Abstract
Cellulose is a major component of the extracellular matrices formed during development of the social amoeba, Dictyostelium discoideum. We isolated insertional mutants that failed to accumulate cellulose and had no cellulose synthase activity at any stage of development. Development proceeded normally in the null mutants up to the beginning of stalk formation, at which point the culminating structures collapsed onto themselves, then proceeded to attempt culmination again. No spores or stalk cells were ever made in the mutants, with all cells eventually lysing. The predicted product of the disrupted gene (dcsA) showed significant similarity to the catalytic subunit of cellulose synthases found in bacteria. Enzyme activity and normal development were recovered in strains transformed with a construct expressing the intact dcsA gene. Growing amoebae carrying the construct accumulated the protein product of dcsA, but did not make cellulose until they had developed for at least 10 hr. These studies show directly that the product of dcsA is necessary, but not sufficient, for synthesis of cellulose.
Although Dictyostelium amoebae do not make cellulose while they are growing, developing cells accumulate cellulose after the mound stage. Cellulose microfibrils are found in the extracellular matrix that surrounds the slugs as well as in the trails they leave behind. During culmination, cellulose is deposited in the stalk tube and in the spore coats and stalk cell walls. An in vitro biochemical assay for cellulose synthase activity has been established and the enzyme has been partially purified from developing cells of Dictyostelium (1).
Dictyostelium presents many advantages as a model for exploring the mechanism of cellulose synthesis in a eukaryote, including the applicability of a variety of molecular genetic techniques, including restriction enzyme-mediated insertional mutagenesis (REMI), which allows the rapid cloning and sequencing of a disrupted gene (2). Because Dictyostelium can be grown and developed as haploids, it is possible to screen directly for morphological mutants that develop within the clonal plaques formed by mutagenized cells grown on bacterial lawns. Because development is an induced event, strains with deleterious developmental mutations can be propagated as amoebae. REMI mutagenesis has allowed the identification of a large number of developmentally regulated genes in Dictyostelium. To that list can now be added the gene for the catalytic subunit of cellulose synthase.
Materials and Methods
Development and Transformation.
Cells were grown in HL5 medium and induced to develop synchronously by depositing them at 5 × 107 cells/cm2 on nitrocellulose filters supported on buffer-saturated pads (3). Strain AX-4 was used as the host for a DpnII REMI mutagenesis with the pBSR1 vector (2, 4). Cells were transformed by electroporation and transferred to tissue culture plates with HL5 medium, and Blasticidin S-resistant transformants were selected by adding 5 μg/ml Blasticidin S (ICN) a day after electroporation and culturing for 1 week (5). Morphological mutants were recognized by the structures formed within plaques generated by the transformed cells. Filter development and slug migration on agar plates were as described (6).
DNA Analyses.
Regions flanking the plasmid insertion site were isolated by plasmid rescue (2). Genomic DNA from strains DG1099 and DG1128 was digested with EcoRI and ligated and electroporated into Escherichia coli SURE cells (Stratagene). Plasmid DNA was isolated from the transformants and sequenced on an ABI 377 automated sequencer (Perkin–Elmer). The disruption of dcsA was recapitulated by homologous recombination (2) with linearized HindIII plasmid, where 4.1 kb of the gene flanks the insertion site.
Northern Blots.
RNA was isolated from 108 developed cells by using Trizol (GIBCO/BRL). Electrophoretic separations of RNA (20 μg per sample) and transfers to nylon membranes (MagnaGraph, Osmonics, Minnetonka, MN) were as described (6). Probes for dcsA mRNA were generated by random hexamer labeling of a DNA fragment containing the 5′ 2.2 kb of the gene (ATG to the BglII site).
Western Blots.
Proteins separated by SDS/PAGE were transferred to nitrocellulose and incubated with primary and secondary antibodies as described (7), with the exception that the blots were blocked with 1 mg/ml bovine hemoglobin (Sigma) before antibody incubation. The primary antibody was raised to the U2 peptide (TKADYEFLGLLDADQQPHPDC) linked to keyhole limpet hemocyanin, affinity-purified to the same peptide on a SulfoLink column (Pierce), and diluted 10-fold before use. The secondary antibody was goat anti-rabbit IgG-conjugated to alkaline phosphatase (Sigma) that was visualized by 5-bromo-4-chloro-3-indolyl phosphate and nitroblue tetrazolium.
Microscopy.
