Abstract
Peripheral ganglion neurons confer sensory information including touch, pain, temperature, and proprioception. Sensory modality is linked to specific neurotrophin (NTF) requirements. NT-3 supports survival of neurons that differentiate primarily into proprioceptors whereas nerve growth factor and brain-derived neurotrophic factor (BDNF) support subpopulations that transmit nociception and mechanoreception, respectively. We examined sensory neurons of gene-targeted mouse mutants at the NT-4, BDNF, NT-3, and TrkA loci. We show that NT-4 functions early in gangliogenesis, upstream of BDNF. In the absence of NT-4 function, BDNF-dependent, TrkB-expressing neurons fail to appear. The results are consistent with the model that precursor cells intended to become BDNF-dependent mechanoreceptors instead differentiate into NT-3-dependent proprioceptive neurons.
Peripheral sensory ganglia are composed of morphologically similar but functionally diverse neurons. Dorsal root and trigeminal ganglia project to multiple targets and mediate information including mechanoreception, nociception, proprioception, and thermal reception. Little is known of the process that specifies formation of these diverse neuronal phenotypes in the peripheral ganglia, whereas more is known about the trophic requirements that emerge as neurons differentiate. The neurotrophin (NTF) family of growth factors is essential for supporting the survival of peripheral neurons during the period of target innervation (1). The classical “Neurotrophic Hypothesis” suggests a role of target-derived NTFs in the sculpting and molding of the peripheral nervous system. Recent studies have implicated NTFs earlier in sensory ganglion development, preceding target innervation (2). It now is proposed that neurotrophin-3 (NT-3) and brain-derived neurotrophic factor (BDNF) regulate neuronal precursor numbers early in gangliogenesis (3–5).
Neurotrophin interactions are mediated through three high-affinity receptor tyrosine kinases (TrkA, TrkB, and TrkC) that are differentially expressed throughout sensory ganglion development. NT-3 primarily binds and activates TrkC; however, strong evidence suggests that NT-3 may also function through TrkA and TrkB receptors (6, 7). Nerve growth factor (NGF) functions through the TrkA receptor, whereas both BDNF and NT-4 equally bind and activate TrkB.
The earliest requirement for NTFs has been reported in cultured chick neural crest cells, where NT-3 functions as a mitogenic factor (8, 9), whereas BDNF can direct pluripotent neural crest cells to differentiation along the sensory neuron lineage (10, 11). The earliest mammalian requirements are thought to occur during the precursor stage of sensory gangliogenesis. Analysis of gene-targeted knockouts in mice has suggested that NT-3 and BDNF function in establishing precursor numbers (5, 12, 13). However, it still remains to be determined whether NTFs function as trophic factors for neuronal precursors (12) or, rather, function indirectly to regulate neuronal differentiation (13).
It is well established that early NTF requirements correlate with differentiated sensory neuron populations that transmit specific modalities, i.e., pain, temperature, or touch. For example, NT-3 via TrkC functions to support Ia (reflex)-associated proprioceptive neurons (2–5, 14). In addition, NGF and BDNF support nociceptive and mechanoreceptive populations (5, 15, 16), respectively, whereas the function of NT-4 is less well understood. Culture studies have demonstrated that both BDNF and NT-4 function through a common TrkB receptor to support the survival of distinct subsets of peripheral sensory neurons (17). However, initial examination of NT-4-deficient mice revealed no significant losses in neural crest-derived sensory neurons (18, 19). To examine further the role of NT-4 in the developing dorsal root ganglion (DRG), we evaluated NT-4, BDNF, NT-3, and TrkA gene-targeted mutants alone and in combination for neurotrophic requirement and phenotypic alterations. Our genetic and cell culture data indicate that NT-4 functions early in development to specify the emergence and differentiation of DRG neuron precursors as BDNF-dependent TrkB-expressing neurons.
Methods
Targeting Construct for Generation of NT-4 and TrkA Mutant Mice.
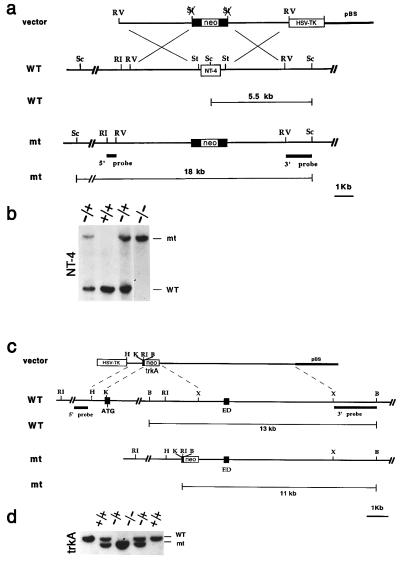
Targeting Vector electroporation and selection. For the NT-4 gene, a replacement-type targeting vector was constructed from a 7.0-kb 129Sv (Stratagene) mouse genomic fragment containing the NT-4 coding sequence. The vector for targeting the TrkA gene was generated by using a HindIII/KasI 0.8-kb fragment containing the first 53 bases of the exon with the translation start codon (ATG). A 7-kb fragment containing the extracellular domain (ED) exon corresponding to nucleotides 918-1247 of the TrkA cDNA sequence was used as 3′ fragment for the targeting vector. The neo gene with the phosphoglycerate kinase 1 promoter and the bovine growth hormone polyadenylation sequence (pGKneobpA) was employed as a positive selectable marker; the pGK-thymidine kinase cassette was used as a negative selectable marker (5). Electroporation and selection were performed by using the CJ7 embryonic stem (ES) cell line as described (2). For the NT-4 gene, DNAs derived from G418/1-(2-deoxy-2-fluoro-β-d-arabinofuranosyl)-5-iodouracil-resistant ES clones were screened by using a diagnostic ScaI restriction enzyme digestion using the 5′ and 3′ probes external to the targeting vector sequence indicated in Fig. 1a. Recombinant clones containing the predicted 18-kb rearranged band were obtained at a frequency of 1/10. Screening of the TrkA clones was performed with the 3′ external probe by detecting a reduction of the 13-kb BamHI restriction fragment to 11-kb band (Fig. 1c). Recombination at the 5′ end of the targeting vector was detected by the change of the 25-kb EcoRI restriction fragment to the predicted 18-kb rearranged band (not shown). Homologous recombinant clones at the TrkA locus were observed at the frequency of 1/250.
