Abstract
NRSF/REST is a protein that silences transcription of a number of genes that contain a DNA element called the neuron-restrictive silencer element (NRSE). During embryogenesis, REST is expressed ubiquitously in nonneural cells, but is down-regulated during differentiation of neural progenitors into neurons. REST is also up-regulated in adult neurons by activity, suggesting a possible role for the protein in synaptic plasticity. To understand mechanisms that control expression of REST, we identified and characterized the promoter region of the mouse REST gene (mREST). A 4.5-kb DNA segment containing three exons (A, B, and C) that correspond to alternatively spliced 5′ untranslated regions (5′UTRs) was isolated and its DNA sequence was determined. Reverse transcription-PCR analyses of fibroblasts, astrocytes, and neural progenitors identified variants in which these exons were spliced to exon D, suggesting that exons A, B, and C may each have a promoter. Consistent with this hypothesis, primer extension and in vitro transcription experiments revealed clusters of RNA transcription initiation sites upstream of exons A, B, and C. Tests of REST/luciferase reporter constructs in Neuro2A and NIH 3T3 cells revealed promoters upstream of exons A and B that were active in both cell lines, and a promoter upstream of exon C that was weakly active only in NIH 3T3 cells. Six enhancer and two repressor regions were found to overlap each of the three promoters, and some of these were found to be cell type-specific. Combinatorial arrangements of these promoters with enhancer and repressor regions may allow modulation of REST expression in particular contexts.
The neuron-restrictive silencer factor (NRSF), also known as RE-1 silencing transcription factor (REST) is a 116-kDa C2H2 zinc finger protein that is related to the Gli-Krüppel family of transcriptional repressors. It was originally identified as a factor that binds to a particular 21-bp DNA regulatory element, called the neuron-restrictive silencer element (NRSE; ref. 1), also known as repressor element 1 (RE-1; ref. 2). NRSF is a potent repressor of genes containing the NRSE (3). Several of the genes containing the NRSE are essential for establishment and maintenance of the neuronal phenotype; these include neuron-specific cytoskeletal proteins, neurotransmitters and their biosynthetic enzymes, synaptic vesicle components, neurotrophins, and cell adhesion molecules (3–9).
In rodents, NRSF is expressed ubiquitously in developing nonneural tissues between embryonic days 8.5 and 9.5 (E8.5–9.5) (10). At the onset of neuronal differentiation, however, only low levels of NRSF mRNA and protein are found in neural progenitors within the ventricular zone, and NRSF is further down-regulated in neurons (1, 2). Recent phenotypic analyses of REST knockout mice and experiments involving ectopic expression of NRSF in chicken embryos (10) showed that perturbation of REST expression and function results in cellular apoptosis, aberrant differentiation and morphogenesis, and lethality. These experiments indicated that NRSF function is essential for normal embryogenesis.
Although NRSF is down-regulated in postmitotic neurons, detectable levels of NRSF mRNA and protein are still found in the adult central nervous system (11). In addition, REST is induced in hippocampal and cortical neurons by the glutamate analog, kainic acid (11), suggesting that its expression may be modulated by neuronal activity. Moreover, a neuronal-specific splice variant called REST4 is induced by protein kinase A and functions as a dominant-negative by binding to full-length NRSF/REST, derepressing two genes that are involved in cholinergic neurotransmission (12). Thus, in addition to its role during early development, NRSF/REST may modulate the expression of NRSE-containing genes in mature neurons and play an important role in activity-dependent synaptic plasticity.
Previous analyses of rat REST genomic DNA identified three exons designated A, B, and C, which correspond to 5′ untranslated regions (5′UTRs) (11). REST mRNA transcripts having one or another of these UTRs were found in the rat and all were spliced to the second exon (called exon D) containing the ATG initiator. Thus, transcription of the REST gene is likely to be initiated upstream of each of the three alternative 5′UTRs. This raises the possibility that there may be at least three promoters within REST, the usage of which might be context-dependent.
In the present study, we identify and characterize three clusters of transcription initiation sites and three promoters within the 5′ end of the mouse REST gene (mREST), investigate the expression in different cell types of exons A, B, and C encoding alternative 5′ untranslated regions (UTRs), and delineate certain positive and negative regulatory regions that overlap with the multiple promoter activities of mREST.
Materials and Methods
Characterization of mREST Clones.
