Abstract
The nonstructural NSm protein of tomato spotted wilt tospovirus (TSWV) represents a putative viral movement protein involved in cell-to-cell movement of nonenveloped ribonucleocapsid structures. To study the molecular basis of NSm function, we expressed the protein in Escherichia coli and investigated protein–protein and protein–RNA interactions of NSm protein in vitro. NSm specifically interacts with TSWV N protein and binds single-stranded RNA in a sequence-nonspecific manner. Using NSm as a bait in a yeast two-hybrid screen, we identified two homologous NSm-binding proteins of the DnaJ family from Nicotiana tabacum and Arabidopsis thaliana.
Tomato spotted wilt tospovirus (TSWV) is a member of the plant-infecting tospovirus genus of the arthropod-borne Bunyaviridae family. Tospoviruses are severe plant pathogens with a broad host range and are transmitted by thrips as vectors. An estimated annual crop loss of over one billion United States dollars puts tospoviruses among the 10 most detrimental plant viruses worldwide (1, 2). The tripartite genome of TSWV, using a negative/ambisense coding strategy, contains five ORFs. The L protein, encoded by the L RNA, represents the putative viral RNA-dependent RNA polymerase, an essential component of virions and nucleocapsids (3–5). The S RNA encodes the nonstructural protein NSs of unknown function and the N protein, the main constituent of the nucleocapsid, which is known to bind RNA and to form homooligomeric structures (6–8). The spike glycoproteins G1 and G2 and the putative movement protein NSm are encoded by the ambisense M RNA (9).
Plant viruses have to overcome the barrier of cellulose-containing cell walls to establish a successful systemic infection of the plant host. Specialized viral movement proteins evolved that facilitate transport of infectious material through plasmodesmata, the intercellular connections of plant cells (10–12). Recent findings suggest that these proteins, which recognize and transport the viral genomes as naked nucleic acids or in complex with other viral proteins, resemble plant proteins that are involved in selective trafficking of protein and protein–nucleic acid complexes through plasmodesmata as part of fundamental transport and signaling processes (13, 14).
Tospoviruses are RNA viruses with a segmented single-stranded genome using negative and ambisense coding strategies. Thus, the RNA-dependent RNA polymerase has to be cotransported with the viral RNA to allow transcription and replication within newly infected cells. A model proposed for TSWV movement predicts a tubule-guided mechanism to transport nucleocapsids, infectious subviral particles of viral genomic RNA complexed with nucleocapsid protein (N), and the viral polymerase (15). Involvement of tubular structures has been described for como-, nepo-, and caulimoviruses (16–22). The NSm protein shows some typical characteristics of viral movement proteins such as expression only at early stages of infection, intracellular localization of NSm close to plasmodesmata and nucleocapsid associations in the cytoplasm, and a tubule-forming capacity (15, 23).
Thus far, little is known about molecular properties of NSm that enable it to specifically recognize and transport nucleocapsids and to interact with factors of the host cell. Here we characterize the role played by the NSm protein with respect to protein–RNA and protein–protein interactions in vitro. Using NSm as a bait in the yeast two-hybrid system, we identify host proteins that may be involved in the TSWV movement process by interacting with NSm in plants.
Materials and Methods
Expression and Purification of Recombinant Proteins.
The NSm gene [TSWV isolate L3 (24)] was cloned into the vector pET-28a (Novagen) and expressed with an N-terminal (His)6-fusion in E. coli HMS174(DE3). To enhance the NSm translation, the argU gene encoding a rare tRNAarg4 [(25); S. Leclair, personal communication] was coexpressed in Escherichia coli. Cells were lysed in 8 M urea/300 mM NaCl/50 mM Tris⋅HCl, pH 8.0, and the protein was purified by affinity chromatography on Ni-nitrilotriacetate agarose (Qiagen) according to the manufacturer's manual. Apparent homogeneity of the eluted protein was evaluated by SDS/PAGE, and these samples were dialyzed against 300 mM NaCl/50 mM Tris⋅HCl, pH 8.0. (His)6-tagged TSWV N protein was expressed and purified as described previously (8).
Retardation Gel Electrophoresis.
PCR products containing 198 bp from the 5′- and the 3′-ends of TSWV S RNA, respectively, and a T7 promoter introduced by the respective PCR primers were transcribed in vitro by using T7 RNA polymerase (Roche Molecular Biochemicals) and [α-33P]UTP (Amersham Pharmacia Biotech). Primers: 5′-TAATACGACTCACTATAAGAGCAATTGTGTCAGAATTTTGTTCATAATCA-3′ and 5′-GTACCAAGTTCATGAAT-3′ for the 5′-end and 5′-TAATACGACTCACTATAGGGCAAAGCAACAATGCTTTCCTTAGTGAGCTT-3′ and 5′-AGAGCAATCGTGTCAATTTTGTGTTCATACCTT-3′ for the 3′-end. The contaminating cDNA templates were removed by digestion with RNase free DNase (Roche Molecular Biochemicals). Transcripts were purified by gel filtration (G-50 Nick Columns, Amersham Pharmacia Biotech) and checked on sequencing gels. A control transcript was generated by using a cDNA of barley yellow mosaic bymovirus (BaYMV) carrying a region between nucleotides 3050 and 3222 of RNA 2 (26). To generate dsRNA, transcripts were generated from pBluescript KS and SK vectors (Stratagene), which contain a T7 promotor site and had been digested with KpnI or SacI, respectively. To obtain double-stranded RNA, the transcripts were combined in equal amounts. The RNA transcripts were combined with purified (His)6-tagged NSm in a total volume of 10 μl of 1× TBP buffer (90 mM Tris base/90 mM boric acid/150 mM KCl, pH 8.0), incubated on ice for 15 min, and separated by electrophoresis on a native 4% polyacrylamide gel in 0.5× TB (90 mM Tris base/90 mM boric acid, pH 8.0). The gels were dried and analyzed by using a Storm 860 beta imager (Molecular Dynamics). Recombinant expressed and purified (His)6-tagged GFP, (His)10-tagged ARF1 from Catharanthus roseus (GenBank accession no. AF005238), and unmodified BSA (New England Biolabs) were used as control proteins. For RNA competition experiments, 16S and 23S ribosomal RNA from E. coli MRE600 (Roche Molecular Biochemicals) were used.