Figure 1.
Generation of NT-4 and TrkA mutant mice. (a) Schematic diagram showing the replacement vector and strategy used to inactivate the NT-4 locus. The NT-4 coding region is indicated. Restriction enzyme sites are as indicated: Sc, ScaI; RI, EcoRI; St, StuI; and RV, EcoRV. (b) Southern blot analysis of tail DNA from a litter obtained by intercrossing two NT-4 +/− mice. ScaI restriction enzyme digestion and the 3′ probe indicated in a were employed to detect rearrangement in the mouse NT-4 locus. The 5.5-kb wild-type (WT) and 18-kb rearranged (mt) DNA bands are indicated. (c) Schematic diagram showing the replacement vector and strategy used to inactivate the TrkA locus. Solid boxes indicate the TrkA exons containing the translation start site (ATG) and the extracellular domain exon (ED) corresponding to nucleotide sequence 918-1247 of the TrkA cDNA. Restriction enzyme sites are as indicated: B, BamHI; RI, EcoRI; X, XhoI; and H, HindIII. pBS indicates the pBluescript cloning vector (Stratagene). (d) Southern blot analysis of tail DNA from a litter obtained by intercrossing two TrkA +/− mice. BamHI restriction enzyme digestion and the 3′ probe indicated in a were used to detect rearrangements in the mouse TrkA locus. The 13-kb wild-type (WT) and 11-kb rearranged (mt) DNA fragments are indicated.
Generation of Mutant Mice.
Two independent targeted ES cell clones for both the NT-4 and the TrkA recombinant clones were injected into C57BL/6 blastocysts to generate chimeras that transmitted the mutated allele to the progeny. Breeding of two NT-4 or TrkA heterozygous (+/−) mice gave rise to homozygous mutant mice at a frequency of 25% (Fig. 1 b and d). Mice consisting of a mixed 129Sv and C57BL/6 background were bred in a specific pathogen-free facility with food and water ad libitum, and wild-type (wt) littermates were employed as controls.
Histology and Cell Counts.
Embryos were collected at embryonic day 13 (E13) and fixed in Bouin's solution [71.4% picric acid solution (1.2% wt/vol)/23.8% formalin/4.8% glacial acetic acid] for 24 hr and washed for 2 days in 70% ethanol. Neonatal mice (P0) were perfused with 4% paraformaldehyde in 0.1 M phosphate buffer saline (PBS), pH 7.4, followed by Bouin's postfixation. The tissues were dehydrated in ethanol, embedded in paraffin, serially sectioned at 5 μM, and stained with hematoxylin/eosin (H&E). DRG were counted at the fourth lumbar segment every 40 μM at P0 and every 20 μM at embryonic stages. No correction was made for split nuclei.
Quantitative Immunohistochemistry, Histochemistry, and Counts.
Paraffin-embedded neonatal lumbar spinal cords (n = 6) were serially sectioned at a thickness of 5 μM, and every 10th section (50 μM) was stained with H&E and counted. The remaining sections were immunostained (5) every 50 μM with antibodies directed against the following proteins: somatostatin (SS; 1:150; Zymed), substance P (SP; 1:150; Zymed), SSEA-3 (1:300; Hybridoma Bank, Iowa City, IA) (20, 21), calcitonin gene-related protein (cGRP; 1:50; Biogenesis, Kingston, NH), and parvalbumin (PA; 1:250; Swant, Bellinzona, Switzerland). Immunopositive neurons were counted and compared with the total number of H&E-stained neurons. Neuronal differentiation was examined by using the neuronal marker antiperipherin (1:300; Chemicon) in wt and mutant mice at E11, E12, and E13. Cutaneous merkel endings and cells were identified by using anti-PGP9.5 antibody (1:300; Biogenesis) (22, 23). TrkB-positive DRG neurons were stained with anti-Trk B (1:100) (6) in wt and NT-4 mutant mice. Finally, to determine muscle fiber types, serial cross-sections were stained for myosin ATPase activity by using preincubation pH values of 4.30, 4.50, and 10.30, as described previously (24). Tissues were visualized by using an Olympic Optical BX50 microscope.
Terminal Deoxynucleotidyltransferase (TdT)-Mediated dUTP-Biotin Nick End Labeling (TUNEL) Staining.
Apoptotic cell death was evaluated by using TUNEL assay in wt and mutant mice at E11, E12, and E13 (n = 5–6 for each time point). Paraffin sections (5 μM) were digested with 10 mg/ml proteinase K for 10 min and quenched in 3% hydrogen peroxide. Tissues were treated with TdT solution containing 40 mM biotinylated dUTP and 0.3 unit/ml TdT in 1× TdT buffer for 1 hr at 37°C. Tissues were incubated with avidin-biotin-peroxidase complex and developed with diaminobenzidine. Tissues were counterstained with H&E, and lumbar DRG cells were counted for the percentage of TUNEL-positive cells.
In Situ Hybridization.
