Abstract
Familial Alzheimer’s disease (AD [MIM 104300]) has been a focus of intense investigation, primarily in Caucasian families from Europe and North America families. Although the late-onset form of familial AD, beginning after age 65 years, has been linked to regions on chromosomes 10q and 12p, the specific genetic variants have not yet been consistently identified. Using a unique cohort of families of Caribbean Hispanics ancestry, we screened the genome using 340 markers on 490 family members from 96 families with predominantly late-onset AD. We observed the strongest support for linkage on 18q (LOD=3.14). However, 17 additional markers (chromosomes 1-6, 8, 10, 12, and 14) exceeded a two-point LOD score of 1.0 under the affecteds-only autosomal dominant model or affected sibpair model. As we previously reported the fine-mapping effort on 12p showing modest evidence of linkage, we focused our fine-mapping efforts on two other candidate regions in the current report, namely 10q and 18q. We added 31 family members and eight additional Caribbean Hispanic families to fine map 10q and 18q. With additional microsatellite markers, the evidence for linkage for 18q strengthened near 112 cM, where the two-point LOD score for D18S541 was 3.37 and the highest NPL score in that region was 3.65 (P=0.000177). This narrow region contains a small number of genes expressed in the brain. However, at 10q (134-138 cM), the NPL score decreased from 3.15 (P=0.000486) to 2.1 (P=0.0218), but two broad peaks remained overlapping with previously reported peaks. Our results provide modest support for linkage on 10q and 12p in this cohort of Caribbean Hispanic families with familial Alzheimer’s disease, and strong evidence for a new locus on 18q.
Keywords: Alzheimer’s disease, genome scan, linkage analysis
The ε4 variant of APOE (APOE [MIM 107741]) is the only known gene associated with increased risk of late-onset Alzheimer’s disease (AD),1 but this variant explains only 20% of the attributable risk.2Genetic linkage studies suggest additional putative loci for late-onset AD.3-8 A locus on chromosome 12p11 (AD5 [MIM 602096]) that confers modest susceptibility to AD was found several years ago,5,9,10 and a locus on chromosome 10q24 (AD6 [MIM 605526]) segregating with either the diagnosis of AD6,8 or with elevated plasma amyloid β has also been identified.7 In families where AD has been confirmed by postmortem examination, Pericak-Vance and associates4 also reported a susceptibility locus on chromosome 9p22. All of these studies have been conducted in Caucasian families from North America or Europe.
The frequency of AD in Caribbean Hispanics was observed to be two- to three-fold higher compared with non-Hispanics living in a New York community,11 but the association between APOE-ε4 and sporadic AD in a population-based study including Caribbean Hispanics was weak.12,13 To investigate the genetics of AD in this population, we initiated a linkage study. Our initial results included a strong association between APOE-ε4 and late-onset familial AD,14 a founder mutation in presenilin 1 (PSEN1 [MIM 104311]) among families with early-onset AD,15 and modest evidence for linkage to chromosome 12p for late-onset AD in this population.16 In this report, we describe the results of fine mapping on chromosomes 10q and 18q based on the results of the first genome scan for familial AD using families of Caribbean Hispanic descent.
Materials and methods
Families
We initially studied 490 individuals (394 relatives and 96 probands) of 96 Caribbean Hispanic families. In the second-stage fine mapping, we added eight families for a total of 521 individuals (417 relatives and 104 probands) of 104 families. The sampling design and detailed characteristics of the participants have been described elsewhere.14 Briefly, multiple sources were used to recruit families. We recruited from clinics in the Dominican Republic and Puerto Rico, as well as the Alzheimer’s Disease Research Center Memory Disorders Clinic at Columbia University in the New York City. To augment family recruitment, we advertized in local newspapers and media in Dominican Republic, Puerto Rico, and New York. In addition, we recruited probands identified in an epidemiological study in northern Manhattan11 when the informant reported family members with dementia.
All patients and participating family members were examined and blood was obtained. Patients with AD met the National Institute of Neurological and Communicative Disorders and Stroke and the Alzheimer’s Disease and Related Disorders Association (NINCDS-ADRDA).17 The Clinical Dementia Rating Scale (CDR) was used to rate disease severity.18 Brain imaging and other laboratory studies were reviewed when available, and offered when medically required for diagnosis. The battery of neuropsychological tests used had been developed and evaluated extensively in Hispanics.19-21 Age at onset was based on the age at which the memory complaint was first reported, or if unavailable the age at first examination. Patients from four families with AD had autopsies confirming the clinical diagnoses of AD. In the current genome scan, we conducted linkage analysis using possible AD as affected. The affection status for family members with other forms of dementia or mild cognitive impairment was classified as unknown or ambiguous for the linkage analyses.
The Institutional Review Board of Columbia Presbyterian Medical Center and Columbia University Health Sciences and the Bioethics National Committee for Research in the Dominican Republic approved the study. Informed consent was obtained either from the participant or from a family member (surrogate) when the individual was demented.
Genotyping
First-stage mapping Using the first set of 96 families, we genotyped 340 autosomal microsatellite markers with an average inter-marker distance of 10.2 cM. Genotypes were generated as described in our earlier report.16 APOE genotypes were performed using a standard method22 with slight modification.12Genotype data were stored in LABMAN,23 and then output in standard LINKAGE file format for analysis. Maps from the Marshfield Medical Research Foundation24 and the Genome Database were used for locus order and inter-marker distance. Order was confirmed by comparison with the Human Genome Browser.25 Two or three individuals from CEPH pedigrees were included on each PCR plate. Discrepant genotypes were observed in 176 of 8607 genotypes for an error rate of 2.04%. This is a maximum error rate as no families were spread over more than one PCR plate, reducing plate-to-plate variation. Errors that caused mendalization errors in a family with greater than two samples were corrected. Data were completed for 96.1% of the sample-marker combinations.
