DNA-less daughters. (ii) Freed from the cell surface and stabilizing selection for the now useless function of marking the surface septation site, FtsZ genes triplicated, yielding γ-tubulin for centrosomes and α- and β-tubulins for microtubules to push them apart by polymerization. 10 nm septin filaments evolved from a eubacterial GTPase to position actin rings. (iii) Kinesin evolved from an early myosin to cross-link spindle microtubules and actively slide them to separate centrosomes and attached chromosomes. (iv) Dynein evolved from an AAA ATPase to pull chromosomes (attached to microtubule minus ends by protocentromeres) and protonuclei along microtubules towards centrosomes and to separate astral microtubules. (v) Ciliary transition fibres evolved, laterally attaching a microtubule ring to the surface membrane, so their polymerization evaginated it as a protocilium; further duplications generated δ- and ϵ-tubulins, making centriolar triplets to rigidify the ciliary base. A novel kinesin (II) transported precursors into the ciliary compartment, helped by intraciliary transport particles of proteins related to vesicle coat and nuclear pore proteins; homologous α-helical-solenoid/β-sheet-propeller proteins of these three macromolecular complexes, never found in bacteria, refute theories of symbiogenetic origins of nuclei or cilia (Jekély & Arendt 2006). Numerous dynein duplications generated ciliary doublet arms, causing sliding and ciliary bending. Cytoplasmic dyneins, kinesins and myosins evolved to move vesicles and organelles along the new interphase cytoskeleton and develop cell polarity. Once centromere-based DNA segregation was efficient, mutation pressure linearized the chromosome and made multiple chromosomes and replicons per chromosome (Cavalier-Smith 1981, 1987b), while meiosis arose to correct polyploidization from residual segregation errors (Cavalier-Smith 2002b,c). The first eukaryote probably inherited cell differentiation programmes and resistant walled exospores (called cysts in protozoa) from actinobacteria; sexual cell fusion evolved prior to encystment to provide more resources to survive starvation (Cavalier-Smith 2002b).
As the eukaryotic morphological fossil record is incomparably better than that of bacteria, eukaryote phylogeny, the position of the root of the eukaryote subtree, and the nature of the first eukaryote are of utmost importance for properly interpreting it and dating the history of life. Recently, misunderstandings on these scores fostered by biased single-gene trees have been corrected by the use of exceptionally rare, practically irreversible shared-derived genetic characters like gene fusions and gene replacements and by multi-gene trees. That the root of the eukaryote tree is between animals and fungi on the one hand and the plant and chromist kingdoms on the other (figure 5) was proposed on grounds of ciliary and cytoskeletal evolution (Cavalier-Smith 2002b), and is now strongly supported by such molecular cladistic evidence (Stechmann & Cavalier Smith 2002, 2003; Richards & Cavalier-Smith 2005). Plants, chromists and three protozoan infrakingdoms (Alveolata, Excavata, Rhizaria) were grouped together as bikonts—a clade defined by evolution in their common ancestor of centriolar and ciliary transformation, in which at cell division the new centriole of each daughter cell bears the anterior cilium, but this reorients at the next cell division to become the older posterior centriole and cilium, often with different ultrastructure and beat pattern (Cavalier-Smith 2002b). This unique pattern of ciliary differentiation was known in all bikont groups, except Rhizaria. Now it has been found also in the rhizarian phylum Cercozoa (Karpov et al. 2006), so it was already present in the cenancestor of bikonts and helps define them.
The best available multi-gene tree based on 149 proteins strongly corroborates the bipartition between unikonts and bikonts and the monophyly of major groups (figures 5 and 7), including opisthokonts, unikonts, Plantae, Excavata and chromalveolates (Rodriguez-Ezpeleta et al. 2005). For parsimony in evolution of protein-targeting machinery in secondarily enslaved chloroplasts, Cavalier-Smith (1999) proposed that all chromophyte (chlorophyll c-containing) algae arose by a single enslavement of a red alga, and aplastidic chromalveolates evolved by chloroplast loss. This major phylogenetic simplification was rapidly compellingly supported by the serendipitous discovery of two gene replacements that must have occurred in the common ancestor of all chromalveolates. The original red algal plastid versions of the enzymes glyceraldehyde-phosphate dehydrogenase and fructose-bisphosphate aldolase were replaced by duplicates of the host cytosolic version of these enzymes (Patron et al. 2004). Chromalveolate monophyly means that the whole chromalveolate clade must be younger than red algae. There are many abundantly fossiliferous chromalveolates: haptophytes, diatoms, silicoflagellates, chrysomonads, dinoflagellates, and others that rarely fossilize—ciliates, in amber only—and, more dubiously, brown algae—I am sceptical of almost all fossil identifications of brown algae. Multi-gene trees show chromalveolates as younger than the primary divergence within reds: between (thermophilic) Cyanidiophyceae and the rest (Yoon et al. 2002). Not only symbiogenesis, but also LGT, can give relative (not absolute) dates independently of the fossil record, because of their rarity and the certainty that the donor cannot have evolved after the last common ancestor of all descendants of the recipient (disallowing time reversal). Thus, shared lateral transfers of bacterial genes by the metamonad superclass Eopharyngia (e.g. Giardia and Trichomonas) prove that the primary divergence within this group was after the evolution of all the bacterial donors. They also prove that Eopharyngia are a derived secondarily anaerobic clade within eukaryotes and excavates (Cavalier-Smith 2003a), and therefore that the root of the eukaryote tree cannot be between Giardia and Trichomonas, as 18S rRNA trees once suggested, corroborating evidence from many protein trees that rRNA trees were seriously misleading, with overall topology dominated by long-branch artefacts when both bacteria and eukaryotes are included.
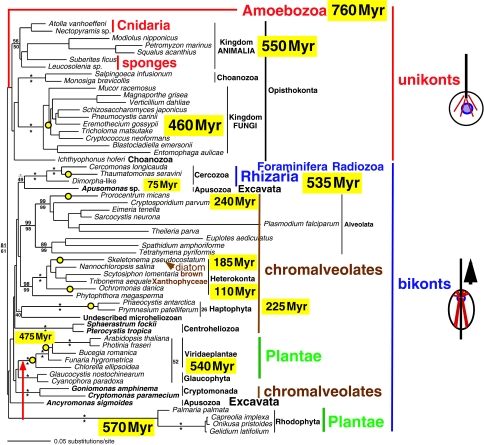
Figure 7.
Distance tree for 58 eukaryotes based on 2994 nucleotides of 28S rRNA. This weighted least squares power 2 (GTR+Γ+I model: a=0.59231, i=0.24389) tree is from von der Heyden (2004). Numbers at major nodes only from 500 wl2 bootstrap samplings (limited to 10 min per replicate) (upper) and from 1000 BioNJ replicates (lower) are percentages of recovery of that clade (asterisks indicate 100%). Dates are for the earliest unambiguous fossils; if not otherwise obvious, dated clades are marked by yellow-filled open circles. The great variability in branch length shows that rRNA evolution is extremely non-clock-like; the still-longer-branch Amoebozoa, Foraminifera, Euglenozoa and Metamonada were omitted to reduce tree distortion and increase legibility; the position of Amoebozoa based on other evidence is shown in red. Foraminifera and Radiozoa (non-phaeodarian radiolaria) constitute the phylum Retaria, which is probably sister to Cercozoa (Cavalier-Smith 1999, 2003b). As in most single-gene trees, the monophyly of Plantae, chromalveolates and Apusozoa is not recovered. Red algae (Rhodophyta) are too low on this tree because of their very long branch, and are really sisters of Viridaeplantae (red arrow); as previously suggested for 18S rRNA (Cavalier-Smith et al. 1996), the gross acceleration in 28S rRNA evolutionary rate could have been entirely in the stem of the florideophyte red algal tree (bangiophytes have relatively normally short branches on 18S rRNA; also true for recent 28S sequences, unavailable for this analysis) as its branches have a phylogenetic depth comparable to their green-plant sisters. Thus, the ancestral florideophyte ribosome probably underwent marked episodic quantum evolution, somewhat less extreme than in the long stems at the base of the eukaryote and neomuran clades that gave rise to the misleading three-domain concept of the tree of life (Cavalier-Smith 2002a). The thumbnail sketches show that in addition to ciliary transformation from younger anterior to dissimilar older posterior organelles, bikonts have a microtubular cytosketeton (red) of cortical band ciliary roots, not a simple cone of separate microtubules like unikonts.
An important consequence of this root of the eukaryote tree and strong evidence that extant anaerobic eukaryotes probably all have organelles (hydrogenosomes or mitosomes) evolutionarily derived from aerobic mitochondria (Embley 2006) is that the last common ancestor of all eukaryotes was an aerobic amoeboflagellate with mitochondria. As this cenancestor arose by enslaving an α-proteobacterium, all eukaryotes must be younger than the oldest α-proteobacteria. Their origin must postdate the primary diversification of eubacteria into phyla and later diversification of proteobacteria into classes. Eukaryotes are younger than eubacteria, as morphological fossils showed long ago. This illustrates how phylogenetic analysis can order evolutionary events logically in time, independently of fossils. But only fossils give absolute dates.
4. Mapping the tree onto the fossil record
(a) The age of eukaryote taxa
The oldest unambiguously eukaryotic fossils are vase-shaped (e.g. Melanocyrillium), going back at least 760 Myr and probably slightly earlier (Porter & Knoll 2000; Porter et al. 2003). All are probably arcellinid lobose testate amoebae (phylum Amoebozoa), which originated relatively early within class Lobosea (Nikolaev et al. 2005). Those forms sometimes suggested to be euglyphid filose testate amoebae could also be agglutinated tests of arcellinids; I do not accept them as evidence for Cercozoa. Undoubted euglyphid fossils occur much later in amber (Foissner & Schiller 2001), but most Cercozoa fossilize poorly; exceptions are ebriids, first in the Palaeocene and throughout the Tertiary (Tappan 1980), and Phaeodaria, from ca 75 Myr (Takahashi 2004). Table 1 references the earliest fossils reasonably attributable to specific protist phyla (see also figure 7). Most groups apparently originate close to or a little before the Cambrian animal explosion or at the beginning of the Mesozoic or Caenozoic. Thus, protist phyla mainly radiated 570–500 Myr ago, soon after snowball earth unfroze, whereas many now important classes originated later during recovery from the end Permian and end Cretaceous mass extinctions (Mundil et al. 2004). Protist diversification broadly resembles the familiar pattern for animal phyla: sudden origins of many phyla near the Precambrian boundary, and novel classes and/or orders in the Early Mesozoic and Cenozoic to exploit niches or whole adaptive zones emptied by the greatest mass extinctions.
Table 1.
Earliest reasonably confident fossil dates for the major eukaryote groups.
| taxa | date (Myr ago) | reference |
|---|---|---|
| unikonts | ||
| opisthokonts | ||
| Animalia | 550 | Conway Morris (2006) |
| Fungi | 460a | Padovan et al. (2005) |
| Choanozoa | no fossils | |
| Amoebozoa (Lobosea: Arcellinida) | 760 | Porter & Knoll (2000) |
| bikonts | ||
| Rhizaria | ||
| Retaria | ||
| Foraminifera | ||
| unilocular | 535 | McIlroy et al. (2001) |
| multilocular | 390 | Armstrong & Brasier (2005) |
| Radiozoa | 505 | Won & Below (1999) |
| Cercozoa | ||
| Euglyphida | 15 | Foissner & Schiller (2001) |
| Ebriida | 60–65 | Tappan (1980) |
| Phaeodaria | 75 | Takahashi (2004) |
| Excavata | no certain fossils; claims for euglenoids and kinetoplastids all post-Permian | |
| Plantae | ||
| Viridaeplantae | ||
| Chlorophyta | 540 | Tappan (1980) |
| Embryophyta | 475 | Wellman et al. (2003) |
| Rhodophyta | 570 | Xiao et al. (2004) |
| Glaucophyta | no fossils | |
| chromalveolates | ||
| Alveolata | ||
| Ciliophora | 100 | Acaso et al. (2005) |
| Myzozoa | ||
| Dinozoa | ||
| Peridinea | 240 | Fensome et al. (1993) |
| Chromista | ||
| Cryptista | no fossils | |
| Heterokonta | ||
| Ochrophyta | ||
| Silicoflagellata | 105–110 | McCartney (1993) |
| Chrysomonadea | 75 | Cornell (1972) |
| Diatomea | 185 | Tappan (1980) |
| Haptophyta | ||
| coccolithophorids | 225 | Bown et al. (2004) |
A claim for 600 Myr old ‘lichen-like’ fossils (Yuan et al. 2005) is doubly confusing. Lichens are large fungi that cultivate cyanobacteria within their tissues. These fossils are entirely different; cyanobacteria (less likely green algae) are permeated by filaments so slender that they are probably actinobacteria not fungi; analogy with actinobacterial endophytes of cereal plants is more apposite (Conn & Franco 2004; Tian et al. 2004).
The marked discrepancy between these late morphological records of all fossils unambiguously attributable to specific eukaryotic phyla and the very early occurrence of steranes often interpreted as eukaryotic derivatives (Brocks et al. 1999) is explained if the 2.7 Gyr ago steranes actually came from eubacteria, at least four groups of which can make sterols (Cavalier-Smith 2002a; Pearson et al. 2003). Of these, Mycobacteria make cholesterol like eukaryotes, and belong to actinobacteria, the probable ancestors of eukaryotes. Sterol biosynthesis evolved polyphyletically by modifying universal eubacterial isoprenoid metabolism after atmospheric oxygenation made it mechanistically possible, long before eukaryotes. It was probably vertically inherited by the first eukaryote from an actinobacterium. Claims that mycobacterial sterol synthesis enzymes were laterally transferred from eukaryote DNA have not been substantiated by phylogenetic analysis (Cavalier-Smith & Chao 2003b), and reflect the false assumption that the ancestors of eukaryotes were archaebacteria, not modified derivatives of actinobacteria that generated the eukaryote/archaebacterial cenancestor. Fossil steranes are not sound evidence for eukaryotes (Cavalier-Smith 2002b); even were they specific for eukaryotes, the potential for downward mobility of petroleum fractions containing them is a perpetual worry for age authenticity—this problem is absent for body fossils, though their identification as eukaryotic or bacterial is also sometimes practically impossible, sometimes overoptimistic.
Some palaeontologists, rightly I think, do not accept Grypania (1.8 Gyr old; Han & Runnegar 1992; redated by Knoll et al. 2006) as eukaryotic rather than a giant cyanobacterial sheath. Identity of the largest, most complex fossils ca 1.5 Gyr ago often called eukaryotic (Javaux et al. 2001, 2003) is problematic. While some might be stem eukaryotes (Javaux et al. 2003), this is not compellingly demonstrated. I consider it more likely that most, probably all, are simply unusually large and complex prokaryotes. Unlike Javaux et al. (2001), I do not think their morphology demands the presence of an internal cytoskeleton or endomembranes; it is within the morphogenetic capabilities of bacteria. I agree with Butterfield (2005) that the morphologically most complex of these fossils is probably not an alga as originally assumed, but a sporangial entity broken from a branching trophic hyphal network, as are his beautiful 0.8–0.9 Gyr fossils (mostly Tappania). In 2001, I microscopically examined the Harvard Tappania plana specimen shown in fig. 4 of Javaux et al. (2003); I told Javaux that I thought it was not a single algal cell and that the structure labelled there by an arrow was the broken base of a branched hypha passing through the outer ‘wall’ towards the dense central mass. However, I disagree with Butterfield's suggestion that these fossils are probably fungi. They could instead be actinobacteria similar to modern Amycolatopsis decaplanina and Kibdelosporangium that generate large pseudosporangia from mycelial networks (Wink et al. 2004); their pseudosporangia are very variable in size, but usually somewhat smaller than in the fossils, but the thickness of their hyphae is indistinguishable. Confusion of actinomycetes and fungi was long-standing even for extant cultivated species; actinomycetes were treated as fungi in my undergraduate mycology textbook (Alexopoulos 1952); it is markedly more difficult to differentiate between them in fossils.
I also do not accept Bangiomorpha (Butterfield et al. 1990; Butterfield 2000) as a red alga or eukaryote; it could be a mixture of two cyanobacterial species, possibly stigonematalean; I was mistaken in earlier calling it Oscillatoria-like (Cavalier-Smith 2002a). It is certainly not like the red alga Bangia, as it lacks the characteristic elongated projections of cells in its holdfast that result from intrusive growth, which had they been present would have supported a eukaryote nature. As some filamentous red algae lack such intrusive cells and are effectively indistinguishable at low resolution from Bangiomorpha, we cannot be sure it is not a red alga. However, the holdfast and all other morphological features are no more complex than in some cyanobacteria. Unfortunately, it lacks characters that require presence of a eukaryotic cytoskeleton or endomembrane system that would firmly make it eukaryotic or that prove that it cannot simply be a complex cyanobacterium. Assuming Bangiomorpha to be correctly dated at 1.2 Gyr (currently unclear; perhaps younger), Bangiomorpha is 600 Myr older than any fossil reasonably confidently a red alga (Xiao & Knoll 1999) and ca 400 Myr older than unambiguously eukaryotic fossils.
Another Late Proterozoic fossil type sometimes called eukaryotic are broad filaments, e.g. Palaeovaucheria, conventionally assigned to the heterokont Xanthophyceae and dating to ca 1 Gyr ago (Butterfield 2004). These lack features unambiguously identifying them as xanthophytes, heterokonts, or even eukaryotes; they could be large filamentous cyanobacteria. Although we cannot rule out their being a stem eukaryotic alga unattributable to phylum or kingdom, their early date makes it virtually impossible that they are xanthophytes, given the well-established heterokont molecular trees and recency of all well-fossilized heterokont groups (diatoms, haptophytes, chrysomonads, silicoflagellates); none go even into the Palaeozoic (table 1), and trees show xanthophytes as no older (figure 7; Cavalier-Smith & Chao 2006).
There are many technically inadequate attempts to date eukaryotes assuming a universal molecular clock (Graur & Martin 2004), even though this assumption is false and grossly misleading, as evolutionary rates of all molecules can change idiosyncratically across a molecular tree, sometimes by many orders of magnitude (Cavalier-Smith 2002a). Yet some stability in evolutionary rates of nucleotide or amino acid substitution exists in local parts of the tree for a given molecule, and assuming a local ‘clock’ is sometimes useful for interpolating between known fossil dates. Extrapolation backwards earlier than fossil dates is considerably more hazardous; nonetheless, with a statistical model that allows rates to change across the tree, reasonably realistic models of substitution, and calibration of many points on the tree by known fossil dates (not one as in many studies), it is worth attempting to try to assess congruence between the fossil record and molecular phylogenetic trees, and to improve interpretation of both if they mismatch; Roger & Hug (2006) discuss some of the problems.