Protein Overlay Assay.
A protein overlay assay was performed according to ref. 27 by using recombinant TSWV N, NSm, and gluthatione S-transferase (GST) from Schistosoma japonicum. Proteins separated on SDS/PAGE were transferred to nitrocellulose membrane and renatured after removal of SDS (28). The membrane was blocked in 20 mM Tris⋅HCl, pH 7.6/1% milk powder and incubated for 1 hr in 20 mM Tris⋅HCl, pH 7.6, containing 10 μg/ml (His)6-tagged N protein that had been purified under native conditions as previously described (8). N protein purified in the stated manner forms homomultimeric structures in vitro in the absence of viral genomic RNA (8). The membrane was extensively washed and incubated overnight in 0.3% glutardialdehyde (Merck) to crosslink the protein complexes. After subsequent washing, the N protein was detected by using specific antibodies (anti-BR01-IgG; Loewe Biochemica, Sauerlach, Germany) in a standard protocol (29). Membranes were stained with Ponceau S (Sigma/Aldrich).
Construction of a Nicotiana tabacum cDNA Library in pAD-GAL4–2.1.
Poly(A)+-RNA from leaf tissues of N. tabacum cv. Samsun was prepared by using the guanidine thiocyanate method (30) and an Oligotex mRNA Kit (Qiagen), and a cDNA library was synthesized with a Stratagene HybriZAP-2.1 cDNA synthesis kit (Stratagene). The library contained 1.55 × 106 recombinant clones with an average insert size of 1.5 kb. The phage library was converted into a plasmid library by in vivo excision (31).
Yeast Two-Hybrid Screen.
A cDNA encoding TSWV NSm protein (24) was cloned in plasmid pAS2 (CLONTECH) to generate a NSm fusion with the GAL4 DNA binding domain in the yeast strain PJ69–4A (32, 33). The N. tabacum cDNA library in plasmid pAD-GAL4–2.1 was transformed into the yeast strain Y187 (34). Colonies were rinsed off the plates and stored in 25% glycerol at −70°C. Aliquots were used to perform exhaustive mating with the PJ69–4A/pAS2-NSm bait described above according to refs. 35 and 36. Zygotes were plated onto synthetic dropout medium lacking leucine, tryptophan, and histidine and supplemented with 5 mM 3-aminotriazole (LWH5) and incubated at 30°C for 5 to 10 days. Growing colonies were tested for β-galactosidase activity (37) followed by preparation of plasmids from positive clones. In control experiments, rescued plasmids were cotransformed with plasmids pAS2, pBD-SNF1 (38), and pBD-p53 (39). Inserts were analyzed by DNA sequencing. 5′ ends of incomplete cDNA clones were obtained by using the Marathon cDNA Amplification Kit (CLONTECH) and Advantage cDNA Polymerase [CLONTECH; (40, 41)].
In Vitro Pull-Down Assay.
The coding region of cDNA from plasmid pAD-NtDnaJ_M541 was cloned as a BamHI-SmaI fragment into the vector pGEX-2T (Amersham Pharmacia Biotech), expressed in E. coli HMS174(DE3), and purified as a GST-fusion protein. The TSWV NSm gene and a luciferase gene used as a control were PCR amplified with primers containing a T7 promoter. PCR products were used as a template for TNT Quick Coupled Transcription/Translation System (Promega) with l[35S]Met (Amersham Pharmacia Biotech) as a label. The protein products were separated on SDS/PAGE and detected by phosphorimaging.
For a GST-pull-down assay (42), gluthatione Sepharose 4B matrix (Amersham Pharmacia Biotech) was loaded with either purified GST-NtDnaJ_M541 fusion protein, or with a control (GST) or was incubated without proteins in 150 mM NaCl/20 mM Tris⋅HCl, pH 7.5/0.1% IGEPAL CA-630 (Sigma). These matrices were incubated with either l[35S]Met NSm protein or with l[35S]Met luciferase as control. After extensive washing, the proteins bound to the beads were denatured by heating, resolved on an SDS/PAGE, and detected by phosphorimaging.
Results
Expression of Recombinant TSWV NSm Protein and Characterization of Its RNA-Binding Capacity.