Second-stage mapping Following the first-stage mapping, we observed that 18q, 10q, and 12p showed the strongest support for linkage either in two-point or multipoint analyses. We previously reported our results on the fine-mapping analysis for 12p.16 Thus, we focused our fine-mapping efforts on 18q and 10q using 104 families (96 families from the first-stage mapping plus eight new families). For 18q, we genotyped 18 additional markers from 96 to 126 cM, with an average inter-marker distance in the second-stage mapping of 1.8 cM. For 10q, we genotyped 12 additional markers between 90 and 160 cM, where our peaks were observed, resulting in an average inter-marker distance of 3.0 cM. We also genotyped eight additional markers between 42 and 94 cM, where other researchers found peaks, resulting in an average inter-marker distance of 4.1 cM. Following this, we then genotyped additional markers near D10S677 (117.4 cM) and D10S1230 (142.8 cM), where we had previously observed suggestive peaks.
Prior to the genome scan, we used 100 markers to examine for relationships among family members using the programs PEDCHECK26 and RELATIVE.27 As RELATIVE uses allele sharing among siblings (as well as non-sibling relatives) to test for nonpaternity, it is well suited for late-onset diseases like AD. Based on the results, we corrected the relationships (eg, full sibling to half-sibling), and we excluded individuals who were determined to be biologically unrelated.
Statistical analysis
In the first-stage scan, we performed a two-point affecteds-only analysis and multipoint affected relative pair analysis. For two-point linkage analysis, we carried out an affecteds-only analysis assuming autosomal dominant inheritance and also an affected sibpair analysis using ANALYZE.28 For multipoint nonparametric linkage analysis, we used GENEHUNTER2.29 When necessary, uninformative nonfounders were excluded (eg, unaffected children) to circumvent the computational limitation of the software. For all linkage analyses, we assumed a susceptibility allele frequency of 0.001. Allele frequencies for all markers were based on data derived from all participating family members.23,30 This method gives asymptotically unbiased estimates of the allele frequencies, thereby reducing the risk of false positives.31 As linkage analysis is sensitive to allele frequency, we reanalyzed the three candidate regions after re-computing allele frequency by randomly selecting one individual from each family. None of the markers in the three candidate regions showed significant differences in allele frequency by either method of estimating allele frequency (data not shown).
In the second-stage mapping, we carried out a series of linkage and family-based association analyses with additional markers using 104 families. First, two-point and multipoint linkage analyses were completed using the same models as in the first-stage mapping. Second, a family-based association analysis was implemented in Sib-TDT using the AD phenotype. Allelic association is expressed as a Z-score, and P-values are computed based on the normal distribution approximation.32 Third, we conducted a joint analysis of linkage and linkage disequilibrium, as implemented in PSEUDOMARKER.33 This method approximates a model-free affected relative pair approach, improves the power by jointly analyzing linkage and linkage disequilibrium, and does not break pedigrees into separate relative pairs. For 10q only, we tested allelic association for a series of markers between D10S1239 and D10S1237 using SNPs and age at onset as the phenotype implemented in QTDT to confirm the findings by Li et al.34,35 Lastly, because we16 and others3,9,36,37 have shown that there may be gene-gene interaction with APOE, we conducted two-point and multipoint linkage analyses, conditional on the APOE ε4 allele. For this purpose, we considered an individual with AD and at least one APOE ε4 allele affected. An individual was considered unknown otherwise (ε4-positive conditional analysis). To determine the independent effect of APOE, we conducted an analogous conditional linkage analysis, in which an individual with AD was considered affected in the absence of an APOE ε4 allele (ε4-negative conditional analysis). By carrying out the conditional analysis, it is not necessary to arbitrarily stratify families into APOE ε4-positive or -negative families. However, we did not test for family-based allelic association because we treated unaffected individuals as well as affected individuals without a copy of ε4 as unknown.
Results
Subjects and families
In the first-stage mapping, we studied 96 families with at least two individuals with AD, which included 490 family members. The demographic and clinical characteristics of the participating family members are presented in Table 1. In addition to probands, approximately four relatives were examined per family, which included affected as well as unaffected individuals. The mean age at onset for affected individuals was 73.6 years. The majority of the families (85.7%) were from the Dominican Republic, 10.2% were from Puerto Rico, and 4.1% came from elsewhere in the Caribbean. Previously, we have shown that the study population is relatively homogeneous.15
Table 1.
Description of Caribbean Hispanic families
| Characteristics | First stage | Second stage |
|---|---|---|
| Number of families | 96 | 104 |
| Number of relatives examined | 394 | 417 |
| Individuals per family | 5.1 | 5.1 |
| Proportion of women (%) | 66.7% | 66.8% |
| Mean age at onset of AD (years) | 73.6 | 73.5 |
| Affection status | ||
| Probable AD | 60.2% | 60.1% |
| Unaffected | 35.3% | 36.3% |
| Unknown | 4.5% | 3.6% |
| Families with: | ||
| ≥5 affecteds | 12 (82:54) | 12 (82:55) |
| 4 affecteds | 12 (48:24) | 13 (52:27) |
| 3 affecteds | 21 (63:38) | 21 (63:42) |
| 2 affecteds | 51 (102:57) | 58 (116:65) |
| Totala | 96 (295:173) | 104 (313:189) |
| APOE—ε4 allele frequency | ||
| Probable AD | 48.8% | 47.0% |
| Unaffected | 35.8% | 35.4% |
In all, 22 individuals were classified as unknown.
Overall, 237 individuals met the criteria for probable AD, 157 were unaffected. The study included families with affected sibpairs as well as families with multiple individuals with AD who were distantly related (eg, avunculars, cousins). There were 12 families with five or more affected family members and 12 families with four affected members. Approximately one-half of the families included two affected members. Eight families had predominantly earlyonset (defined as the majority of affected individuals having onset before age 65 years) disease and 88 families had late-onset AD.
In the second-stage mapping, we added eight families for a total of 104 families and 521 family members. We found the clinical characteristics for the additional families to be similar to those of the families used in the first-stage mapping (Table 1).
The frequency of APOE-ε4 among affected individuals was significantly higher than expected under the null hypothesis of no association (Z = 4.26, P= 1.01 × 10-5, allele frequencies in Table 1). We were unable to compare allele frequencies for highly polymorphic microsatellite markers between individuals from the Puerto Rico and Dominican Republic because there were too few individuals from the Puerto Rico to make such comparisons meaningful.