Only two such studies, using a Bayesian ‘relaxed clock’, attain the highest current standards. One based on 129 proteins, 36 eukaryotes and six Phanerozoic fossil calibration points based on macro-organisms (three animals, two land plants, one fungal) deduced 950–1259 Myr ago (mean 1085) for the divergence of Amoebozoa from other eukaryotes (Douzery et al. 2004); given the difficulty of resolving the branching order of Amoebozoa and bikonts on trees (Cavalier-Smith 2004b), this probably approximates to an estimate for the cenancestral eukaryote. Berney (2005) and Berney & Pawlowski (2006), using 18S rRNA (1465 nucleotides) from 83 eukaryotes and 26 Phanerozoic microfossil time constraints, similarly dated the cenancestral eukaryote at 948–1357 Myr ago (mean 1126). Using microfossils to constrain trees has the advantage that such organisms as diatoms, radiolaria and foraminifera are superabundant, with billions of clearly identifiable fossils, so a group's first appearance is more accurately dated than for sparsely recorded macro-organisms and unlikely to be underestimated. However, Bayesian algorithms are unlikely to model episodic evolution (e.g. dramatic short-term changes in evolutionary rate) well. For rRNA, the evolutionary rate probably massively increased in the stem eukaryote lineage (possibly by approx. four orders of magnitude; Cavalier-Smith 2002a) then subsequently greatly slowed again. If slowing finished before the initial radiations, the earlier fast rates should not distort Berney's conclusion (though probably fatal for similar studies of the whole tree). If higher rates lasted after the primary eukaryotic divergence but declined before fossil calibration dates, this gene tree would overestimate the age of the cenancestral eukaryote. As many proteins used by Douzery et al. (2004) were ribosomal (coevolving with rRNA), their dates also could be overestimates. Many other proteins used in their dataset were novel eukaryotic proteins, expected to have evolved faster early on when function was freshly established than later when stabilizing and purifying selection would predominate; thus, the protein dates may be overestimates. This is supported by reanalysis of these data by a technically superior method; calculating trees separately for each gene and then combining likelihoods (Roger & Hug 2006) gave a younger estimate of ca 900 Myr ago. Based on numerous proteins, this should be more reliable than the 200 Myr earlier date from 18S rRNA (Berney 2005; Berney & Pawlowski 2006), especially if hyper-fast rRNA evolution in the pre-eukaryote stem (Cavalier-Smith 2002a) persisted somewhat after the earliest divergences.
Berney used a clever method to test identifications of contentious Proterozoic fossils like Palaeovaucheria, Tappania, Pterocladus (claimed to be a cladophoran green alga) and Bangiomorpha. He assumed palaeontologists correctly identified them and used their dates as constraints for Bayesian calculations, deducing a hypothetical origin time for groups with easily identified fossils, e.g. diatoms, dinoflagellates. Invariably, this dated these groups with a continuous unambiguous fossil record grossly earlier than their actual fossil dates (typically overestimating ages of rhizosolenid diatoms, pennate diatoms and coccolithophorid haptophytes by 4–5 times). This strongly supports the view that these fossils were misidentified as crown eukaryotes (Cavalier-Smith 2002b). Although some might be stem eukaryotes, all are likely simply complex bacteria. Likewise, assuming that some vase-like fossils are euglyphids (Porter & Knoll 2000; Porter et al. 2003) overestimates these dates 4–8 times (because euglyphids nest relatively shallowly in the cercozoan tree; Cavalier-Smith & Chao 2003a); by contrast arcellinids nest relatively deeply in the amoebozoan tree (Nikolaev et al. 2005); their date of more than 760 Myr ago (Porter & Knoll 2000; Porter et al. 2003) is consistent within the error range with estimates of ca 950 Myr ago for Amoebozoa (Berney 2005; Berney & Pawlowski 2006) and ca 0.9 Gyr ago for eukaryotes (Roger & Hug 2006).
My objection to identifying these fossils as eukaryotes is primarily on morphological grounds; morphology inadequately supports it. Their conflict with the phylogenetic evidence for approximately equal ages for bikonts and unikonts, and thus with plants and chromists being not dramatically older than animals, made me examine them critically, but I should be sceptical even were they much younger. The relaxed Bayesian estimates for red algae (ca 730 Myr ago from rRNA; less than 928 Myr ago from proteins) and Bangiophyceae (ca 680 Myr ago from rRNA) are consistent with ca 570 Myr old fossils being genuine florideophytes (Xiao et al. 2004). These estimates should not be called ‘molecular’. They synthesize fossil and molecular data with a model for rate change across the tree, i.e. fossil dates modulated and integrated by extensive comparative molecular evidence. Molecular trees and substitution models help integrate the partial fossil evidence with more representative molecular data and apply dates to groups lacking fossils. Molecular sequences alone give no dates.
Inferred cenancestral dates for chromalveolate groups (haptophytes 560 Myr ago; heterokonts 580 Myr ago; alveolates 600 Myr ago) are consistent with the requirement that chomalveolates postdate red algae (having originated by one red algal enslavement), but the mean unseparated protein date for chromalveolates (919 Myr ago, Douzery et al. 2004) is only just consistent with the red algal protein date and markedly earlier than the rRNA red algal date, suggesting an overestimate. Estimates for the red/green algal divergence of ca 930 (rRNA) and 1010 Myr ago (proteins, unseparated; Douzery et al. 2004) should be slightly younger than for the origin of Plantae as glaucophytes diverge earlier. However, as rRNA gives 812 Myr ago for the primary animal radiation and proteins 695 Myr ago, early backwardly extrapolated dates of these studies may be 100–300 Myr too old, it being unlikely that animals have been missed in the fossil record (Peterson & Butterfield 2005; Conway Morris 2006)—their true age is probably ca 555 Myr ago (if Vendobiota are not animals) or ca 570 Myr ago if Vendobiota are animals.
Taking fossils and molecular evidence together, 900±100 Myr ago is the most likely origin date for eukaryotes. Dating the origin of chloroplasts and Plantae is harder, but most likely 570–850 Myr ago, whereas ca 570 Myr ago is reasonable for the origin of chromalveolates, opisthokonts, Rhizaria and excavates, when snowball earth melted.
(b) Archaebacteria are the youngest prokaryotes
Archaebacteria were named when all known archaebacteria were methanogens: Woese (1977) assumed that metabolism using H2 to reduce CO2 to methane was very ancient. His primary reason for supposing archaebacteria to be ancient was that their rRNA seemed as divergent from that of eukaryotes and eubacteria as they were from each other; he implicitly assumed that rRNA evolved at the same rate in all three groups and that all three were equally old (Balch et al. 1977; Woese & Fox 1977). Fossil dates (eukaryotes much younger than eubacteria) were ignored. Thus, the widespread belief that archaebacteria are ancient stems from dubious assumptions not from direct evidence. Stackebrandt & Woese (1981) explicitly stated there was no evidence that rRNA evolved at the same rate in the three groups, but that important caveat has been generally ignored; numerous interpretations have assumed that substitution rates are uniform for rRNA, especially bacterial, despite this being false and misleading. Even that caveat overlooked the likelihood that quantum evolution during transitions between the three groups mainly caused the differences, not long elapsed time (Cavalier-Smith 1981, 1987b, 1991a), as critical interpretation of the trees in the light of fossils effectively proves (Cavalier-Smith 2002a). We cannot use phyletic depth on rRNA trees to determine age independently of fossils. But we can combine tree data with fossil evidence for relatives to estimate dates for groups without fossils. As archaebacteria are sisters of eukaryotes, they cannot be dramatically older. Thus, they are probably only ca 0.9 Gyr old. Eubacteria are probably 2.2–2.6 Gyr older.
It is unlikely that neomura are much older than either eukaryotes or archaebacteria. Probably eukaryotes and archaebacteria each originated almost immediately after neomura (Cavalier-Smith 2002a). It is especially unlikely that neomura can be as old as eubacteria. If they were, bacteria with a neomuran glycoprotein envelope, DNA-handling enzymes and ribosomes, but lacking specific archaebacterial properties (e.g. isoprenoid ether lipids, archaeosine, flagella), must have existed for 2.5 Gyr before eukaryotes without leaving any descendants not converted to eukaryotes or archaebacteria; it is incredible that a major niche for such an intermediate bacterium persisted for more than 2 Gyr, but was suddenly wiped out coincidentally with the origin of eukaryotes.
There is no morphological fossil record for archaebacteria and scant prospect of ever finding any. One potentially important biomarker unique to archaebacteria is the double-length C40 isoprenoid lipids that span the entire CM of hyperthermophilic (not other) archaebacteria. As hyperthermophily is probably the ancestral state (Cavalier-Smith 2002b), these probably first evolved in the cenancestral archaebacterium and would mark their origin. Unfortunately, they are unknown earlier than the Mesozoic, suggesting relative instability (they ought to date from the Early Neoproterozoic (ca 0.9 Gyr), like eukaryotes) that underestimates the age of archaebacteria (Cavalier-Smith 2002a). Linear C20–C30 isoprenoids ca 1.6 Gyr are proposed as biomarkers for halophilic archaebacteria, which typically make membrane lipid isoprenoids of C20 and C25 (Summons et al. 1998). However, all eubacteria have long-chain isoprenols of C50 or C55 lengths as membrane carriers, and one wonders whether partial degradation products of these or other eubacterial isoprenoids might persist and be hard to distinguish from halobacterial isoprenoids. R. E. Summons (2005, personal communication) noted that polyunsaturation of these eubacterial isoprenols makes them highly susceptible to degradation by light and oxidation, unlike saturated archaebacterial lipids. Given that atmospheric oxygen was substantially sparser ca 1.6 Gyr ago (Holland 2006) and the deep ocean might have been anoxic, possibly anoxic burial of eubacteria occurred rapidly enough for degradation to yield the observed spectrum of chain lengths.
Organic carbon samples 2.8 Gyr ago (and somewhat later) are markedly lighter isotopically than achievable by one-step CO2 fixation by Rubisco (Δ13C −38‰). Previously, assuming that archaebacterial methanogenesis was ancient, this was interpreted as the result of recycling of biogenic methane made by archaebacteria (archaebacterial methanogenesis has exceptionally low 13C : 12C ratios: Δ13C −30–50‰) by methanotrophic bacteria into organic carbon (Hayes 1983, 1994). Hayes suggested that the rise of oxygen restricted methanogen habitats sufficiently to make their hypothetical contribution quantitatively undetectable less than 2.2 Gyr ago. As phylogenetic and morphological fossil evidence in combination strongly indicate that archaebacteria are much younger than 2.0–2.8 Gyr, explanations for these light carbon deposits cannot involve archaebacteria. Straus et al. (1992) suggested that chemosynthetic bacteria or anoxygenic photosynthesis might participate in two-step fractionation that could produce lighter organic carbon than Rubisco alone. A specific suggestion was a chemotroph (presumably using Rubisco) taking as substrate CO2 already made light by photosynthetic fixation and released by respiration. The lightness of the final product would depend on local recycling efficiency compared with mixing in the atmospheric pool. Section 5d develops an analogous explanation involving greater phototroph diversity generated by the glycobacterial revolution 2.8 Gyr ago.
Ultralight reduced carbon is also made by autotrophic acetogenesis; some Clostridiales thus make acetate from CO2 and hydrogen, with similar 13C depletion to methanogenesis (Δ13C −59‰; Gelwicks et al. 1989). As they use the same input materials as methanogens but are undoubtedly much older, they are a potentially more plausible explanation for the isotopic data. As anaerobes, their relative contribution to fixed carbon compared with aerobic phototrophs would have become insignificant just when the ultralight signal went. However, Clostridiales are Endobacteria, possibly not present as early as 2.7 Gyr ago: §4d suggests they arose 2.3–1.5 Gyr ago, but does not firmly exclude that they evolved earlier and contributed to the ultralight signal.
Another potential explanation is abiotic sources of methane plus aerobic biological methanotrophy. Some abiotic methane is not ultralight. Iron- and nickel-rich alloys, common in some minerals, catalyse production from bicarbonate at 200–300 °C of abiotic methane indistinguishably ultralight (Δ13C−35–50‰) from archaebacterial methane. However, as §5d argues, biotic depletion is more likely. But, given the potential of three very different kinds of explanation not involving archaebacteria for the 2.8–2.1 Gyr ago carbon isotopic data, they are not specific evidence for Archaean or Palaeoproterozoic archaebacteria, especially as all other evidence strongly contradicts that. Hayes (1994) recognized that ‘the antiquity of methanogens… is more speculative than proven’. I consider it now disproved (Cavalier-Smith 2006a).
(c) Antiquity of negibacterial phyla, the oldest prokaryotes
Sound evidence for life more than 3.5 Gyr ago is wanting (Fedo et al. 2006). Diverse evidence makes it highly probable that life, negibacteria in particular, existed in the Late Archaean, 2.5–2.9 Gyr; no evidence absolutely requires the presence of Unibacteria (Posibacteria, Archaebacteria) or eukaryotes. Evidence for life from 3.5 to 2.9 Gyr is less compelling, coming from only three sources: stromatolites and possible/probable body fossils (both reviewed by Schopf 2006) and isotopically light organic carbon suggestive of fractionation by Rubisco (Hayes & Waldbauer 2006). Inorganic processes can produce both organic carbon and carbon isotopic fractionation of the order observed but are insufficiently understood. Carbon isotope modelling has concentrated on the known biotic world rather than on attempting to predict what the carbon cycle would have been like prior to life to provide a proper baseline for comparison. We need to understand whether abiotic forces could have caused the consistent light organic carbon from 2.8 to 3.5 Gyr ago and to seek biomarkers in that period. For stromatolites of certain types inorganic causes have been proposed; although such causes can be hard to rule out in individual cases (Brasier et al. 2002, 2006), the overall pattern of the evidence (Schopf 2006) and lack of abiogenic explanations for conical stromatolites suggests that negibacteria had originated by ca 3.1 Gyr ago and quite possibly ca 3.5 Gyr ago. However, putative body fossils over 2.8 Gyr old (Schopf 2006) are less convincingly biogenic than later ones; none older than 2.8 Gyr is obviously cyanobacterial and biogenicity of some is problematic (Brasier et al. 2002, 2006). No putative morphological fossils more than 2.1 Gyr old can unambiguously be assigned to a phylum.
Biomarkers for hopanoids suggest that glycobacteria, cyanobacteria in particular, existed more than 2.7 Gyr ago (Summons et al. 1999, 2006), assuming that these potentially mobile organics are truly endogenous to these rocks, which is uncertain (Schopf 2001). Because of this worry, the extensive evidence that oxygenic photosynthesis had arisen by the Late Archaean (Holland 2006) is probably stronger evidence for negibacteria and cyanobacteria. If 2.7 Gyr hopanes are endogenous, this is a minimum age, earlier deposits being unstudied. I suggest that the glycobacterial revolution and origin of cyanobacteria occurred 2.8–2.9 Gyr ago and that all indications of life from 3.5 to 2.9 Gyr ago can be attributed to Eobacteria, with ecosystems entirely dependent on photosynthetic Chlorobacteria for primary production, or to stem bacteria. I suggest ca 2.9 Gyr ago for the origin of oxygenic photosynthesis because the limited and temporary oxidative removal of abiogenic atmospheric methane by biogenic oxygen seems a simpler explanation of the 2.9 Gyr ago glaciation than indirect depletion of hydrogen and methane by biological sulphate reduction as Kasting & Ono (2006) suggest. If oxygenic photosynthesis originated in the common ancestor of Hadobacteria and cyanobacteria (Cavalier-Smith 2006a), biogenic oxygen first came not from cyanobacteria but from an immediately preceding stem lineage. However, there is no reason to postulate significant delay after photosystem duplication before the origin of phycobilisomes. A 100 Myr delay could have occurred only if a well-adapted oxygenic bacterium existed that long in a well-defined adaptive zone. No such organisms exist, consistent with quantum evolutionary principles that they were evolutionarily unstable, short-lived transitional intermediates in perfecting oxygenic photosynthesis. It is unlikely that cyanobacteria postdated oxygenic photosynthesis by even 1 Myr, so if the 2.9 Gyr glaciation was caused by methane oxidation glycobacteria and cyanobacteria probably originated ca 2.9 Gyr ago. This increased biotic complexity of microbial mats, with at least one new group that used only Rubisco for carbon fixation, would have made carbon fixed within mats still lighter—directly by increasing the Rubisco : hydroxypropionate ratio and indirectly by increasing the scope for repeated recycling of light CO2 within the mat, and might therefore have caused the ca 2.8 Gyr ago ultralight carbon isotopic spike (Hayes 1983, 1994; Hayes & Waldbauer 2006). Carotenoid biomarkers demonstrate proteobacteria 1.6 Gyr ago (Brocks et al. 2005), and R. E. Summons (personal communication) found them in 2.7 Gyr old samples. If confirmed, the radiation into all four photosynthetic glycobacterial phyla plus the heterotrophic spirochaetes was over by 2.7 Gyr ago.
If Chlorobacteria are the earliest diverging phylum, as I argue, they must be older than the divergence of Hadobacteria (i.e. 2.9 Gyr ago, if oxygenic photosynthesis is that age, as just suggested, and did evolve immediately prior to that node on the tree; figure 5). The marked separation of chlorobacteria on rRNA and some other trees from other photosynthetic groups suggests that they may have arisen at least 10% earlier than that (i.e. ca 3.2 Gyr ago); an origin 20% earlier (ca 3.5 Gyr ago) would be not unreasonable, assuming some substitutional saturation reducing the good resolution on sequence trees that such a difference in age should otherwise cause. Thus, chlorobacteria probably originated ca 3.2±0.3 Gyr ago. Although Chlorobacteria are probably the most ancient phylum, the first life would have been a stem lineage preceding the divergence of chlorobacteria and other organisms. Even though the bacterial cenancestor was probably already complex, with probably at least 2000 genes and a complex envelope with peptidoglycan and lipoprotein and fully complete metabolic and bioenergetic (probably both photosynthetic and anaerobic respiratory) repertoires, bacterial generations can be so rapid, selective forces so strong, and bacterial populations so huge, that bacterial evolution can be extremely fast on a geological time-scale. The time from the first life to the cenancestral negibacterium need not have exceeded 1–10 Myr. As Eobacteria include photoautotrophs, photoheterotrophs, non-photosynthetic anaerobic respirers and fermenters, thermophiles and mesophiles, they could have formed a diversified photosynthesis-based ecosystem for hundreds of millions of years before oxygenic photosynthesis and glycobacteria. As photosynthetic chlorobacteria are typically filamentous gliders, like cyanobacteria, they could have helped generate all genuinely biogenic Archaean stromatolites. Contributions from cyanobacteria could have been largely, perhaps entirely, Proterozoic.
Flagella probably evolved in the common ancestor of Gracilicutes (Proteobacteria, Sphingobacteria, Planctobacteria, Spirochaetes) and Eurybacteria. As this ancestrally photosynthetic assemblage is probably sister to cyanobacteria, it should be approximately equal in age. As multigene trees do not clearly resolve the branching order of these phyla, deduced mainly by molecular cladistics, they probably diverged in a rapid early radiation of glycobacteria. The often facultatively aerobic proteobacteria, which include purple photosynthetic bacteria and an immense variety of chemotrophs and heterotrophs, e.g. sulphate reducers and H2S- and iron-oxidizers, and the Sphingobacteria that comprise anaerobic green-sulphur bacteria (e.g. Chlorobium) and aerobic Flavobacteria (including predatory cytophagas) are probably almost as old as cyanobacteria. Thus, rapid diversification of photosynthetic machineries and antenna pigments followed the glycobacterial revolution. Photoreceptors allowing physiological chromatic adaptation and light-oriented movements probably also stem from the glycobacterial ancestor; sensory rhodopsins occur in cyanobacteria and proteobacteria (Jung et al. 2003; Ruiz-Gonzalez & Marin 2004; Venter et al. 2004; Vogeley et al. 2004). One green non-S bacterium photosynthesizes using geothermal radiation in deep-sea vents (Beatty et al. 2005); like sulphur-oxidizers there, they are glycobacteria and so younger than chlorobacteria. If deep ocean chlorobacteria do not include autotrophs, vent autotrophy probably postdates the glycobacterial revolution and is irrelevant to early life.