Oligonucleotide primers (nucleotides 252–420, GenBank accession no. U13878) were used in a PCR-based screening of a P1 mouse genomic library (Genome Systems, St. Louis). The mREST clones were digested, electrophoresed, and transferred to membranes (Hybond-N; Amersham Pharmacia) as described (13). 32P-labeled probes corresponding to a 169-bp PCR product from the third zinc finger of mouse NRSF (bp 252–420, see above), and a 219-bp product derived from exon A of rat REST (bp 21–239; GenBank accession no. AF037200) were prepared by using the High Prime DNA Labeling kit (Roche Molecular Biochemicals). The mREST fragments identified by probes were subcloned into pBluescript SK− (Stratagene). The sequence of a 4.5-kb SmaI to BamHI segment of mREST was determined on both strands by using a Perkin–Elmer Biosystems sequencer.
Cell Culture.
Neuro2A, HeLa, and NIH 3T3 cells were maintained in DMEM supplemented with 10% FBS. Cortical astrocytes were prepared from brains of postnatal day 2–6 rat pups and maintained (14). Neural progenitors and neuronal cultures were prepared from the hippocampus and cortex, respectively, of embryonic day 17 rat embryos as described (15). Hippocampal progenitors were grown in neurobasal medium with B27 supplement (Life Technologies, Grand Island, NY) and 20 ng/ml of basic fibroblast growth factor (Sigma). Cortical neurons were plated in NB/B27 media supplemented with 10% FBS for 24 hr, switched to NB/B27 for 3 additional days, and plated on a poly-l-lysine/laminin substrate (Sigma) in 100-mm dishes.
Analyses of REST Transcript Expression.
Total RNA was isolated from NIH 3T3 cells, cortical neurons, astrocytes, and neural progenitor cells by extraction with Trizol (Life Technologies). Reverse transcription-PCR (RT-PCR) consisted of 30 cycles of 1 min at 95°C, 1 min at 60°C, and 1 min at 72°C, with final extension of 10 min at 72°C (16). Sense primers used in PCR reactions were as follows: exon A, 5′-tgtgctggaatgtgcggctc-3′; exon B: 5′-tcgcagcgaccgggggccg-3′; exon C, 5′-gcctatttcagttaggaatgccgc-3′. Two antisense primers within exon D were used: D1, 5′-ggttaaggccatgcccatgttggcac-3′and D2, 5′-ctcctccagaagactgccccatcacctg-3′. The sizes of PCR products were as follows: (exon A/D1, 356 bp; exon A/D2, 234 bp; exon B/D2, 123 bp). Glyceraldehyde-3-phosphate dehydrogenase mRNA levels were used as an internal control. Primers for these reactions were G1, 5′-gcatccactggtgctgccaaggctgtgggc-3′, and G2, 5′-catgtgggccatgaggtccaccac-3′.
Primer Extension and in Vitro Transcription Analyses.
Primer extension products were generated by using the following primers corresponding to regions within exons A, B, and C: A2, 5′cttcggcggcgctcctggtcttctgctgttg-3′; A4, 5′-tcgcggagcgggatcagaccgccggccctg-3′; B2, 5′- gatcgggcctgtaccggggctccgcggcgc-3′; B3, 5′-ctcgctacccgcgtccgatgcgggcctgta-3′; B5, 5′-cagccccctccccctgccgcccgcccacta-3′; C2, 5′-cagcccgagagcaacgtgttgcaccgaattc-3′; and C3, 5′-cactcccaaggaaatctccacgtg-3′. Primers were labeled with [γ-32P]ATP (Dupont/NEN) using polynucleotide kinase. A total of 5 × 104 dpm of each mREST-specific primer was hybridized to 10 μg of total RNA at 65°C for 90 min in a buffer containing 150 mM KCl, 10 mM Tris⋅HCl (pH 8.3), 1 mM EDTA. Reverse transcription reactions were performed with Superscript II (Life Technologies), treated with RNase A (20 μg/ml), and electrophoresed on a 6% sequencing gel. Sequencing reactions of mREST DNA using the same primers were run in adjacent lanes to determine the sizes of extension products, which were visualized by autoradiography.
RNA polymerase II run-off transcription assays were performed (17) by using nuclear extracts prepared from NIH 3T3 and HeLa cells (18). To analyze start sites upstream of exon A, two templates were used: a BglII/SacI fragment (−591 bp to +280 bp of mREST) and BglII/XhoI (obtained by XhoI digestion of construct ABC at +411). To analyze exon B, three templates were used: a 637-bp PCR product (+521 to +1158), a 793-bp PCR product (+521 to +1214), and a HindIII-linearized B1 construct. To analyze exon C, three templates were used: a 361-bp PCR product (+1157 to +1518), a C1 construct that was linearized at the EcoRI site (+1431 bp), and a BC1 construct linearized with EcoRI.