First-stage mapping
Two-point analysis The positive results of two-point analyses for 340 autosomal microsatellite markers with an average inter-marker distance of 10.2 cM under the genetic homogeneity and affected sibpair models are shown in Table 2 (see supplementary graphs). We observed 17 markers exceeding an LOD score of 1.0 using either model. The three highest two-point LOD scores were 3.26 for D18S541 in the affected sibpair model, 2.44 for D18S844 in the homogeneity model, and 2.0 for D14S587 in the affected sibpair model. When restricted to late-onset families, the LOD score increased to 3.62 for D18S541 and to 2.98 for D18S844 under the homogeneity model, but decreased to 1.69 for D14S587 under the affected sibpair model.
Table 2.
Summary table for the two-point linkage analysisa results
|
Marker information |
All families |
Late onset families |
||||
|---|---|---|---|---|---|---|
| Chr | Marker | cM | Model | Lod score | Model | Lod score |
| 1 | D1S551 | 113.7 | ASP | 1.00 | ASP | 1.12 |
| 2 | D2S1777 | 99.4 | LOD | 1.49 | LOD | 1.33 |
| ASP | 1.51 | ASP | 1.32 | |||
| D2S1326 | 149.9 | b | ASP | 1.15 | ||
| D2S1353 | 164.5 | LOD | 1.52 | LOD | 1.59 | |
| D2S2944 | 210.4 | ASP | 1.31 | ASP | 1.31 | |
| 3 | D3S2418 | 215.8 | LOD | 1.07 | b | |
| 4 | D4S2366 | 12.9 | LOD | 1.59 | LOD | 1.31 |
| ASP | 1.10 | ASP | 1.03 | |||
| D4S2382 | 57.0 | LOD | 1.25 | LOD | 1.26 | |
| D4S3335 | 195.1 | LOD | 1.10 | LOD | 1.25 | |
| 5 | D5S2488 | 0.0 | LOD | 1.14 | LOD | 1.12 |
| ASP | 1.57 | ASP | 1.76 | |||
| 6 | GAAT3A06 | 36.4 | LOD | 1.65 | LOD | 1.31 |
| ASP | 1.47 | ASP | 1.11 | |||
| D6S1280 | 73.1 | LOD | 1.58 | LOD | 1.58 | |
| 8 | D8S1136 | 82.3 | b | LOD | 1.08 | |
| 10 | D10S1230 | 142.8 | LOD | 1.28 | LOD | 1.15 |
| ASP | 1.19 | |||||
| 12 | D12S1042 | 48.7 | LOD | 1.42 | LOD | 1.34 |
| ASP | 1.30 | ASP | 1.27 | |||
| 14 | D14S587 | 55.8 | LOD | 1.73 | LOD | 1.62 |
| ASP | 2.00 | ASP | 1.69 | |||
| D14S592 | 66.8 | LOD | 1.52 | LOD | 1.28 | |
| ASP | 1.32 | ASP | 1.16 | |||
| 18 | D18S541 | 106.8 | LOD | 3.14 | LOD | 3.62 |
| ASP | 3.26 | ASP | 3.40 | |||
| D18S844 | 116.4 | LOD | 2.44 | LOD | 2.98 | |
| ASP | 1.51 | ASP | 2.01 | |||
The results from the affecteds-only dominant model under the assumption of genetic homogeneity (LOD) and affected sibpair (ASP) model are shown.
The cells with lod scores less than one for both models are left empty.
Multipoint analysis Two of the highest multipoint NPL scores were observed on 18q and 10q (see supplementary material). On 18q, the strongest support was observed for D18S541 (NPL=2.70, P=0.0022; 112.3 cM), and the NPL scores for the adjacent markers were 1.42 (P=0.0646) for D18S1270 and 1.82 (P=0.026; 96.5 cM) for D18S844 (119.7 cM). The peak spanned over 20 cM. For 10q, the major peak was at D10S1230 with an NPL score of 3.15 (P=0.000487), and a minor peak was observed at D10S677 with an NPL score of 1.54 (P=0.048502; 117.4 cM). Although the major peak spans approximately 35 cM, the two peaks combined span as much as 60 cM.
Second-stage mapping
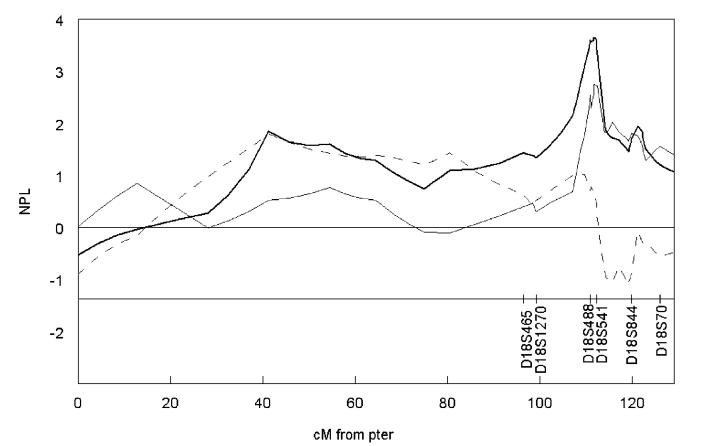
18q We re-examined the entire region between D18S1270 (96.5 cM) and D18S70 (126.0 cM) in detail with nine additional markers, because the initial peak was broad and the distal end of the peak remained elevated. Two-point analysis supported linkage for D18S541 (LOD=3.37) and D18S844 (LOD=2.06). With additional markers, the peak came downto the right of D18S844. The two markers to the distal end of D18S844, namely D18S1122 (122.8 cM) and D18S70 (126.0 cM), had LOD scores of 0.76 and 0.18, respectively, suggesting a boundary of the peak. In the multipoint analysis, the support for linkage was strongest at D18S1106 (NPL=3.65, P=0.000177; 111.7 cM), which is located less than 1cM away from D18S541; further, the peak was much narrower than in the first-stage scan (Figure 1). As the peak was quite narrow, we conducted an additional analysis to assess the likelihood of genotyping errors leading to a sharp false-positive peak, by repeating the multipoint linkage analysis of the peak region excluding one marker at a time. If the peak were due to genotyping errors, it is expected that the linkage at the peak region would diminish; however, the magnitude of the NPL score change for each marker was minimal (data not shown).