Slug slime trails were lifted from the agar surface on a glass coverslip and prepared for FBA28 [fluorescent brightening agent 28 (color index no. 40622; trade names: Calcofluor White ST or Tinopal LPW)] epifluorescence microscopy as described (8). Developing structures on filters were prepared for sectioning and light microscopy by rapid freezing in liquid nitrogen-cooled liquid propane using a freeze-plunging device (9), freeze-substituted in 1% (wt/vol) OsO4 in acetone at −80°C for 3 days, warmed through −40°C, −20°C, and 4°C to room temperature, infiltrated with Spurr's epoxy resin, polymerized, sectioned (0.5 μm), stained with 0.5% (wt/vol) toluidine blue, and mounted in Permount (Fisher Scientific). Cells developing on filters were prepared for scanning electron microscopy as described (10). Slime trails were collected onto 100-mesh Formvar-coated specimen grids (Pelco) and were prepared for direct shadowing transmission electron microscopy as described (8).
In Vivo Labeling.
Filters bearing developing structures at midculmination (or the equivalent time of development for dcsA− strains) were transferred to 25-μl drops of KK2 buffer (1) containing 5 μCi of [14C]glc (NEN) for 2 hr, returned to the original filter pads for 24 hr, collected, and subjected to either (i) 0.5 M KOH for 30 min at 100°C or (ii) acetic-nitric reagent (11) for 30 min at 100°C. The insoluble materials were collected onto Whatman GF/C glass fiber filters in a filtration manifold (Millipore) after first adding equal volumes of 0.5 M HCl or 0.5 M NaOH to the respective treatments. The filters were washed three times with H2O, once with methanol, air-dried, and counted.
Cellulose Synthase Assay.
Cellulose synthase activity was determined as described (8). Amylase sensitivity of in vitro products was determined by adding an equal volume of salivary amylase (12) to the reaction tube, incubating at 22°C for 4 hr, boiling, and processing the samples as described above. Whole-cell lysates were prepared by sonication in 10 mM Na-Hepes, pH 7.0, or by a filter-lysis method (13) and assayed as described (1). The in vitro product used in the solubility tests was generated as follows. Cells were plated for development on buffered agar, collected at early culmination, and disrupted as described (1), and crude membranes were prepared and protein concentrations were determined as described (8). Assay conditions were as above for the whole-cell lysates. A small quantity of unlabeled carrier cellulose (Sigmacell) was added to each reaction tube, and the tube contents were treated with one of the following: (i) 66% ethanol for 30 min at −20°C; (ii) 0.5 M KOH for 30 min at 100°C; or (iii) acetic-nitric reagent for 30 min at 100°C. The ethanol precipitates were collected as above and washed three times with 66% ethanol and once with methanol. The other samples were treated, and the retained radioactivity on all of the filters was determined as described above.
Results
Isolation of Mutant Strains that Fail to Synthesize Cellulose.
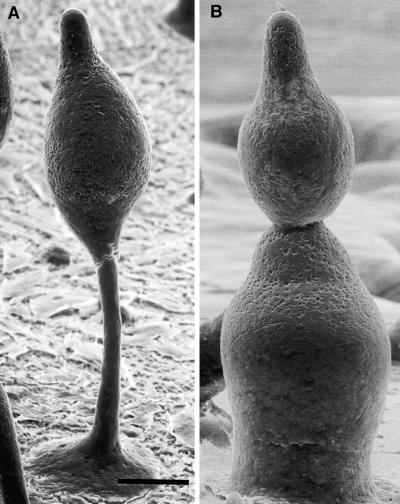
REMI mutagenesis was used to generate several hundred independent morphological mutants of Dictyostelium (2). Mutant strains that aggregated normally but failed to make proper fruiting bodies were screened with FBA28. Wild-type strains fluoresced blue when illuminated with ultraviolet light because of the cellulose deposited in the sheath surrounding cell mounds and slugs and from the cellulose in the stalk and spores (14). Two mutants, DG1099 and DG1128, did not bind FBA28 at any stage of development, although they proceeded through the first 20 hr of development and initiated culmination in a manner indistinguishable from wild-type strains. However, the mutant culminants collapsed on themselves to form structures resembling snowmen (compare Fig. 1), and these eventually “melted” as the cells in the structure lysed. No spores, stalk cells, or stalk tubes were visible in these structures (data not shown).
Figure 1.
Scanning electron micrographs of developing wild-type and cellulose-deficient strains. (A) Developing fruiting body of the wild type after 18 hr of development. Note the well-defined stalk and basal disk. (B) Developing fruiting body of the cellulose-deficient strain DG1099 after 18 hr of development. Note the absence of a stalk or basal disk. The apical-most mass is similar in appearance to the cell mass in A. The multiple lobes seen here are characteristic of the mutant strains, giving them the appearance of snowmen. (Bar = 50 μm.)
The Gene for the Dictyostelium Cellulose Synthase Catalytic Subunit.