Although some suggest that bacterial sulphate reduction (BSR) is 3.5 Gyr old, sulphate deposits more than 3.2 Gyr ago, but not for the next 900 Myr, imply that it evolved less than 3.2 Gyr ago (Kasting & Ono 2006). Phylogenetic evidence fits the widespread consensus that isotopic sulphur fractionation began only ca 2.7 Gyr ago and sharply increased quantitatively ca 2.3 Gyr ago when atmospheric oxygenation would have increased sulphate supplies (Holland 2006; Kasting & Ono 2006). Heterotrophic anaerobic iron and sulphate reducers probably evolved early in diversification of Proteobacteria (sensu Cavalier-Smith 2002a) in lineages that lost photosynthesis. Probably BSR was first exclusively by Proteobacteria, making its onset a marker for their origin; possibly all Archaean BSR was proteobacterial. However, metabolism of most Chlorobacteria is unknown; if their huge uncharted diversity (Morris et al. 2004) included sulphate reducers, BSR could be 3.2 Gyr old. Carotenoid biomarker and phylogenetic evidence fit an origin of Proteobacteria and Sphingobacteria ca 2.8 Gyr ago. As the distinctively motile heterotrophic spirochaetes adapted for corkscrewing through soft media like microbial mats or sloppy organics-rich mud, probably diverged slightly before, they are marginally older. Thus, by ca 2.8 Gyr ago seven of the eight negibacterial phyla had already evolved. Only Planctobacteria, apparently sisters of (or derived from) Proteobacteria, which mostly lost murein and are often aerobic flagellates or parasites, may be somewhat younger. Possibly Planctobacteria evolved 2.1–2.5 Gyr ago during the great oxygenation event, becoming significant heterotrophic bacterioplankton; a substantially later origin is unlikely, or else they would nest more definitely and more shallowly within Proteobacteria in sequence trees.
Eurybacteria are probably at least as old as Proteobacteria; although including photosynthetic Heliobacteria, which do not fix CO2, and the thermophilic or hyperthermophilic Thermotogales, they contribute little to biogeochemical cycles, having low diversity and narrow metabolic capabilities. Antiquity of Thermotogales is hard to estimate as hyperthermophily of Thermotoga probably makes it branch artefactually early on many trees; nonetheless, Thermotogales probably did diverge relatively early and may be nearly as old as Proteobacteria. As hyperthermophilic Thermotoga species nest relatively shallowly among thermophilic species, transfer of hyperthermophilic genes (notably reverse gyrase) from archaebacteria (Nesbo et al. 2001; Nesbo & Doolittle 2003) is much more recent, not contradicting the late origin of archaebacteria. The evolutionary importance of eurybacteria is the evidence they give of early diversification of flagellate glycobacteria and the likelihood that they were ancestral to Posibacteria and thus ultimately to neomura and eukaryotes (Cavalier-Smith 2006a).
(d) Probable intermediate age of posibacteria
Despite the usual (not universal) failure of Actinobacteria/neomura to group with Endobacteria, Posibacteria are most probably monophyletic (§2b and Cavalier-Smith 2006a). Therefore, as Endobacteria nest within Eurybacteria in virtually all sequence trees, Posibacteria must be substantially younger. If Eurybacteria are ca 2.75 Gyr old, Posibacteria can hardly be older than the 2.3 Gyr ago oxygenation event or else they would not nest so reliably within Eurybacteria on trees. I think they cannot be younger than ca 1.5 Gyr old or they would nest even more securely and distinctly more shallowly within Eurybacteria. Therefore, Posibacteria probably evolved ca 2.0–1.5 Gyr ago. While Actinobacteria are predominantly aerobic, endobacteria exploit aerobic and anaerobic niches to the full and include early diverging sulphate reducers. The extra-thick walls of Posibacteria and preadapted resistant endospores made them dominant bacteria in soils that became increasingly developed as a favourable microbial habitat after an ozone layer developed (and geographically more extensive by continental accretion). Endospores and thick walls protected from drying during wind dispersal and drought.
Although Actinobacteria are related to Endobacteria, it is uncertain if they are sisters to Endobacteria, and roughly equally old (previously assumed; Cavalier-Smith 2002), or derived from them, and thus younger. Many actinobacteria use the thick posibacterial walls to become morphologically highly complex; actinomycetes can be macroscopic and visible to the naked eye, like some cyanobacteria that they rival and sometimes exceed in morphological complexity. Actinomycetes are throughout the world in soils and sediments, even in deepest oceanic trenches. Surprisingly, palaeontologists almost never consider them possible candidates for more complex fossils, especially those conventionally seen as candidate stem eukaryotes. 1.5 Gyr ago is when distinctly larger than previously fossil thick-walled cysts begin to occur at low frequency (Schopf & Klein 1992; Knoll et al. 2006). Butterfield (2005) broke with the tradition that most if not all of these are algal, for which there was never compelling evidence; in his words, micropalaeontologists adopted ‘a search image weighted excessively in favour of unicellular plant protists’. He reasonably suggests that Tappania and possibly also Germinosphaera, Foliomorpha, Trachyhystrichosphaera, Shuiyousphaeridium and Dictyosphaera are not single cells but multi-genome marine benthic mycelial, osmotrophic, heterotrophs. He thought they were fungi, but such as Tappania could be actinomycetes similar to Amycolatopsis (Wink et al. 2004) and Kibdelosporangium. If they are, as I suspect, actinomycetes are probably at least 1.5 Gyr old if any Roper fossils (Javaux et al. 2001, 2003; Knoll et al. 2006) are actinomycetes (I agree with Knoll et al. (2006) that Roper T. plana may differ from Butterfield's Neoproterozoic fossils).
Since actinomycetes are just one derived subclade within the actinobacterial tree, unless Tappania is an extinct derivative of an earlier branching clade now represented only by morphologically simpler species, then actinobacteria are probably distinctly older. I suggest that Actinobacteria are no younger than ca 1.8 Gyr. Thus, we have a rough estimate of the maximum age of Endobacteria as 2.3 Gyr (2 Gyr more likely) and a minimum age for Actinobacteria of ca 1.8 Gyr. These estimates are compatible with their being sisters and both 2–1.8 Gyr old or with Actinobacteria being younger than Endobacteria and derived from them. The closeness of these estimates implies that even if Actinobacteria evolved from Endobacteria, they should nest deeply within Endobacteria, less than or equal to 10% of the distance from their base with perfect phylogenetic reconstruction. In the real world of imperfect phylogenetic algorithms and pervasive systematic biases, e.g. from the exceptionally high-GC nucleotide composition of actinobacterial DNA and unusually low GC in Endobacteria, we should not expect Actinobacteria and Endobacteria to group together on most single-gene or multigene trees, even if Posibacteria are monophyletic. If they are sisters, mild systematic biases—or sampling error on single-gene trees—could falsely separate them.
Similar considerations and more accurately dating Actinobacteria are important for interpreting sequence trees in relation to neomuran origins. If eukaryotes and neomura are only 0.9 Gyr old and Actinobacteria are 1.8 Gyr old, perfect trees ought to nest neomura within Actinobacteria. Previously, as some eukaryote-like properties are phylogenetically restricted, I assumed that actinobacteria are paraphyletic ancestors of neomura, and thus older (Cavalier-Smith 2002a). If instead neomura and Actinobacteria are sisters (not disproved) and ca 1.5 Gyr old, as in some fossil interpretations, neomura should NOT nest within Actinobacteria on perfect trees but should be their sisters (as they are weakly on some trees, especially if the often closer, probably artefactual, position near neomura of Thermotoga/Aquifex is set aside). Systematic bias from accelerated evolution of most neomuran genes may be sufficient to exclude neomura from Actinobacteria artefactually on most sequence trees, even if they differ in age by 800 Myr; thus we cannot use their normal exclusion to argue that their ages are markedly closer than that, or for their being sisters.
5. Synthesis: how microbial quantum evolution changed the world
(a) Overview: the three ages of life
The preceding analyses and synthesis yield a simple picture of the history of the biosphere and its impact on the atmosphere and earth's surface (figure 8). Three great eras of unequal length were punctuated by two short periods of dramatic change: the glycobacterial and neomuran revolutions. First, more than 2.8–2.9 Gyr ago, when only eobacteria existed, ecosystems and biogeochemical cycles were very simple. I call this first stable era the age of chlorobacteria, as life depended fundamentally on energy and fixed carbon provided by the single photosystem of chlorobacteria—the most primitive form of anoxygenic photosynthesis using hydrogen or H2S as hydrogen donor. Ecosystems were anaerobic (Pavlov & Kasting 2002). Heterotrophs were present and probably included anaerobic fermenters (like Anaerolinea; Sekiguchi et al. 2003) and a limited range of anaerobic respirers (probably not sulphate reducers more than 3.2 Gyr ago) but no aerobic respirers. Next was the immensely long age of cyanobacteria, when cyanobacterial photosynthesis predominantly fed ecosystems and the biosphere differentiated into aerobic and anaerobic zones (2.8–0.9 Gyr ago). Eobacteria expanded into both, but became minor contributors to biogeochemistry compared with the four ancestrally photosynthetic phyla stemming from the glycobacterial revolution. Of these, cyanobacteria were the dominant aerobic autotrophs and proteobacteria the dominant heterotrophs and anaerobes The versatility of proteobacteria produced the greatest diversity of physiological mechanisms, helping them colonize almost every bacterial biogeochemical adaptive zone except oxygenic photosynthesis and methanogenesis. The glycobacterial revolution and ensuing physiological diversification of the six glycobacterial phyla sparked 0.5 Gyr of instability, with snowball Earth global freezing and atmospheric and crustal oxidation. Thereafter Earth entered a relatively uneventful phase—the ‘boring billion’ (ca 1.2 Gyr)—when major bacterial metabolic diversification ceased and the biosphere and Earth's surface were in a relatively stable steady state.
Figure 8.
Key events in global history, integrating evidence from palaeontology and cell phylogeny. The Late Archaean and Late Proterozoic saw momentous global changes caused fundamentally by two biological revolutions—the glycobacterial and neomuran. Most phyla probably originated in bursts of adaptive radiation and quantum evolution relatively soon after these major innovations. Several new physiologies thereby created dramatically changed biogeochemical cycles. Except for a probable delay in eukaryote diversification caused by the Neoproterozoic snowball Earth episodes, their timing was probably not affected by environmental factors but by their inherent evolutionary difficulty; adaptive zones for the new phyla were created for the first time by their novel body plans, e.g. water splitting (oxygenic) photosynthesis by cyanobacteria and their immediate ancestors, phagotrophy by eukaryotes. By contrast the two major mass extinctions led to substantial lower level (class or order) innovations by removing competitors from swathes of previously occupied niches. Extinction of acritarchs probably allowed dinoflagellates, haptophytes, and later diatoms to evolve. The time of origin of Chlorobacteria (and therefore when the age of Eobacteria began) is very uncertain—any time from ca 2.9 to 3.5 Gyr, with a date within 3.1–3.5 Gyr ago being slightly preferred (see text).
During this quasi-equilibrium posibacteria evolved, paving the way for its disruption by the neomuran revolution near the Meso/Neoproterozoic boundary and the immediately ensuing eukaryote and archaebacterial revolutions that generated the modern world. This third phase of Earth history I call ‘the age of eukaryotes’ as it has been dominated by greater morphological complexity made possible by the eukaryote cell and phagotrophy. As for the age of cyanobacteria, there was early instability for at least 100 Myr and global glaciation while new groups diversified to generate the major novel types. Invention of archaebacterial methanogenesis and methanotrophs, origin of the chloroplast and secondary symbiogeneses to form more complex algae, and the origin of fungal hyphal heterotrophy were probably all between 850 and 570 Myr ago, amidst the Neoproterozoic snowball Earth episodes. All major forms of neomuran matter and energy processing evolved by 570–545 Gyr, when the origin of epithelial and mesenchymal connective tissue generated the animal kingdom and immediately following Cambrian explosion of animals.
(b) Methane and the Precambrian biosphere
After water vapour and CO2, methane is the most important greenhouse gas; per molecule its warming effect is ca 21 times that of CO2. Today methane made by free-living and symbiotic archaebacteria probably greatly exceeds abiotic methane. The Archaean sun was weaker; climate modellers argue that more greenhouse gas was thus necessary to prevent global oceanic freezing. It was once thought that substantially higher levels of CO2 would suffice (Kasting & Pollack 1984), but palaeosol evidence suggested atmospheric CO2 levels below those that on their own would prevent global freezing (Kasting 1987). Following Lovelock's (1988) suggestion of methane as an important Archaean greenhouse gas, the assumption of palaeoclimatic models was that biogenic methane could solve the faint Archaean sun problem (Pavlov et al. 2000; Kasting & Siefert 2002; Kasting & Ono 2006). However, as discussed above, archaebacteria probably did not exist in the Archaean or Early Proterozoic. Some geologists argue that CO2 levels were much higher than palaeosol data would allow (Ohmoto 2004; Ohmoto et al. 2004), and thus potentially sufficient to solve the faint sun problem, but arguments against that scenario seem convincing (Kasting 2004). Thus, at least some role for inorganic methane seems inescapable unless net heating by water vapour and clouds was proportionally higher than currently assumed or a fourth factor is overlooked. Abiotic methane (Horita & Berndt 1999; Foustoukos & Seyfried 2004; Scott et al. 2004a) is found, e.g. in deep mines in Canada and South Africa and in mid-oceanic rifts. Was there enough to solve the faint sun problem, with presently assumed levels of water vapour and the maximum CO2 allowable by palaeosol data? Before the 2.3 Gyr ago oxygenation and origin of biological methanotrophy, methane would have been destroyed by oxidation more slowly than today. According to fig. 4 of Kasting & Ono (2006), the suggested maximum level of abiogenic methane of 1000 p.p.m. would be amply sufficient to solve the problem in conjunction with higher CO2 levels, below the palaeosol limit, as would 10–20 times lower methane levels. Therefore an early origin of archaebacteria need not be postulated to save the Archaean earth from perpetual freezing. Phylogenetic evidence for the absence of archaebacteria from the Archaean usefully narrows the range of allowable possibilities for the levels of CO2, methane, and temperature.
Methanotrophy and biological methanogenesis are both done by a suite of 15–16 different enzymes related to those that mediate other C1-compound conversions, some using unusual cofactors made by enzymes encoded by ca 10 other genes (Chistoserdova et al. 2004). Belief that these 25–26 genes were restricted to methanogenic archaebacteria and proteobacteria of subphylum Rhodobacteria (purple bacteria plus non-photosynthetic descendants) fostered hypotheses of LGT from archaebacteria to proteobacteria (Chistoserdova et al. 1998; Boucher et al. 2003; Martin & Russell 2003) or vice versa (Cavalier-Smith 2002a). Lateral transfer from archaebacteria, the more popular, was temporally impossible because methanogens are probably about three times younger than Rhodobacteria (ca 2.7 Gyr old), and practically highly improbable because their C1-related genes are scattered all over the chromosome and could hardly all be cotransferred. Reverse transfer from Proteobacteria to Archaebacteria is mechanistically more plausible as many are closely clustered in operons in eubacteria and could theoretically be cotransferred. However, discovery of related genes in Planctobacteria, probably sisters of Proteobacteria, and in non-methanogenic archaebacteria and one even in the actinobacterium Streptomyces, plus recent phylogenetic analysis argues strongly that almost all these genes have simply been inherited vertically, being repeatedly lost by lineages without them (Chistoserdova et al. 2004). Lateral transfer need be invoked only for one Fae homologue of unknown function from a proteobacterium to the planctomycete Pirellula. Phylogeny of six genes with straightforward history (three C1 pathway and three cofactor genes) is entirely congruent, if rooted between Proteobacteria/Planctobacteria (collectively Exoflagellata; Cavalier-Smith 2002a) and Archaebacteria, with my rooted universal tree (figure 5), favouring vertical inheritance. Gene phylogenies contradict suggestion D of Chistoserdova et al. (2004) of lateral transfer from Planctobacteria to Proteobacteria and Archaebacteria.
The simplest interpretation of evolution of methylotrophy and methanogenesis is not scenario E of Chistoserdova et al. (2004), which makes the usual incorrect assumption that the tree's root is between neomura and eubacteria, but that these C1 enzymes first evolved in the common ancestor of Gracilicutes and Eurybacteria, were inherited vertically by Planctobacteria, Rhodobacteria and Archaebacteria, and lost by eurybacteria, eukaryotes, most Posibacteria, Sphingobacteria and Spirochaetes. In Planctobacteria they do not mediate methylotrophy or methanogenesis; they originally probably oxidized C1 compounds, e.g. formaldehyde. Aerobic methylotrophy and methanotrophy occur only in α- and γ-proteobacteria, probably originating after they diverged from planctobacteria. If vertically inherited, these enzymes must date approximately to the ancestral rhodobacterium (2.7 Gyr; see above); C1 enzymes generally must be as old. Thus, methylotrophy extends back to the 2.78 ultralight C-isotope spike discussed above. Methanotrophy need not be that old; adding only one enzyme, methane monooxygenase, more easily transferred by LGT than the whole pathway, would convert a methylotroph to a methanotroph. However, the closest relative of proteobacterial methane monooxygenase is ammonium monooxygenase of β-proteobacteria, somewhat more divergent from methane oxygenases of α- and γ-proteobacteria than they are from each other, suggesting that divergence in function of the two paralogues occurred in the ancestral rhodobacterium and methanooxygenase and methanotrophy also originated ca 2.7 Gyr ago, as suggested for methylotrophy. Some aerobic methanotrophs can scavenge methane even from the atmosphere (Henckel et al. 2000). Aerobic methanotrophy needs more than 20 p.p.m. methane; atmospheric abiotic methane levels needed to solve the faint sun problem would have provided enough. The initial limiting factor was oxygen (Hayes 1994).
It is reasonable that C1 oxidation originated at virtually the same time as oxygenic photosynthesis providing the oxygen (2.8–2.9 Gyr ago). The initial impetus could have been protection against harmful molecules like formaldehyde by converting them to CO2. Abiotic methane destruction was probably markedly accelerated ca 2.7 Gyr ago by aerobic methanotrophy. Lovelock (1988) and Pavlov et al. (2000) suggested that the origin of oxygenic photosynthesis would have accelerated methane removal by oxygenating the atmosphere and could thus have caused the 2.45–2.25 Gyr ago global snowball Earth episodes (probably two). Section 5d discusses this further.