(His)6-tagged NSm protein was expressed in E. coli, purified to apparent homogeneity, and tested for RNA-binding properties. For this, in vitro transcripts corresponding to the 3′- and 5′-ends of the viral S RNA, respectively (43), were used. A 172-base transcript of RNA 2 of BaYMV (26) was used as a control. TSWV transcripts as well as the control BaYMV RNA formed complexes with the recombinant TSWV NSm protein, resulting in bands of lower electrophoretic mobility (Fig. 1). Although both in vitro transcripts derived from the TSWV RNA bound to NSm in a very similar manner, the affinity of NSm to the control BaYMV transcript appeared to be slightly lower, as concluded from the higher amount of protein needed to achieve a complete retardation of the RNA. Both 3′- and 5′-end probes derived from the TSWV RNA were retarded when 3.4 pmol or more NSm was present in the samples, whereas protein amounts of 1.7 pmol had no effect on the mobility of the RNA. With 6.8 pmol of NSm protein, the band of free RNA was quantitatively shifted to high molecular mass complexes. Increasing the NSm amount to 13.6 pmol did not alter the mobility of these complexes. The presence of 32 pmol of NSm prevented the RNA from entering into the gel, indicating the formation of very high molecular mass complexes (data not shown). Similar results were obtained with the BaYMV RNA transcript. Here, the appearance of bands with lower electrophoretic mobility and a complete shift of the free RNA took place when 6.8 pmol and 13.6 pmol NSm were used, respectively. Again radioactive RNA was hindered from entering into the gel when 32 pmol or more was applied to the samples. When single-stranded and double-stranded transcripts derived from pBluescript were used, binding to single-stranded RNA was observed but no binding to double-stranded RNA was found with the same amounts of NSm (data not shown). As additional controls, (His)6-tagged GFP, (His)10-tagged ARF1, and unmodified BSA in comparable concentrations were tested for their capacity to retard the different RNA transcripts in a gel shift assay. No retardation of electrophoretic mobility of RNA transcripts occurred for either of the proteins. Thus, the observed RNA binding was not caused by unspecific binding of the (His)6-tag or by unspecific protein-binding properties of the RNA transcripts (data not shown).
Figure 1.
Gel retardation assay of RNA–NSm complexes. (His)6-tagged NSm protein was incubated with [α-33P]UTP-labeled RNA, and the RNA-protein complexes were resolved on native gels. The gels were evaluated by phosphorimaging. Free RNA, representing either 198 bases from the 3′-end of TSWV S-RNA (A) or a 172-base fragment from the RNA 2 of BaYMV (B), is marked by arrows. The amounts of NSm protein used in the binding assays are indicated.
Unlabeled E. coli rRNA was used to compete the NSm-RNA binding, and the extent of retardation of radioactive RNA was quantified by phosphorimaging and subsequent densitometric evaluation (Fig. 2). The half titration points of BaYMV and TSWV RNA were determined by using logarithmic fit and were found to be of the same order of magnitude (about 10 and 16 rRNA/[α-33P]UTP-labeled RNA, respectively), exhibiting again a slightly lower affinity of NSm to the BaYMV RNA transcript.
Figure 2.
Competition of the RNA–NSm complex with nonlabeled RNA. (A) In vitro-transcribed and [α-33P]UTP-labeled RNA was incubated with 13.6 pmol TSWV NSm protein and subsequently competed with E. coli rRNA. RNA–protein complexes were visualized by phosphorimaging. (B) Extent of RNA retardation was determined by densitometric quantitation of the free RNA in each lane in comparison to the free RNA in lane 1 (indicated with an arrow). Lanes 1–9: 1.6 ng of [α-33P]UTP-labeled RNA; lanes 2–9, 13.6 pmol (His)6-tagged NSm protein; amounts of E. coli rRNA added to the samples were in lane 3: 8 ng; lane 4: 16 ng; lane 5: 32 ng; lane 6: 64 ng; lane 7: 128 ng; lane 8: 256 ng; lane 9: 512 ng.
Interaction of NSm with the Nucleocapsid Protein N.
Purified (His)6-tagged NSm, crude extract of E. coli expressing (His)6-tagged NSm, purified GST protein, and purified (His)6-tagged N protein were separated by SDS/PAGE, blotted onto membrane, and probed with (His)6-N protein. The protein complexes were crosslinked and subsequently probed with anti-N antibodies (Fig. 3A). Position of the proteins was confirmed by staining the membrane with Ponceau S (Fig. 3B). The TSWV N protein was detected as a discrete band in the sample containing the purified NSm protein, as well as in lanes containing a crude extract from E. coli expressing the (His)6-tagged NSm. By comparison with bands on the stained membrane, these N signals colocalized with the band of (His)6-tagged NSm. Neither other proteins of the crude E. coli lysate nor the control GST protein interacted with the N protein in the overlay assay. To exclude possible crossreactivity of the anti-N antibody with NSm, the same assay was performed without the N protein in the overlay buffer. No crossreactions of the antibodies were observed (data not shown).
Figure 3.