In situ hybridization utilized TrkC-specific 35S-labeled UTP antisense probes and NT-4-specific digoxigenin-UTP antisense, and sense probes were performed as described previously (2, 25–27). Tissues were counterstained with H&E for total cell counts and photographed by using an Olympus Optical BX50 microscope.
Axonal Counts.
The number of myelinated axons in the saphenous, common peroneal, and tibial nerves was examined in adult wt and NT-4 mutant mice (n = 5). A 5-mm segment of nerve was excised and fixed in a 3% glutaraldehyde, pH 8.0, solution. Tissues were dehydrated and embedded in JB-4 medium. Semithin sections (0.5 mm) were cut by using an ultramicrotome and stained with toluidine blue. Nerve cross-sections were counted for the total number of myelinated axons.
Culture Studies.
For primary cultures, E13 DRG were isolated (thoracic level 12 to lumbar level 5) from wt and mutant mice (n = 8–12) and cultured in the presence of the following NTFs: 10 ng/ml NGF (7S; GIBCO); 2 ng/ml BDNF; 10 pg/ml NT-3; and 2 ng/ml NT-4 (Regeneron Pharmaceuticals, Tarrytown, NY) (28). Neuron cultures were maintained at 37°C in a humidified incubator with 4% CO2. Neurons were counted every 24 hr in a heated Olympus IMT-2 inverted microscope under phase contrast.
Coordination Studies.
NT-4 mutant and wt littermates at 3 months of age (n = 6) were placed on a stationary rotarod (Columbus Instruments, Columbus, OH); acceleration began at 5 rpm and increased 10 rpm/min. Five trials were performed daily with a 15-min rest period between trials (over 4 consecutive days). Animals were kept on a 12-hr light cycle, and trials were performed in the middle of the light cycle.
Statistics.
Mean and SEM were evaluated between wt and mutant groups, and the significance was determined by Mann–Whitney rank sum test.
Results
NT-4 Mutation Modulates DRG Neuron Number in BDNF and NT-3 Mutant Mice.
Biochemical and cell culture studies have demonstrated that BDNF and NT-4 function through a common TrkB receptor and can mediate the survival of the same subset of peripheral sensory neurons (29–31). Analysis of targeted gene knockouts has revealed early requirements for NGF, BDNF, NT-3, and their cognate receptors TrkA, TrkB, and TrkC in sensory ganglion development (1). These mice die shortly after birth and have reduced number of sensory neurons. In contrast, NT-4 mutant mice appear normal and do not exhibit a reduction in neural crest-derived sensory neurons (18, 19). We previously have described mice that are mutated at the TrkC, BDNF, and NT-3 genes (2, 5). Fig. 1 diagrammatically illustrates the respective recombination cassettes employed (Fig. 1 a and c) and the Southern blots (Fig. 1 b and d) verifying genetic transmission of the mutated NT-4 and TrkA alleles into heterozygous and homozygous mice. In general, our observation of these mutant mice is in agreement with previous reports (16, 18, 19). In particular, the NT-4 mutant homozygotes are viable, fertile, and do not exhibit overt behavioral disorders under laboratory conditions.
To better analyze the role of each of the NTFs and Trk receptors in DRG development, each of the single mutant mice and combinations thereof were examined for development of DRG neurons. Table 1 shows DRG neuron counts from NT-4, NT-3, BDNF, and TrkA mutant mice at E13 and P0. In accord with previous studies, TrkA, BDNF, and NT-3 mutants exhibit reduction in sensory neuron number by E13, whereas NT-4 mutant DRG are similar to wt (Table 1, E13) (2–5, 18, 19, 32, 33). At P0, NT-4 mutant DRG have increased neuron numbers as compared with wt (Table 1, P0). When BDNF/NT-4 double mutants were analyzed, the neuronal losses present in the single BDNF mutant were abolished (Table 1). Thus, loss of NT-4 function resulted in apparent rescue of the sensory neurons lost in BDNF-deficient ganglia. We next examined NT-3/NT-4 double mutants and observed a significantly greater loss of DRG neurons (91%) as compared with those seen in the NT-3 mutant alone (78%). DRG neuron numbers in TrkA/NT-4 double mutants did not differ from the TrkA mutant alone (Table 1). These latter results indicate that the NT-4 mutation does not interact genetically with all neurotrophins and their receptors but, rather, specifically with BDNF and NT-3. Similar findings were observed in trigeminal sensory neurons for each of the mutant mice (Table 1). Thus, in vivo, absence of NT-4 function results in an increase in NT-3-dependent neurons as manifested in the NT-3/NT-4 double mutants. In addition, the absence of DRG neuronal loss in the BDNF/NT-4 double mutant indicates that these ganglia lack BDNF-dependent neurons. In lumbar DRG, the onset of BDNF and NT-3 dependence occurs as early as E11 and is completed by E13 (2, 5, 12, 13), indicating that NT-4 exerts its effect on DRG early in ganglion development.
Table 1.