Figure 1.
The multipoint nonparametric analysis for 18q from the second-stage fine mapping using all families. The heavy solid line represents the linkage analysis using all families; the light solid line the Apolipoprotein E ε4-positive conditional linkage analysis; and the dotted line the Apolipoprotein E ε4-negative conditional linkage analysis. Map distances are based on the Marshfield genetic map. (Marshfield Medical Research Foundation, Center for Medical Genetics, http://research.marshfieldclinic.org/genetics/).
Using the same set of 104 families, we then conducted a family-based association analysis using Sib-TDT. Multiple markers on 18q met minimal criteria for statistically significant (P<0.05) allelic association as shown in Table 3, namely D18S465, D18S1092, D18S1106, D18S541, D18S1358, D18S870, and D18S58. For D18S541 and D18S870, two alleles were associated with AD (P<0.05 uncorrected for multiple testing). Joint linkage and association analysis using PSEUDOMARKER indicated that, for D18S541, linkage in the presence of linkage disequilibrium did not differ significantly from the linkage model alone (P=4.4 × 10-5 vs 3.1 × 10-5).
Table 3.
Fine-mapping family-based association analysis of 18q and 10q
| Marker | Allele | cM | Z′ | P-valuea | Marker | Allele | cM | Z′ | P-valuea |
|---|---|---|---|---|---|---|---|---|---|
| 18q | 10q | ||||||||
| D18S465 | 7 | 99.2 | 1.731 | 0.0417 | D10S1432 | 6 | 93.9 | 1.683 | 0.04619 |
| D18S1092 | 1 | 107.0 | 1.688 | 0.0457 | D10S580 | 5 | 96.7 | 2.187 | 0.01437 |
| D18S488 | 3 | 110.9 | 1.643 | 0.0502 | D10S2327 | 6 | 100.9 | 1.702 | 0.04438 |
| D18S1106 | 11 | 111.7 | 1.693 | 0.0452 | rs13595 | 1 | 113.9 | 1.828 | 0.03378 |
| D18S541 | 22 | 112.3 | 1.804 | 0.0356 | rs4925 | 2 | 130.2 | 1.692 | 0.04532 |
| 17 | 1.801 | 0.0359 | 7 | 133.1 | 1.718 | 0.04290 | |||
| D10S610 | |||||||||
| D181358 | 1 | 112.4 | 2.125 | 0.0168 | D10S1230 | 5 | 142.8 | 1.704 | 0.04419 |
| D18S870 | 1 | 113.8 | 2.407 | 0.0080 | 7 | 3.228 | 0.00062 | ||
| D18S58 | 7 | 115.6 | 2.047 | 0.0203 | D10S1213 | 4 | 148.2 | 1.865 | 0.03109 |
| D18S1122 | 8 | 122.8 | 2.113 | 0.0173 | D10S1222 | 7 | 156.3 | 2.042 | 0.02058 |
| D10S1223 | 7 | 156.3 | 2.744 | 0.00304 |
Only the alleles with a nominal P-value <0.05 are listed. P-values are not corrected for multiple testing.
Lastly, we conducted a conditional linkage analysis based on the APOE genotype (Figure 1). This analysis revealed that the peak on the 18q largely derived from individuals who were APOE4-positive, and the peak essentially disappeared when we considered APOE4-negatives.
10q With 12 additional markers near D10S1230, the location narrowed somewhat. The two markers that gave the strongest signals from the two-point analysis were D10S190 (LOD=1.81) and D10S1230 (LOD=1.34). The multipoint analysis revealed the strongest peak at 138 cM near D10S190 (NPL=2.02; P=0.0157) (Figure 2). With saturation of 10q, our peak shifted slightly proximal compared with our initial mapping, and yet the candidate region remains broad, spanning over 50 cM.
Figure 2.
The multipoint nonparametric analysis for 10q from the second-stage fine mapping using all families. The locations and markers of reported suggestive linkage findings by other groups on 10q are shown below. To estimate confidence intervals, we subtracted one from the highest reported LOD Score. The number next to each bar or dot indicates the reference (1. Bertram et al., 2000; 2. Ertekin-Taner et al., 2002; 3. Myer et al., 2000; 4. Kehoe et al., 1999; 5. Blacker et al., 2003; 6. Myer et al., 2002; 7. Farrer et al., 2003; 8. Li et al., 2003; 9. Ertekin-Taner et al., 2003). Map distances are based on the Marshfield genetics map (Marshfield Medical Research Foundation, Center for Medical Genetics, http://research.marshfieldclinic.org/genetics/).
Our subsequent family-based association analysis further supported the findings from the linkage analysis. Although multiple markers in this region supported allelic association as shown in Table 3, the strongest allelic association was observed with D10S1230 (Z=3.23, P=0.000623). This is the most likely cause of the linkage from our sample. In addition, D10S610 and rs4925 at around 130 cM also showed modest allelic association.
We examined 10q using the two-point joint linkage and association approach. In this set of analyses, D10S190 (at 138 cM; LOD=1.95 under the autosomal recessive model) and D10S1230 (at 143 cM; LOD=1.28 under the autosomal dominant model) showed only modest linkage as well as linkage in the presence of linkage disequilibrium. The autosomal recessive model showed stronger support than the autosomal dominant model. In the smaller peak at around 115 cM, D10S583 showed a significant result for linkage disequilibrium given linkage under the autosomal recessive model (P=0.003869), as well as for a joint linkage and linkage disequilibrium test under the autosomal recessive model (P=0.004188). No other markers showed support for this region.
As our peak for the AD phenotype centered on D10S190 and Li et al35 reported allelic association between glutathione S-transferase omega-1 (GSTO1) and GSTO2 with age at onset of AD and Parkinson’s disease (PD), we examined allelic association for seven markers (D10S583, D10S1671, D10S670, rs4925, D10S1429, D10S610, D10S187) near GSTO1 using the age at onset as the phenotype. We did not find a significant association with rs4925 (P=0.4); further, the age at onset was slightly lower for the individuals with more common allele compared with those with rare allele (73.4 vs 75.6 for common vs rare allele, respectively). However, we did observe a weak association with D10S610 (F=5.51, P 0.02), which is approximately 3 cM away from the rs4925 SNP. The strongest association with AD was with marker D10S1230 (P=0.0006), approximately 12 cM distal.