For each transcription reaction, between 200 and 500 ng of a mREST/pGL3-Basic construct or 20 ng of a purified promoter fragment were incubated with nuclear extract (50–100 μg). Transcription was initiated by addition of 0.5 μCi [α-32P]UTP (Dupont/NEN) and a mixture of 0.5 mM ATP, CTP,GTP, and 0.01 mM UTP. Reactions were electrophoresed on a 6% sequencing gel adjacent to DNA sequencing reactions and visualized by autoradiography.
Cellular Transfection of mREST Reporters.
The mREST fragments shown in Fig. 5 were inserted into the promoterless luciferase reporter vector pGl3-Basic (Promega); Plasmids were prepared by using Maxi-Prep columns (Qiagen, Chatsworth, CA). The DNA (0.5 μg of each reporter) was transfected into Neuro2A and NIH 3T3 cells at a density of 1.5 × 105 cells/well in 6-well plates using Fugene reagent (Roche Molecular Biochemicals). The reporter plasmid pCMVβ (CLONTECH) was cotransfected to provide an internal standard of β-galactosidase activity to which luciferase activities were normalized (19). The luciferase activities shown were obtained from 3–12 independent experiments each performed in triplicate.
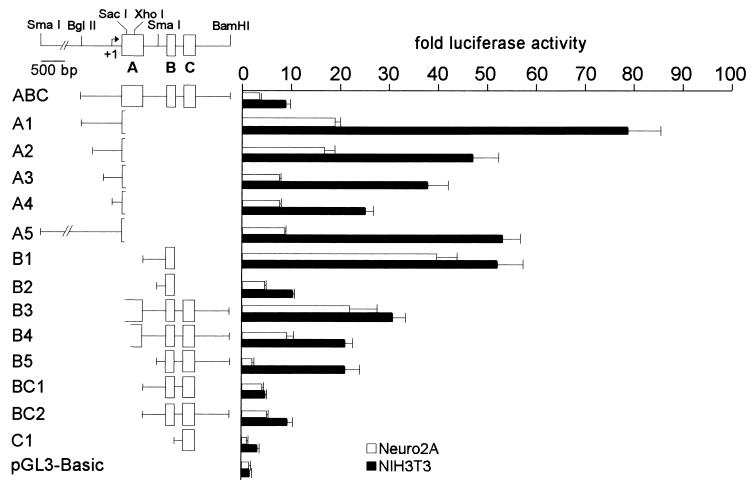
Figure 5.
Schematic representation of 14 mREST/luciferase reporter constructs and corresponding activities after transfection into Neuro2A or NIH 3T3 cells. The activity levels are expressed as the fold increase in relative light units above the level produced by the promoterless reporter, pGL3-Basic.
Results
Isolation and Characterization of the 5′ End of mREST.
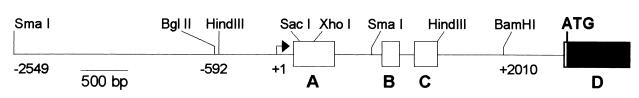
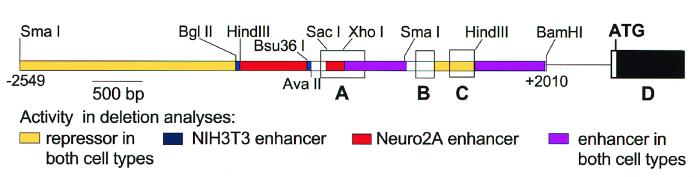
P1 clones for mREST were isolated by PCR screening using primers that were complementary to a cDNA segment encoding the third zinc finger of mouse NRSF. Restriction fragments containing the 5′ end of the mREST gene were then identified by Southern hybridization using a cDNA probe corresponding to the rat A 5′UTR (11). Further mapping of restriction sites and positions of the first four exons (A, B, C, and D) were performed to provide a detailed picture of the genomic organization of the 5′ end of mREST (Fig. 1). Comparison of mREST sequences of exons A, B, and C that encode the alternatively spliced 5′UTRs displayed a high degree of similarity (85%, 99%, and 82%, respectively) to those previously described for the rat REST gene (11).
Figure 1.
Schematic representation of the 5′ end of mREST. Restriction sites used to prepare reporter constructs are shown. Exons A, B, and C encoding 5′ UTR segments, which are alternatively spliced to exon D, are shown in open boxes. The arrow labeled +1 denotes the transcription start site that was located 30 bp downstream of the TATA box.