Protein overlay assay. (A) Proteins were separated on SDS/PAGE, blotted on poly(vinylidene difluoride) membrane, and probed with a (His)6-tagged TSWV N protein. After extensive washing, the protein complexes were crosslinked with 0.3% glutaraldehyde. TSWV N protein bound to the membrane was detected by using specific antibodies. Lane 1: purified GST-protein (27 kDa) used as a negative control; lane 2: extract from a culture of E. coli HMS174(DE3) expressing a (His)6-tagged NSm protein; lane 3: purified (His)6-tagged NSm protein (35.7 kDa); lane 4: (His)6-tagged TSWV N protein (31.1 kDa). (B) Proteins were visualized by staining with Ponceau S. Lanes as described in A.
Yeast Two-Hybrid Interaction Screen Using TSWV NSm Protein as a Bait.
Using an interaction mating procedure, 8.1 × 106 zygotes containing the NSm protein as a bait and a GAL4-activation domain fused cDNA-library from N. tabacum leaf material were screened. Interaction was monitored by histidine prototrophic growth and β-galactosidase activity. Plasmids were rescued and verified by double transformation into yeast in combination with the original bait plasmid pBD-NSm, control bait plasmids, and empty vectors (44). Two identical clones (designated NtDnaJ_M541 and M551) were found to interact specifically with NSm fused to the GAL4-DNA-binding domain (Fig. 4). They encoded a region of C-terminal 240 aa of a DnaJ-like protein, which showed the highest homology to Arabadopsis thaliana genes (GenBank accession nos. AC005489, AC010871, AC007258, AC007109, AL021749, and AL049658). A full-length clone was obtained by 5′-RACE with gene-specific primers on a cDNA adapter ligated RACE library from N. tabacum leaves (GenBank accession no. of the full-length sequence: AF191497).
Figure 4.
Protein interactions of NtDnaJ_M541 in the yeast two-hybrid system. Yeast cells transformed with bait and prey vectors were plated on selective medium lacking leucine, tryptophan, and histidine and supplemented with 5 mM 3-aminotriazole. Empty vector constructs and a combination of SNF1/SNF4 were used as negative and positive controls, respectively (A). Aliquots containing 107 cells of the respective colonies were spotted on a filter and tested for β-galactosidase activity (B). Combinations of transformed plasmid: 1, pBD-NSm/pAD-NtDnaJ_M541; 2, pAS2/pAD-NtDnaJ_M541; 3, pBD-SNF1/pAD-NtDnaJ_M541; 4, pBD-SNF1/pAD-SNF4; 5, pAS2/pACT2.
In a similar approach, we used a pACT2 cDNA library from A. thaliana (45) and screened 6 × 106 yeast clones for interaction with the NSm bait construct. A clone (denoted AtA39) could be identified to interact specifically with TSWV NSm, revealing no unspecific binding to unrelated proteins. AtA39 was found to be identical with one of the Arabidopsis orthologs [GenBank accession no. AL021749 (46)] to NtDnaJ_M541. The partial cDNA of AtA39 encoded 314 aa with 70% identity (78% similarity) to NtDnaJ_M541 (Fig. 5).
Figure 5.
N. tabacum and A. thaliana DnaJ-like proteins. Multiple sequence alignment by using pileup (Wisconsin Package Ver. 9.1, Genetics Computer Group). NtDnaJ_M541, N. tabacum DnaJ-like protein (GenBank accession no. AF191497), AtA39, A. thaliana DnaJ-like protein, (GenBank accession no. AL021749). For both NtDnaJ_M541 and AtA39, amino acids identified to interact in the two-hybrid screens are in bold letters. The DnaJ domain is depicted, and dots indicate gaps.
In Vitro Binding of a N. tabacum DnaJ-Like Protein NtDnaJ_M541 to TSWV NSm.
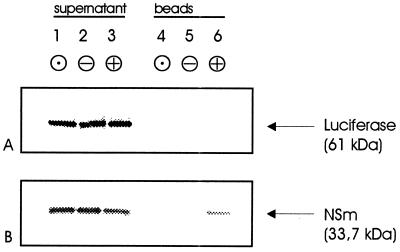
The specificity of the NSm-NtDnaJ_M541 protein interaction was tested by using a GST-pull-down assay (42). A GST fusion of NtDnaJ_M541 protein was expressed in E. coli, purified to homogeneity, and bound to gluthatione-Sepharose beads. The beads were incubated with l[35S]Met-labeled NSm protein, extensively washed, and analyzed by SDS/PAGE. As negative controls, unloaded and GST-loaded beads were used. To investigate possible unspecific binding activities of GST-NtDnaJ_M541, l[35S]Met-labeled luciferase was assayed in the same way.
The l[35S]Met-labeled proteins in the supernatant were monitored after separation on SDS/PAGE (Fig. 6, lanes 1–3). Matrix-bound l[35S]Met-labeled protein could be detected only in beads loaded with GST-NtDnaJ_M541 and l[35S]Met-NSm (Fig. 6, lanes 4–6). No binding to unloaded or GST-loaded beads was observed, and no l[35S]Met luciferase was found to bind in any of the samples, demonstrating the specificity of in vitro binding of NSm to NtDnaJ_M541.
Figure 6.
In vitro binding of N. tabacum DnaJ-like protein NtDnaJ_M541 to TSWV NSm. Gluthatione beads were loaded with purified GST-NtDnaJ_M541 protein (⊕), with GST (⊝), or were incubated only with buffer (⊙). The beads were used to test interaction with in vitro-transcribed and -translated l[35S]Met-labeled proteins. l[35S]Met luciferase (A) and l[35S]Met NSm (B) were incubated with the beads, which were then extensively washed. The bound proteins were eluted by denaturing, separated on an SDS/PAGE, and the labeled proteins were detected by using a phosphorimager. Lanes 1–3: supernatants after incubation with l[35S]Met-labeled proteins; lane 4–6: matrix-bound proteins eluted by denaturation.