For both 18q22 and 10q, we reanalyzed the two-point and multipoint linkage analyses previously described using the allele frequencies estimated from one randomly selected individual from each family to reduce the influence that large extended pedigrees may have on allele frequency estimation. However, the allele frequencies were comparable and the LOD and NPL scores remained unchanged.
Discussion
In this unique group of Caribbean Hispanics with familial AD, we observed a modest peak at chromosome 10q near 138 cM, which spans approximately 15 cM and a weaker peak at 115 cM. The major peak on 10q was more distal than those reported by some groups.6-8 A new peak was identified at 18q22, and with additional fine mapping the region surrounding the peak narrowed considerably, and there existed evidence of an epistatic effect with APOE ε4.
Overall, 17 markers had an LOD score of greater than or equal to one in the first stage of the genome scan reported here. Other genome-wide searches3,9,37-39 have reported many of these loci, for example, 1q, 10q, 12p, 14q, while some are new. For example, Kehoe et al3 and Blacker et al38 reported a modest peak on 1q23 (178 cM). We do observe weak linkage in the region, but observed a stronger finding in a more proximal location at ∼114 cM. For 14q, we observed a peak on D14S587 (55 cM) as Blacker et al38 did in ‘early/mixed’ families; however, our samples, based predominantly on late-onset AD individuals, showed linkage to the same region. For 10q, 12p, and 18q, we chose to fine map these candidate regions because we observed strong evidence for linkage, and others reported strong linkage.
The observed variability in linkage findings is not surprising given the genetic complexity of AD. Specifically, there are likely to be multiple susceptibility genes, incomplete penetrance, and putative environmental risk factors. In addition, these studies use different genetic models, multiple testing of markers, and different study samples from different ethnic backgrounds. The primary goal of our study was to identify candidate regions that may contain AD susceptibility genes. According to the criteria suggested by Lander and Kruglyak,40 loci on chromosomes 10q26, 12p12, and 18q21 linked to AD would fulfill the criteria for suggestive of linkage. Some of the loci we observed from our genome-wide search will likely be false-positives. Thus, the findings here, as with any other genome scan, will need independent confirmation.
At least four loci have been linked to AD on chromosomes 9p, 10q, 12p, and 20p. However, though several putative loci have been linked to familial AD, only a few reports of allelic variation have been reported, but these reports need to be confirmed in independent samples. Pericak-Vance et al9 identified a locus on chromosome 12p conferring susceptibility to AD. Subsequent work by her group, ourselves, and others has suggested that there may be two separate loci on chromosome 12 depending on the presence of APOE-ε4 and clinical heterogeneity related to the presence of Lewy bodies.5,10,16,36,38,39,41 In the initial scan, Kehoe et al3 observed a peak at 12p only for the APOE-ε4 negative families, not for the APOE-ε4-positive families. However, when Blacker et al38 followed up the earlier genome scan using a larger sample, the support for linkage on 12p weakened. Previously, we reported our fine-mapping results that showed two peaks near D12S1623 (16 cM) and D12S1042 (49 cM).16 When we restricted the analysis to late-onset AD families, both markers showed strong linkage. When we conducted the analysis stratified by the APOE status, the linkage for D12S1623 was observed only in the APOE ε4-negative families. Two candidate genes in the region, alpha-2-macro-globulin (A2M [MIM 103950]) and the low-density lipoprotein receptor-related protein (LRP1 [MIM 107770]), have been the target of investigations, but without consistent results.16,36,42-45 Saunders et al,46 using the larger NIMH samples, identified several haplotypes that showed an association with AD, suggesting that there may be a gene—either A2M or one nearby—that may confer a modest effect toward AD.
Linkage findings on 10q also provide an illustration of the difficulties in replicating linkage and in estimating the location of gene underlying complex disorders.47,48 Six studies based on three different but correlated phenotypes (familial AD;3,6,8,38 variation in plasma amyloid β levels;7 and age at onset of AD34) showed evidence of linkage to 10q. The peaks in these studies cover a wide chromosomal region from 81 to 135 cM; however, it is not possible to determine whether these represent one or multiple loci.48 Three studies3,6,7 found the strongest support for linkage at ∼81 cM (D10S1227-D10S1211), while Bertram et al8 observed a peak between 115 and 127 cM, and later Blacker et al38 reported a peak at 135 cM (D10S1237) in a two-point analysis (θ=0.25). Using age at onset as the phenotype, Li et al34 reported a peak at 135 cM using the same marker (D10S1237) as Blacker et al, and subsequently reported an association with GSTO1. As we observed a peak nearby at 138 cM with the AD phenotype, we examined allelic association using both AD and age at onset phenotypes. For the age at onset phenotype, we did not find an association with rs4925 (P=0.4), but we did observe a weak association with D10S610 (F=5.51, P=0.02), which is approximately 3 cM away from the rs4925 SNP. For the AD phenotype, however, we observed weak allelic associations for rs4925 (Z=1.692; P=0.045323) as well as D10S610 (Z=1.718; P=0.042898). Although we did observe a weak association in the region, our finding is somewhat different from that of Li et al. The mean age at onset observed in our samples was comparable to that reported by Li et al. Yet, our association with rs4925 is weak for the AD phenotype and not significant for the age at onset phenotype. Further, while Li et al found the less common allele to be associated with later age at onset, we did not observe any difference in the mean age at onset between the two alleles. Further studies are needed to understand the allelic association since our finding is based on a relatively small sample of Caribbean Hispanics, while that of Li et al’s findings is based on a larger set of US Caucasians. Thus, at this point, it is not possible to conclude whether GSTO1 or some other gene is responsible for our linkage and association finding in this region.