Genomic DNA was isolated from strains DG1099 and DG1128 and digested with EcoRI for cloning by plasmid rescue in E. coli (15). Partial sequencing showed that the same gene had been disrupted in both mutant strains. A 600-bp portion of the sequence was used to screen a cDNA library prepared from wild-type cells at the slug stage. The largest cDNA clone recognized by the probe was sequenced and found to have a single, long ORF of 3.2 kb that terminated in a poly(A) tract. Genomic DNA covering the cDNA was sequenced from the original EcoRI clones and a KpnI clone that extended upstream of the plasmid insertion. Comparisons of the genomic and cDNA sequences showed that the ORF is flanked by high A+T regions characteristic of noncoding regions in Dictyostelium and that there are no introns in this gene.
To confirm that the gene was essential for accumulation of cellulose, we generated a new allele by homologous recombination with a construct in which the cloned gene was disrupted at the BglII site (Fig. 2A). This new mutant was indistinguishable from DG1099 and DG1128 in development and also never produced FBA28-positive material.
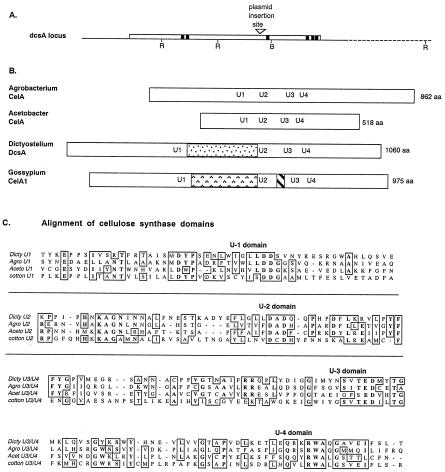
Figure 2.
Structure of the dcsA gene and comparison of its product, DcsA, with related proteins. (A) The region flanking DpnII restriction site (▿), where plasmids were found to have inserted in strains DG1099 and DG1128, contains a single, large ORF of 3.2 kb (open box). Pertinent restriction sites (RI, EcoRI; B, BglII) are marked. Sequences encoding hydrophobic domains of ≥20 aa are shown as solid boxes. (B) The predicted product, DcsA, is most similar to the cellulose synthases of A. tumefaciens and A. xylinum and could be aligned on the basis of conserved motifs U2–U4, which are marked at their position within each protein. An insertion occurred between the U1 and U2 motifs in both DcsA and the putative cellulose synthase of cotton, G. hirsutum, CelA1. However, the sequence of the insert is not conserved between these organisms. (C) Detailed comparison of the amino acid sequences surrounding the conserved motifs shows extended similarities especially between the established bacterial cellulose synthases and DcsA.
The predicted product of the gene disrupted in strains DG1099 and DG1128 is a protein of 1,060 aa that shows significant sequence similarity to the catalytic subunit of cellulose synthases reported from Acetobacter xylinum (16–18) and Agrobacterium tumefaciens (19) as well as putative cellulose synthases from higher plants including cotton, Gossypium hirsutum (20) (Fig. 2B). There is considerable sequence conservation surrounding the conserved aspartates in the U1, U2, and U3 motifs and the presence of the QXXRW signature sequence (21) in all these cellulose synthase sequences (Fig. 2C). The conserved motifs KAG and QTP identified by comparisons of other glycosyltransferases to cellulose synthases (22, 23) also are found in the Dictyostelium sequence.
An insert relative to the bacterial proteins follows the U1 domain in the Dictyostelium and higher plant proteins, although the Dictyostelium insert bears no sequence similarity to the plant insert. The insert between U2 and U3 that is found in the plant cellulose synthases is not encountered in the Dictyostelium protein. The degree of sequence similarity of the Dictyostelium protein to previously characterized catalytic subunits of cellulose synthase and the fact that disrupting the gene encoding this protein precludes cellulose synthesis prompted us to name the gene dcsA, for Dictyostelium cellulose synthase.
Hybridization of dcsA to genomic DNA digested with a variety of restriction enzymes showed no evidence for a family of cross-hybridizing genes (data not shown). Furthermore, we localized dcsA to a single locus at the distal end of chromosome 1 by probing an ordered set of large inserts carried in yeast artificial chromosomes (24) (data not shown).
Cloned dcsA Restores the Wild-Type Phenotype.
We cloned a full-length copy of dcsA into a plasmid in which the expression of the gene would be controlled by the regulatory region from the actin 15 gene, which is active in growing cells before the initiation of development and throughout the first 12 hr of development (25). Transformation of dcsA− cells with the act15∷dcsA+ construct completely overcame the mutant defects such that normally proportioned fruiting bodies were formed (data not shown).
Expression of dcsA.
RNA was prepared at intervals during development of wild-type cells, separated electrophoretically, and probed for dcsA mRNA. A single band of apparent molecular mass of 3.6 kb accumulated after 12 hr of development and continued to increase until culmination at 22 hr of development (Fig. 3A). This mRNA was completely missing in strain DG1128 (dcsA−) but was abundant during early development in the transformant TL128 that carries the act15∷dcsA+ construct (Fig. 3A).