Methanogenesis is probably younger than archaebacteria, as methanogens nest within one archaebacterial subphylum, Euryarchaeota (Gribaldo & Brochier 2006). As archaebacterial sequence trees are substantially non-clock like (Gribaldo & Brochier 2006), estimating the age of methanogens is somewhat hazardous. In the absence of a Bayesian relaxed clock analysis using an arbitrary age for the archaebacterial cenancestor to estimate how much younger methanogens may be, a rough estimate can be made: inspection of the concatenated ribosomal protein tree (Gribaldo & Brochier 2006) and a crude clock suggest that the cenancestral methanogen was 13–32% younger than the cenancestral archaebacterium. A rounded middle 20% and date of 0.9 Gyr ago for archaebacteria (as for eukaryotes; see above) gives ca 720 Myr ago for the origin of archaebacterial methanogenesis. As this coincides with onset of Neoproterozoic near-global freezing, I suggest this atmospheric infusion of biogenic methane caused snowball Earth indirectly by the mechanism of Schrag et al. (2002). Eventually stabilization occurred as the biosphere adapted to the innovation. One important adaptation would have been the origin of anaerobic methanotrophy, done only by still-uncultivated archaebacteria (Orphan et al. 2002). Methanotrophic archaebacteria all nest within the methanogens (Gribaldo & Brochier 2006), so must be younger. They probably evolved from methanogens by reversing the metabolic pathway and adding methane oxidase; small innovations, given prior evolution of methanogenesis (Chistoserdova et al. 2005). Anaerobic methanotrophs are most related to Methanosarcinales, which from the same concatenated tree (Gribaldo & Brochier 2006) are 24–57% younger; if methanogens are 720 Myr old and Methanosarcinales ca 30% younger, Methanosarcinales would be ca 500 Myr old. If anaerobic methanogens are their sisters, they are similarly aged or a little older. I suggest they evolved less than 570 Gyr ago, when snowball Earth episodes ceased, and by depleting methane close to its source (their methanogenic congeners) they have helped prevent excessive global warming by biogenic methane ever since.
(c) A brief history of nitrogen fixation
After life began, global biomass was limited by the initial stock of reduced nitrogen and carbon. Once photosynthetic carbon fixation evolved, reduced nitrogen became a key limitation. Unsurprisingly therefore, biological nitrogen fixation is very ancient, dating back to the oldest phylum, Chlorobacteria (Cavalier-Smith 2006a). Contrary to what is widely supposed, inheritance of N-fixation genes has probably been largely vertical, with LGT being rare (Raymond et al. 2004). The most ancient N-fixation enzymes are group II enzymes that work best under anaerobic conditions, having evolved from photosynthetic pigment biogenesis genes before the great oxygenation event. Type I genes, associated with mechanisms to circumvent the potentially inhibitory effects of oxygen, probably originated just prior to the glycobacterial revolution when the ancestors of cyanobacteria and proteobacteria diverged, immediately after the photosystem duplication that made oxygenic photosynthesis (Cavalier-Smith 2006a). The great antiquity of nitrogen fixation means that the inferred reduction of lightning during the Archaean posed no problems for biology; any crisis was before the cenancestor, not 2.2 Gyr ago as Navarro-Gonzalez et al. (2001) postulated.
(d) The glycobacterial revolution, biotic and abiotic lags and the Palaeoproterozoic snowball
Oxygenic photosynthesis provided most atmospheric oxygen and oxidized the oceans and surface rocks (Holland 2006). The striking coincidence of this great oxidation with the first snowball Earth episodes suggests they are causally connected. I argued above that Archaean climatic stability depended on greenhouse effects of both CO2 and abiotic methane. Oxygenic photosynthesis by using a more abundant hydrogen source (water, not H2 or H2S) allowed life to expand immensely. By increasing photosynthetic flux, it would reduce CO2 levels directly. Rising oxygen would reduce methane levels by abiotic atmospheric oxidation and enabling evolution of aerobic methanotrophy. Thus, it would reduce both major greenhouse gases, eventually bringing on snowball Earth. Unlike others proposing methane removal by atmospheric oxidation as the prime cause of Palaeoproterozoic global freezing (Pavlov et al. 2000; Kopp et al. 2005; Kasting & Ono 2006), I think the methane was not biogenic; nor should the roles of CO2 draw-down by expanded phototrophy and of proteobacterial methanotrophy be ignored. If there was no biogenic methane, the drop in methane would have been even faster than they assume.
Kopp et al. (2005) argue that cyanobacterial expansion and oxygen rise were so fast that cyanobacteria must have evolved only ca 2.4 Gyr ago. But if the 2.7 Gyr bitumen biomarkers are truly endogenous (uncertain) that cannot be true. Apart from possible past downward mobility, Kopp et al. (2005) give three reasons for discounting that 2.7 Gyr hopanoid evidence for cyanobacteria: (i) some methylotrophs (e.g. Methylobacterium) make low levels of 2-methyl-bacteriohopanepolyol; (ii) other hopanols are not restricted to aerobes but present in Geobacter; (iii) methyl-bacteriohopanepolyol might have been made by ancestral cyanobacteria before oxygenic photosynthesis arose. All are refuted by phylogenetic arguments: Methylobacterium and other aerobic methylotrophs, and Geobacter are all proteobacteria, thus part of the Gracilicute clade, whose common ancestor must have had two photosystems like cyanobacteria (Cavalier-Smith 2006a). They are also part of a larger ancestrally flagellate eubacterial clade that is sister to the non-flagellate cyanobacteria. They cannot therefore be older than cyanobacteria and are probably somewhat younger. If any of these bacteria were present 2.7 Gyr ago, cyanobacteria must have been also. Hopanols evolved no later than the common ancestor of cyanobacteria and proteobacteria; as cyanobacteria are holophyletic (Gupta et al. 2003), their common ancestor with Gracilicutes had both hopanols and two contrasting photosystems. Unless a reason other than the origin of oxygenic photosynthesis can be found for divergence of two photosystems in one cell, then this glycobacterial common ancestor already had at least primitive oxygenic photosynthesis. As there is no evidence for hopanoids or two photosystems in Eobacteria, they probably evolved in this common ancestor immediately before it diverged to cyanobacteria and gracilicutes/eurybacteria. Thus, hopanols in general, not just 2-methyl-bacteriohopanepolyol, are probably good proxies for the age of cyanobacteria and (insignificantly earlier) glycobacteria as a whole. If my argument (based on catalase evolution) that oxygenic photosynthesis evolved in the common ancestor of glycobacteria and Hadobacteria is correct (Cavalier-Smith 2006a), it probably even slightly preceded the origin of glycobacteria. Even though cyanobacteria probably do not make sterols (Summons et al. 2006), their biosynthesis requires oxygen, suggesting that biotic oxygen was made in microbial mats as early as 2.715 Gyr ago. It is unwise to seek escape from this refutation in an unknown anaerobic mechanism for adding oxygen (Kopp et al. 2005).
Hayes & Waldbauer (2006) point out that the reservoir of reduced iron and sulphur able to mop up early biogenic oxygen was so huge that one expects a substantial lag after oxygenic photosynthesis before atmospheric oxygen levels rose. If oxygen was not generated faster than these minerals could remove it, a 400 Myr lag can easily be explained by that reservoir size. Kopp et al. (2005) calculated biogenic oxygen fluxes compared with removal by Fe, assuming that cyanobacterial population expansion was limited only by total oceanic P or N nutrients, and concluded that atmospheric oxygen should rise enough to deplete methane within a few million years. But this has two problems: it is the available stock of reduced Fe and S that matters initially not its regeneration rate, which Kopp et al. assumed limited O2 sequestration; secondly, it is unrealistic that cyanobacteria were limited only by nutrients. Before an ozone layer they would have been seriously restricted by harmful UV radiation (Cockell 2000; Cockell & Horneck 2001).
Consider the isotopic evidence for the state of the carbon cycle at the putative time (2.8 Gyr) of the glycobacterial revolution. Rothman et al. (2003) and Hayes & Waldbauer (2006) showed that traditional steady-state carbon cycle models of the past 30 years have been oversimplified in two respects, making predictions disagree with the data. First, it is unrealistic to treat all oceanic/atmospheric carbon as one pool; one must differentiate between dissolved organic carbon and CO2 (Rothman et al. 2003). Secondly, the crust/oceans/atmosphere is not self-contained; as geothermal activity continually injects CO2 from the mantle, one must also weigh export of oxidizing power to the mantle in the balance (Hayes & Waldbauer 2006). Hayes & Waldbauer (2006) also note that as crustal burial processes important for redox mass balance calculations operate on a much slower time-scale than biological processes that directly generate or consume oxygen, isotopic fractionation of buried carbon need not correlate with atmospheric changes; large negative excursions of Δ13C in organic carbon are not mirrored by complementary changes in inorganic carbonate from 2.8 to 2.0 Gyr ago, so they argue that changes in burial rates were not a major factor—in marked contrast to Neoproterozoic perturbations to the isotope record affecting both organic and inorganic deposits.
A further complication not considered previously is heterogeneity of the environment where primary production occurs. For Phanerozoic oceans dominated by plankton a homogeneous well-mixed model is reasonable. But in the Archaean and Early Proterozoic more than 2.3 Gyr ago with no ozone layer, UV irradiation probably largely excluded photosynthetic bacterioplankton from the upper photic zone. Phototrophs were probably concentrated in shallow water where iron-impregnated mineral grains or snow overlying thin ice offered enough protection from UV radiation. Global productivity was much less than today; life concentrated in stratified microbial mats (fossilizable as stromatolites) rather than as well-mixed phytoplankton freely communicating with the atmospheric CO2 pool. Mat complexity increased greatly with the glycobacterial revolution (figure 9) offering increased opportunity for internally recycling CO2. Figure 9b shows a simplified model with only two strata, which allows two-stage fractionation by Rubisco. First, by cyanobacteria in the upper layer. Then by anaerobic bacteria, e.g. chlorobacteria or purple bacteria (proteobacteria) such as Chromatium in the lower layer. Fermentative and respiring heterotrophs would have been in both layers using organics generated by the phototrophs. CO2 generated by these heterotrophs or phototrophs at night would be already depleted of 13C by Rubisco carbon fixation. Recycling it within the mat could deplete it again—and again, producing increasingly light organic carbon for burial. For a strictly two stage recycling (figure 9b) the depletion in buried organic carbon would be: ΔC+forrΔC, where ΔC is the depletion by a single stage, r is the fraction of CO2 fixed by the second stage (assumed for simplicity to be solely purple bacteria, but in practice it could be any Rubisco-using phototroph in the mat) and for is the fraction of buried carbon that comes from the purple bacteria after recycling (1−for would come from cyanobacteria).
Figure 9.
Ecosystems and the carbon cycle before and after the glycobacterial revolution. (a) In the age of chlorobacteria, life was mainly restricted to low diversity microbial mats protected from UV radiation by mineral particles. (b) After cyanobacteria evolved, mats became more complex with upper aerobic and lower anaerobic layers increasing the scope for internal CO2 recycling, with serial 13C isotope depletion by Rubisco. Isotopic composition of buried organic carbon depends on the fraction, r, of once depleted CO2 used by the second stage and on the fraction for of total C buried coming from second-stage organisms. (c) After flagella and proteobacteria evolved, they displaced chlorobacteria from most habitats and began to form a facultatively aerobic photosynthetic plankton, whose quantitative contribution greatly expanded with the ozone layer as UV inhibition declined and cyanobacterial phytoplankton evolved, especially prochlorophytes adapted to higher light intensities. As the fraction of buried organic carbon coming from plankton, fop, increased, that from recycling within the mat (for) became quantitatively insignificant from Mid-Proterozoic to Phanerozoic, thus abolishing the ultralight carbon signal of stage (b).
If cyanobacterial photosynthesis and CO2 trapping were very efficient, the upper layer could absorb most unfractionated CO2 diffusing from the atmosphere, leaving only a small proportion for the lower layer. Thus, values for r could probably easily exceed 0.5 and could be as high as 1. Assuming a value of 0.9 and a value of 0.5 for for gives a total fractionation 1.45 times that of one stage. Note that for is determined by the relative biomass of the two layers, not by relative rates of synthesis. This is important because if photosynthesis in the lower layer were CO2-limited, it would occur at a lower rate. But if the lower layer were thicker it could have a higher biomass, even if turnover was slower. If it were three times as thick and as dense, for would be 0.75 and total fractionation 1.65 times that of a single stage. In fact, mats can have much greater anaerobic than aerobic biomass (Sorensen et al. 2005). If one Rubisco stage can achieve −30‰, two could yield −45‰ and three could yield Δ13C −63‰. I propose that a sudden increase in biological complexity of microbial mats, favouring multistage recycling, caused the negative Δ13C spike 2.77 Gyr ago and that this negative spike is the best way of dating the glycobacterial revolution. There is no need to invoke methanogenesis or acetogenesis. Logan et al. (1999) found that in terminal Proterozoic samples those from microbial mats were more depleted in 13C than planktonic samples, fitting the idea that mats can favour recycling already lighter CO2 more than possible in suspension and thus serial light-biased fractionations by Rubisco. Mats often have three layers, with green-sulphur bacteria at the bottom, but in some habitats these largely replace the purple bacterial layer of figure 9c (Sorensen et al. 2005).
This stratified mat model explains timing of the onset and end of this unusual depletion. The negative spike would disappear completely when the photic zone was fully colonized by glycobacterial plankton (mainly cyanobacteria and proteobacteria) after the ozone layer was formed. If it is correct, the isotope record suggests that this expansion into the plankton was complete by 2.1 Gyr in the later stages of the great oxidation event. However, the spike is greatly reduced by 2.55 Gyr, well before even the slight oxidation that ended mass-independent oxygen 2.45 Gyr ago and the likely onset of the ozone layer (2.3 Gyr). This early decline of the negative spike clearly contradicts the methanogenesis explanation, which suggested that termination was caused by atmospheric oxidation of methane, which could not have occurred till after 2.45 Gyr. It is no problem for the stratification theory if a partial shift into the plankton occurred prior to the ozone layer. A sound phylogenetic reason expects such a partial shift: the origin of flagella. Flagella arose after the glycobacterial revolution, but before the origin of proteobacteria (Cavalier-Smith 2006a). I suggest that the spike peak represents the brief period when cyanobacteria had evolved, but proteobacteria had not. At that time the only phototrophs were chlorobacteria and cyanobacteria, both with gliding motility and no flagella. If that is correct, the second anaerobic stage of CO2 recycling (figure 9b) must then have been by Rubisco-using chlorobacteria, not proteobacteria. If flagella evolved ca 2.75 Gyr ago, they would have enabled facultatively aerobic photosynthetic purple bacteria to swim above the mat and photosynthesize as plankton, reducing the recycling fraction r. If they kept in the lowest photic zone region they would suffer little UV damage.
This scenario is evolutionarily sounder than the original methanogenesis/methylotrophy as there is direct evidence from biomarkers that cyanobacteria and purple bacteria had both evolved by that date. If the overlying water column were also stratified, impeding downward flux of CO2 from the atmosphere, this would have favoured preferential reuse of respired CO2 compared with that from the atmospheric pool, increasing fractionation. However, this scenario does not require two different types of photosynthesis. It could occur by repeatedly recycling previously fixed and respired CO2 by one kind of photosynthesizer alone, e.g. purple bacteria in an entirely anaerobic mat or as suggested by Straus et al. (1992) by a chemotroph within the mat recycling previously phototrophically fixed CO2. The key thing is restriction of free mixing with the atmospheric CO2 pool, i.e. the physical conditions not the precise organisms involved. Thus, extra-light carbon cannot be used to infer the presence of a particular kind of bacterium; rather it tells us either that special physical conditions to allow local recycling of gaseous products without global mixing were present in the particular habitats where the ultralight carbon was laid down or that little-known abiotic processes contributed to such a signature. However, although §4b cited some able to do so, there is no obvious rationale why abiotic causes should have been globally effective just then, neither before nor later, so they are probably irrelevant. Stratified mats, composed of the same globally distributed bacteria could give a consistent global isotopic signal without mat CO2 having to mix freely globally. As Hayes & Waldbauer (2006) stress, kinetic effects can dominate the short term, and the spike was definitely short term compared with rock burial cycles. Microbial mats are not the only situation where isotopic fractionation can be higher than the standard model predicts. Biomass stemming from proteobacterial chemotrophs densely packed within animals is markedly more depleted (Δ13C −30–35‰; much more than in phytoplankton) than the 24‰ caused in one step by the corresponding Rubisco (Scott et al. 2004b). This is partly because the symbiotic system recycles already depleted inorganic carbon from the local environment; but CO2 recycling within the animal's chemotroph mass could also occur.
The above is oversimplified. Like earlier models (Hayes 1983, 1994; Hayes & Waldbauer 2006) it ignores the fact that Δ13C by Rubisco varies evolutionarily, being typically lower in bacteria than eukaryotes, usually ca −20‰ not −30‰ as in spinach (Guy et al. 1993). This is a problem for the classical assumption that pre-2.77 Gyr ago Archaean Δ13C of 35‰ reflects a single-stage Rubisco fractionation. There was no spinach in the Archaean. I suggest that two-stage recycling in mats is also needed to explain the more than 2.77 Gyr ago data—unless they were produced purely abiotically and the 2.77 Gyr spike is a marker not for glycobacteria but for the origin of life and photosynthesis, which cannot be conclusively rejected, but I think is unlikely. The problem for the chlorobacterial Early Archaean is even greater, because most chlorobacteria, except Oscillochloris that uses Rubisco with Δ13C of ca 20‰ (Ivanovsky et al. 1999), do not use Rubisco, but the 3-hydroxypropionate pathway, showing less favouritism for light carbon (e.g. Chloroflexus). We currently cannot infer which carbon fixation pathway was ancestral for chlorobacteria and whether both or only one were present in the Archaean. Assuming an equal mixture gives Δ13C of only −16‰, so much recycling would be needed to produce the observed values; some is unavoidable even if they only used Rubisco like that of Oscillochloris (Ivanovsky et al. 1999). However, there is a vast unexplored diversity of chlorobacteria, the most neglected bacterial phylum; other mechanisms may exist with higher fractionation. Immensely more research is needed on Chlorobacteria if we are to understand Archaean ecosytems.
Another complication is the unpublished evidence for a temporary pulse of oxygen sufficient to abolish mass-independent sulphur isotope fractionation around 2.9 Gyr ago (Kasting & Ono 2006). Was the glycobacterial revolution, therefore, earlier than suggested? Not necessarily. One possibility is that this pulse came from pre-cyanobacterial oxygenic photosynthesizers, which probably evolved immediately before Cyanobacteria and Hadobacteria diverged (Cavalier-Smith 2006a). This could be a signal from that node in the tree, and could have caused the glaciation 2.9 Gyr ago, as suggested above. If it were, why did oxygen not remain high enough to prevent mass-independent fractionation? One possibility is that aerobic respiration evolved and reduced oxygen concentrations at source sufficiently for abiotic hydrogen and methane to accumulate enough to scavenge atmospheric oxygen, keeping it below the critical level more than 2.45 Gyr ago. Thereafter continued oxygenic photosynthesis produced the ozone layer, allowing cyanobacterial populations to expand rapidly and oxidize the earth. Once oxygen reached a certain threshold and the ozone layer began, positive feedback through population expansion would make oxygen levels rise explosively. Snowball Earth itself would have favoured phototroph expansion into the plankton through UV protection from snow on sea ice (Cockell et al. 2002; Cockell & Cordoba-Jabonero 2004), which would probably not have been too thick for light penetration (McKay 2000) and thus synergistic with ozone rise. As cytochrome oxidase probably evolved from an oxygen-independent oxidase previously used in anaerobic respiration, and the earlier part of the respiratory chain was just taken over from anaerobic respiration, aerobic respiration could have evolved rapidly after oxygen levels in mats became appreciable. This is important, as it must have happened fast enough to prevent oxygen rising above the threshold for oxidizing minerals like uraninite (only 10 times that which abolishes mass-independent fractionation: Kasting & Ono 2006) which remained reduced until 2.4 Gyr ago.