Expression of mREST Transcripts Containing Exons A, B, and C in Various Differentiated Cells.
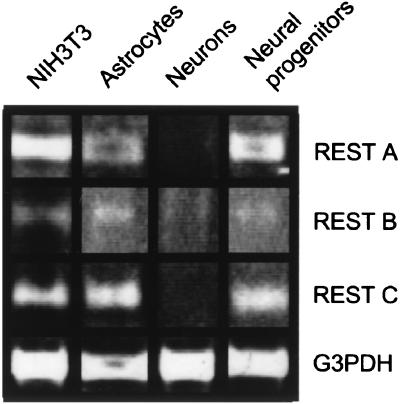
To examine the expression in different cell types of REST transcripts containing the alternative 5′UTRs, RT-PCR analyses were performed on RNA isolated from NIH 3T3 cells, and from primary cultures of rat astrocytes, neural progenitors, and cortical neurons. As shown in Fig. 2, REST mRNA transcripts containing exon A, B, or C linked to exon D were detected in RT-PCR reactions of NIH 3T3 cells, cultured astrocytes, and neural progenitors. The mRNAs containing exons A and C were not detected in cultured cortical neurons when less than 100 ng of cDNA was used in RT-PCR reactions. These data suggest that only a low level of mRNAs containing exons A, B, and C are expressed by cortical neurons, with transcripts containing exon B being the most abundant. No amplification products were obtained in absence of reverse transcriptase, indicating that the observed PCR products were amplified from the cDNA and not from contaminating genomic DNA (data not shown).
Figure 2.
Ethidium bromide staining of an agarose gel containing products of RT-PCR. RT-PCR used 5′ primers from exons A, B, and C and a 3′ primer within exon D on RNA from murine NIH 3T3 cells and rat astrocytes, cortical neurons, and neural progenitors. Glyceraldehyde-3-phosphate dehydrogenase RT-PCR was performed as a positive control.
Transcription of the mREST Initiates Upstream of Each of the Exons Encoding the Alternative 5′UTRs.
The presence of each of the three 5′UTR exons in mREST mRNA transcripts suggested that transcription might initiate upstream of each of the three exons A, B, and C. To address this possibility, transcription initiation sites were mapped using both primer extension and in vitro transcription assays. Examples of experiments used to determine RNA start sites occurring at the 5′ end of exon A are shown in Fig. 3; similar results were also used to map start sites that occurred upstream of exons B and C (data not shown).
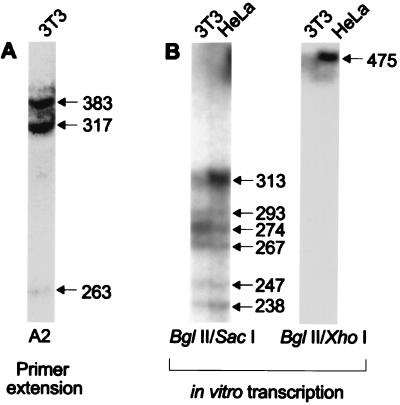
Figure 3.
Analysis of start sites for mREST transcripts in exon A. Primer extension (A) generated three products; the sizes of these RNA transcripts (in nucleotides, nt) were determined by comparison with adjacent DNA sequencing reactions (not shown). The 317-nt product was arbitrarily assigned as +1. (B) Nuclear run-off transcription assays using BglII/SacI (Left) and BglII/XhoI (Right) templates.
As shown in Fig. 3A, extension using the A2 primer gave products of 383, 317, and 263 nt, respectively. The 317-nt product initiated at a position 30 bp downstream of the TATA box within mREST (Fig. 4). This initiation site was arbitrarily assigned the position of +1. The 383-nt product initiated 11 bp downstream of a CAAT box, which is located approximately 50 bp upstream of the TATA box (see Fig. 4). The 263-nt product and extension products from the A4 primer (data not shown) were found in a separate cluster of initiation sites located 17 bp downstream of an Sp1 site that was located at +115.