In contrast, Bertram et al8 reported significant peaks at 115 cM (D10S583) and 127 cM (D10S1671), and observed allelic association at D10S583, located near the gene insulin degrading enzyme (IDE [MIM 146680]). Ait-Ghezala et al49 supported this finding in a follow-up case-control study. However, Abraham et al50 could not confirm the association with D10S583 or with SNPs flanking IDE. In a subsequent genome scan, Blacker et al38 reported two modest and broad peaks on 10q: one more proximal (80-100 cM) than the previously reported peaks (115 and 127 cM) and another at 135 cM. In our data, we did observe a modest peak at 138 cM. Given that multiple data sets using multiple correlated phenotypes show modest support for linkage on 10q covering a wide region, a susceptibility gene, if present, may only confer a weak effect. Alternatively, there may be more than one gene involved and they may interact. In our conditional analysis of APOE 4, we did find a slightly stronger evidence of linkage at the D10S190, but it was not statistically significant. Lastly, Ertekin-Taner et al7 reported linkage to 10q (∼80 cM) for plasma amyloid β protein levels as an intermediate risk factor phenotype for AD, then they subsequently reported the QTL locus to be the α-T catenin gene.51 In our data set, we did not observe any signal in this region when we used the AD phenotype.
An additional locus on chromosome 9p22 has been linked to AD from different samples: two studies in the US Caucasians;4,38 and in inbred Israeli Arabs.39 On chromosome 20p, AD8 [MIM607116]), a locus that includes the gene encoding cystatin-C (CST3 [MIM 604312]), was linked to very late-onset AD,37 and appeared to interact with the amyloid precursor protein. In contrast, we did not observe any support for linkage in these two regions to the AD phenotype.
It is important to note that, with the exception of the studies by three groups (Ertekin-Taner et al,7 Farrer et al,39 and the current study), all other studies include some subsets of the NIMH samples; thus, overlapping linkage peaks from different studies need to be interpreted with caution.
Our most robust linkage finding on 18q was previously reported by Pericak-Vance et al4 to have an LOD score of 1.1. With additional markers, the linkage peak sharpened, and the peak centers around D18S541, since both the joint linkage and association analysis and family-based association analysis strongly support D18S541. As with any genome scan, it is difficult to exclude the possibility of a false-positive finding. However, the addition of more markers narrowed the chromosomal region considerably, and the NPL score attained greater statistical significance. Further, the distal peak tapered off, arguing against the possibility of a spurious finding that occurs at the distal ends of the chromosome. Subsequent re-analyses of the region—first by sequentially excluding one marker at a time and second by using the allele frequencies based on a single randomly selected individual per family, rather than using all family members—did not change the LOD or NPL scores, suggesting that the observed linkage is less likely to be due to either genotyping errors or due to inaccurate allele frequency estimates.
This region on 18q contains approximately 4 000 000 base pairs including several candidate genes for which mRNA expression in brain has been observed. These include NETO1, a neuropilin (NRP) and tolloid (TLL)-like protein, and the predicted RefSeq genes for HSPC154 and MGC39671. There are also approximately 50 genes predicted using the Genscan Gene prediction track of the UCSC Genome Browser in the region.
We did not observe significant linkage to APOE in either the two-point or multipoint analyses. Similarly, we did not observe an association between APOE-ε4 and sporadic Alzheimer disease in the randomly selected Caribbean Hispanics, suggesting that APOE confers a weak effect in this population.12,13 Among familial AD, however, we reported14 the APOE-ε4 allele to be strongly associated (Z=4.26, P=1.01 × 10-5). In the current study, the joint linkage and association analysis of APOE yielded a P-value of 0.00802 for linkage disequilibrium under the dominant model, a P-value of 0.04863 for linkage, and a P-value of 0.00408 for a joint test of linkage and linkage disequilibrium. As expected, the linkage to APOE was weak. This illustrates the difficulties in identifying relatively common alleles, with weak to modest effects when studying late-onset disorders.
In sum, this was the first genome scan focused on familial AD in Caribbean Hispanics. We observed suggestive evidence for linkage to chromosome 18q and continue to support evidence for linkage to 10q.
Acknowledgements
Support to this work was provided by Federal Grants AG15473, AG08702, AG07232, Alzheimer’s Association grant IIRG-02-4387, the Charles S Robertson Memorial Gift for Alzheimer’s Disease Research from the Banbury Fund and the Blanchette Hooker Rockefeller Foundation. We also thank the members of Estudio Familiar de Influencia Genetica en Alzheimer, The Sociedad Dominicana de Geriatria y Gerontologia, The Sociedad Dominicana de Neurologia y Neurocirugia, The Sociedad Dominicana de Psiquiatria and the Associacion Dominicana Alzheimer y Similares, Inc, and The Bioethics National Committee for Research in the Dominican Republic.