Figure 3.
Expression of dcsA. (A) RNA was prepared at 4-hr intervals during early development and 2-hr intervals throughout late development of wild type (AX4), the dcsA− strain (DG1128), and the rescued strain (TL128). (B) An antibody was prepared against a peptide corresponding to the U2 region of DcsA. Total cellular proteins were prepared at 4-hr intervals during early development and 2-hr intervals throughout late development of wild type (AX4), the dcsA− strain (DG1128), and the rescued strain (TL128).
Antibodies prepared to a peptide from the U2 region of DcsA recognized a pair of proteins of 100 kDa and 120 kDa that accumulated rapidly after 10 hr of development in wild-type cells, but did not recognize any proteins in extracts of dcsA− cells of strain DG1128 (Fig. 3B). In the rescued strain, TL128, the pair of proteins was detected early in development and found to persist throughout development (Fig. 3B). Because both of the proteins recognized by the anti-DcsA antibodies depended on the dcsA gene, it is likely that they represent differently modified forms of DcsA.
Phenotypic Consequences of the Disruption of dcsA.
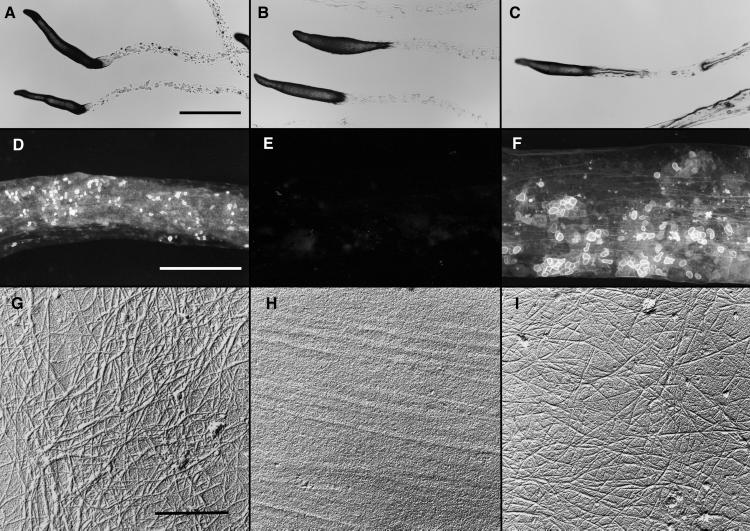
Wild-type, dcsA−, and act15∷dcsA+ cells all made slugs that were positively phototactic, were similarly shaped and sized, and left trails as they migrated that could be readily seen in the light microscope (Fig. 4 A–C). Wild-type trails fluoresced when treated with FBA28 (Fig. 4D) and contained microfibrils of cellulose (Fig. 4G). The dcsA− trails did not fluoresce (Fig. 4E) and contained no microfibrils (Fig. 4H). Those of the act15∷dcsA+ slugs fluoresced (Fig. 4F) and contained cellulose microfibrils (Fig. 4I).
Figure 4.
Slime trails produced by slugs of wild-type, dcsA−, and the dcsA rescued strains. Migrating slugs were phototactically directed to migrate across 2% water agar dishes. They were photographed in place (A–C) or the trails were collected onto coverslips (D–F) or electron microscope specimen grids (G–I). (A–C) All strains formed normal-appearing slugs that migrated phototactically and left behind slime trails. (Bar = 1 mm.) (D–F) The trails left behind wild-type slugs (D), and those of the rescued strain TL128 (F) fluoresced brightly whereas there was only background fluorescence from the trails left behind the dcsA− strain DG1128 (E). (Bar = 100 nm.) (G–I) Slime trails collected on electron microscope specimen grids were treated with Proteinase K (to remove obscuring proteins) and shadowed unidirectionally from 17° with Pt/C and with C from 85°. Microfibrils were seen clearly in the trails left by the wild type (G) and rescued strain, TL128 (I). Microfibrils were absent from the trails of the dcsA− strain, DG1128 (H). (Bar = 50 nm.)
The dcsA−-developing culminants were distinctively different from those of the wild type (Fig. 1). The developing culminants of the act15∷dcsA+ strain appeared normal. The dcsA− null mutant culminants had from one to four lobes stacked on each other, the topmost lobe having the morphology of a normal, early culminant (Fig. 1B).