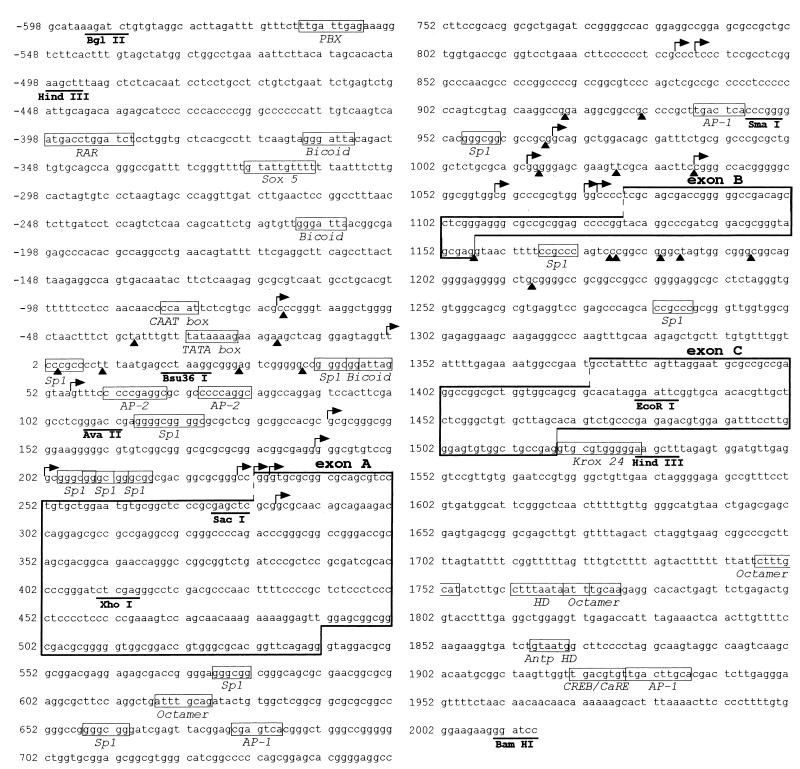
Figure 4.
DNA sequence of a 2.6-kb BglII–BamHI fragment from mREST. The sequences for exons A, B, and C that show a high degree of similarity with comparable exons in rat REST (11) are boxed. The transcription start sites determined by primer extension and nuclear run-off assays are indicated with rightward pointing arrows and filled triangles, respectively. Putative binding sites for trans-factors are indicated with small open boxes.
The in vitro transcriptions of two different DNA templates indicated multiple start sites upstream of exon A (Fig. 3B). The 313- and 475-nt transcription products initiated at approximately the same location as that indicated by the largest primer extension product (Fig. 3A). Five other minor products were produced; these mapped RNA start sites within the region between −35 and +39.
Primer extension and in vitro transcription identified several clusters of start sites that mapped to particular positions upstream of each of the three exons A, B, and C (see Fig. 4). Only RNA start sites that were corroborated in extensions with multiple primers, that were produced by in vitro transcriptions of more than one template, or that showed correlation between extension and transcription were included. These data indicate that several start sites occur upstream of exons A, B, and C and support the hypothesis that there are promoters upstream of each of these exons.
Three Promoters, Six Enhancer Regions, and Two Repressor Regions within the 5′ End of mREST.
To demonstrate that promoters exist upstream of each of the three exons and to examine regions that enhance or repress the activity, 14 different reporter constructs were analyzed after transfection of NIH 3T3 or Neuro2A cells (Fig. 5). NIH 3T3 and Neuro2A cells endogenously express the full-length NRSF/REST and truncated REST 4 proteins, respectively (11). The mRNAs encoding both of these proteins are expressed as variants that contain exons A, B, or C (11). Our constructs were designed to examine promoter activity upstream of these exons either individually or in combination.
A construct designated ABC, containing exons A, B, and C and spanning the region between −592 and + 2010 of the mREST, was 3-fold more active than the promoterless control vector in Neuro2A cells and 8-fold more active than the control in NIH 3T3 cells, indicating that this region could exhibit promoter activity in both neural and nonneural cell types. To analyze the region upstream of exon A for promoter activity, five different constructs (A1–A5) were tested (Fig. 5). A1 (−592/+280) was considerably more active than ABC in both cell types, indicating that there is a promoter upstream of exon A (designated promoter A), and that the +280 to +2010 region is likely to contain sequences that repress transcription of promoter A. Sequential 5′ deletion of sequences upstream of exon A revealed regions that differentially enhanced the activity of promoter A in the two cell lines. Deletion from −592 to −498, the A2 construct, did not appreciably reduce activity in Neuro2A cells, but reduced activity to 59% of A1 in NIH 3T3 cells, suggesting that the deleted region between −592 and −498 enhances the activity of promoter A in NIH 3T3 cells. Deletion of sequences from −498 to +21 (construct A3) resulted in a significant loss of activity in Neuro2A cells, but not in NIH 3T3 cells, indicating that this region enhances the activity of promoter A in Neuro2A cells. Further deletion to +109 (construct A4) resulted in a loss of activity in NIH 3T3 cells, but no appreciable reduction of activity in Neuro2A cells, indicating that +21 to +109 enhances activity of promoter A in NIH 3T3 cells. A5, which contained an additional segment of 1,957 base pairs upstream of −592, showed a reduced level of activity as compared with that of A1, indicating that the added region represses the activity of promoter A.