References
- 1.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 2.Slooter AJ, Cruts M, Kalmijn S, Hofman A, Breteler MM, Van Broeckhoven C, et al. Risk estimates of dementia by apolipoprotein E genotypes from a population-based incidence study: the Rotterdam Study. Arch Neurol. 1998;55:964–968. doi: 10.1001/archneur.55.7.964. [DOI] [PubMed] [Google Scholar]
- 3.Kehoe P, Wavrant-De Vrieze F, Crook R, Wu WS, Holmans P, Fenton I, et al. A full genome scan for late onset Alzheimer’s disease. Hum Mol Genet. 1999;8:237–245. doi: 10.1093/hmg/8.2.237. [DOI] [PubMed] [Google Scholar]
- 4.Pericak-Vance MA, Grubber J, Bailey LR, Hedges D, West S, Santoro L, et al. Identification of novel genes in late-onset Alzheimer’s disease. Exp Gerontol. 2000;35:1343–1352. doi: 10.1016/s0531-5565(00)00196-0. [DOI] [PubMed] [Google Scholar]
- 5.Wu WS, Holmans P, Wavrant-DeVrieze F, Shears S, Kehoe P, Crook R, et al. Genetic studies on chromosome 12 in late-onset Alzheimer disease. JAMA. 1998;280:619–622. doi: 10.1001/jama.280.7.619. [DOI] [PubMed] [Google Scholar]
- 6.Myers A, Holmans P, Marshall H, Kwon J, Meyer D, Ramic D, et al. Susceptibility locus for Alzheimer’s disease on chromosome 10. Science. 2000;290:2304–2305. doi: 10.1126/science.290.5500.2304. [DOI] [PubMed] [Google Scholar]
- 7.Ertekin-Taner N, Graff-Radford N, Younkin LH, Eckman C, Baker M, Adamson J, et al. Linkage of plasma Abeta42 to a quantitative locus on chromosome 10 in late-onset Alzheimer’s disease pedigrees. Science. 2000;290:2303–2304. doi: 10.1126/science.290.5500.2303. [DOI] [PubMed] [Google Scholar]
- 8.Bertram L, Blacker D, Mullin K, Keeney D, Jones J, Basu S, et al. Evidence for genetic linkage of Alzheimer’s disease to chromosome 10q. Science. 2000;290:2302–2303. doi: 10.1126/science.290.5500.2302. [DOI] [PubMed] [Google Scholar]
- 9.Pericak-Vance MA, Bass MP, Yamaoka LH, Gaskell PC, Scott WK, Terwedow HA, et al. Complete genomic screen in late-onset familial Alzheimer disease. Evidence for a new locus on chromosome 12. JAMA. 1997;278:1237–1241. [PubMed] [Google Scholar]
- 10.Rogaeva E, Premkumar S, Song Y, Sorbi S, Brindle N, Paterson A, et al. Evidence for an Alzheimer disease susceptibility locus on chromosome 12 and for further locus heterogeneity. JAMA. 1998;280:614–618. doi: 10.1001/jama.280.7.614. [DOI] [PubMed] [Google Scholar]
- 11.Tang MX, Cross P, Andrews H, Jacobs DM, Small S, Bell K, et al. Incidence of AD in African-Americans, Caribbean Hispanics, and Caucasians in northern Manhattan. Neurology. 2001;56:49–56. doi: 10.1212/wnl.56.1.49. [DOI] [PubMed] [Google Scholar]
- 12.Maestre G, Ottman R, Stern Y, Gurland B, Chun M, Tang MX, et al. Apolipoprotein E and Alzheimer’s disease: ethnic variation in genotypic risks. Ann Neurol. 1995;37:254–259. doi: 10.1002/ana.410370217. [DOI] [PubMed] [Google Scholar]
- 13.Tang MX, Stern Y, Marder K, Bell K, Gurland B, Lantigua R, et al. The APOE-epsilon4 allele and the risk of Alzheimer disease among African Americans, whites, and Hispanics. JAMA. 1998;279:751–755. doi: 10.1001/jama.279.10.751. [DOI] [PubMed] [Google Scholar]
- 14.Romas SN, Santana V, Williamson J, Ciappa A, Lee JH, Rondon HZ, et al. Familial Alzheimer disease among Caribbean Hispanics: a reexamination of its association with APOE. Arch Neurol. 2002;59:87–91. doi: 10.1001/archneur.59.1.87. [DOI] [PubMed] [Google Scholar]
- 15.Athan ES, Williamson J, Ciappa A, Santana V, Romas SN, Lee JH, et al. A founder mutation in presenilin 1 causing early-onset Alzheimer disease in unrelated Caribbean Hispanic families. JAMA. 2001;286:2257–2263. doi: 10.1001/jama.286.18.2257. [DOI] [PubMed] [Google Scholar]
- 16.Mayeux R, Lee JH, Romas SN, Mayo D, Santana V, Williamson J, et al. Chromosome-12 mapping of late-onset Alzheimer disease among Caribbean Hispanics. Am J Hum Genet. 2002;70:237–243. doi: 10.1086/324773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan E. Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA work group under the auspices of the department of health and human services task force on Alzheimer’s disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 18.Hughes CP, Berg L, Danziger WL, Coben LA, Martin RL. A new clinical scale for the staging of dementia. Br J Psychiatry. 1982;140:566–572. doi: 10.1192/bjp.140.6.566. [DOI] [PubMed] [Google Scholar]
- 19.Pittman J, Andrews H, Tatemichi T, Link B, Struening E, Stern Y, et al. Diagnosis of dementia in a heterogeneous population. A comparison of paradigm-based diagnosis and physician’s diagnosis. Arch Neurol. 1992;49:461–467. doi: 10.1001/archneur.1992.00530290043010. [DOI] [PubMed] [Google Scholar]
- 20.Stern Y, Andrews H, Pittman J, Sano M, Tatemichi T, Lantigua R, et al. Diagnosis of dementia in a heterogeneous population. Development of a neuropsychological paradigm-based diagnosis of dementia and quantified correction for the effects of education. Arch Neurol. 1992;49:453–460. doi: 10.1001/archneur.1992.00530290035009. [DOI] [PubMed] [Google Scholar]
- 21.Stricks L, Pittman J, Jacobs DM, Sano M, Stern Y. Normative data for a brief neuropsychological battery administered to English- and Spanish-speaking community-dwelling elders. J Int Neuropsychol Soc. 1998;4:311–318. [PubMed] [Google Scholar]
- 22.Hixson JE, Vernier DT. Restriction isotyping of human apolipoprotein E by gene amplification and cleavage with HhaI. J Lipid Res. 1990;31:545–548. [PubMed] [Google Scholar]
- 23.Adams P. LABMAN and LINKMAN: a data management system specifically designed for genome searches of complex diseases. Genet Epidemiol. 1994;11:87–98. doi: 10.1002/gepi.1370110109. [DOI] [PubMed] [Google Scholar]
- 24.Broman KW, Murray JC, Sheffield VC, White RL, Weber JL. Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am J Hum Genet. 1998;63:861–869. doi: 10.1086/302011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goring HH, Ott J. Relationship estimation in affected sib pair analysis of late-onset diseases. Eur J Hum Genet. 1997;5:69–77. [PubMed] [Google Scholar]