Finally, why did global glaciations cease for 1.6 Gyr? This is unexplained. Had the sun perhaps warmed enough to avoid global freezing altogether? Perhaps biology helped by invasion of land by cyanobacteria that the ozone layer allowed after 2.25 Gyr. Vast desert areas and parts of the Arctic are now covered by a thin dark cryptogamic crust of cyanobacteria, lichens and fungi (Johansen 1993). Prominent therein is the black cyanobacterium Scytosiphon, but actinobacteria and proteobacteria also abound (Nagy et al. 2005). They reduce albedo and in the Arctic can warm surfaces by 8–12 °C and soil by 4–5 °C (Gold 1998). Cyanobacterial blackening of Early-Mid-Proterozoic continents, albeit initially perhaps only half their present extent, possibly saved the day, until quantum evolution again intervened, starting the Neoproterozoic snowball.
In discussing carbonate isotopic levels 2.0–2.3 Gyr ago, Hayes & Waldbauer (2006) wrote ‘In all likelihood, the diagenetic alternative has failed to win popularity because an alternative does not appear to be needed.’ Likewise the biogenic methane explanation for the organic carbon negative spike and the increased 13C in 2.0–2.3 Gyr carbonates remains popular because the necessity for an alternative is widely unrecognized. If the present explanation is found wanting, another must be found, not invoking methanogens, which were almost certainly absent more than 1.5 Gyr ago, and probably also more than 0.75 Gyr.
(e) Neoproterozoic snowball Earth
The explanation offered here for the onset of the Neoproterozoic glaciations is essentially that of Schrag et al. (2002), with one crucial difference. The extra source of methane that they suggested indirectly caused CO2 levels to drop (and thus cause cooling) was the invention for the first time of biological methanogenesis, here inferred (§5b) as ca 720 Myr ago, not a large-scale release of oceanic methane hydrates. A biogenic source is more plausible, as methane-hydrate release is often catastrophic. Slow release (as by spread of newly-evolved methanogens around the world) is crucial to their model so it did not cause sudden global warming itself. If biological methane were the cause, it could accumulate again after one glaciation to repeat the process. So, what terminated snowball Earth episodes? I am sceptical that changes in continental disposition alone were sufficient (Schrag et al. 2002). Continents seem not to have been concentrated greatly more near the Equator at the onset of the snowball than ca 250 Myr ago when there was no glaciation (Torsvik 2003). I inferred above that anaerobic methanotrophy probably arose ca 570 Gyr ago, after which snowball Earth did not occur. I suggest these methanotrophs reduced biotic methane emission close to source, dropping atmospheric levels below those critical for the Schrag et al. (2002) scenario. Thus, some archaebacteria caused snowball Earth, then others saved the world from recurrences—so far.
The age estimated above of the plant kingdom and the cyanobacterial enslavement that first generated eukaryotic algae is 850 Myr, when atmospheric oxygen is inferred to have risen to present levels (Holland 2006). Holland (2006) attributes this rise to burial of fixed carbon and the origins of new types of eukaryote algae and protozoa. I suggest that larger cell sizes and more recalcitrant cell walls (both vegetative and of cysts) of the newly evolved eukaryotic algae and of cysts of protozoa that fed on them were the key causes of greater burial. Thus, cyanobacteria probably first oxygenated the atmosphere to moderate levels. Then eukaryotic algae increased the concentration further soon after their origin. Survival of eukaryotic algae through global freezing is sometimes raised as an objection to or problem for snowball Earth (Hoffman & Schrag 2002). But it is not. Equatorial sea ice was probably thin enough for sub-ice photosynthesis and survival of some unicellular eukaryotic algae (McKay 2000).
(f) Evolution of eukaryote algae
The dominant eukaryotic marine phytoplankton since the Permian extinction have been chromalveolates. As reasoned above, their origin by red algal enslavement was much earlier, in the Late Proterozoic. Discounting an unlikely shift in adaptive capacity among major algal lineages, the often spinose or morphologically complex marine acritarchs relatively common from 570 to 250 Myr ago were probably mostly chromalveolates, not green, red or glaucophyte algae, or euglenoids or chlorarachneans, but perhaps extinct classes. It is reasonable to equate the rise in acritarch complexity and speciosity ca 570 Gyr ago (Knoll 1994; Xiao & Knoll 1999; Peterson & Butterfield 2005) immediately after the snowball melt, when 57 new morphotypes appear (Grey et al. 2003), with the origin of chromalveolates. Gammacerane biomarkers are often cited as evidence for ciliates ca 750 Gyr (Anbar & Knoll 2002), but they are not even specific for eukaryotes, being made by proteobacteria. Dinosterols, often cited as evidence for dinoflagellates back to 1.1 Gyr (Anbar & Knoll 2002), are not specific for dinoflagellates, being found in some other chromalveolates. Although dinosterol is unknown in bacteria, 1.1 Gyr seriously contradicts other evidence for dinoflagellate age (250 Myr only; §4a and figure 8 legend) and is more than 600 Myr older than better evidence for chromalveolates. Thus, it is unwise to accept their specificity even for chromalveolates. However, the marked rise of dinosterane to modern proportions (after being undetectable during the preceding 200 Myr) in the Mid-Triassic coincidentally with the very first dinoflagellate fossils favours that date as the real time of origin of peridinean dinoflagellates. The earlier sporadic occurrences of dinosteranes (Late Ordovician/Silurian and Mid-Late Proterozoic) may have been produced by different chromalveolates now extinct. It is unsound to regard these early records as evidence for Palaeozoic, still less Precambrian dinoflagellates (Molowan et al. 2001), given the likelihood of convergence and the contradiction with the recency of morphological evidence and its congruence with molecular trees showing Peridinea as much younger than chromalveolates. Very likely eukaryotic marine phytoplankton from 850 to 570 Myr ago was exclusively prasinophyte green algae. Some putatively eukaryotic sphaeromorph acritarchs in this period have been identified as prasinophytes, but I am unsure this is correct. Organic carbon and biomarkers reveal extensive bacterial photosynthesis 740–700 Myr ago during the first snowball glaciation (Olcott et al. 2005), but their claim that biomarkers also reveal eukaryote algae is invalid.
Stanier (1970) remarked that chloroplasts ought to have arisen before mitochondria as cyanobacteria long preceded the rise in oxygen that would make mitochondria possible. His environmental determinism reflects widespread misconceptions that environment drives megaevolution and ignores endogenous evolutionary limitations that I think mainly govern its timing. Chloroplasts were not enslaved for ca 2 Gyr after cyanobacteria arose for potential enslavement. Enslavement was not possible until after eukaryote cells evolved, which was itself late because of the lateness and improbability of the neomuran revolution and the billions of years that eubacteria were prevented by their murein corset from taking up other cells.
6. Explaining megaevolution: preadaptation and quantum evolution
(a) The neomuran revolution and eukaryogenesis
The neomuran revolution allowed the origin of archaebacteria, and was a crucial prerequisite for the origin of eukaryotes. Its radical nature and probable extreme difficulty explain why eukaryotes emerged so late. Once rigid murein was replaced by flexible glycoprotein, evolution of phagotrophy became relatively easy—but still complex and not inevitable. Almost certainly the actinobacterium that generated it secreted scores, maybe hundreds, of digestive enzymes, so it was pre-adapted for digesting prey. The ancestral neomuran was preadapted for evolving addition of oligosaccharides containing N-acetylglucosamine (GlcNAc) to surface proteins as its actinobacterial ancestor already had an isoprenol carrier for exporting bulky hydrophilic GlcNAc-linked molecules (muramopeptide wall precursors) across the CM, plus enzymes for attaching them to the carrier and for joining GlcNAc to peptidoglycan amino acids that could be adapted to make glycoprotein instead. Minor changes in adhesive properties of its oligosaccharides could enable them to bind prey; softening the surface MreB skeleton would allow prey to be partially wrapped by membrane and eventually engulfed. Most likely this occurred within a few thousand years of the substitution of murein by glycoprotein. Once evolution of phagotrophy started, it would have gone to completion and generated a complete early eukaryote with endomembrane system, endoskeleton, nucleus, mitosis, cilium, peroxisomes, mitochondria and sex, probably in much less than a million years (based on considerations like those suggesting that mitochondria probably only took 10–100 000 years to evolve; Cavalier-Smith 2006b). A neomuran lacking special advantages like phagotrophy (eukaryotes) or hyperthermophily (archaebacteria) over eubacteria could not have spread around the world, still less persist for millions, or even billions of years as some scenarios assume, without leaving even one prokaryotic lineage. Assumptions that such a neomuran prokaryote existed for hundreds of millions of years are science fiction.
That glycoproteins are independent molecules free to diffuse in the fluid CM, and not covalently bonded into a three-dimensional rigid meshwork as in murein, preadapted the first neomuran to evolve phagocytosis. Substitution of murein by glycoproteins was a prerequisite for evolution of phagocytosis by folding the CM around the prey and forming the endomembrane system by evolving coated vesicles. Food vacuoles could not possibly have formed with a thick murein wall covalently attached to the plasma membrane as in Posibacteria. Having only one bounding membrane, not two as in Negibacteria, was also a prerequisite for phagotrophy and eukaryotes; the independent loss of murein by Planctobacteria could not have led to the origin of phagotrophy if Posibacteria had never evolved, though it seems convergently to have allowed a different type of membrane invagination from other Negibacteria that some misinterpret as related to those of eukaryotes (Fuerst & Webb 1991; Lindsay et al. 2001; Fuerst 2005). If Posibacteria evolved only ca 2 Gyr ago, eukaryotes could not have evolved earlier, because of the structural limitations of the negibacterial envelope. As mycoplasmas probably evolved only after eukaryotes gave them a protective habitat (Cavalier-Smith 2002a), the successful transition from posibacterial to neomuran walls was uniquely successful in generating a novel kind of free-living bacterium from a posibacterial ancestor. Its delay of probably 1 Gyr after the origin of Posibacteria is unlikely to have been through environmental limitations. It probably simply reflects the extreme difficulty of this change in wall organization. This explains the lack of innovation for the ‘boring billion’ years: all major metabolic innovations except methanogenesis had already evolved; the only possible major evolutionary advance was by fundamental change in the cell envelope, the mechanistically and selectively difficult replacement of murein by glycoprotein.
Seeking to explain why eukaryotes delayed for a billion years before becoming complex (Anbar & Knoll 2002) is pointless if they originated only ca 900 Myr ago and there was no such delay. A delay of diversification by snowball Earth till ca 570 Gyr is a comprehensible environmental brake. But thereafter protist diversification and the origins of macro-organisms probably took place without significant exogenous limitations, being limited merely by the inherent difficulties of the transitional stages. Three of these were evolutionarily unusually complex cases of quantum evolution: origins of chloroplasts, choanoflagellates, and sponges. Even though origin of plastids was very complex, probably involving tens of thousands of mutations (Cavalier-Smith 1982, 2000), it apparently happened early in bikont and eukaryote evolution and was thus probably not exceptionally difficult, given the prior occurrence of aerobic eukaryotes with good protection against O2 toxicity, and probably not a major retarding factor.
(b) The Cambrian explosions
Origin of choanoflagellate feeding by a periciliary microvillar collar involves complex coordination between probable derivatives of filose pseudopodia; its relative difficulty and the low biodiversity of suitable precursor amoeboflagellates could have been why animals did not evolve immediately after eukaryotes arose. Most Amoebozoa have lobose or discoid pseudopods entirely unsuitable as precursors. Most groups with well-developed filopodia have lost cilia (e.g. nucleariid Choanozoa, euglyphid Cercozoa) or do not make them at the same time in the life cycle as filopodia. Since animals originated via choanoflagellates and sponges, and I think any other route (e.g. evolving a metazoan gut and bilateral muscular motility directly without a filter-feeding sessile intermediate) would have been mechanistically far more difficult and perhaps impossible, the low probability of evolving choanoflagellates and the difficult trophic/skeletal tradeoffs needed to evolve non-feeding cells to support a large sponge body are probably sufficient explanation of the delay, though snowball Earth perhaps lengthened the lag.
Once sponges arose, and the spongocoel later became the coelenterate gut, and nematocysts and synaptic transmission evolved, the only remaining major innovation was the anus. Very likely this anal breakthrough ca 550 Gyr ago stimulated the long-puzzling Cambrian explosion. Peterson & Butterfield (2005) imply that the anus evolved at the Early Ediacara (570 Myr). However, like Conway Morris (2006), I am unconvinced that any Ediacara fauna (Xiao et al. 2005) are bilateria; nor can I place them in any protist or plant group. Most likely they are early animals related to sponges and cnidaria, but fossils have too little detail to show if they belong within either or are a distinct phylum; whichever makes little difference to the suddenness of the animal Cambrian radiation and its contemporaneity with the origin of most protist phyla. The Ediacaran/Cambrian explosion of protists and animals is a fundamental feature of eukaryote evolution, not a preservational artefact (Peterson & Butterfield 2005; Conway Morris 2006). Given the initial bilaterian pattern of a continuous-flow through gut and associated developmental mechanisms, all feasible animal bilateral body plans would inevitably be tried out without further delay by minor developmental modifications to basic animality, leading to an immediate explosive production of all bilateral animal phyla within a few tens of millions of years, each involving rapid quantum evolution and the entry to a major animal adaptive zone. The anus was a prerequisite for intelligence; without it, heads and brains would not have evolved. But after it evolved many primitively sessile groups remained headless. Contrary to Hyman's (1940) postulated multiple decephalizations, I consider that Brachiozoa, bivalves, Bryozoa, crinoids, pterobranchs, were not secondarily beheaded, but primitively acephalic tentaculate feeders, and that heads evolved polyphyletically (but using ancestral bilaterian antero-posterior polarity determinants) in the most cephalized groups (vertebrates, radulate molluscs, arthropods, polychaetes) in lineages that took to active burrowing, creeping or directional swimming rather than sedentary filter feeding or entrapment, the ancestral animal state (sponges) that required only limited neural development (cnidaria).
This burgeoning variation is fascinating from an anthropocentric perspective, but from that of biogeochemistry and global processes the most significant innovation was the later evolution of embryophytes that clothed the land with cooling greenness, enabling complex terrestrial life and greater stability to soil biomes (Wellman et al. 2003). This key innovation was probably delayed merely by the developmental and evolutionary difficulties of the transition from a charophyte alga to an embryophyte, not by environmental factors like the possibly insufficient ozone layer as sometimes speculated; oxygen had probably reached present levels 375 Myr before land plants arose and 300 Myr before animals (Holland 2006), so frequent suggestions that the origin of either was triggered by oxygen are erroneous. Both directly and by altering cloud cover, forest growth affected global processes but relatively far less than bacterial metabolic innovations.
I do not agree that animal origin was inevitable (Conway Morris 2006). Our presence owes much to rare and unique historical and evolutionary accidents. The neomuran revolution was delayed 2.6 Gyr after the origin of life; why could it not have been delayed 4 Gyr, or till after the sun runs out of fuel? Conversely, if on another planet cell walls were not first covalently bonded corsets, but made of modules like neomuran glycoproteins linked by weaker forces, then phagotrophy and its ability to generate internal cellular complexity would probably evolve much earlier. Since complex behaviour depends on internal cell complexity and on morphological combinations of basically similar neurons, morphological complexity, not basic chemical and genic complexity, makes intelligent life. Thus, if many different planets evolved life, wall chemistry of the first cells might be the primary determinant of when, if ever, intelligent life might evolve. Were they bonded covalently in three dimensions or by weaker forces allowing flexibility to phagocytose and make dendrites?
(c) The pattern of Earth history: quantum evolution and punctuated equilibrium versus uniformism
Darwin was puzzled by the sudden Cambrian appearance of animals, thinking it perhaps an artefact of preservation or missing rocks. Half a century of micropalaeontology (Schopf 2001; Knoll 2004) has shown that this suddenness is real; for almost the first 3 Gyr of life only microbes were present. Thus, animal origin was by radical quantum evolution, followed by exceptionally rapid radiation of novel phyla/body plans, as Simpson (1944) correctly argued is the rule for all higher taxa: orders, classes and phyla (megaevolution). It is even more dramatically true of kingdoms, subkingdoms and the only two superkingdoms: Bacteria and Eukaryota. At these highest levels megaevolutionary timing is not stimulated by environmental change, as is much low-level evolution. What matters primarily is the degree of difficulty of the evolutionary changes making the new body plan, and the availability of suitable phylogenetic precursors. Simpson argued that a major transition can occur only if the organism is in some way preadapted and if the novel adaptive zone thus created (e.g. phagotrophy for eukaryotes) was both empty and mutationally and ecologically accessible to it. He stressed that the transition was all-or-none; all intermediate stages invariably die out. Thus, endogenous, not exogenous, factors are the drivers. A novel body plan creates an adaptive zone and changes the environment; its origin is not responsive. It always changes the biotic environment for other life: the origin of vascular plants simultaneously provided food and habitats for the later most speciose groups: beetles and Lepidoptera.
At the largest scales, the phrase punctuated equilibrium aptly describes evolution and Earth history, even though stasis between punctuating episodes of quantum evolution is neither a thermodynamic equilibrium nor a true steady state, since evolution of a more modest kind necessarily continues throughout, as do irreversible inorganic processes like continental drift, orogenesis and erosion, slowing the Earth's rotation, and warming of the Sun. Quantum evolution and punctuated equilibria conceptually correct the deep-seated idea of uniformism: the false assumption that rates and types of change are spread uniformly through time and across lineages. Concepts of quantum evolution and punctuated equilibria are essential for a realistic description of what happened in history. But Gould (1994) confused things by contrasting punctuated equilibria not with uniformism, but with gradualism: the view that evolution proceeds gradually by cumulative spreading within populations of numerous relatively small mutations, not by a sudden ‘macromutation’ making a new organ or species overnight. Stated thus, gradualism is generally correct, but it must be qualified, as hybridization and polyploidy together have created thousands of new species overnight (allopolyploidy), albeit only a minority of all speciations and mostly in hermaphrodites. More dramatically, symbiogenesis wrought far more dramatic sudden changes that contradict overstated versions of gradualism, but only ca 6–7 times in all history (notably origins of mitochondria, plants, chromalveolates). But even these do not undermine the philosophic core of gradualism; the initiating changes were not DNA macromutations, but trivial accidents in the now immensely common process of internalization of foreign cells by phagocytosis: rare failures of digestion. Next was a key mutation that made the symbiont useful to the host in one step. This key mutation—for mitochondria, arguably, inserting an inner membrane carrier—radically changed the selective forces on the host, so an almost inevitable cascade of thousands of mutations (many newly beneficial, but some initially harmful) then led to organelle formation (Cavalier-Smith in press). Key mutational innovations that radically change selective forces on an organism are the fundamental initiating factors in quantum evolution; many examples are given in my writings on cell evolution, and above. But they need not be classical macromutations (Goldschmidt 1940) that were rightly anathema to population geneticists because proponents seemed to ignore the need to explain how mutations drastically changing phenotypes could survive and spread within sexual populations.