Sensory neuron counts in the dorsal root and trigeminal ganglia of wt and mutant mice
| DRG
|
Trigeminal
|
|||||
|---|---|---|---|---|---|---|
| E13 | Reduction, % | P0 | Reduction, % | P0 | Reduction, % | |
| wt | 16,245 ± 651c | 4,434 ± 124g | 27,005 ± 640b | |||
| NT-4 −/− | 15,689 ± 646c | NS | 4,986 ± 236f | φ* | 26,407 ± 1,059a | NS |
| BDNF −/− | 11,274 ± 134c,h | 31* | 3,235 ± 177f | 27*** | 21,418 ± 786a | 21** |
| NT-4 and BDNF −/− | 16,437 ± 915c | NS | 4,311 ± 182c | NS | 24,628 ± 1,142b | 9* |
| NT-3 −/− | 10,459 ± 265b,h | 36** | 960 ± 51d | 78*** | 9,501 ± 1,287b | 65*** |
| NT-3 and NT-4 −/− | 7,820 ± 407a,h | 52*** | 412 ± 26c | 91*** | 6,133 ± 271a | 77*** |
| NT-3 and BDNF −/− | 8,789 ± 625c | 46** | 687 ± 63d | 84*** | 6,932 ± 573c,h | 74*** |
| TrkA −/− | ND | 706 ± 23b | 84*** | ND | ||
| TrkA and NT-4 −/− | ND | 791 ± 55c | 82*** | ND | ||
Neuron numbers (mean ± SEM) of dorsal root and trigeminal ganglia were counted in E13 and P0 mouse spinal cords and heads of wt, NTF/Trk receptor mutant mice. Tissues were fixed in Bouin's solution, embedded in paraffin, serially sectioned (5 μm), and stained with H&E. The tissue slides were counted every 40 μm at P0 and every 20 μm at embryonic stages. No correction was made for split nuclei. Mann–Whitney rank sum test determined the significance levels. φ, A significant increase of 12% (P < 0.05); NS, not significant; ND, not determined. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
n = 2.
n = 3.
n = 4.
n = 5.
n = 6.
n = 9.
n = 10.
Previously described in Liebl et al. (5).
NT-4 −/− Sensory Neurons Do Not Exhibit Increased Cell Death.
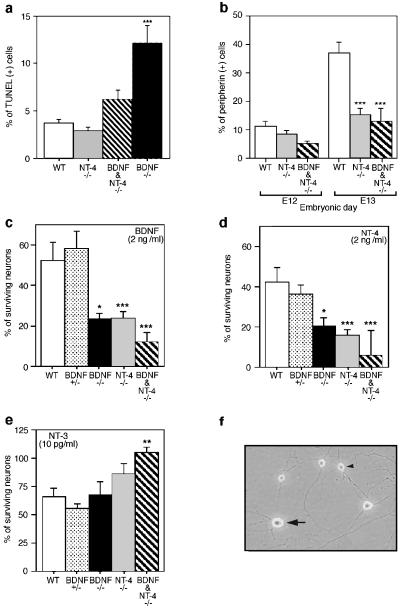
In DRG, targeted mutation of the NT-3 gene causes marked increase in apoptosis, accounting for the observed loss in sensory neurons (12, 13). To address whether NT-4 or BDNF functions in the trophic support of DRG neurons, wt, NT-4, and BDNF mutant DRG were examined for apoptosis by TUNEL staining (Fig. 2a). At E11, the percentage of apoptotic cells in NT-4 mutant ganglia was the same as in wt controls (Fig. 2a). Similar findings were obtained at E12 and E13 (not shown). BDNF/NT-4 double mutants showed a significant reduction in TUNEL staining compared with the BDNF mutation alone (Fig. 2a). In addition, we observed a reduced number of differentiated neurons in the same period as measured by peripherin staining (Fig. 2b). Thus, in the absence of NT-4, a portion of DRG neuronal precursors undergo delayed neuronal differentiation. We conclude that unlike the other NTFs (BDNF, NT-3, and NGF), NT-4 does not appear to function as a trophic factor in developing DRG. In the absence of NT-4, no measurable increase in apoptosis occurs, and the period of differentiation of some neurons is prolonged until after E13.
Figure 2.
Neurotrophin requirements in developing DRG. (a) TUNEL staining was examined at E11 in wt, BDNF, NT-4, and BDNF/NT-4 mutant mice. (b) Peripherin-immunostained cells in wt and mutant DRG. (c– e) Primary culture of wt, BDNF, NT-4, and BDNF/NT-4 mutant E13 DRG neurons incubated with 2 ng/ml BDNF (c), 2 ng/ml NT-4 (d), and 10 pg/ml NT-3 (e). (f) Primary DRG neurons cultured in the presence of NGF, BDNF, and NT-3. Small-soma-diameter neurons (arrowhead) are NGF-dependent, whereas large-soma-diameter neurons (arrow) are BDNF- or NT-3-dependent.
NT-4 −/− DRG Lack BDNF-Dependent Neurons.
To examine whether the effects of the NT-4 mutation could be extended in vitro, we cultured DRG neurons from E13 BDNF, NT-4, and BDNF/NT-4 mutants. In our hands, cultured DRG neurons can be divided into two morphological classes by soma diameter (Fig. 2f). Small-diameter neurons require NGF for survival, whereas large-diameter neurons require BDNF or NT-3 (D.J.L., L.J.K., and L.F.P., unpublished results). At E13, survival of approximately 40–50% of large-diameter wt neurons can be supported by BDNF or NT-4 (Fig. 2 c and d), whereas approximately 65% require NT-3 (Fig. 2e). Neuronal cultures from BDNF mutants exhibit reduced numbers of BDNF- and NT-4-responsive neurons (≈50%; Fig. 2 c and d) but not of NT-3-responsive neurons (Fig. 2e). Neurons cultured from NT-4-deficient embryos also exhibit selective loss of BDNF- and NT-4-responsive neurons (Fig. 2 c and d). Thus, mutation in either the BDNF or NT-4 genes results in loss of DRG neurons that can be cultured in the presence of these NTFs. DRG cultures from NT-4 mutants showed increased numbers of NT-3-dependent neurons as compared with wt controls (Fig. 2e). Reduction of BDNF- and NT-4-responsive neurons coupled with increased NT-3-responsive neurons also was observed in BDNF/NT-4 mutant mice (Fig. 2 c–e). Consistent with examination of the TrkA mutant mice in which no interaction was noted with the NT-4 mutation, NGF had no differential effect on survival of wt or mutant cultures (not shown). Coaddition of BDNF and NT-4 gave results indistinguishable from either factor alone (not shown), suggesting that direct competition between the NTFs is not a component of this phenomenon. These data are consistent with the in vivo results indicating that the NT-4 mutation results in a loss of BDNF-responsive DRG neurons and a compensatory gain in NT-3-dependent neurons.