- 28.Goring HH, Terwilliger JD. Linkage analysis in the presence of errors III: marker loci and their map as nuisance parameters. Am J Hum Genet. 2000;66:1298–1309. doi: 10.1086/302846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES. Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet. 1996;58:1347–1363. [PMC free article] [PubMed] [Google Scholar]
- 30.Terwilliger JD, Ott J. Handbook of Human Genetic Linkage. The Johns Hopkins University Press; Baltimore: 1994. [Google Scholar]
- 31.Ott J. Strategies for characterizing highly polymorphic markers in human gene mapping. Am J Hum Genet. 1992;51:283–290. [PMC free article] [PubMed] [Google Scholar]
- 32.Spielman RS, Ewens WJ. A sibship test for linkage in the presence of association: the sib transmission/disequilibrium test. Am J Hum Genet. 1998;62:450–458. doi: 10.1086/301714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goring HH, Terwilliger JD. Linkage analysis in the presence of errors IV: joint pseudomarker analysis of linkage and/or linkage disequilibrium on a mixture of pedigrees and singletons when the mode of inheritance cannot be accurately specified. Am J Hum Genet. 2000;66:1310–1327. doi: 10.1086/302845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li YJ, Scott WK, Hedges DJ, Zhang F, Gaskell PC, Nance MA, et al. Age at onset in two common neurodegenerative diseases is genetically controlled. Am J Hum Genet. 2002;70:985–993. doi: 10.1086/339815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li YJ, Oliveira SA, Xu P, Martin ER, Stenger JE, Scherzer CR, et al. Glutathione S-transferase omega-1 modifies age-at-onset of Alzheimer disease and Parkinson disease. Hum Mol Genet. 2003;12:3259–3267. doi: 10.1093/hmg/ddg357. [DOI] [PubMed] [Google Scholar]
- 36.Scott WK, Grubber JM, Conneally PM, Small GW, Hulette CM, Rosenberg CK, et al. Fine mapping of the chromosome 12 late-onset Alzheimer disease locus: potential genetic and phenotypic heterogeneity. Am J Hum Genet. 2000;66:922–932. doi: 10.1086/302828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olson JM, Goddard KA, Dudek DM. A second locus for very-late-onset Alzheimer disease: a genome scan reveals linkage to 20p and epistasis between 20p and the amyloid precursor protein region. Am J Hum Genet. 2002;71:154–161. doi: 10.1086/341034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blacker D, Bertram L, Saunders AJ, Moscarillo TJ, Albert MS, Wiener H, et al. Results of a high-resolution genome screen of 437 Alzheimer’s disease families. Hum Mol Genet. 2003;12:23–32. doi: 10.1093/hmg/ddg007. [DOI] [PubMed] [Google Scholar]
- 39.Farrer LA, Bowirrat A, Friedland RP, Waraska K, Korczyn AD, Baldwin CT. Identification of multiple loci for Alzheimer disease in a consanguineous Israeli-Arab community. Hum Mol Genet. 2003;12:415–422. doi: 10.1093/hmg/ddg037. [DOI] [PubMed] [Google Scholar]
- 40.Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 41.Scott WK, Grubber JM, Abou-Donia SM, Church TD, Saunders AM, Roses AD, et al. Further evidence linking late-onset Alzheimer disease with chromosome 12. JAMA. 1999;281:513–514. doi: 10.1001/jama.281.6.513. [DOI] [PubMed] [Google Scholar]
- 42.Blacker D, Wilcox MA, Laird NM, Rodes L, Horvath SM, Go RC, et al. Alpha-2 macroglobulin is genetically associated with Alzheimer disease. Nat Genet. 1998;19:357–360. doi: 10.1038/1243. [DOI] [PubMed] [Google Scholar]
- 43.Dodel RC, Du Y, Bales KR, Gao F, Eastwood B, Glazier B, et al. Alpha2 macroglobulin and the risk of Alzheimer’s disease. Neurology. 2000;54:438–442. doi: 10.1212/wnl.54.2.438. [DOI] [PubMed] [Google Scholar]
- 44.Wavrant-DeVrieze F, Perez-Tur J, Lambert JC, Frigard B, Pasquier F, Delacourte A, et al. Association between the low density lipoprotein receptor-related protein (LRP) and Alzheimer’s disease. Neurosci Lett. 1997;227:68–70. doi: 10.1016/s0304-3940(97)00304-2. [DOI] [PubMed] [Google Scholar]
- 45.Martin-Rehrmann MD, Cho HS, Rebeck GW. Lack of association of two lipoprotein lipase polymorphisms with Alzheimer’s disease. Neurosci Lett. 2002;328:109–112. doi: 10.1016/s0304-3940(02)00511-6. [DOI] [PubMed] [Google Scholar]
- 46.Saunders AJ, Bertram L, Mullin K, Sampson AJ, Latifzai K, Basu S, et al. Genetic association of Alzheimer’s disease with multiple polymorphisms in alpha-2-macroglobulin. Hum Mol Genet. 2003;12:2765–2776. doi: 10.1093/hmg/ddg310. [DOI] [PubMed] [Google Scholar]
- 47.Suarez BK, Hampe CL, Van Eerdewegh P. Problems of replicating linkage claims in psychiatry. In: Gershon ES, Cloninger CR, editors. Genetic Approaches to Mental Disorders. American Psychiatric Press, Inc; Washington, DC: 1994. pp. 23–46. [Google Scholar]
- 48.Roberts SB, MacLean CJ, Neale MC, Eaves LJ, Kendler KS. Replication of linkage studies of complex traits: an examination of variation in location estimates. Am J Hum Genet. 1999;65:876–884. doi: 10.1086/302528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ait-Ghezala G, Abdullah L, Crescentini R, Crawford F, Town T, Singh S, et al. Confirmation of association between D10S583 and Alzheimer’s disease in a case-control sample. Neurosci Lett. 2002;325:87–90. doi: 10.1016/s0304-3940(02)00243-4. [DOI] [PubMed] [Google Scholar]
- 50.Abraham R, Myers A, Wavrant-DeVrieze F, Hamshere ML, Thomas HV, Marshall H, et al. Substantial linkage disequilibrium across the insulin-degrading enzyme locus but no association with late-onset Alzheimer’s disease. Hum Genet. 2001;109:646–652. doi: 10.1007/s00439-001-0614-1. [DOI] [PubMed] [Google Scholar]
- 51.Ertekin-Taner N, Ronald J, Asahara H, Younkin L, Hella M, Jain S, et al. Fine mapping of the alpha-T catenin gene to a quantitative trait locus on chromosome 10 in late-onset Alzheimer’s disease pedigrees. Hum Mol Genet. 2003;12:3133–3143. doi: 10.1093/hmg/ddg343. [DOI] [PMC free article] [PubMed] [Google Scholar]