To assay exon B and sequences immediately upstream of it for promoter activity, two constructs, B1 (+521 to +1158) and B2 (+948 to +1158), were tested. The data indicated that the region upstream and including exon B is a potent promoter (designated promoter B). B1 showed the most potent activity in Neuro2A cells of all constructs tested. B2, in which a 427-bp segment was deleted, showed much less activity than B1 in both Neuro2A and NIH 3T3 cells. These data indicated that the +521 to +948 region contains an enhancer for promoter B that functions in both neural and nonneural cells. In support of this conclusion, constructs in which this region was inserted upstream of an SV40 early promoter had 9- and 8-fold greater activity in Neuro2A and NIH 3T3 cells, respectively, than did the promoter alone (data not shown).
A construct called C1 was tested to examine whether exon C and its immediate upstream region had promoter activity. C1 showed only background levels of activity in Neuro2A cells, but had some promoter activity in NIH 3T3 cells (3-fold over the activity of pGL3-Basic). Because this promoter (designated promoter C) had only minimal activity, we examined the activities of additional constructs that included both promoter B and promoter C together with downstream and upstream segments normally found in native mREST. Comparison of the activities of B1 and BC1 indicated that when promoter C and exon C were added to B1, activities were significantly reduced, indicating that promoter C and exon C repress the activity of promoter B and its enhancer in both cell types.
Further addition to BC1 of the region between the 3′ end of exon C and the 3′ terminal BamHI site (+1518 to +2010) (BC2) increased the activity in Neuro2A and NIH 3T3 cells, indicating that the added segment acts as a distal enhancer in both cell types. An independent test of this region as an enhancer of a heterologous promoter supported this conclusion (data not shown). The region between +1518 and +2010 thus mitigates the repressor activity of the +1157 to +1518 region (C1) on promoter B and its upstream enhancer.
To analyze the function of this distal enhancer in combination with the sequences upstream of promoter B, three additional constructs: B5, B4, and B3 were tested. Construct B5 deleted the +521 to +948 region upstream of exon B, but retained all of the other sequences present in construct BC2. As seen in comparisons of constructs B1 and B2, the region upstream of promoter B had potent enhancer activity in both cell types when presented in the context of promoter B and exon B alone. However, deletion of this region in the context of promoter B, exon B, promoter C, exon C, and the distal enhancer uncovered differential activities of this region in Neuro2A and NIH 3T3 cells (Fig. 5). B5 had 40% of the activity of BC2 in Neuro2A cells, but had 2.2-fold more activity than BC2 in NIH 3T3s; these comparisons suggested that in the context of BC2, the +521 to +948 region functions as an enhancer in Neuro2A cells, but as a repressor in NIH 3T3 cells.
Additional enhancer activities for promoter B were also found immediately upstream of +521. Addition of a region from the 3′ end of exon A (+411 to +521) to BC2 resulted in construct B3, which showed increases in activity in both Neuro2A and NIH 3T3 cells as compared with BC2. Further addition of an adjacent 5′ segment of exon A (+280 to +411) to B3 resulted in construct B4, which showed a 2.5-fold increase in Neuro2A cells, but no appreciable increase in NIH 3T3 cells, as compared with B3. These additional data suggest that the sequences between +280 and +411 enhance promoter B in Neuro2A cells.
The relative activities of promoters A, B, and C in neural progenitor cells were also compared. Constructs A1, B1, and C1 showed 38-, 70-, and 11-fold activity over that of the promoterless pGL3-Basic vector (data not shown). These experiments suggest that although all three mREST promoters are active in neural progenitors, promoter B shows the highest levels of activity.
Discussion
We have characterized the 5′ end of mREST, determined that exons in three different 5′ UTRs (exons A, B, and C) are expressed differentially in neural and nonneural cells, identified clusters of RNA start sites upstream of the exons A, B, and C, and identified three promoters, six enhancer regions, and two repressor regions that might modulate the expression of mREST in neural and nonneural cells. These various activities are summarized schematically in Fig. 6.
Figure 6.
Schematic representation of the three promoters, six enhancer regions, and two repressor regions within mREST. Enhancer regions are categorized by their function in the particular cell types: in Neuro2A cells (red), in NIH 3T3 cells (blue), and in both cell types (purple). Repressor regions (that function in both cell types) are shown in yellow.