NT4 −/− DRG Lack TrkB-Expressing Neurons.
To examine whether the altered NTF requirements seen in NT-4 mutant DRG were reflected in altered NTF receptor expression, we employed a TrkB-specific antibody and TrkC-specific mRNA in situ hybridization to determine the number of expressing neurons in P0 DRG (5, 6). Consistent with the preceding results, NT-4 mutant ganglia are depleted of TrkB-expressing neurons (Table 2 and Fig. 3 a–d). In addition, NT-4 mutant ganglia exhibited increased numbers of TrkC-expressing neurons (Table 2 and Fig. 3 e– j). Taken together, the in vivo and culture studies indicate a loss of BDNF-dependent DRG neurons in BDNF-, NT-4-, and BDNF/NT-4-deficient mice coupled with an increase of NT-3-dependent neurons in DRG of NT-4- and BDNF/NT-4-deficient mice.
Table 2.
Percentage of P0 DRG neurons expressing TrkB or TrkC in wt and NT-4 mutant mice
| % of TrkB (+) neurons | % of TrkC (+) neurons | |
|---|---|---|
| wt | 11.3 ± 0.3% | 7.8 ± 0.8% |
| NT-4 −/− | 1.2 ± 0.4% | 16.8 ± 0.5% |
| P value | P < 0.0001 | P < 0.0001 |
Percentage of TrkB- and TrkC-expressing neurons of the lumbar DRG were counted in sections of the midganglia in wt and NT-4 mutant mice. The number of Trk-positive neurons (Fig. 3 a–j) was compared with total neuron counts (≈2,000–4,000 neurons) in the same lumbar sections (10 sections per animal). Student's t test determined the significance levels. n = 4.
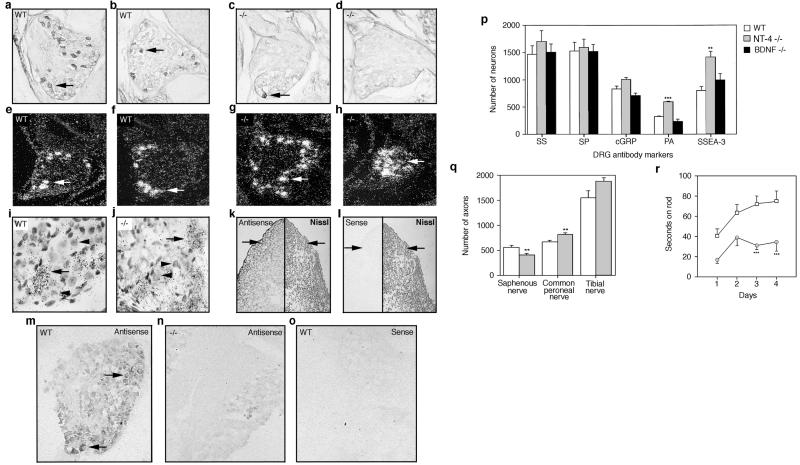
Figure 3.
Anti-TrkB antibody staining (arrows) of neonatal DRG in wt (a and b) and NT-4 mutant (c and d) mice. Dark-field view of TrkC mRNA expression in neurons (arrows) of neonatal DRG in wt (e and f) and NT-4 mutant (g and h) mice. (i and j) Higher, bright-field magnification of g and h. (i and j) Arrows indicate “positive,” TrkC-expressing neurons whereas arrowheads indicate “negative,” TrkC-expressing neurons. Digoxigenin-labeled NT-4-specific antisense and sense probes (k– o) were hybridized to E10 embryo sections of P0, wt, and NT-4 mutant DRG. (k and l) Transverse thoracic section of E10 embryo showing hybridized (Left) and Nissl-stained tissue (Right). Arrows point to region of forming DRG. NT-4 mRNA expression in neonatal wt (m and o) and NT-4 mutant (n) DRG using antisense (m and n) and sense (o) probes. (p) DRG sections from neonatal wt, NT-4, and BDNF mutant mice were immunostained with antibodies for somatostatin (SS), substance P (SP), calcitonin gene-related protein (cGRP), parvalbumin (PA), and SSEA-3. (q) Number of myelinated axons innervating cutaneous tissue (saphenous nerve) and muscle (common peroneal and tibial nerves) in wt and NT-4 mutant mice. (r) Coordination of wt and NT-4 mutant on a rotating rod over 4 consecutive days.
The preceding results in NT-4 and BDNF mutants are most consistent with a function for NT-4 in early sensory gangliogenesis. NT-4 is expressed throughout the E10 embryo (Fig. 3k Left), including the cells that give rise to the DRG (Fig. 3k, arrow), myotome premuscle mass, and the developing mantle and ependymal layers of the spinal cord (Fig. 3k). No signal was observed with a sense probe (Fig. 3l Left). As further controls, the sense and antisense NT-4 probes were hybridized to P0 DRG (Fig. 3 m–o). Antisense probe hybridized only to wt DRG including within discernible neurons (Fig. 3m, arrow). Sense probe again afforded no signal (Fig. 3o). Thus, NT-4 is expressed at the appropriate location and timing to exert an effect on DRG neuronal precursors.