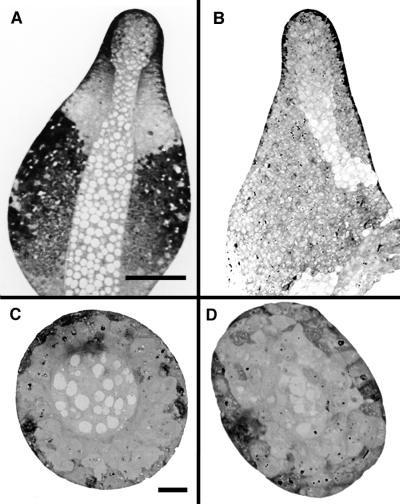
Longitudinal and cross-sections through rapidly frozen and freeze-substituted wild-type and mutant culminants showed that the dcsA− cells formed a central core of vacuolated cells (Fig. 5 B and D), just as occurs in wild type (Fig. 5 A and C).
Figure 5.
Comparative sections of developing culminants of wild-type and dcsA− strains. (A and B) Bright-field light micrographs of longitudinal sections of the wild-type (A) and dcsA− strain (B) reveal that the mutant forms the characteristic central core of vacuolized cells but that these do not have the rigid, cellulose-containing walls of normal stalk cells. Moreover, the cellulosic stalk tube is missing such that a functional stalk cannot be made. A first attempt at forming a stalk is apparent at the bottom of the longitudinal section of the mutant (B). (Bar = 50 μm.) (C–D) Bright-field light micrographs of cross-sections of the wild-type (C) and dcsA− strain (D) show the well-defined stalk tube in the wild type (C). A central core of vacuolated cells is present in the mutant, and there is the suggestion of a defining border (D), but the region is not nearly as well defined as in the wild type. (Bar = 10 μm.)
In Vivo and in Vitro Cellulose Synthase Activity.
To detect cellulose synthase activity in vivo, we radiolabeled developing culminants with [14C]glc, allowed them to complete development, and then looked for radiolabeled glucans that were insoluble in either 0.5 M KOH or acetic-nitric reagent (16). Although all forms of cellulose (and some noncellulosic glucans) are insoluble after heating in 0.5 M KOH (although some may be lost because of end degradation), only crystalline cellulose is insoluble in acetic-nitric (11). Considerable amounts of label were recovered in the alkali-insoluble polymers from wild-type fruiting bodies, and about half of it had the properties of crystalline cellulose (Table 1). Fruiting bodies of the dcsA− strain DG1099 incorporated less than 1% as much label into insoluble glucans, and this may have been background contamination with polymers other than cellulose (Table 1).
Table 1.
Relative solubility of metabolically labeled developing culminants of wild-type and dcsA− strains
| Treatment |
14C-insoluble material, dpm
|
||||
|---|---|---|---|---|---|
| Wild type (AX4) | dcsA− (DG1099) | dcsA− (DG1128) | dcsA− (TL127) | Rescued strain (TL128) | |
| 0.5 M KOH | 402,013 ± 221,742 | 487 ± 689 | 649 ± 918 | 0 | 1,100,790 ± 228,989 |
| Acetic-nitric | 193,849 ± 73,451 | 1,953 ± 90 | 843 ± 498 | 1,855 ± 1,175 | 634,873 ± 59,127 |
The values represent the means of triplicate reactions of a single experiment and the SD of those means.
Cellulose synthase activity was assayed in crude membrane preparations from wild-type, mutant, and rescued strains. In the wild-type and rescued strains, cellulose synthase activity first appeared at 12 hr of development and then increased throughout the remainder of development. No measurable activity was found at any time in development of the dcsA− mutant strains DG1128 (Fig. 6) or DG1099 (data not shown). Likewise, no activity was found during development of the dcsA− strain generated by homologous recombination, TL127 (data not shown). The amoebae often showed low levels of incorporation, which had not been seen in any of the strains used in our previous studies of cellulose synthase activity during development (1, 8). Aggregating cells did not incorporate any glucose from UDP-Glc. Therefore, we suspected the incorporation in amoebae was by residual glycogen synthase activity. This was confirmed when the product formed by amoebae was solubilized with α-amylase, which had no effect on the product formed by the differentiating cells (data not shown).
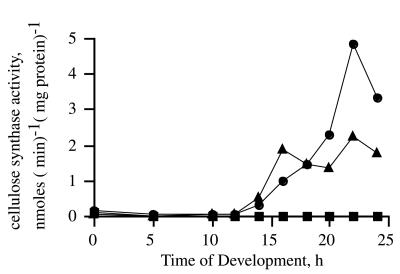
Figure 6.
Developmental time course of cellulose synthase activity. Membranes were prepared from the wild type (●), the dcsA− mutant strain DG1128 (■), and the dcsA+ rescue strain TL128 (▴) at various times during development, and the specific activity of cellulose synthase was determined. The specific activity in cells of the dcsA− strain was at or below background at all times. This was true as well for the DG1099 insertional mutant and for TL127, the strain with the new dcsA− allele created by homologous recombination (data not shown). No activity was detected in amoebae or early-developing stages of TL128, even though the gene was being expressed, as shown by Northern blot analysis, and the protein was present, as shown by Western blot analysis. The low level of incorporation seen in membranes from amoebae was shown to be amylase-sensitive (see Results).