In nonneural NIH 3T3 cells, we found that the relative strengths of the three promoters was promoter A>promoter B>promoter C, consistent with previous data on the relative abundance of mRNAs containing exon A, B, and C in the rat (approximately 80% of REST transcripts contained exon A, 20% contained exon B, and <1% contained exon C; ref. 11). Promoter A contains two distinct classes of elements and clusters of transcription initiation sites. The 5′ portion of promoter A contains TATA and CAAT boxes that appear to be associated with a cluster of RNA start sites initiating immediately downstream of these sequences. The 3′ end of promoter A is particularly GC-rich, contains multiple binding sites for the transcription factor Sp1, and contains a separate cluster of transcription start sites that is adjacent to these Sp1 sites. These observations indicate that promoter A is complex and might itself be partitioned into two promoters (TATA box- and Sp1-containing), suggesting that each may have an accompanying set of start sites and regulatory elements with which it functions. Two other promoters were found in mREST, one upstream of exon B and another upstream of exon C. Neither of these promoters were found to contain any TATA box or related sequences, but were GC-rich and contained binding sites for Sp1. Of all three promoters, promoter B was the most active in neural cells. Moreover, a strong enhancer was also found immediately upstream of promoter B. This enhancer functioned in both neural and nonneural cells in the context of promoter B and exon B, but when combined with additional downstream regions of mREST that included promoter C and exon C, it was found to function only in neural cells. These findings suggest that the cell-specific regulation of any of the three mREST promoters by adjacent or distal enhancer or repressor regions is likely to depend on the complement of trans-factors that bind to these regions in particular cell types. Database analyses of the enhancer regions within mREST have already provided clues (Fig. 4) as to the types of proteins that might regulate these promoters. These include homeodomain proteins of the bicoid, POU, PBX, and Antennapedia class, and members of the retinoic acid receptor, Sox, Krox, AP-1, and CREB families of transcription factors.
RT-PCR analyses of cortical neurons showed that transcripts containing exon B were most abundant. Moreover, in transfection experiments of neural progenitors, promoter B had the most activity. These data suggest that promoter B plays a key role in regulating mREST in neural cells.
Promoter C was found to be the least active mREST promoter—it showed no activity in Neuro2A cells, and was weakly active in NIH 3T3 and neural progenitor cells. Moreover, addition of promoter C and exon C to segments of mREST containing promoters A and B repressed activities of these stronger promoters, indicating that this region is a potent silencer of these upstream mREST promoters in both neural and nonneural cell types.
Multiple promoters have been identified in a number of genes, particularly those that have complex tissue-specific patterns of regulation and multiple contexts of activation by different signals. For instance, the gene encoding the brain-derived neurotrophic factor (7) contains at least four distinct promoters that are responsible for different contexts of regulation. The promoter upstream of exon III, for example, is the one of the four that is regulated by calcium influx and neuronal activity (20). It will be important to determine which of the three mREST promoters is regulated by neuronal activity. Based on our findings, promoter B is the most attractive candidate.
Other examples of multiple promoters and their complex usage in different biological contexts include those regulating expression of the fibroblast growth factors (FGFs) and their receptors (21). Alternative promoter usage in FGF genes generates alternative 5′UTRs that have been shown to differentially regulate mRNA stability and translation (22, 23). Because each of the three mREST promoters encodes a different 5′UTR, it will be informative to examine the half-lives of mREST transcripts having the different 5′UTRs and the ability of each to be differentially localized or translated in the cells in which they are expressed.
Our studies of the regulatory regions of mREST provide a basis for investigation of the contexts in which each of the promoters is used and how they interact with enhancer and repressor elements within mREST. An understanding of the factors that down-regulate mREST expression at the onset of neuronal differentiation would provide additional insights into gene control events that immediately follow neural induction. Moreover, identifying mechanisms that fine-tune mREST expression in neurons during adult life should help us understand the role of NRSF/REST in mature neurons (24).
Acknowledgments
We thank Nicole Son and Judy Yen for excellent technical assistance. We are also grateful to Drs. Bruce Cunningham, Robyn Meech, and J. A. Gally for critical reading of the manuscript. G.M.E. is a consultant to Becton Dickinson and Company. This work was supported by a U.S. Public Health Service grant to G.M.E. (HD33576), a grant from the National Science Foundation to F.S.J. (IBN-9816896), and a grant from the G. Harold and Leila Y. Mathers Foundation.