Discussion
Plant viruses achieve systemic spread by cell-to-cell movement of viral infectious material through plasmodesmata, followed by long-distance transport through the vascular system. For this cell-to-cell movement, they encode specialized movement proteins and develop different movement strategies (reviewed in refs. 12, 47).
One mechanism, which is used by tobamoviruses and dianthoviruses, uses the transport of viral genomic RNA complexed with a movement protein through plasmodesmata that do not undergo extensive structural modifications (48–50). Another mechanism involves the movement of viral genomes in the form of complete virions along the newly formed tubular structures, as shown for comoviruses, caulimoviruses, and nepoviruses (16–22).
Several molecular features of movement proteins of positive sense RNA viruses have been established, including RNA-binding capacities, association with the cytoskeleton, and ability to form dimers (51–53). The TSWV NSm protein has been proposed to be a viral movement protein, because the NSm gene is expressed transiently in infected plants during a short early period of systemic infection (23). Immunogold labeling shows an association of NSm protein with nucleocapsid aggregates and plasmodesmata of infected plant cells (23). NSm forms tubular structures emerging from the cell surface of plant protoplasts and penetrating plasmodesmata in infected plant tissues (15). On the basis of these data, TSWV movement fits best to the tubule model (15, 23).
To investigate the molecular basis of NSm function, we cloned and expressed the NSm gene in E. coli. The results presented in this study provide evidence for multiple in vitro molecular interactions of the TSWV NSm protein. Gel mobility shift assays, by using purified NSm protein and radioactively labeled in vitro transcribed RNA, revealed a sequence-unspecific ssRNA-binding property of NSm (Fig. 1). In protein–RNA binding experiments by using unlabeled E. coli rRNA as a competitor, the bound RNA was effectively superseded from the complex (Fig. 2). Because TSWV genomic RNA is thought to form so-called panhandle structures (1), the binding of NSm to a model RNA substrate has been investigated. This model substrate resembles the precise ends of the 5′ (96 bases) and 3′ (96 bases) terminal sequences of TSWV S RNA linked by six artificial bases and forms stable secondary structure in vitro, indicating a panhandle structure (43). Here, similar reversible binding of NSm was found (T.-R.S., unpublished results). The question whether the panhandle secondary structure of TSWV genomic RNA is involved in specific binding to NSm awaits further investigation.
Similar competition assays by using purified TSWV N protein have revealed a virtually irreversible RNA binding of this protein (7). The fact that NSm binds RNA much less efficiently suggests a transient NSm–RNA interaction during the transport process.
The RNA-binding capability is a common characteristic of movement proteins of positive sense RNA viruses, irrespective of whether they move naked genomic RNA or virions (51, 53–55). Our results provide evidence that the movement protein of a negative-sense RNA virus may have similar properties.
A prerequisite for tubule-guided movement of the TSWV nucleocapsid is a specific recognition of its nucleocapsid components by NSm. Our results from gel-overlay assays (Fig. 3) suggest that this recognition is provided by an interaction of NSm with the N protein, the main constituent of the nucleocapsid. This observation supports data indicating that NSm colocalizes with nucleocapsid (23).
In summary, our data define two molecular properties of the TSWV NSm protein: interaction with the N protein and with ssRNA. These data thus support a tubule-guided mechanism of TSWV nucleocapsid movement proposed by ref. 15.
To answer the question concerning the involvement of host factors, we performed a yeast two-hybrid screen by using NSm as a bait that resulted in the isolation of DnaJ-homologues from both N. tabacum and A. thaliana (Fig. 4). The interaction in yeast was confirmed by a biochemical assay in vitro by using a GST fusion of tobacco DnaJ (NtDnaJ_M541) expressed in E. coli to pull down a radioactively labeled NSm protein transcribed and translated in vitro (Fig. 6).
Sequence alignment showed a high degree of homology between the two DnaJ-homologues interacting with NSm (Fig. 5). Computation of a phylogenetic tree omitting the J-domain shows that they, together with four additional DnaJ-like sequences found in A. thaliana, form a distinct group, more closely related to certain bacterial, fungal, and mammalian DnaJ proteins than to other plant DnaJ proteins.
Proteins of the DnaJ family are characterized by an N-terminal J-domain. In the best described subgroup of this family, the Hsp-40 family, including the E. coli DnaJ protein, four domains were characterized: an N-terminal J-domain, a glycine- and phenylalanine-rich region, a cysteine-rich zinc-finger domain, and a less well conserved C-terminal domain that is thought to be involved in substrate binding. The NSm interacting DnaJ homologues from A. thaliana and N. tabacum belong to a subclass of the DnaJ family that only contain the J-domain. Proteins of this subclass have been implicated to take part in diverse cellular processes, including protein translocation into mammalian endoplasmic reticulum (56), mitochondrial protein import (57), protein import into plant peroxisomes (58), and microtubule formation (59). Generally, DnaJ proteins are key regulators of the Hsp-70 chaperone. The DnaJ-domain binds to Hsp-70, changing its conformation and stimulating ATP hydrolysis (reviewed in ref. 60).