Some key mutations, however, e.g. murein hypertrophy and loss of OM or loss of murein, are phenotypically macromutations, but could be micro at the genetic level, being in principle causable by a single protein-inactivating nucleotide substitution or gene deletion (and could spread in bacteria which are non-sexual, lacking syngamy). Overstatements of gradualism asserting that megaevolution is simply the accumulation of large numbers of small mutations of the same kind as predominate at lower evolutionary levels are simply wrong and must be abandoned. The clearest counter example involves new gene formation by chimaerization; the mutational fusion of two unrelated genes to code for one, more complex, protein of novel function; many involve novel combinations of protein domains, e.g. origins of archaebacterial reverse gyrase or solenoid/propeller proteins vital for eukaryogenesis (figure 6a legend), but novel domains must sometimes evolve thus themselves. Such statistically rare ‘macromutations’ were crucial for many megaevolutionary episodes, but could be totally absent in many cases of microevolution and speciation. Thus, one cannot simply extrapolate from microevolution to megaevolution. New genes are more commonly made by gene duplication and extreme divergence, also non-essential for microevolution but fundamental to megaevolution (e.g. MreB and FtsZ duplications generated the eukaryote cytoskeleton, figure 6). Ancestors of thousands of novel eukaryotic genes cannot be identified by sequence bioinformatics, which misses the most important part of the quantum evolutionary picture because of extreme episodic sequence divergence during eukaryogenesis. With less marked divergence sister protein paralogues can be put on the same sequence tree, but episodic basal divergences so violate substitution models of uniform or only gradually changing divergence that most paralogue trees are very confusing, and seriously misleading conclusions can be drawn (Cavalier-Smith 2002b, 2006a). Mutations of large phenotypic effect are not Goldschmidt's (1940) macromutations by wholesale genomic reorganization, but simple gene mutations, duplications, deletions and chimaerizations. Transposable elements can cause major genome reorganization, but it is typically dissociated from and irrelevant to major phenotypic change (Cavalier-Smith 1993). The largest genomic reorganization in history converted bacterial circular single replicons with numerous operons (cotranscribed gene clusters) and virtually no non-coding DNA to eukaryotic linear chromosomes with multiple replicons and separately transcribed genes during eukaryogenesis. This caused no dramatic phenotypic changes, but was an adaptively trivial consequence of two of them (origins of mitotic spindle and nuclear envelope; Cavalier-Smith 1993, 2005). Goldschmidt's view that big phenotypic changes need big genetic causes was wrong; but in focusing on how mutations effect radical phenotypic change he was far ahead of others.
Population genetic paradigms that focus on changing allele frequencies for existing genes, or phylogenetic trees that chart their divergence, sidestep many key evolutionary innovations and like sequence bioinformatics see only part of the evolutionary picture. These approaches, though vital, must be supplemented. Understanding organismal evolution requires focus on specific phenotypic effects of different types of mutation in specific kinds of organisms, and their ecological consequences, not just treating mutations as generalized abstractions for mathematical manipulation. To comprehend megaevolution of macro-organisms, we must understand developmental biology and how it makes new forms (here genetically small mutations can have huge phenotypic effects; they should not be unthinkingly dismissed as ‘hopeful monsters’—origins of phyla are exceedingly rare events that may involve inherently unlikely intermediates, and key mutations acting early in embryogenesis to reform body plans grossly). Understanding microbial megaevolution requires cell biology—not just biochemistry. To link either with palaeontology we must have the correct phylogenetic tree and map it properly onto the fossil record. Attempts to reconstruct Earth history in a phylogenetic vacuum ignore important constraints, causing mistakes.
Concluding remarks. Gould (1994) also confused us by linking punctuated equilibrium to a mistaken macroevolutionary theory grossly overemphasizing species selection and lineage sorting. Therefore, I must stress that I am neither attacking nor ignoring population genetics or sequence phylogenetics, or the basically sound Neodarwinian paradigm, in advocating the merits of concepts of punctuated equilibrium, quantum evolution, and—especially—key mutations that dramatically change selective forces (and sometimes unusually substantially change phenotypes). These and other special requirements for understanding unique historical events that cannot be treated statistically were part of the synthetic ‘theory’ as seen by Simpson (1953), one of its leading architects, but have been partially forgotten and need restating each generation. Quantum evolution and punctuated equilibria are not mechanisms to replace mutation and selection, but descriptive devices to characterize large-scale evolutionary patterns. Truly synthetic evolutionary biology requires a balanced synthesis of reconstructed historic patterns and mechanistic explanations of the changes. Its must draw on and attempt to integrate all relevant disciplines, none preeminent—not easy in an age of overspecialization. My efforts here to portray a three-stage view of life with only two really major punctuations in more than 3 Gyr may stimulate others to do better. A possible simplification to just two stages, only hinted at above, arises if the origin of life was actually much closer to the glycobacterial revolution than widely assumed, e.g. ca 2.9–3.1 Gyr, making the origins of Chlorobacteria and Hadobacteria only slightly before that of glycobacteria. If so, the glycobacterial revolution would simply be a later part of one explosive early negibacterial radiation, as argued previously (Cavalier-Smith 1987a, 1992, 2002b). In that case the neomuran revolution would be the only really major punctuation in all evolution. Better understanding of the Early Archaean is needed to decide.
Acknowledgments
I thank NERC for research grants, and the Canadian Institute for Advanced Research and NERC for fellowship support.
Footnotes
One contribution of 14 to a Discussion Meeting Issue ‘Major steps in cell evolution’.
References
- Acaso C, Wierzchos J, Speranza M, Gutiérrez J.C, González A.M, de Los Ríos A, Alonso J. Fossil protists and fungi in amber and rock substrates. Micropalaeontology. 2005;51:59–62. 10.2113/51.1.59 [Google Scholar]
- Alexopoulos C.J. Wiley; New York, NY: 1952. Introductory mycology. [Google Scholar]
- Anbar A.D, Knoll A.H. Proterozoic ocean chemistry and evolution: a bioinorganic bridge? Science. 2002;297:1137–1142. doi: 10.1126/science.1069651. 10.1126/science.1069651 [DOI] [PubMed] [Google Scholar]
- Armstrong H.A, Brasier M.D. 2nd edn. Blackwell Scientific Publications; Oxford, UK: 2005. Microfossils; p. 296. [Google Scholar]
- Balch W.E, Magrum L.J, Fox G.E, Wolfe R.S, Woese C.R. An ancient divergence among the bacteria. J. Mol. Evol. 1977;9:305–311. doi: 10.1007/BF01796092. 10.1007/BF01796092 [DOI] [PubMed] [Google Scholar]
- Bartlett M.S, Thomm M, Geiduschek E.P. Topography of the euryarchaeal transcription initiation complex. J. Biol. Chem. 2004;279:5894–5903. doi: 10.1074/jbc.M311429200. 10.1074/jbc.M311429200 [DOI] [PubMed] [Google Scholar]
- Beatty J.T, Overmann J, Lince M.T, Manske A.K, Lang A.S, Blankenship R.E, Van Dover C.L, Martinson T.A, Plumley F.G. An obligately photosynthetic bacterial anaerobe from a deep-sea hydrothermal vent. Proc. Natl Acad. Sci. USA. 2005;102:9306–9310. doi: 10.1073/pnas.0503674102. 10.1073/pnas.0503674102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berney, C. 2005 Contributions to the molecular phylogeny of eukaryotes, with focus on amoeboid protists and environmental diversity. DSc thesis 3631, University of Geneva.
- Berney C, Pawlowski J. A molecular time-scale for eukaryote evolution recalibrated with the continuous microfossil record. Proc. R. Soc. B. 2006;273 doi: 10.1098/rspb.2006.3537. 10.1098/rspb.2006.3537 (Published online 20 April 2006.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G. Intracellular protein topogenesis. Proc. Natl Acad. Sci. USA. 1980;77:1496–1500. doi: 10.1073/pnas.77.3.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher Y, Douady C.J, Papke R.T, Walsh D.A, Boudreau M.E, Nesbo C.L, Case R.J, Doolittle W.F. Lateral gene transfer and the origins of prokaryotic groups. Annu. Rev. Genet. 2003;37:283–328. doi: 10.1146/annurev.genet.37.050503.084247. 10.1146/annurev.genet.37.050503.084247 [DOI] [PubMed] [Google Scholar]
- Bown P.R, Lees J.A, Young J.R. Calcareous nannoplankton evolution and diversity through time. In: Thierstein H.R, Young J.R, editors. Coccolithophores from cellular process to global impact. Springer; Berlin: 2004. pp. 481–508. [Google Scholar]
- Brasier M.D, Green O.R, Jephcoat A.P, Kleppe A.K, Van Kranendonk M.J, Lindsay J.F, Steele A, Grassineau N.V. Questioning the evidence for Earth's oldest fossils. Nature. 2002;416:76–81. doi: 10.1038/416076a. 10.1038/416076a [DOI] [PubMed] [Google Scholar]
- Brasier M, McLoughlin N, Green O, Wacey D. A fresh look at the fossil evidence for early Archaean cellular life. Phil. Trans. R. Soc. B. 2006;361:887–902. doi: 10.1098/rstb.2006.1835. 10.1098/rstb.2006.1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretschneider T, Diez S, Anderson K, Heuser J, Clarke M, Muller-Taubenberger A, Kohler J, Gerisch G. Dynamic actin patterns and Arp2/3 assembly at the substrate-attached surface of motile cells. Curr. Biol. 2004;14:1–10. doi: 10.1016/j.cub.2003.12.005. 10.1016/j.cub.2003.12.005 [DOI] [PubMed] [Google Scholar]
- Brocks J.J, Logan G.A, Buick R, Summons R.E. Archean molecular fossils and the early rise of eukaryotes. Science. 1999;285:1033–1036. doi: 10.1126/science.285.5430.1033. 10.1126/science.285.5430.1033 [DOI] [PubMed] [Google Scholar]
- Brocks J.J, Love G.D, Summons R.E, Knoll A.H, Logan G.A, Bowden S.A. Biomarker evidence for green and purple sulphur bacteria in a stratified Palaeoproterozoic sea. Nature. 2005;437:866–870. doi: 10.1038/nature04068. 10.1038/nature04068 [DOI] [PubMed] [Google Scholar]
- Butterfield N.J. Bangiomorpha pubescens n. gen., n. sp.: implications for the evolution of sex, multicellularity, and the Mesoproterozoic/Neoproterozoic radiation of eukaryotes. Paleobiology. 2000;26:386–404. [Google Scholar]
- Butterfield N.J. A vaucheriacean alga from the middle Neoproterozoic of Spitzbergen: implications for the evolution of Proterozoic eukaryotes and the Cambrian explosion. Paleobiology. 2004;30:231–252. [Google Scholar]
- Butterfield N.J. Probable Proterozoic fungi. Paleobiology. 2005;31:165–182. [Google Scholar]
- Butterfield N.J, Knoll A.H, Swett K. A bangiophyte red alga from the Proterozoic of arctic Canada. Science. 1990;250:104–107. doi: 10.1126/science.11538072. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. The origin and early evolution of the eukaryotic cell. In: Carlile M.J, Collins J.F, Moseley B.E.B, editors. Molecular and cellular aspects of microbial evolution. Cambridge University Press; Cambridge, UK: 1981. pp. 33–84. [Google Scholar]
- Cavalier-Smith T. The origins of plastids. Biol. J. Linn. Soc. 1982;17:289–306. [Google Scholar]
- Cavalier-Smith T. The origin of cells: a symbiosis between genes, catalysts, and membranes. Cold Spring Harb. Symp. Quant. Biol. 1987a;52:805–824. doi: 10.1101/sqb.1987.052.01.089. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. The origin of eukaryotic and archaebacterial cells. Ann. N. Y. Acad. Sci. 1987b;503:17–54. doi: 10.1111/j.1749-6632.1987.tb40596.x. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. The evolution of cells. In: Osawa S, Honjo T, editors. Evolution of life. Springer; Tokyo: 1991a. pp. 271–304. [Google Scholar]
- Cavalier-Smith T. Intron phylogeny: a new hypothesis. Trends Genet. 1991b;7:145–148. [PubMed] [Google Scholar]
- Cavalier-Smith T. Origins of secondary metabolism. Ciba Found. Symp. 1992;171:64–80. doi: 10.1002/9780470514344.ch5. discussion 80-7. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Evolution of the eukaryotic genome. In: Broda P, Oliver S.G, Sims P, editors. The eukaryotic genome. Cambridge University Press; Cambridge, UK: 1993. pp. 333–385. [Google Scholar]
- Cavalier-Smith T. Principles of protein and lipid targeting in secondary symbiogenesis: euglenoid, dinoflagellate, and sporozoan plastid origins and the eukaryotic family tree. J. Eukaryot. Microbiol. 1999;46:347–366. doi: 10.1111/j.1550-7408.1999.tb04614.x. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Membrane heredity and early chloroplast evolution. Trends Plant Sci. 2000;5:174–182. doi: 10.1016/s1360-1385(00)01598-3. 10.1016/S1360-1385(00)01598-3 [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Obcells as proto-organisms: membrane heredity, lithophosphorylation, and the origins of the genetic code, the first cells, and photosynthesis. J. Mol. Evol. 2001;53:555–595. doi: 10.1007/s002390010245. 10.1007/s002390010245 [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. The neomuran origin of archaebacteria, the negibacterial root of the universal tree and bacterial megaclassification. Int. J. Syst. Evol. Microbiol. 2002a;52:7–76. doi: 10.1099/00207713-52-1-7. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int. J. Syst. Evol. Microbiol. 2002b;52:297–354. doi: 10.1099/00207713-52-2-297. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Origins of the machinery of recombination and sex. Heredity. 2002c;88:125–141. doi: 10.1038/sj.hdy.6800034. 10.1038/sj.hdy.6800034 [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. The excavate protozoan phyla Metamonada Grassé emend. (Anaeromonadea, Parabasalia, Carpediemonas, Eopharyngia) and Loukozoa emend. (Jakobea, Malawimonas): their evolutionary affinities and new higher taxa. Int. J. Syst. Evol. Microbiol. 2003a;53:1741–1758. doi: 10.1099/ijs.0.02548-0. 10.1099/ijs.0.02548-0 [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Protist phylogeny and the high-level classification of Protozoa. Eur. J. Protistol. 2003b;39:338–348. 10.1078/0932-4739-00002 [Google Scholar]
- Cavalier-Smith T. The membranome and membrane heredity in development and evolution. In: Hirt R.P, Horner D.S, editors. Organelles, genomes and eukaryote phylogeny. Taylor & Francis; London: 2004a. pp. 335–351. [Google Scholar]
- Cavalier-Smith T. Only six kingdoms of life. Proc. R. Soc. B. 2004b;271:1251–1262. doi: 10.1098/rspb.2004.2705. 10.1098/rspb.2004.2705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. Economy, speed and size matter: evolutionary forces driving nuclear genome miniaturization and expansion. Ann. Bot. 2005;95:147–175. doi: 10.1093/aob/mci010. 10.1093/aob/mci010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith, T. 2006a Rooting the tree of life by transition analysis. Biol. Direct [DOI] [PMC free article] [PubMed]
- Cavalier-Smith T. 2006b Origin of mitochondria by intracellular enslavement of a photosynthetic purple bacterium. Proc. R. Soc. B (10.1098/rspb.2006.3531). (Published online 11 April 2006.) [DOI] [PMC free article] [PubMed]
- Cavalier-Smith, T. In press. The chimaeric origin of mitochondria: photosynthetic cell enslavement, gene-transfer pressure, and compartmentation efficiency. In Origins of mitochondria and hydrogenosomes (ed. W. Martin). Berlin: Springer.