Cells transformed with the act15∷dcsA+ rescue vector expressed the gene from the start of development, but no cellulose synthase activity was detected until the usual time in development in crude membranes prepared, assayed, and processed by the standard method (Fig. 6). To test whether the cellulose synthase activity might be present in a soluble form, we assayed whole-cell lysates (prepared both by sonication and filter lysis) from vegetative cells and cells in early aggregation. No activity was detected in reactions treated by the standard method or when ethanol precipitation and washing were used to precipitate all glucans (data not shown). Thus, even though the act15∷dcsA+ rescued cells had accumulated DcsA protein recognizable on Western blots of vegetative and aggregating cells, there was no cellulose synthase activity until the mound stage after 12 hr of development (Fig. 6).
The solubility characteristics of the products generated in the in vitro assays of the various strains were compared to ensure that expression of dcsA+ was sufficient to generate authentic cellulose. Extracts from either wild-type or the rescued strain generated labeled products that were insoluble in 66% ethanol, 0.5 M KOH, or acetic-nitric reagent (Table 2). Extracts from the dcsA− mutant strains incorporated less than 1% as many counts into insoluble material. Because most glucan polymers, including the (1→3)-β-glucan callose, are insoluble in 66% ethanol, it is clear that membrane preparations from developing dcsA− cells fail to make any glucose polymer at all.
Table 2.
Relative solubility of 14C product formed in in vitro enzyme assays
| Treatment |
14C-insoluble material, dpm
|
||||
|---|---|---|---|---|---|
| Wild type (AX4) | dcsA− (DG1099) | dcsA− (DG1128) | dcsA− (TL127) | Rescued strain (TL128) | |
| 66% EtOH | 17,167 ± 386 | 244 ± 26 | 165 ± 26 | 177 ± 7 | 13,314 ± 525 |
| (100%) | (100%) | ||||
| 0.5 M KOH | 10,166 ± 368 | 94 ± 64 | 0 | 0 | 9,742 ± 141 |
| (59%) | (73%) | ||||
| Acetic-nitric | 13,209 ± 1,118 | 4 ± 18 | 1 ± 22 | 0 | 9,165 ± 167 |
| (77%) | (69%) | ||||
The values represent the means of triplicate reactions of a single experiment and the SD of those means. The numbers in parentheses represent the percentage of the retained activity for the given treatment compared with the retained activity after the 66% ethanol treatment.
Discussion
dcsA Is a Bona Fide Cellulose Synthase.
Several lines of evidence suggest that dcsA encodes the catalytic subunit of the cellulose synthase. (i) DcsA shows the D… D… D… QXXRW pattern that is found in other cellulose synthases (21). Although overall sequence similarity is low between the various cellulose synthases, it is much higher in the region around these conserved residues. Interestingly, some plant and bacterial genes with the D… D… D… QXXRW pattern are not cellulose synthases (22, 26–30), so the presence of this pattern itself is not sufficient to declare that a gene is a cellulose synthase (29, 30). (ii) Cellulose microfibrils are found in wild-type slug trails but are absent from the trails of dcsA− slugs, which are still sufficiently well formed that they can be used to lift the attached slug off the substratum. (iii) The cellulosic walls formed by wild-type spores and stalk cells as well as the cellulose-containing stalk tube are missing in the fruiting bodies of dcsA− strains. (iv) There is no in vivo incorporation of [14C]glc into a glucan polymer in dcsA− culminants, and there is no in vitro incorporation of [14C]glc from UDP-Glc into a glucan polymer by membrane preparations from dcsA− culminants. Moreover, the product solubility studies showed that there is no glucose incorporation into a low crystallinity cellulose, which would have been precipitated by the 66% ethanol. (v) The patterns of dcsA mRNA and protein accumulation in normal development are consistent with the known developmental patterns of cellulose synthase activity (1, 8, 31). (vi) Introducing the cloned dcsA gene into dcsA− cells rescues the wild-type phenotype: microfibrils are found in the slug slime trail; fruiting bodies complete with spores and stalk cells that stain with FBA28 are produced; and glucose is incorporated (in vivo and in vitro) into a polymer that is alkali-insoluble and resistant to acetic-nitric reagent, which is indicative of the production of crystalline cellulose (11).