The specific interaction between NSm and proteins from the DnaJ family suggests a possible involvement of a Hsp-70-dependent mechanism in the TSWV movement. Interestingly, Hsp-70 activity has been implicated in viral movement of BYV, a member of the Closteroviridae. A Hsp-70 homolog encoded by the viral genome was shown to escort virions to their destination in infected cells and to participate in the intercellular translocation of BYV (61). In complementation assays, the BYV-encoded Hsp-70 homolog can substitute potexvirus or hordeivirus movement proteins, providing a direct evidence for its role in cell-to-cell movement (62). An additional feature of the BYV Hsp-70 protein is its ability to bind to microtubules (63). The cytoskeleton is thought to be involved in viral intracellular and possibly intercellular movement. Besides a direct association between a viral movement protein and cytoskeleton, which is shown for the TMV P30 protein (52, 64), an indirect contact via a Hsp-70 protein, either as an intrinsic viral component or recruited by viral structures, is also conceivable. The fact that Hsp-70 has been detected in phloem exudates (65) indicates that it might have some intrinsic properties that facilitate its intercellular transport through plasmodesmata. This leads to speculation that the protein itself may be involved in transport processes. In the case of protein import to mitochondria, Hsp-70 in fact acts as a molecular motor providing a driving force for the translocation process (66).
The transport of RNA–protein complexes through plasmodesmata involves major structural modifications on the side of plasmodesmata by changing the size exclusion limit and on the side of transported complexes by changing their size via disassembly or unfolding. Therefore, it seems reasonable to discuss an Hsp-70-dependent mechanism. Hsp-70 may be recruited via the NSm interacting DnaJ protein to the TSWV transport complex.
Summarizing, interaction of NSm with the TSWV N protein and RNA may provide a molecular basis for specific recognition of nucleocapsid structures, and the interaction with a plant DnaJ protein could link the viral structures to elements of a plant machinery directing intercellular transport through plasmodesmata.
Acknowledgments
We gratefully acknowledge Jeff Schell for his constant support and for helpful discussions. We thank Christoph Minke for providing cDNA clones for the 5′- and 3′-end of the TSWV S RNA. Edith Oehmen, BAYER AG, Monheim, Germany, provided the N. tabacum 5′-RACE-library, and the pACT2 cDNA library from A. thaliana was donated by Klaus Salchert and Csaba Koncz (Max Plancke Institut, Cologne), who also carefully read this manuscript. Many thanks to both of them. The cDNA of the RNA 2 of BaYMV and the E. coli argU gene encoding tRNAarg4 were provided by Hans-Henning Steinbiss and Stephane Leclair (Max Planck Institute, Cologne). Klaus Richter contributed to the construction of the GAL4 activation domain fusion cDNA library during a practical course in our lab. We gratefully acknowledge Christina Philipp for her contribution as a technician in our laboratory. This work was supported by a Max-Planck-Gesellschaft fellowship to T.-R.S., a fellowship of the BAYER AG, Germany, to J.F.U., a fellowship from the Boehringer Ingelheim Fonds to G.L.B., and a grant of the Deutsche Forschungsgemeinschaft (Ke 690/1–1) to J.-W.K.
Abbreviations
- BaYMV
barley yellow mosaic bymovirus
- TSWV
tomato spotted wilt tospovirus
- GST
gluthatione S-transferase
Footnotes
The sequence reported in this paper has been deposited in the GenBank database (accession no. AF191497).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.030548397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.030548397
References
- 1.Goldbach R, Peters D. In: The Bunyaviridae. Elliot R M, editor. New York: Plenum; 1996. pp. 129–157. [Google Scholar]
- 2.Prins M, Goldbach R W. Trends Microbiol. 1998;6:31–35. doi: 10.1016/S0966-842X(97)01173-6. [DOI] [PubMed] [Google Scholar]
- 3.de Haan P, Kormelink R, de Oliveira Resende R, van Poelwijk F, Peters D, Goldbach R W. J Gen Virol. 1991;72:2207–2216. doi: 10.1099/0022-1317-72-9-2207. [DOI] [PubMed] [Google Scholar]