- Cavalier-Smith T, Chao E.E. Phylogeny and classification of phylum Cercozoa (Protozoa) Protist. 2003a;154:341–358. doi: 10.1078/143446103322454112. 10.1078/143446103322454112 [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T, Chao E.E. Phylogeny of Choanozoa, Apusozoa, and other Protozoa and early eukaryote megaevolution. J. Mol. Evol. 2003b;56:540–563. doi: 10.1007/s00239-002-2424-z. 10.1007/s00239-002-2424-z [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T, Chao E.E. Phylogeny and megasystematics of phagotrophic heterokonts (kingdom Chromista) J. Mol. Evol. 2006;62:388–420. doi: 10.1007/s00239-004-0353-8. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T, Allsopp M.T.E.P, Chao E.E, Boury-Esnault N, Vacelet J. Sponge phylogeny, animal monophyly and the origin of the nervous system: 18S rRNA evidence. Can. J. Zool. 1996;74:2031–2045. [Google Scholar]
- Chistoserdova L, Vorholt J.A, Thauer R.K, Lidstrom M.E. C1 transfer enzymes and coenzymes linking methylotrophic bacteria and methanogenic Archaea. Science. 1998;281:99–102. doi: 10.1126/science.281.5373.99. 10.1126/science.281.5373.99 [DOI] [PubMed] [Google Scholar]
- Chistoserdova L, Jenkins C, Kalyuzhnaya M.G, Marx C.J, Lapidus A, Vorholt J.A, Staley J.T, Lidstrom M.E. The enigmatic planctomycetes may hold a key to the origins of methanogenesis and methylotrophy. Mol. Biol. Evol. 2004;21:1234–1241. doi: 10.1093/molbev/msh113. 10.1093/molbev/msh113 [DOI] [PubMed] [Google Scholar]
- Chistoserdova L, Vorholt J.A, Lidstrom M.E. A genomic view of methane oxidation by aerobic bacteria and anaerobic archaea. Genome Biol. 2005;6:208. doi: 10.1186/gb-2005-6-2-208. 10.1186/gb-2005-6-2-208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke G.D, Beiko R.B, Ragan M.A, Charlebois R.L. Inferring genome trees by using a filter to eliminate phylogenetically discordant sequences and a distant matrix based on mean normalised BLASTP scores. J. Bacteriol. 2002;184:2072–2080. doi: 10.1128/JB.184.8.2072-2080.2002. 10.1128/JB.184.8.2072-2080.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockell C.S. Ultraviolet radiation and the photobiology of earth's early oceans. Orig. Life Evol. Biosph. 2000;30:467–499. doi: 10.1023/a:1006765405786. 10.1023/A:1006765405786 [DOI] [PubMed] [Google Scholar]
- Cockell C.S, Cordoba-Jabonero C. Coupling of climate change and biotic UV exposure through changing snow-ice covers in terrestrial habitats. Photochem. Photobiol. 2004;79:26–31. 10.1562/0031-8655(2004)79%3C26:COCCAB%3E2.0.CO;2 [PubMed] [Google Scholar]
- Cockell C.S, Horneck G. The history of the UV radiation climate of the earth—theoretical and space-based observations. Photochem. Photobiol. 2001;73:447–451. doi: 10.1562/0031-8655(2001)073<0447:thotur>2.0.co;2. 10.1562/0031-8655(2001)073%3C0447:THOTUR%3E2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Cockell C.S, Rettberg P, Horneck G, Wynn-Williams D.D, Scherer K, Gugg-Helminger A. Influence of ice and snow covers on the UV exposure of terrestrial microbial communities: dosimetric studies. J. Photochem. Photobiol. B. 2002;68:23–32. doi: 10.1016/s1011-1344(02)00327-5. 10.1016/S1011-1344(02)00327-5 [DOI] [PubMed] [Google Scholar]
- Comfort D, Clubb R.T. A comparative genome analysis identifies distinct sorting pathways in gram-positive bacteria. Infect. Immun. 2004;72:2710–2722. doi: 10.1128/IAI.72.5.2710-2722.2004. 10.1128/IAI.72.5.2710-2722.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn V.M, Franco C.M. Analysis of the endophytic actinobacterial population in the roots of wheat (Triticum aestivum L.) by terminal restriction fragment length polymorphism and sequencing of 16S rRNA clones. Appl. Environ. Microbiol. 2004;70:1787–1794. doi: 10.1128/AEM.70.3.1787-1794.2004. 10.1128/AEM.70.3.1787-1794.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway Morris S. Darwin's dilemma: the realities of the Cambrian ‘explosion’. Phil. Trans. R. Soc. B. 2006;361:1069–1083. doi: 10.1098/rstb.2006.1846. 10.1098/rstb.2006.1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell W.C. Late Cretaceous chrysomonad cysts. Palaeogeogr. Palaeoclimatol. Palaeoecol. 1972;12:33–47. 10.1016/0031-0182(72)90005-3 [Google Scholar]
- Cubonova L, Sandman K, Hallam S.J, Delong E.F, Reeve J.N. Histones in crenarchaea. J. Bacteriol. 2005;187:5482–5485. doi: 10.1128/JB.187.15.5482-5485.2005. 10.1128/JB.187.15.5482-5485.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos D, Dokudovskaya S, Alber F, Williams R, Chait B.T, Sali A, Rout M.P. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol. 2004;2:e380. doi: 10.1371/journal.pbio.0020380. 10.1371/journal.pbio.0020380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douzery E.J, Snell E.A, Bapteste E, Delsuc F, Philippe H. The timing of eukaryotic evolution: does a relaxed molecular clock reconcile proteins and fossils? Proc. Natl Acad. Sci. USA. 2004;101:15 386–15 391. doi: 10.1073/pnas.0403984101. 10.1073/pnas.0403984101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Embley T.M. Multiple secondary origins of the anaerobic lifestyle in eukaryotes. Phil. Trans. R. Soc. B. 2006;361:1055–1067. doi: 10.1098/rstb.2006.1844. 10.1098/rstb.2006.1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedo C.M, Whitehouse M.J, Kamber B.S. Geological constraints on detecting the earliest life on Earth: a perspective from the Early Archean (older than 3.7 Gyr) of southwest Greenland. Phil. Trans. R. Soc. B. 2006;361:851–867. doi: 10.1098/rstb.2006.1836. 10.1098/rstb.2006.1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fensome, R. A., Taylor, F. J. R., Norris, G., Sargeant, W. A. S., Wharton, D. I. & Williams, G. L. A. 1993 A classification of living and fossil dinoflagellates. Micropalaeontology, Special Publication no. 7
- Foissner W, Schiller W. Stable for 15 million years: scanning electron microscope investigation of Miocene euglyphid thecamoebians from Germany, with description of the new genus Scutieuglypha. Protistology. 2001;37:167–180. 10.1078/0932-4739-00012 [Google Scholar]
- Foustoukos D.I, Seyfried W.E., Jr Hydrocarbons in hydrothermal vent fluids: the role of chromium-bearing catalysts. Science. 2004;304:1002–1005. doi: 10.1126/science.1096033. 10.1126/science.1096033 [DOI] [PubMed] [Google Scholar]
- Fuerst J.A. Intracellular compartmentation in planctomycetes. Annu. Rev. Microbiol. 2005;59:299–328. doi: 10.1146/annurev.micro.59.030804.121258. 10.1146/annurev.micro.59.030804.121258 [DOI] [PubMed] [Google Scholar]
- Fuerst J.A, Webb R.I. Membrane-bounded nucleoid in the eubacterium Gemmata obscuriglobus. Proc. Natl Acad. Sci. USA. 1991;88:8184–8188. doi: 10.1073/pnas.88.18.8184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelwicks J.T, Risatti J.B, Hayes J.M. Carbon isotope effects associated with autotrophic acetogenesis. Org. Geochem. 1989;14:441–446. doi: 10.1016/0146-6380(89)90009-0. 10.1016/0146-6380(89)90009-0 [DOI] [PubMed] [Google Scholar]
- Gentle I, Gabriel K, Beech P, Waller R, Lithgow T. The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J. Cell Biol. 2005;164:19–25. doi: 10.1083/jcb.200310092. 10.1083/jcb.200310092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille C, Goede A, Schloetelburg C, Presissner R, Kloetzel P.M, Gobel U.B, Frommel C. A comprehensive view on proteasomal sequences: implications for the evolution of the proteasome. J. Mol. Biol. 2003;326:1437–1448. doi: 10.1016/s0022-2836(02)01470-5. 10.1016/S0022-2836(02)01470-5 [DOI] [PubMed] [Google Scholar]
- Gitai Z, Dye N, Shapiro L. An actin-like gene can determine cell polarity in bacteria. Proc. Natl Acad. Sci. USA. 2004;101:8643–8648. doi: 10.1073/pnas.0402638101. 10.1073/pnas.0402638101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitai Z, Dye N.A, Reisenauer A, Wachi M, Shapiro L. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell. 2005a;120:329–341. doi: 10.1016/j.cell.2005.01.007. 10.1016/j.cell.2005.01.007 [DOI] [PubMed] [Google Scholar]
- Gitai Z, Thanbichler M, Shapiro L. The choreographed dynamics of bacterial chromosomes. Trends Microbiol. 2005b;13:221–228. doi: 10.1016/j.tim.2005.03.006. 10.1016/j.tim.2005.03.006 [DOI] [PubMed] [Google Scholar]
- Gold W.G. The influence of cryptogamic crusts on the thermal environment and temperature relations of plants in a high arctic polar desert, Devon Island, NWT, Canada. Arctic Antarct. Alpine Res. 1998;30:108–120. [Google Scholar]
- Goldschmidt R. Yale University Press; New Haven, CT: 1940. The material basis of evolution. [Google Scholar]
- Gould S.J. Tempo and mode in the macroevolutionary reconstruction of Darwinism. Proc. Natl Acad. Sci. USA. 1994;91:6764–6771. doi: 10.1073/pnas.91.15.6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graur D, Martin W. Reading the entrails of chickens: molecular timescales of evolution and the illusion of precision. Trends Genet. 2004;20:80–86. doi: 10.1016/j.tig.2003.12.003. 10.1016/j.tig.2003.12.003 [DOI] [PubMed] [Google Scholar]
- Grey K, Walter M.R, Calver C.R. Neoproterozoic biotic diversification: snowball earth or an aftermath of the Acraman impact? Geology. 2003;31:459–462. 10.1130/0091-7613(2003)031%3C0459:NBDSEO%3E2.0.CO;2 [Google Scholar]
- Gribaldo S, Brochier-Armanet C. The origin and evolution of Archaea: a state of the art. Phil. Trans. R. Soc. B. 2006;361:1007–1022. doi: 10.1098/rstb.2006.1841. 10.1098/rstb.2006.1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribaldo S, Philippe H. Ancient phylogenetic relationships. Theor. Popul. Biol. 2002;61:391–408. doi: 10.1006/tpbi.2002.1593. 10.1006/tpbi.2002.1593 [DOI] [PubMed] [Google Scholar]
- Gupta R.S, Pereira M, Chandrasekera C, Johari V. Molecular signatures in protein sequences that are characteristic of cyanobacteria and plastid homologues. Int. J. Syst. Evol. Microbiol. 2003;53:1833–1842. doi: 10.1099/ijs.0.02720-0. 10.1099/ijs.0.02720-0 [DOI] [PubMed] [Google Scholar]
- Guy R.D, Fogel M.L, Berry J.A. Photosynthetic fractionation of the stable isotopes of oxygen and carbon. Plant Physiol. 1993;101:37–47. doi: 10.1104/pp.101.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han T.M, Runnegar B. Megascopic eukaryotic algae from the 2.1-billion-year-old Negaunee iron-formation, Michigan. Science. 1992;257:232–235. doi: 10.1126/science.1631544. [DOI] [PubMed] [Google Scholar]
- Hayes J.M. Geochemical evidence bearing on the origin of aerobiosis, a speculative hypothesis. In: Schopf J.M, editor. Earths' earliest biosphere: its origin and evolution. ch. 12. Princeton University Press; Princeton, NJ: 1983. pp. 291–301. [Google Scholar]
- Hayes J.M. Global methanotrophy at the Archean–Proterozoic transition. In: Bengtson S, editor. Early life on Earth, Nobel Symposium 84. Columbia University Press; New York, NY: 1994. pp. 220–236. [Google Scholar]
- Hayes J.M, Waldbauer J.R. The carbon cycle and associated redox processes through time. Phil. Trans. R. Soc. B. 2006;361:931–950. doi: 10.1098/rstb.2006.1840. 10.1098/rstb.2006.1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henckel T, Jackel U, Schnell S, Conrad R. Molecular analyses of novel methanotrophic communities in forest soil that oxidize atmospheric methane. Appl. Environ. Microbiol. 2000;66:1801–1808. doi: 10.1128/aem.66.5.1801-1808.2000. 10.1128/AEM.66.5.1801-1808.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman P.F, Schrag D.P. The snowball earth hypothesis: testing the limits of global change. Terra Nova. 2002;14:129–155. 10.1046/j.1365-3121.2002.00408.x [Google Scholar]
- Holland H.D. The oxygenation of the atmosphere and oceans. Phil. Trans. R. Soc. B. 2006;361:903–915. doi: 10.1098/rstb.2006.1838. 10.1098/rstb.2006.1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horita J, Berndt M.E. Abiogenic methane formation and isotopic fractionation under hydrothermal conditions. Science. 1999;285:1055–1057. doi: 10.1126/science.285.5430.1055. 10.1126/science.285.5430.1055 [DOI] [PubMed] [Google Scholar]
- Hyman L.H. McGraw-Hill; New York, NY: 1940. The invertebrates. [Google Scholar]
- Ingouff M, FitzGerald J.N, Guerin C, Robert H, Sorensen M.B, Van Damme D, Geelen D, Blanchoin L, Berger F. Plant formin AtFH5 is an evolutionarily conserved actin nucleator involved in cytokinesis. Nat. Cell Biol. 2005;7:374–380. doi: 10.1038/ncb1238. 10.1038/ncb1238 [DOI] [PubMed] [Google Scholar]
- Ivanovsky R.N, Fal Y.I, Berg I.A, Ugolkova N.V, Krasilnikova E.N, Keppen O.I, Zakharchuc L.M, Zyakun A.M. Evidence for the presence of the reductive pentose phosphate cycle in a filamentous anoxygenic photosynthetic bacterium, Oscillochloris trichoides strain DG-6. Microbiology. 1999;145:1743–1748. doi: 10.1099/13500872-145-7-1743. [DOI] [PubMed] [Google Scholar]
- Javaux E.J, Knoll A.H, Walter M.R. Morphological and ecological complexity in early eukaryotic ecosystems. Nature. 2001;412:66–69. doi: 10.1038/35083562. 10.1038/35083562 [DOI] [PubMed] [Google Scholar]
- Javaux E.J, Knoll A.H, Walter M. Recognizing and interpreting the fossils of early eukaryotes. Orig. Life Evol. Biosph. 2003;33:75–94. doi: 10.1023/a:1023992712071. 10.1023/A:1023992712071 [DOI] [PubMed] [Google Scholar]
- Jekély G, Arendt D. Evolution of intraflagellar transport from coated vesicles and autogenous origin of the eukaryotic cilium. BioEssays. 2006;28:191–198. doi: 10.1002/bies.20369. 10.1002/bies.20369 [DOI] [PubMed] [Google Scholar]
- Johansen J.R. Cryptogamic crusts of semiarid and arid lands of North America. J. Phycol. 1993;29:140–147. 10.1111/j.0022-3646.1993.00140.x [Google Scholar]
- Jung K.H, Trivedi V.D, Spudich J.L. Demonstration of a sensory rhodopsin in eubacteria. Mol. Microbiol. 2003;47:1513–1522. doi: 10.1046/j.1365-2958.2003.03395.x. 10.1046/j.1365-2958.2003.03395.x [DOI] [PubMed] [Google Scholar]
- Karpov S.A, Bass D, Mylnikov A.P, Cavalier-Smith T. Molecular phylogeny of Cercomonadidae and kinetid patterns of Cercomonas and Eocercomonas gen. nov. (Cercomonadida, Cercozoa) Protist. 2006;157 doi: 10.1016/j.protis.2006.01.001. (Published online 27 April 2006.) [DOI] [PubMed] [Google Scholar]
- Kasting J.F. Theoretical constraints on oxygen and carbon dioxide concentrations in the Precambrian atmosphere. Precambrian Res. 1987;34:205–229. doi: 10.1016/0301-9268(87)90001-5. 10.1016/0301-9268(87)90001-5 [DOI] [PubMed] [Google Scholar]
- Kasting J.F. Palaeoclimatology: Archaean atmosphere and climate. Nature. 2004;432 doi: 10.1038/nature03166. 10.1038/nature03166 1 p following 460. [DOI] [PubMed] [Google Scholar]
- Kasting J.F, Ono S. Palaeoclimates: the first two billion years. Phil. Trans. R Soc. B. 2006;361:917–929. doi: 10.1098/rstb.2006.1839. 10.1098/rstb.2006.1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasting J.F, Pollack J.B. Effects of high CO2 levels on surface temperature and atmospheric oxidation state of the early Earth. J. Atmos. Chem. 1984;1:403–428. doi: 10.1007/BF00053803. 10.1007/BF00053803 [DOI] [PubMed] [Google Scholar]
- Kasting J.F, Siefert J.L. Life and the evolution of Earth's atmosphere. Science. 2002;296:1066–1068. doi: 10.1126/science.1071184. 10.1126/science.1071184 [DOI] [PubMed] [Google Scholar]
- Knoll A.H. Proterozoic and early Cambrian protists: evidence for accelerating evolutionary tempo. Proc. Natl Acad. Sci. USA. 1994;91:6743–6750. doi: 10.1073/pnas.91.15.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll A.H. Princeton University Press; Princeton, NJ: 2004. Life on a young planet: the first three billion years of evolution on earth. [Google Scholar]
- Knoll A.H, Javaux E.J, Hewitt D, Cohen P. Eukaryotic organisms in Proterozoic oceans. Phil. Trans. R. Soc. B. 2006;361:1023–1038. doi: 10.1098/rstb.2006.1843. 10.1098/rstb.2006.1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga Y, Kyuragi T, Nishihara M, Sone N. Did archaeal and bacterial cells arise independently from noncellular precursors? A hypothesis stating that the advent of membrane phospholipid with enantiomeric glycerophosphate backbones caused the separation of the two lines of descent. J. Mol. Evol. 1998;46:54–64. doi: 10.1007/pl00006283. 10.1007/PL00006283 [DOI] [PubMed] [Google Scholar]
- Kopp R.E, Kirschvink J.L, Hilburn I.A, Nash C.Z. The Paleoproterozoic snowball Earth: a climate disaster triggered by the evolution of oxygenic photosynthesis. Proc. Natl Acad. Sci. USA. 2005;102:11 131–11 136. doi: 10.1073/pnas.0504878102. 10.1073/pnas.0504878102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunisawa T. Dichotomy of major bacterial phyla inferred from gene arrangement comparisons. J. Theor. Biol. 2006;239:367–375. doi: 10.1016/j.jtbi.2005.08.004. 10.1016/j.jtbi.2005.08.004 [DOI] [PubMed] [Google Scholar]
- Lerat E, Daubin V, Ochman H, Moran N.A. Evolutionary origins of genomic repertoires in bacteria. PLoS Biol. 2005;3:e130. doi: 10.1371/journal.pbio.0030130. 10.1371/journal.pbio.0030130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay M.R, Webb R.I, Strous M, Jetten M.S, Butler M.K, Forde R.J, Fuerst J.A. Cell compartmentalisation in planctomycetes: novel types of structural organisation for the bacterial cell. Arch. Microbiol. 2001;175:413–429. doi: 10.1007/s002030100280. 10.1007/s002030100280 [DOI] [PubMed] [Google Scholar]
- Logan G.A, Calver C.R, Gorjan P, Summons R.E, Hayes J.M, Walter M.R. Terminal Proterozoic mid-shelf benthic microbial mats in the Centralian Superbasin and their environmental significance. Geochim. Cosmochim. Acta. 1999;63:1345–1358. doi: 10.1016/s0016-7037(99)00033-2. 10.1016/S0016-7037(99)00033-2 [DOI] [PubMed] [Google Scholar]