Prior analysis of the in vitro products of the Dictyostelium cellulose synthase indicated the presence of (1→3)-β linkages characteristic of the polymer callose in addition to the expected (1→4)-β linkages of cellulose (1). Yet, Dictyostelium accumulates no measurable callose in vivo. Membrane preparations from plants synthesize in vitro almost exclusively callose (produced abundantly as a wound response in vivo) in preference to cellulose (32), perhaps because the same enzyme is responsible for the synthesis of both and that cell disruption results in a change in the linkage formed (33, 34). This hypothesis is supported by our observation that membrane preparations from dcsA− cells synthesize neither (1→4)-β- nor (1→3)-β-linked glucans nor any glucan polymer for that matter. Thus, the cellulose synthase catalytic subunit of Dictyostelium appears to be responsible for synthesis of both of the linkage types detected previously in the in vitro products (1).
DcsA Is Necessary but Not Sufficient for Cellulose Synthesis.
Rescue of normal development and cellulose synthesis by transformation of dcsA− cells with the cloned dcsA gene demonstrated further that dcsA is necessary for cellulose synthesis. The rescue vector used the regulatory sequences from the act15 gene (25), which resulted in the expression of dcsA mRNA (detected by Northern blots) and DcsA protein (detected by Westerns) in growing amoebae and throughout early development. In the wild type, the mRNA and protein do not appear until 12 hr of development. However, in the act15∷dcsA+ cells that precociously accumulate DcsA, no cellulose synthesis was detected by FBA28 fluorescence and no in vitro activity was detectable until after 12 hr of development. These data indicate that dcsA alone is not sufficient for cellulose synthesis, that other components are necessary for it to occur, and that these components are not synthesized until aggregation is completed.
Developmental Consequences of the Absence of Cellulose.
Culmination began normally in the dcsA− strains, with a central core of enlarged cells defining the nascent stalk, the prestalk cells surrounding the core assuming their usual positions and orientations, and the cell mass assuming the characteristic shape of an early culminant. However, at the point when the cell mass should have begun to rise off of the surface, the absence of cellulose had its effect; the central core of vacuolating cells could not support the cell mass in the absence of the strengthening cellulose microfibrils in a stalk tube. In normal development, stalk cells vacuolate and lay down thick cellulose walls within the stalk tube, providing strength to the stalk. In the mutants, these cells vacuolated and died, but left no support structure behind. The cell masses appeared to reattempt to culminate, a second central core of cells forming and the prestalk cells surrounding that core assuming their proper positions. However, each attempt was doomed to failure as the structure collapsed upon itself. The mutant prestalk cells showed no evidence of recognizing that cellulose was not being deposited. However, none of the dcsA− cells in the sorus had the characteristic ellipsoid shape of spores, and exocytosis of the prespore vesicles (which, in normal development results in the formation of the spore coat) did not occur, suggesting that the absence of cellulose was somehow communicated to the prespore cells.
Acknowledgments
We thank Debbie Delmer for discussions. This work was supported by grants from the National Institutes of Health (HD30892) to W.F.L. and from the Office of Basic Energy Sciences, U.S. Department of Energy (DE-FG03-99ER20335) to R.L.B. and by funds from the Offices of the Provost and the Vice President for Research and the Research Enhancement Fund of the College of Arts and Sciences, Texas Tech University to R.L.B., and departmental and university support to the Department of Biological Sciences Electron Microscopy Laboratory at Texas Tech.
Abbreviations
- FBA 28
fluorescent brightening agent 28
- REMI
restriction enzyme-mediated insertional mutagenesis
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database [accession no. AF163835 (4.1-kb genomic sequence containing the dcsA sequence and upstream region)].