- 4.van Poelwijk F, Boye K, Oosterling R, Peters D, Goldbach R. Virology. 1993;197:468–470. doi: 10.1006/viro.1993.1614. [DOI] [PubMed] [Google Scholar]
- 5.Adkins S, Quadt R, Choi T J, Ahlquist P, German T. Virology. 1995;207:308–311. doi: 10.1006/viro.1995.1083. [DOI] [PubMed] [Google Scholar]
- 6.de Haan P, Wagemakers L, Peters D, Goldbach R W. J Gen Virol. 1990;71:1001–1007. doi: 10.1099/0022-1317-71-5-1001. [DOI] [PubMed] [Google Scholar]
- 7.Richmond K E, Chenault K, Sherwood J L, German T L. Virology. 1998;248:6–11. doi: 10.1006/viro.1998.9223. [DOI] [PubMed] [Google Scholar]
- 8.Uhrig J F, Soellick T-R, Minke C J, Philipp C, Kellmann J-W, Schreier P H. Proc Natl Acad Sci USA. 1999;96:55–60. doi: 10.1073/pnas.96.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kormelink R, de Haan P, Meurs C, Peters D, Goldbach R W. J Gen Virol. 1992;73:2795–2804. doi: 10.1099/0022-1317-73-11-2795. [DOI] [PubMed] [Google Scholar]
- 10.Lucas W J, Gilbertson R L. Annu Rev Pythopathol. 1994;32:387–411. [Google Scholar]
- 11.Ghoshroy S, Lartey R, Sheng J S, Citovsky V. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:25–48. doi: 10.1146/annurev.arplant.48.1.27. [DOI] [PubMed] [Google Scholar]
- 12.Lazarowitz S G. Curr Opin Plant Biol. 1999;2:332–338. doi: 10.1016/S1369-5266(99)80058-2. [DOI] [PubMed] [Google Scholar]
- 13.Lucas W J, Wolf S. Curr Opin Plant Biol. 1999;2:192–197. doi: 10.1016/S1369-5266(99)80035-1. [DOI] [PubMed] [Google Scholar]
- 14.Xoconostle-Cazares B, Yu X, Ruiz-Medrano R, Wang H L, Monzer J, Yoo B C, McFarland K C, Franceschi V R, Lucas W J. Science. 1999;283:94–98. doi: 10.1126/science.283.5398.94. [DOI] [PubMed] [Google Scholar]
- 15.Storms M M H, Kormelink R, Peters D, vanLent J W M, Goldbach R W. Virology. 1995;214:485–493. doi: 10.1006/viro.1995.0059. [DOI] [PubMed] [Google Scholar]
- 16.Linstead P J, Hills G J, Plaskitt K A, Wilson I G, Harker C L, Maule A J. J Gen Virol. 1988;69:1809–1818. [Google Scholar]
- 17.Kasteel D T J, Wellink J, Verver J, vanLent J W M, Goldbach R W, van Kammen A. J Gen Virol. 1993;74:1721–1724. doi: 10.1099/0022-1317-74-8-1721. [DOI] [PubMed] [Google Scholar]
- 18.Wieczorek A, Sanfacon H. Virology. 1993;194:734–742. doi: 10.1006/viro.1993.1314. [DOI] [PubMed] [Google Scholar]
- 19.Ritzenthaler C, Schmit A C, Michler P, Stussi-Garaud C, Pinck L. Mol Plant–Microbe Interact. 1995;8:379–387. [Google Scholar]
- 20.Perbal M C, Thomas C L, Maule A J. Virology. 1993;195:281–285. doi: 10.1006/viro.1993.1375. [DOI] [PubMed] [Google Scholar]
- 21.van Lent J, Wellink J, Goldbach R. J Gen Virol. 1990;71:219–223. [Google Scholar]
- 22.van Lent J, Storms M, van der Meer F, Wellink J, Goldbach R W. J Gen Virol. 1991;72:2615–2623. doi: 10.1099/0022-1317-72-11-2615. [DOI] [PubMed] [Google Scholar]
- 23.Kormelink R, Storms M, Van Lent J, Peters D, Goldbach R. Virology. 1994;200:56–65. doi: 10.1006/viro.1994.1162. [DOI] [PubMed] [Google Scholar]
- 24.Kellmann J W, Schreier P H. Plant Mol Biol. 1996;30:1075. doi: 10.1007/BF00020121. [DOI] [PubMed] [Google Scholar]
- 25.Schenk P M, Baumann S, Mattes R, Steinbiss H H. BioTechniques. 1995;19:196–200. [PubMed] [Google Scholar]
- 26.Reichel C, Maas C, Schulze S, Schell J, Steinbiss H H. J Gen Virol. 1996;77:587–592. doi: 10.1099/0022-1317-77-4-587. [DOI] [PubMed] [Google Scholar]
- 27.Owen D J, Vallis Y, Noble M E M, Hunter J B, Dafforn T R, Evans P R, McMahon H T. Cell. 1999;97:805–815. doi: 10.1016/s0092-8674(00)80791-6. [DOI] [PubMed] [Google Scholar]
- 28.Glenney J R, Jr, Weber K. Methods Enzymol. 1983;102:204–210. doi: 10.1016/s0076-6879(83)02021-2. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 30.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 31.Vekris A. Nucleic Acids Res. 1994;22:4842–4843. doi: 10.1093/nar/22.22.4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.James P, Halladay J, Craig E A. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gietz R D, Schiestl R H. Methods Mol Cell Biol. 1995;5:255–269. [Google Scholar]
- 34.Harper J W, Adami G R, Wie N, Keyomarsi K, Elledge S J. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 35.Bendixen C, Gangloff S, Rothstein R. Nucleic Acids Res. 1994;22:1778–1779. doi: 10.1093/nar/22.9.1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fromont-Racine M, Rain J-C, Legrain P. Nat Genet. 1997;16:277–282. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]