Altered Neuronal Markers in NT4 −/− DRG.
For DRG, definitive markers that specifically identify mature somatosensory neuronal subclasses do not exist currently. However, the use of several antibodies permits discrimination of subsets of neurons (20, 34, 35). NGF-responsive DRG neurons transmit nociceptive information and express somatostatin (SS), substance P (SP), and calcitonin gene-related protein (cGRP). In contrast, large soma neurons that transmit mechanoreceptive and proprioceptive information are either BDNF- and/or NT-3-dependent and express parvalbumin (PA), SSEA-3, and, to a small extent, cGRP. We examined the relative proportions of SS, SP, cGRP, PA, and SSEA-3 in wt, NT-4, and BDNF mutant DRG (Fig. 2p). These data confirm that nociceptive (TrkA), SS-, and SP-expressing neurons remain unchanged in BDNF and NT-4 ganglia. In contrast, NT-4 mutant DRG have increased PA- and SSEA-3-positive neurons, which is most consistent with an increased presence of NT-3-dependent large soma neurons (Fig. 2p).
Altered Sensory Afferents in NT-4 Mutants.
NT-3-dependent DRG neurons innervate cutaneous tissue with mechanoreceptors (saphenous nerve) and skeletal muscle with proprioceptors (common peroneal and tibial nerves) via fast-conducting myelinated axons. To examine whether the increase in NT-3-dependent large soma neurons in NT-4 mutant DRG was reflected in target innervation, the saphenous, common peroneal, and tibial nerves were excised and the myelinated fibers were counted (Fig. 3q). Consistent with previous reports (36), the number of myelinated axons was reduced (16%) in the saphenous nerve of NT-4 mutants as compared with wt littermates (Fig. 3q). Examination of the common peroneal and the tibial nerves yielded different results. The number of myelinated tibial nerve axons in NT-4 mutants increased 19% over wt, and an 18% increase was observed in the common peroneal nerve (Fig. 3q). These results support the model that the increase of NT-3-dependent neurons seen in NT-4 mutant DRG represents an increase in proprioceptive muscle afferents and not mechanoreceptive cutaneous neurons. In light of our preceding data, the loss in saphenous axons seen in NT-4 mutants (Fig. 3q; ref. 36) likely reflects loss of BDNF-dependent mechanoreceptors that normally form a component of this nerve (37, 38).
We also examined cutaneous mechanoreceptor targets of NT-3-dependent neurons in wt, BDNF, and NT-4 mutants. The mystical pad has a specific organization of mechanoreceptors that are dependent only on NT-3 and not on BDNF (22, 23, 39). In particular, Merkel cell–neurite complexes (MCNC) terminate on the outer-root sheath of the vibrissal follicle sinus complex (VFSC), and neurons expressing these specialized endings are dependent on NT-3 for survival and development of the sensory ending. We employed anti-PGP 9.5 immunofluorescence to identify Merkel innervation of the VFSC in wt, NT-4, and BDNF mutants. We found no significant difference in the numbers or distribution of MCNC between wt, NT-4, and BDNF mutants (not shown). These results support further the model that NT-4 mutants have no increased number of cutaneous mechanoreceptors.
Reduced Coordination in NT4 −/− Mice.
Somatosensory proprioception is a higher-order function that integrates the relay of information from muscle directly through reflex arcs at the spinal cord and also to the brain via ascending spinal projections. The role of NT-3 in development and survival of proprioceptive sensory neurons has been well established (2, 3, 5, 14). To investigate further whether the observed increase in common peroneal and tibial axons in NT-4 mutant mice affects target muscle innervation and proprioceptive coordination, these mice were evaluated for their ability to maintain balance on a rotating rod. Throughout the study, NT-4 mutants were impaired as compared with their wt littermates (Fig. 3r). Because the NT-4 mutants exhibited a learning curve parallel to controls, we interpret the results to indicate that the reduced ability to balance on the rod was not directly related to learning impairment. Alterations in sensory modality with increased proprioceptive input and decreased mechanoreception likely would result in incorrect wiring and aberrant motility and/or balance. We examined muscle fiber composition in the NT-4 mutants and observed no differences compared with wt (not shown); thus, muscle malfunction cannot explain the observed loss of balance (24). The formal possibility that NT-4 mutants may have alterations in motor neuron conductivity, neurotransmitter release, and other centers that affect coordination (i.e., motor cortex and cerebellum) is not ruled out by the present study. However, we have not observed defects in central nervous system motor centers, and studies by others on independently derived NT-4 mutant mice have not reported such defects.
Discussion
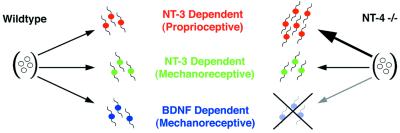
The Neurotrophic Hypothesis implicates target-derived NTFs in the sculpting of the peripheral nervous system through competition by NTF-dependent neurons for limiting amounts of trophic factors (40, 41). Recent studies have also implicated NTFs earlier in the development of sensory ganglia, suggesting roles for NTFs in cell proliferation (2, 5, 12, 13, 25, 42–44). Thus, considerable insight has been gained about the differentiation and death of sensory neurons, but less is known about the mechanisms that regulate cell fate and determination in these cells. The present study indicates a regulatory function for NT-4 in the differentiation pathway of DRG precursors for BDNF- and NT-3-dependent neurons. NT-4 is a required upstream effector for the normal development of BDNF-dependent mechanoreceptors (Fig. 4). Because migratory neural crest cells (NCC) are pluripotent, it is unlikely that NT-4 functions before gangliogenesis. However, TrkB activation can induce pluripotent NCC to differentiate along the primary sensory neuron lineage (10, 11) and would suggest that in vivo, NT-4 may function to prime postmigratory NCC.