- Lopez P, Casane D, Philippe H. Heterotachy, an important process of protein evolution. Mol. Biol. Evol. 2002;19:1–7. doi: 10.1093/oxfordjournals.molbev.a003973. [DOI] [PubMed] [Google Scholar]
- Lovelock J. Oxford University Press; Oxford, UK: 1988. The ages of Gaia. [Google Scholar]
- Martin W, Koonin E.V. Introns and the origin of nucleus-cytosol compartmentalization. Nature. 2006;440:41–45. doi: 10.1038/nature04531. 10.1038/nature04531 [DOI] [PubMed] [Google Scholar]
- Martin W, Müller M. The hydrogen hypothesis for the first eukaryote. Nature. 1998;392:37–44. doi: 10.1038/32096. 10.1038/32096 [DOI] [PubMed] [Google Scholar]
- Martin W, Russell M.J. On the origins of cells: a hypothesis for the evolutionary transitions from abiotic geochemistry to chemoautotrophic prokaryotes, and from prokaryotes to nucleated cells. Phil. Trans. R. Soc. B. 2003;358:59–85. doi: 10.1098/rstb.2002.1183. 10.1098/rstb.2002.1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney K. Silicoflagellates. In: Lipps J, editor. Fossil prokaryotes and protists. Blackwell Scientific Publications; Oxford, UK: 1993. pp. 143–154. [Google Scholar]
- McIlroy D, Green O.R, Brasier M.D. Palaeobiology and evolution of the earliest agglutinated Foraminifera: Platysolenites, Spirosolenites and related forms. Lethaia. 2001;34:13–29. 10.1080/002411601300068170 [Google Scholar]
- McKay C.P. Thickness of tropical ice and photosynthesis on a snowball Earth. Geophys. Res. Lett. 2000;27:2153–2156. doi: 10.1029/2000gl008525. 10.1029/2000GL008525 [DOI] [PubMed] [Google Scholar]
- Molowan J.M, Jacobsen S.R, Dahl J, Al-Hajji A, Huizinga B.J, Fago F.J. Molecular fossils demonstrate Precambrian origin of dinoflagellates. In: Zhuralev A, Riding R, editors. Ecology of the Cambrian radiation. Columbia University Press; New York, NY: 2001. pp. 474–493. [Google Scholar]
- Morris R.M, Rappe M.S, Urbach E, Connon S.A, Giovannoni S.J. Prevalence of the Chloroflexi-related SAR202 bacterioplankton cluster throughout the mesopelagic zone and deep ocean. Appl. Environ. Microbiol. 2004;70:2836–2842. doi: 10.1128/AEM.70.5.2836-2842.2004. 10.1128/AEM.70.5.2836-2842.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundil R, Ludwig K.R, Metcalfe I, Renne P.R. Age and timing of the Permian mass extinctions: U/Pb dating of closed-system zircons. Science. 2004;305:1760–1763. doi: 10.1126/science.1101012. 10.1126/science.1101012 [DOI] [PubMed] [Google Scholar]
- Nagy M.L, Perez A, Garcia-Pichel F. The prokaryotic diversity of biological soil crusts in the Sonoran Desert (Organ Pipe Cactus National Monument, AZ) FEMS Microbiol. Ecol. 2005;54:233–245. doi: 10.1016/j.femsec.2005.03.011. 10.1016/j.femsec.2005.03.011 [DOI] [PubMed] [Google Scholar]
- Navarro-Gonzalez R, McKay C.P, Mvondo D.N. A possible nitrogen crisis for Archaean life due to reduced nitrogen fixation by lightning. Nature. 2001;412:61–64. doi: 10.1038/35083537. 10.1038/35083537 [DOI] [PubMed] [Google Scholar]
- Nesbo C.L, Doolittle W.F. Targeting clusters of transferred genes in Thermotoga maritima. Environ. Microbiol. 2003;5:1144–1154. doi: 10.1046/j.1462-2920.2003.00515.x. 10.1046/j.1462-2920.2003.00515.x [DOI] [PubMed] [Google Scholar]
- Nesbo C.L, L'Haridon S, Stetter K.O, Doolittle W.F. Phylogenetic analyses of two “archaeal” genes in Thermotoga maritima reveal multiple transfers between archaea and bacteria. Mol. Biol. Evol. 2001;18:362–375. doi: 10.1093/oxfordjournals.molbev.a003812. [DOI] [PubMed] [Google Scholar]
- Nikolaev S.I, Berney C, Fahrni J.F, Bolivar I, Polet S, Mylnikov A.P, Aleshin V.V, Petrov N.B, Pawlowski J. The twilight of Heliozoa and rise of Rhizaria, an emerging supergroup of amoeboid eukaryotes. Proc. Natl Acad. Sci. USA. 2004;101:8066–8071. doi: 10.1073/pnas.0308602101. 10.1073/pnas.0308602101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaev S.I, Mitchell E.A, Petrov N.B, Berney C, Fahrni J, Pawlowski J. The testate lobose amoebae (order Arcellinida Kent, 1880) finally find their home within Amoebozoa. Protist. 2005;156:191–202. doi: 10.1016/j.protis.2005.03.002. 10.1016/j.protis.2005.03.002 [DOI] [PubMed] [Google Scholar]
- Ohmoto H. The Archaean atmosphere, hydrosphere and biosphere. In: Eriksson P.G, Altermann W, Nelson D.R, Mueller W.U, Catuneanu O, editors. The Precambrian earth: tempos and events. Elsevier; Amsterdam: 2004. pp. 361–388. [Google Scholar]
- Ohmoto H, Watanabe Y, Kumazawa K. Evidence from massive siderite beds for a CO2-rich atmosphere before ∼1.8 billion years ago. Nature. 2004;429:395–399. doi: 10.1038/nature02573. 10.1038/nature02573 [DOI] [PubMed] [Google Scholar]
- Olcott A.N, Sessions A.L, Corsetti F.A, Kaufman A.J, de Oliviera T.F. Biomarker evidence for photosynthesis during Neoproterozoic glaciation. Science. 2005;310:471–474. doi: 10.1126/science.1115769. 10.1126/science.1115769 [DOI] [PubMed] [Google Scholar]
- Orphan V.J, House C.H, Hinrichs K.U, McKeegan K.D, DeLong E.F. Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments. Proc. Natl Acad. Sci. USA. 2002;99:7663–7668. doi: 10.1073/pnas.072210299. 10.1073/pnas.072210299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padovan A.C, Sanson G.F, Brunstein A, Briones M.R. Fungi evolution revisited: application of the penalized likelihood method to a Bayesian fungal phylogeny provides a new perspective on phylogenetic relationships and divergence dates of Ascomycota groups. J. Mol. Evol. 2005;60:726–735. doi: 10.1007/s00239-004-0164-y. 10.1007/s00239-004-0164-y [DOI] [PubMed] [Google Scholar]
- Patron N.C, Rogers M.B, Keeling P.J. Gene replacement of fructose-1,6-bisphosphate aldolase supports the hypothesis of a single photosynthetic ancestor of chromalveolates. Eukaryot. Cell. 2004;3:1169–1175. doi: 10.1128/EC.3.5.1169-1175.2004. 10.1128/EC.3.5.1169-1175.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov A.A, Kasting J.F. Mass-independent fractionation of sulfur isotopes in Archean sediments: strong evidence for an anoxic Archean atmosphere. Astrobiology. 2002;2:27–41. doi: 10.1089/153110702753621321. 10.1089/153110702753621321 [DOI] [PubMed] [Google Scholar]
- Pavlov A.A, Kasting J.F, Brown L.L, Rages K.A, Freedman R. Greenhouse warming by CH4 in the atmosphere of early Earth. J. Geophys. Res. 2000;105:11 981–11 990. doi: 10.1029/1999je001134. 10.1029/1999JE001134 [DOI] [PubMed] [Google Scholar]
- Pearson A, Budin M, Brocks J.J. Phylogenetic and biochemical evidence for sterol synthesis in the bacterium Gemmata obscuriglobus. Proc. Natl Acad. Sci. USA. 2003;100:15 352–15 357. doi: 10.1073/pnas.2536559100. 10.1073/pnas.2536559100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peretó J, López-García P, Moreira D. Ancestral lipid biosynthesis and early membrane evolution. Trends Biochem. Sci. 2004;29:469–477. doi: 10.1016/j.tibs.2004.07.002. 10.1016/j.tibs.2004.07.002 [DOI] [PubMed] [Google Scholar]
- Peterson K.J, Butterfield N.J. Origin of the Eumetazoa: testing ecological predictions of molecular clocks against the Proterozoic fossil record. Proc. Natl Acad. Sci. USA. 2005;102:9547–9552. doi: 10.1073/pnas.0503660102. 10.1073/pnas.0503660102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H, Douady C.J. Horizontal gene transfer and phylogenetics. Curr. Opin. Microbiol. 2003;6:498–505. doi: 10.1016/j.mib.2003.09.008. 10.1016/j.mib.2003.09.008 [DOI] [PubMed] [Google Scholar]
- Porter S.M, Knoll A.H. Testate amoebae of the Chuar Group, Grand Canyon. Paleobiology. 2000;27:345–370. [Google Scholar]
- Porter S.M, Meisterfeld R, Knoll A.H. Vase-shaped microfossils from the Neoproterozoic Chuar Group, Grand Canyon: a classification guided by modern testate amoebae. J. Paleontol. 2003;77:409–429. [Google Scholar]
- Raymond J, Siefert J.L, Staples C.R, Blankenship R.E. The natural history of nitrogen fixation. Mol. Biol. Evol. 2004;21:541–554. doi: 10.1093/molbev/msh047. 10.1093/molbev/msh047 [DOI] [PubMed] [Google Scholar]
- Richards T.A, Cavalier-Smith T. Myosin domain evolution and the primary divergence of eukaryotes. Nature. 2005;436:1113–1118. doi: 10.1038/nature03949. 10.1038/nature03949 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Ezpeleta N, Brinkmann H, Burey S.C, Roure B, Burger G, Loffelhardt W, Bohnert H.J, Philippe H, Lang B.F. Monophyly of primary photosynthetic eukaryotes: green plants, red algae, and glaucophytes. Curr. Biol. 2005;15:1325–1330. doi: 10.1016/j.cub.2005.06.040. 10.1016/j.cub.2005.06.040 [DOI] [PubMed] [Google Scholar]
- Roger A.J, Hug L.A. The origin and diversification of eukaryotes: problems with molecular phylogenetics and molecular clock estimation. Phil. Trans. R. Soc. B. 2006;361:1039–1054. doi: 10.1098/rstb.2006.1845. 10.1098/rstb.2006.1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman D.H, Hayes J.M, Summons R.E. Dynamics of the Neoproterozoic carbon cycle. Proc. Natl Acad. Sci. USA. 2003;100:8124–8129. doi: 10.1073/pnas.0832439100. 10.1073/pnas.0832439100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Gonzalez M.X, Marin I. New insights into the evolutionary history of type 1 rhodopsins. J. Mol. Evol. 2004;58:348–358. doi: 10.1007/s00239-003-2557-8. 10.1007/s00239-003-2557-8 [DOI] [PubMed] [Google Scholar]
- Schopf J.W. Princeton University Press; Princeton, NJ: 2001. Cradle of life: the discovery of earth's earliest fossils. [Google Scholar]
- Schopf J.W. Fossil evidence of Archaean life. Phil. Trans. R. Soc. B. 2006;361:869–885. doi: 10.1098/rstb.2006.1834. 10.1098/rstb.2006.1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopf J.W, Klein C, editors. The Proterozoic biosphere: a multidisciplinary study. Cambridge University Press; Cambridge, UK: 1992. [Google Scholar]
- Schrag D.P, Berner R.A, Hoffman P.F, Halverson G.P. On the initiation of a snowball Earth. Geochem. Geophys. Geosyst. 2002;3:1–21. 10.1029/2001GC000219 [Google Scholar]
- Scott H.P, Hemley R.J, Mao H.K, Herschbach D.R, Fried L.E, Howard W.M, Bastea S. Generation of methane in the Earth's mantle: in situ high pressure–temperature measurements of carbonate reduction. Proc. Natl Acad. Sci. USA. 2004a;101:14 023–14 026. doi: 10.1073/pnas.0405930101. 10.1073/pnas.0405930101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott K.M, Schwedock J, Schrag D.P, Cavanaugh C.M. Influence of form IA RubisCO and environmental dissolved inorganic carbon on the Δ13C of the clam-chemoautotroph symbiosis Solemya velum. Environ. Microbiol. 2004b;6:1210–1219. doi: 10.1111/j.1462-2920.2004.00642.x. 10.1111/j.1462-2920.2004.00642.x [DOI] [PubMed] [Google Scholar]
- Sekiguchi Y, Yamada T, Hanada S, Ohashi A, Harada H, Kamagata Y. Anaerolinea thermophila gen. nov., sp. nov. and Caldilinea aerophila gen. nov., sp. nov., novel filamentous thermophiles that represent a previously uncultured lineage of the domain Bacteria at the subphylum level. Int. J. Syst. Evol. Microbiol. 2003;53:1843–1851. doi: 10.1099/ijs.0.02699-0. 10.1099/ijs.0.02699-0 [DOI] [PubMed] [Google Scholar]
- Simpson G.C. Columbia University Press; New York, NY: 1944. Tempo and mode in evolution. [Google Scholar]
- Simpson G.C. Columbia University Press; New York, NY: 1953. The major features of evolution. [Google Scholar]
- Sorensen K.B, Canfield D.E, Teske A.P, Oren A. Community composition of a hypersaline endoevaporitic microbial mat. Appl. Environ. Microbiol. 2005;71:7352–7365. doi: 10.1128/AEM.71.11.7352-7365.2005. 10.1128/AEM.71.11.7352-7365.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stackebrandt E, Woese C.R. The evolution of prokaryotes. In: Carlile M.I, Collins I.F, Moseley B.E.B, editors. Molecular and cellular aspects of microbial evolution. Cambridge University Press; Cambridge, UK: 1981. pp. 1–32. [Google Scholar]
- Stanier R.Y. Some aspects of the biology of cells and their possible evolutionary significance. In: Charles H.P, Knight J.G, editors. Organization and control in prokaryotic and eukaryotic cells (Society for General Microbiology Symposium 20) Cambridge University Press; Cambridge, UK: 1970. pp. 1–38. [Google Scholar]
- Stechmann A, Cavalier-Smith T. Rooting the eukaryote tree by using a derived gene fusion. Science. 2002;297:89–91. doi: 10.1126/science.1071196. 10.1126/science.1071196 [DOI] [PubMed] [Google Scholar]
- Stechmann A, Cavalier-Smith T. The root of the eukaryote tree pinpointed. Curr. Biol. 2003;13:R665–R666. doi: 10.1016/s0960-9822(03)00602-x. 10.1016/S0960-9822(03)00602-X [DOI] [PubMed] [Google Scholar]
- Straus H, Des Marais D.J, Hayes J.M, Summons R.E. The carbon isotope record. In: Schopf J.M, Klein C, editors. The Proterozoic biosphere: a multidisciplinary study. Cambridge University Press; Cambridge, UK: 1992. pp. 117–134. [Google Scholar]
- Summons R.E, Powell T.G, Boreham C.J. Petroleum geology and geochemistry of the middle Proterozoic McArthur Basin, Northern Australia. III. Composition of extractable hydrocarbons. Geochim. Cosmochim. Acta. 1998;52:1747–1763. 10.1016/0016-7037(88)90001-4 [Google Scholar]
- Summons R.E, Jahnke L.L, Hope J.M, Logan G.A. 2-Methylhopanoids as biomarkers for cyanobacterial oxygenic photosynthesis. Nature. 1999;400:554–557. doi: 10.1038/23005. 10.1038/23005 [DOI] [PubMed] [Google Scholar]
- Summons R.E, Bradley A.S, Jahnke L.L, Waldbauer J.R. Steroids, triterpenoids and molecular oxygen. Phil. Trans. R. Soc. B. 2006;361:951–968. doi: 10.1098/rstb.2006.1837. 10.1098/rstb.2006.1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi O. Phaeodarian radiolaria from the Upper Cretaceous beds of the central Japan. Rev. Micropalaeontol. 2004;47:119–125. 10.1016/j.revmic.2004.06.001 [Google Scholar]
- Tappan H. Freeman; San Francisco, CA: 1980. The palaeobiology of plant protists. [Google Scholar]
- Tian X.L, Cao L.X, Tan H.M, Zeng Q.G, Jia Y.Y, Han W.Q, Zhou S.N. Study on the communities of endophytic fungi and endophytic actinomycetes from rice and their antipathogenic activities in vitro. World J. Microbiol. Biotechnol. 2004;20:303–309. 10.1023/B:WIBI.0000023843.83692.3f [Google Scholar]
- Torsvik T.H. Geology. The Rodinia jigsaw puzzle. Science. 2003;300:1379–1381. doi: 10.1126/science.1083469. 10.1126/science.1083469 [DOI] [PubMed] [Google Scholar]
- Venter J.C, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. 10.1126/science.1093857 [DOI] [PubMed] [Google Scholar]
- Vogeley L, Sineshchekov O.A, Trivedi V.D, Sasaki J, Spudich J.L, Luecke H. Anabaena sensory rhodopsin: a photochromic color sensor at 2.0 A. Science. 2004;306:1390–1393. doi: 10.1126/science.1103943. 10.1126/science.1103943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Heyden, S. 2004 Testing ubiquitous dispersal and freshwater/marine divergence in free-living protist groups. DPhil thesis, University of Oxford, Oxford.
- Wellman C.H, Osterloff P.L, Mohiuddin U. Fragments of the earliest land plants. Nature. 2003;425:282–285. doi: 10.1038/nature01884. 10.1038/nature01884 [DOI] [PubMed] [Google Scholar]
- Wink J, Gandhi J, Kroppenstedt R.M, Seibert G, Straubler B, Schumann P, Stackebrandt E. Amycolatopsis decaplanina sp. nov., a novel member of the genus with unusual morphology. Int. J. Syst. Evol. Microbiol. 2004;54:235–239. doi: 10.1099/ijs.0.02586-0. 10.1099/ijs.0.02586-0 [DOI] [PubMed] [Google Scholar]
- Woese C.R. A comment on methanogenic bacteria and the primitive ecology. J. Mol. Evol. 1977;9:369–371. doi: 10.1007/BF01796101. 10.1007/BF01796101 [DOI] [PubMed] [Google Scholar]
- Woese C.R. The universal ancestor. Proc. Natl Acad. Sci. USA. 1998;95:6854–6859. doi: 10.1073/pnas.95.12.6854. 10.1073/pnas.95.12.6854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C.R, Fox G.E. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl Acad. Sci. USA. 1977;74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C.R, Debrunner-Vossbrinck B.A, Oyaizu H, Stackebrandt E, Ludwig W. Gram-positive bacteria: possible photosynthetic ancestry. Science. 1985;229:762–765. doi: 10.1126/science.11539659. [DOI] [PubMed] [Google Scholar]
- Won M, Below R. Cambrian Radiolaria from the Georgina Basin, Queensland, Australia. Micropaleontology. 1999;45:325–363. [Google Scholar]
- Xiao S, Knoll A.H. Fossil preservation in the Neoproterozoic Doushantuo phosphorite Lagerstatte, South China. Lethaia. 1999;32:219–240. doi: 10.1111/j.1502-3931.1999.tb00541.x. [DOI] [PubMed] [Google Scholar]
- Xiao S, Knoll A.H, Yuan X, Pueschel C.M. Phosphatized multicellular algae in the Neoproterozoic Doushantuo formation, China, and the early evolution of florideophyte red algae. Am. J. Bot. 2004;91:214–227. doi: 10.3732/ajb.91.2.214. [DOI] [PubMed] [Google Scholar]
- Xiao S, Shen B, Zhou C, Xie G, Yuan X. A uniquely preserved Ediacaran fossil with direct evidence for a quilted bodyplan. Proc. Natl Acad. Sci. USA. 2005;102:10 227–10 232. doi: 10.1073/pnas.0502176102. 10.1073/pnas.0502176102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Doolittle R.F, Bourne P.E. Phylogeny determined by protein domain content. Proc. Natl Acad. Sci. USA. 2005;102:373–378. doi: 10.1073/pnas.0408810102. 10.1073/pnas.0408810102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon H.S, Hackett J.D, Pinto G, Bhattacharya D. The single, ancient origin of chromist plastids. Proc. Natl Acad. Sci. USA. 2002;99:15 507–15 512. doi: 10.1073/pnas.242379899. 10.1073/pnas.242379899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X, Xiao S, Taylor T.N. Lichen-like symbiosis 600 million years ago. Science. 2005;308:1017–1020. doi: 10.1126/science.1111347. 10.1126/science.1111347 [DOI] [PubMed] [Google Scholar]