Abbreviations
- NRSF
neuron-restrictive silencer factor
- REST
repressor element 1 silencing transcription factor
- NRSE
neuron-restrictive silencer element
- UTR
untranslated region
- RT
reverse transcription
- mREST
mouse gene encoding NRSF/REST
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the Genbank database (accession no. AF220154).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.050578797.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.050578797
References
- 1.Schoenherr C J, Anderson D J. Science. 1995;267:1360–1363. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- 2.Chong J A, Tapia-Ramirez J, Kim S, Toledo-Aral J J, Zheng Y, Boutros M C, Altshuller Y M, Frohman M A, Kraner S D, Mandel G. Cell. 1995;80:949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- 3.Schoenherr C J, Paquette A J, Anderson D J. Proc Natl Acad Sci USA. 1996;93:9881–9886. doi: 10.1073/pnas.93.18.9881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wood I C, Roopra A, Buckley N J. J Biol Chem. 1996;271:14221–14225. doi: 10.1074/jbc.271.24.14221. [DOI] [PubMed] [Google Scholar]
- 5.Lonnerberg P, Schoenherr C J, Anderson D J, Ibanez C F. J Biol Chem. 1996;271:33358–33365. doi: 10.1074/jbc.271.52.33358. [DOI] [PubMed] [Google Scholar]
- 6.Thiel G, Greengard P, Sudhof T C. Proc Natl Acad Sci USA. 1991;88:3431–3435. doi: 10.1073/pnas.88.8.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Timmusk T, Palm K, Metsis M, Reintam T, Paalme V, Saarma M, Persson H. Neuron. 1993;10:475–489. doi: 10.1016/0896-6273(93)90335-o. [DOI] [PubMed] [Google Scholar]
- 8.Kallunki P, Jenkinson S, Edelman G M, Jones F S. J Biol Chem. 1995;270:21291–21298. doi: 10.1074/jbc.270.36.21291. [DOI] [PubMed] [Google Scholar]
- 9.Kallunki P, Edelman G M, Jones F S. J Cell Biol. 1997;138:1343–1354. doi: 10.1083/jcb.138.6.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Z-F, Paquette A J, Anderson D J. Nat Genet. 1998;20:136–142. doi: 10.1038/2431. [DOI] [PubMed] [Google Scholar]
- 11.Palm K, Belluardo N, Metsis M, Timmusk T. J Neurosci. 1998;18:1280–1296. doi: 10.1523/JNEUROSCI.18-04-01280.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shimojo M, Paquette A J, Anderson D J, Hersh L B. Mol Cell Biol. 1999;19:6788–6795. doi: 10.1128/mcb.19.10.6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Southern E M. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 14.Krushel L A, Sporns O, Cunningham B A, Crossin K L, Edelman G M. Proc Natl Acad Sci USA. 1995;92:4323–4327. doi: 10.1073/pnas.92.10.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hopker V H, Amoureux M C, Varon S. J Neurosci. 1997;49:355–363. [PubMed] [Google Scholar]
- 16.Amoureux M C, Wurch T, Pauwells P J. Biochem Biophys Res Commun. 1995;214:639–645. doi: 10.1006/bbrc.1995.2334. [DOI] [PubMed] [Google Scholar]
- 17.Pugh B F. In: Methods in Molecular Biology: In Vitro Transcription and Translation Protocols. Tymms M J, editor. Vol. 37. Totowa, NJ: Humana; 1995. pp. 349–357. [Google Scholar]
- 18.Dignam J D, Lebovitz R M, Roeder R G. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meech R, Kallunki P, Edelman G M, Jones F S. Proc Natl Acad Sci USA. 1999;96:2420–2425. doi: 10.1073/pnas.96.5.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shieh P B, Hu S-C, Bobb K, Timmusk T, Ghosh A. Neuron. 1998;20:727–740. doi: 10.1016/s0896-6273(00)81011-9. [DOI] [PubMed] [Google Scholar]
- 21.Szebenyi G, Fallon J F. Int Rev Cytol. 1999;185:45–106. doi: 10.1016/s0074-7696(08)60149-7. [DOI] [PubMed] [Google Scholar]
- 22.Myers R L, Payson R A, Chotani M A, Deaven L L, Chiu I-M. Oncogene. 1993;8:341–349. [PubMed] [Google Scholar]
- 23.Chotani M A, Chiu I-M. Cell Growth Differ. 1997;8:999–1013. [PubMed] [Google Scholar]
- 24.Jones F S, Meech R. BioEssays. 1999;21:372–376. doi: 10.1002/(SICI)1521-1878(199905)21:5<372::AID-BIES3>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]