Figure 4.
Schematic diagram for model of NT-4 function in early gangliogenesis to direct precursor cells along a BDNF-dependent lineage. In the absence of NT-4, precursors continue to expand and ultimately differentiate along the NT-3-dependent proprioceptive pathway.
We presume that NT-4 action in early sensory gangliogenesis is mediated through the TrkB receptor. One prediction arising from our model would be that TrkB knockouts should exhibit the same DRG phenotype as NT-4 knockouts (no neuron loss) rather than BDNF knockouts (>30% neuron loss). Richard Smeyne (personal communication) has examined DRG efferents in TrkB knockout mice (45) and detects no afferent loss. Comparative examination of BDNF, NT-4, and TrkB mutant mice will provide further insight into the mechanism of BDNF-dependent sensory neuron differentiation.
Acknowledgments
We thank Dr. Louis Reichardt for kindly providing the TrkB antibody and Dr. Richard Smeyne for communicating unpublished results. We thank the members of the Parada Lab for double-blind cell counts and for helpful discussions and past members of the Parada Lab (S. W. Reid, J. Flynn, M. Mazzula, and M. E. Palko) for assistance in the generation of the knockout mice. We thank Regeneron Pharmaceuticals for providing NTFs. This work was supported by National Institutes of Health Grant R01 NS33199 (to L.F.P.). L.J.K. was partially supported by National Institutes of Health Training Grant T32GM08203.
Abbreviations
- NTF
neurotrophin
- NT-3 and NT-4
neurotrophin-3 and -4, respectively
- BDNF
brain-derived neurotrophic factor
- NGF
nerve growth factor
- E
embryonic day
- DRG
dorsal root ganglion
- wt
wild type
- H&E
hematoxylin/eosin
- TdT
terminal deoxynucleotidyltransferase
- TUNEL
TdT-mediated dUTP-biotin nick end labeling
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.040562597.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.040562597
References
- 1.Snider W D. Cell. 1994;77:627–638. doi: 10.1016/0092-8674(94)90048-5. [DOI] [PubMed] [Google Scholar]
- 2.Tessarollo L, Vogel K S, Palko M E, Reid S W, Parada L F. Proc Natl Acad Sci USA. 1994;91:11844–11848. doi: 10.1073/pnas.91.25.11844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ernfors P, Lee K F, Kucera J, Jaenisch R. Cell. 1994;77:503–512. doi: 10.1016/0092-8674(94)90213-5. [DOI] [PubMed] [Google Scholar]
- 4.Farinas I, Jones K R, Backus C, Wang X Y, Reichardt L F. Nature (London) 1994;369:658–661. doi: 10.1038/369658a0. [DOI] [PubMed] [Google Scholar]
- 5.Liebl D J, Tessarollo L, Palko M E, Parada L F. J Neurosci. 1997;17:9113–9121. doi: 10.1523/JNEUROSCI.17-23-09113.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farinas I, Wilkinson G A, Backus C, Reichardt L F, Patapoutian A. Neuron. 1998;21:325–334. doi: 10.1016/s0896-6273(00)80542-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tessarollo L, Tsoulfas P, Donovan M J, Palko M E, Blair-Flynn J, Hempstead B L, Parada L F. Proc Natl Acad Sci USA. 1997;94:14776–14781. doi: 10.1073/pnas.94.26.14776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalcheim C, Carmeli C, Rosenthal A. Proc Natl Acad Sci USA. 1992;89:1661–1665. doi: 10.1073/pnas.89.5.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pinco O, Carmeli C, Rosenthal A, Kalcheim C. J Neurobiol. 1993;24:1626–1641. doi: 10.1002/neu.480241207. [DOI] [PubMed] [Google Scholar]
- 10.Sieber-Blum M. Neuron. 1991;6:949–955. doi: 10.1016/0896-6273(91)90235-r. [DOI] [PubMed] [Google Scholar]
- 11.Sieber-Blum M, Ito K, Richardson M K, Langtimm C J, Duff R S. J Neurobiol. 1993;24:173–184. doi: 10.1002/neu.480240205. [DOI] [PubMed] [Google Scholar]
- 12.ElShamy W M, Linnarsson S, Lee K F, Jaenisch R, Ernfors P. Development. 1996;122:491–500. doi: 10.1242/dev.122.2.491. [DOI] [PubMed] [Google Scholar]
- 13.Farinas I, Yoshida C K, Backus C, Reichardt L F. Neuron. 1996;17:1065–1078. doi: 10.1016/s0896-6273(00)80240-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klein R, Silos-Santiago I, Smeyne R J, Lira S A, Brambilla R, Bryant S, Zhang L, Snider W D, Barbacid M. Nature (London) 1994;368:249–251. doi: 10.1038/368249a0. [DOI] [PubMed] [Google Scholar]
- 15.Crowley C, Spencer S D, Nishimura M C, Chen K S, Pitts-Meek S, Armanini M P, Ling L H, MacMahon S B, Shelton D L, Levinson A D, et al. Cell. 1994;76:1001–1011. doi: 10.1016/0092-8674(94)90378-6. [DOI] [PubMed] [Google Scholar]