- 37.Bartel P, Fields S. Methods Enzymol. 1995;254:241–263. doi: 10.1016/0076-6879(95)54018-0. [DOI] [PubMed] [Google Scholar]
- 38.Fields S, Song O-K. Nature (London) 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 39.Iwabuchi K, Li B, Bartel P, Fields S. Oncogene. 1993;8:1693–1696. [PubMed] [Google Scholar]
- 40.Frohman M A, Dush M K, Martin G R. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frohman M A. Methods Enyzmol. 1993;218:340–358. doi: 10.1016/0076-6879(93)18026-9. [DOI] [PubMed] [Google Scholar]
- 42.Melcher K, Johnston S A. Mol Cell Biol. 1995;15:2839–2848. doi: 10.1128/mcb.15.5.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Minke C. Ph.D. thesis. Cologne, Germany: Univ. of Cologne; 1998. [Google Scholar]
- 44.Bartel P, Chien C, Sternglanz R, Fields S. BioTechniques. 1993;14:920–924. [PubMed] [Google Scholar]
- 45.Németh K, Salchert K, Putnoky P, Bhalerao R, Koncz-Kálmán Z, Stankovic-Stangeland B, Bakó L, Mathur J, Ökrész L, Stabel S, et al. Genes Dev. 1998;12:3059–3073. doi: 10.1101/gad.12.19.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bevan M, Bancroft I, Bent E, Love K, Goodman H, Dean C, Bergkamp R, Dirkse W, Van Staveren M, Stiekema W, et al. Nature (London) 1998;391:485–488. doi: 10.1038/35140. [DOI] [PubMed] [Google Scholar]
- 47.Carrington J C, Kasschau K D, Mahajan S K, Schaad M C. Plant Cell. 1996;8:1669–1681. doi: 10.1105/tpc.8.10.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deom C M, Lapidot M, Beachy R N. Cell. 1992;69:221–224. doi: 10.1016/0092-8674(92)90403-y. [DOI] [PubMed] [Google Scholar]
- 49.Lucas W J, Ding B, van der Schoot C. New Phytol. 1993;125:435–476. doi: 10.1111/j.1469-8137.1993.tb03897.x. [DOI] [PubMed] [Google Scholar]
- 50.Lazarowitz S G, Beachy R N. Plant Cell. 1999;11:535–554. doi: 10.1105/tpc.11.4.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Citovsky V, Knorr D, Schuster G, Zambryski P. Cell. 1990;60:637–647. doi: 10.1016/0092-8674(90)90667-4. [DOI] [PubMed] [Google Scholar]
- 52.Heinlein M, Epel B L, Padgett H S, Beachy R N. Science. 1995;270:1983–1985. doi: 10.1126/science.270.5244.1983. [DOI] [PubMed] [Google Scholar]
- 53.Jansen K A J, Wolfs J A M, Lohuis H, Goldbach R W, Verduin B J M. Virology. 1998;242:387–394. doi: 10.1006/viro.1997.9000. [DOI] [PubMed] [Google Scholar]
- 54.Kasteel D T J, van der Wel N N, Jansen K A J, Goldbach R W, vanLent J W M. J Gen Virol. 1997a;78:2089–2093. doi: 10.1099/0022-1317-78-8-2089. [DOI] [PubMed] [Google Scholar]
- 55.Osman T A M, Hayes R J, Buck K W. J Gen Virol. 1992;73:223–227. doi: 10.1099/0022-1317-73-2-223. [DOI] [PubMed] [Google Scholar]
- 56.Zimmermann R. Biol Chem. 1998;379:275–282. [PubMed] [Google Scholar]
- 57.Rassow J, Maarse A C, Krainer E, Kubrich M, Muller H, Meijer M, Craig E A, Pfanner N. J Cell Biol. 1994;127:1547–1556. doi: 10.1083/jcb.127.6.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Crookes W J, Olsen L J. J Biol Chem. 1998;273:17236–17242. doi: 10.1074/jbc.273.27.17236. [DOI] [PubMed] [Google Scholar]
- 59.Oka M, Nakai M, Endo T, Lim C R, Kimata Y, Kohno K. J Biol Chem. 1998;273:29727–29737. doi: 10.1074/jbc.273.45.29727. [DOI] [PubMed] [Google Scholar]
- 60.Kelley W L. Trends Biochem Sci. 1998;23:222–227. doi: 10.1016/s0968-0004(98)01215-8. [DOI] [PubMed] [Google Scholar]
- 61.Medina V, Peremyslov V V, Hagiwara Y, Dolja V V. Virology. 1999;260:173–181. doi: 10.1006/viro.1999.9807. [DOI] [PubMed] [Google Scholar]
- 62.Agranovsky A A, Folimonov A S, Folimonova S Y, Morozov S Y, Schiemann J, Lesemann D, Atabekov J G. J Gen Virol. 1998;79:889–895. doi: 10.1099/0022-1317-79-4-889. [DOI] [PubMed] [Google Scholar]
- 63.Karasev A V, Kashina A S, Gelfand V I, Dolja V V. FEBS Lett. 1992;304:12–14. doi: 10.1016/0014-5793(92)80578-5. [DOI] [PubMed] [Google Scholar]
- 64.McLean B G, Zupan J, Zambryski P C. Plant Cell. 1995;7:2101–2114. doi: 10.1105/tpc.7.12.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schobert C, Grossmann P, Gottschalk M, Komor E, Pecsvaradi A, Zurnieden U. Planta. 1995;196:205–210. [Google Scholar]
- 66.Voisine C, Craig E A, Zufall N, von Ahsen O, Pfanner N, Voos W. Cell. 1999;97:565–574. doi: 10.1016/s0092-8674(00)80768-0. [DOI] [PubMed] [Google Scholar]