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.040565697.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.040565697
References
- 1.Blanton R L, Northcote D H. Planta. 1990;180:324–332. doi: 10.1007/BF00198783. [DOI] [PubMed] [Google Scholar]
- 2.Kuspa A, Loomis W F. Proc Natl Acad Sci USA. 1992;89:8803–8807. doi: 10.1073/pnas.89.18.8803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sussman M. Methods Cell Biol. 1987;28:9–29. doi: 10.1016/s0091-679x(08)61635-0. [DOI] [PubMed] [Google Scholar]
- 4.Shaulsky G, Escalante R, Loomis W F. Proc Natl Acad Sci USA. 1996;93:15260–15265. doi: 10.1073/pnas.93.26.15260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adachi H, Hasebe T, Yoshinaga K, Ohta T, Sutoh K. Biochem Biophys Res Commun. 1994;205:1808–1814. doi: 10.1006/bbrc.1994.2880. [DOI] [PubMed] [Google Scholar]
- 6.Shaulsky G, Loomis W F. Dev Biol. 1993;160:85–98. doi: 10.1006/dbio.1993.1288. [DOI] [PubMed] [Google Scholar]
- 7.Knecht D A, Loomis W F. Science. 1987;236:1081–1086. doi: 10.1126/science.3576221. [DOI] [PubMed] [Google Scholar]
- 8.Blanton R L. Development (Cambridge, UK) 1993;119:703–710. [Google Scholar]
- 9.Mollenhauer H M. Microsc Res Tech. 1999;44:195–200. doi: 10.1002/(SICI)1097-0029(19990115/01)44:2/3<195::AID-JEMT11>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 10.Wu L, Franke J, Blanton R L, Podgorski G J, Kessin R H. Dev Biol. 1995;167:1–8. doi: 10.1006/dbio.1995.1001. [DOI] [PubMed] [Google Scholar]
- 11.Updegraff D M. Anal Biochem. 1969;32:420–424. doi: 10.1016/s0003-2697(69)80009-6. [DOI] [PubMed] [Google Scholar]
- 12.Olaitan S A, Northcote D H. Biochem J. 1962;82:509–519. doi: 10.1042/bj0820509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das O P, Henderson E J. Biochim Biophys Acta. 1983;736:45–56. [Google Scholar]
- 14.Harrington B J, Raper K B. J Appl Microbiol. 1968;16:106–113. doi: 10.1128/am.16.1.106-113.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuspa A, Loomis W F. Genetics. 1994;138:665–674. doi: 10.1093/genetics/138.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong H C, Fear A L, Calhoon R D, Eichinger G H, Mayer R, Amikam D, Benziman M, Gelfand D H, Meade J H, Emerick A W, et al. Proc Natl Acad Sci USA. 1990;87:8130–8134. doi: 10.1073/pnas.87.20.8130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saxena I M, Lin F C, Brown R M., Jr Plant Mol Biol. 1990;15:673–683. doi: 10.1007/BF00016118. [DOI] [PubMed] [Google Scholar]
- 18.Saxena I M, Brown R M., Jr J Bacteriol. 1995;177:5276–5283. doi: 10.1128/jb.177.18.5276-5283.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matthysse A G, White S, Lightfoot R. J Bacteriol. 1995;177:1069–1075. doi: 10.1128/jb.177.4.1069-1075.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pear J R, Kawagoe Y, Schrecknegost W E, Delmer D P, Stalker D M. Proc Natl Acad Sci USA. 1996;93:12637–12642. doi: 10.1073/pnas.93.22.12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saxena I M, Brown R M, Jr, Fèvre M, Geremia R A, Henrissat B. J Bacteriol. 1995;177:1419–1424. doi: 10.1128/jb.177.6.1419-1424.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stasinopoulos S J, Fisher P R, Stone B A, Stanisich V A. Glycobiology. 1999;9:31–41. doi: 10.1093/glycob/9.1.31. [DOI] [PubMed] [Google Scholar]
- 23.Wiggins C A R, Munro S. Proc Natl Acad Sci USA. 1998;95:7945–7950. doi: 10.1073/pnas.95.14.7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuspa A, Loomis W F. Proc Natl Acad Sci USA. 1996;93:5562–5566. doi: 10.1073/pnas.93.11.5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cohen S, Knecht D, Lodish H, Loomis W. EMBO J. 1986;5:3361–3366. doi: 10.1002/j.1460-2075.1986.tb04651.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cutler S, Somerville C. Curr Biol. 1997;7:R108–R111. doi: 10.1016/s0960-9822(06)00050-9. [DOI] [PubMed] [Google Scholar]
- 27.Saxena I M, Brown R M. Cellulose. 1997;4:33–49. [Google Scholar]
- 28.Delmer D P. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:245–276. doi: 10.1146/annurev.arplant.50.1.245. [DOI] [PubMed] [Google Scholar]
- 29.Taylor N G, Scheible W-R, Cutler S, Somerville C R, Turner S R. Plant Cell. 1999;11:769–779. doi: 10.1105/tpc.11.5.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arioli T, Burn J E, Betzner A S, Williamson R E. Trends Plant Sci. 1998;3:165–166. [Google Scholar]
- 31.Blanton R L. In: Dictyostelium: A Model System for Cell and Developmental Biology. Maeda Y, Inouye K, Takeuchi I, editors. Tokyo: Universal Academy Press; 1997. pp. 379–391. [Google Scholar]
- 32.Delmer D P. Annu Rev Plant Physiol. 1987;38:259–290. [Google Scholar]
- 33.Jacob S R, Northcote D H. J Cell Sci Suppl. 1985;2:1–11. doi: 10.1242/jcs.1985.supplement_2.1. [DOI] [PubMed] [Google Scholar]
- 34.Delmer D P, Cooper G, Alexander D, Cooper J, Hayashi T, Nitsche C, Thelen M. J Cell Sci Suppl. 1985;2:33–50. doi: 10.1242/jcs.1985.supplement_2.3. [DOI] [PubMed] [Google Scholar]