- 16.Smeyne R J, Klein R, Schnapp A, Long L K, Bryant S, Lewin A, Lira S A, Barbacid M. Nature (London) 1994;368:246–249. doi: 10.1038/368246a0. [DOI] [PubMed] [Google Scholar]
- 17.Davies A M, Horton A, Burton L E, Schmeizer C, Vandlen R, Rosenthal A. J Neurosci. 1993;13:4961–4967. doi: 10.1523/JNEUROSCI.13-11-04961.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conover J C, Erickson J T, Katz D M, Bianchi L M, Poueymirou W T, McClain J, Pan L, Helgren M, Ip N Y, Boland P, et al. Nature (London) 1995;375:235–238. doi: 10.1038/375235a0. [DOI] [PubMed] [Google Scholar]
- 19.Liu X, Ernfors P, Wu H, Jaenisch R. Nature (London) 1995;375:238–241. doi: 10.1038/375238a0. [DOI] [PubMed] [Google Scholar]
- 20.Dodd J, Solter D, Jessell T M. Nature (London) 1984;311:469–472. doi: 10.1038/311469a0. [DOI] [PubMed] [Google Scholar]
- 21.Jessell T M, Dodd J. Philos Trans R Soc London B. 1985;308:271–281. doi: 10.1098/rstb.1985.0027. [DOI] [PubMed] [Google Scholar]
- 22.Fundin B T, Silos-Santiago I, Ernfors P, Fagan A M, Aldskogius H, DeChiara T M, Phillips H S, Barbacid M, Yancopoulos G D, Rice F L. Dev Biol. 1997;190:94–116. doi: 10.1006/dbio.1997.8658. [DOI] [PubMed] [Google Scholar]
- 23.Rice F L, Albers K M, Davis B M, Silos-Santiago I, Wilkinson G A, LeMaster A M, Ernfors P, Smeyne R J, Aldskogius H, Phillips H S, et al. Dev Biol. 1998;198:57–81. [PubMed] [Google Scholar]
- 24.Brooke M H, Kaiser K K. Arch Neurol. 1970;23:369–379. doi: 10.1001/archneur.1970.00480280083010. [DOI] [PubMed] [Google Scholar]
- 25.Birren S J, Lo L, Anderson D J. Development. 1993;119:597–610. doi: 10.1242/dev.119.3.597. [DOI] [PubMed] [Google Scholar]
- 26.Martin-Zanca D, Barbacid M, Parada L F. Genes Dev. 1990;4:683–694. doi: 10.1101/gad.4.5.683. [DOI] [PubMed] [Google Scholar]
- 27.Harland R M. In situ Hybridization: An Improved Whole Mount Method for Xenopus Embryos. New York: Academic; 1991. [DOI] [PubMed] [Google Scholar]
- 28.Klesse L J, Parada L F. J Neurosci. 1998;18:10420–10428. doi: 10.1523/JNEUROSCI.18-24-10420.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barbacid M. J Neurobiol. 1994;25:1386–1403. doi: 10.1002/neu.480251107. [DOI] [PubMed] [Google Scholar]
- 30.Davies A M. Curr Biol. 1994;4:273–276. doi: 10.1016/s0960-9822(00)00064-6. [DOI] [PubMed] [Google Scholar]
- 31.Ip N Y, Yancopoulos G D. Ann Neurol. 1994;35,Suppl.:S13–S16. doi: 10.1002/ana.410350706. [DOI] [PubMed] [Google Scholar]
- 32.Ernfors P, Lee K F, Jaenisch R. Nature (London) 1994;368:147–150. doi: 10.1038/368147a0. [DOI] [PubMed] [Google Scholar]
- 33.Jones K R, Farinas I, Backus C, Reichardt L F. Cell. 1994;76:989–999. doi: 10.1016/0092-8674(94)90377-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carr P A, Nagy J I. Brain Res Bull. 1993;30:209–219. doi: 10.1016/0361-9230(93)90246-8. [DOI] [PubMed] [Google Scholar]
- 35.Snider W D, Wright D E. Neuron. 1996;16(2):229–232. doi: 10.1016/s0896-6273(00)80039-2. [DOI] [PubMed] [Google Scholar]
- 36.Stucky C L, DeChiara T, Lindsay R M, Yancopoulos G D, Koltzenburg M. J Neurosci. 1998;18:7040–7046. doi: 10.1523/JNEUROSCI.18-17-07040.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McMahon S B, Armanini M P, Ling L H, Phillips H S. Neuron. 1994;12:1161–1171. doi: 10.1016/0896-6273(94)90323-9. [DOI] [PubMed] [Google Scholar]
- 38.Wright D E, Snider W D. J Comp Neurol. 1995;351:329–338. doi: 10.1002/cne.903510302. [DOI] [PubMed] [Google Scholar]
- 39.Albers K M, Perrone T N, Goodness T P, Jones M E, Green M A, Davis B M. J Cell Biol. 1996;134:487–497. doi: 10.1083/jcb.134.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hamburger V, Yip J W. J Neurosci. 1984;4:767–774. doi: 10.1523/JNEUROSCI.04-03-00767.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barde Y A. Prog Clin Biol Res. 1994;390:45–56. [PubMed] [Google Scholar]
- 42.Buchman V L, Davies A M. Development. 1993;118:989–1001. doi: 10.1242/dev.118.3.989. [DOI] [PubMed] [Google Scholar]
- 43.Snider W D, Silos-Santiago I. Philos Trans R Soc London B. 1996;351:395–403. doi: 10.1098/rstb.1996.0034. [DOI] [PubMed] [Google Scholar]
- 44.Lefcort F, Clary D O, Rusoff A C, Reichardt L F. J Neurosci. 1996;16:3704–3713. doi: 10.1523/JNEUROSCI.16-11-03704.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klein R, Smeyne R J, Wurst W, Long L K, Auerbach B A, Joyner A L, Barbacid M. Cell. 1993;75:113–122. [PubMed] [Google Scholar]