Abstract
To identify genes required for maintaining neuronal viability, we screened our collection of Drosophila temperature-sensitive paralytic mutants for those exhibiting shortened lifespan and neurodegeneration. Here, we describe the characterization of wasted away (wstd), a recessive, hypomorphic mutation that causes progressive motor impairment, vacuolar neuropathology, and severely reduced lifespan. We demonstrate that the affected gene encodes the glycolytic enzyme, triosephosphate isomerase (Tpi). Mutations causing Tpi deficiency in humans are also characterized by progressive neurological dysfunction, neurodegeneration, and early death. In Tpi-deficient flies and humans, a decrease in ATP levels did not appear to cause the observed phenotypes because ATP levels remained normal. We also found no genetic evidence that the mutant Drosophila Tpi was misfolded or involved in aberrant protein–protein associations. Instead, we favor the hypothesis that mutations in Tpi lead to an accumulation of methylglyoxal and the consequent enhanced production of advanced glycation end products, which are ultimately responsible for the death and dysfunction of Tpi-deficient neurons. Our results highlight an essential protective role of Tpi and support the idea that advanced glycation end products may also contribute to pathogenesis of other neurological disorders.
Keywords: advanced glycation end product
Forward genetic strategies in Drosophila have proven to be a powerful approach for dissecting the molecular mechanisms of neuronal signaling. Unbiased screens for mutants based solely on phenotype enabled the molecular identification and characterization of proteins with key roles in neuronal function that were previously unknown or inaccessible via alternative approaches. The phenotype of temperature-sensitive (ts) paralysis has been particularly potent for discovering genes with crucial roles in neural function (1–3). Over the years, we have amassed a large collection of ts-paralytic mutations and discovered among them genes encoding a variety of ion channels, ion channel regulators, components of the synaptic release machinery, and proteins required for synaptic growth.
One of the important insights derived from investigating these mutants is that in most cases their conditional phenotype is not caused by the temperature sensitivity of the mutant proteins (4–8). Instead, most of the mutations cause unconditional complete or partial loss of activity of the affected proteins, resulting in varying degrees of neural impairment even at nominally permissive temperatures. At permissive temperatures, these impairments are generally not sufficient to cause drastic deficits in locomotor activity or other behaviors. However, when neural function is impaired for any of a variety of reasons, the mutant flies apparently cannot withstand the greater physiological demands of elevated temperature and become paralyzed within seconds to minutes.
Recently, a number of investigators have used the experimental advantages of Drosophila to model human neurodegenerative disorders such as Parkinson's disease, Alzheimer's disease, and triplet repeat disorders (reviewed in ref. 9). Most of these studies have relied on existing knowledge to introduce human disease genes into flies or target the Drosophila counterpart of these genes for experimental manipulation to create flies with phenotypes that mimic the human syndromes. A more open-ended strategy to investigate the basic biology of neurodegeneration is to use a forward genetics approach to identify a collection of genes whose function is necessary to ensure neuronal maintenance and survival. Isolation and characterization of such “neurodegeneration suppressor” genes should provide important insights into the molecular mechanisms that maintain neuronal viability. Although several studies have focused on forward genetic screens for neurodegeneration in Drosophila, the number of mutations identified in this way still remains relatively small (10–16).
We reasoned that our collection of ts-paralytic mutants, originally isolated because they exhibited aberrant behaviors likely to be associated with altered or impaired neural function, included those whose behavioral deficits were associated with neurodegeneration. For example, some of the mutants showed progressive motor impairments even at permissive temperature as a function of age. Indeed, an initial histological survey of our collection demonstrated that it was enriched for mutants with defects in neuronal survival: ≈10% of the mutants examined manifest varying degrees of age-dependent neuropathology in the central nervous system (17). Among the previously identified mutants associated with neurodegeneration are those encoding ion channels, the Na+/K+ ATPase, and components of the synaptic release machinery (17–19).
Here, we demonstrate that one of the previously uncharacterized mutations disrupts the gene encoding the glycolytic enzyme, triosephosphate isomerase (Tpi), resulting in progressive motor impairment, severely reduced lifespan, and neurodegeneration. These phenotypes closely resemble those associated with triosephosphate deficiency in humans (20, 21). Thus, even though our initial mutant screen did not specifically target human disease genes, we have obtained an excellent Drosophila model for a human neurodegenerative disorder. Our results indicate that the neurological phenotypes do not result simply from a reduction in ATP production. Nor do we have any genetic evidence of aberrant protein misfolding or inappropriate associations in wstd mutants. Instead, we favor the idea that loss of Tpi activity, which interconverts dihydroxyacetone phosphate (DHAP) and glyceraldehyde-3-phosphate (GAP), results in elevated levels of DHAP. DHAP, in turn, spontaneously converts to methylglyoxal, a highly reactive compound that modifies proteins and DNA through the formation of advanced glycation end products (AGEs), which are deleterious to neurons and other cells (22–25). We contend that enhanced AGE production is responsible for the dysfunction and death of Tpi-deficient neurons (26). Our results highlight an essential protective role of Tpi and underscore the possibility that AGEs contribute to pathogenesis of other neurological disorders as well.
Results
ts Paralysis and Shortened Lifespan in wstd Mutants.
wstd1 was isolated >25 years ago, in a large-scale screen for ts-paralytic mutants (B.G., unpublished work). It was selected for further investigation based on an initial survey of our ts-paralytic collection for neurodegeneration mutants (17). At 22°C, wstd1 homozygotes exhibit essentially normal locomotor activity. However, when shifted abruptly to 38°C, the mutants lose motor control and fall to the bottom of the vial within 1 min with occasional spasmodic activity and leg twitching. Paralysis becomes complete within 3–5 min at the elevated temperature. In contrast, WT and wstd1/+ flies remain active at 38°C for >30 min.
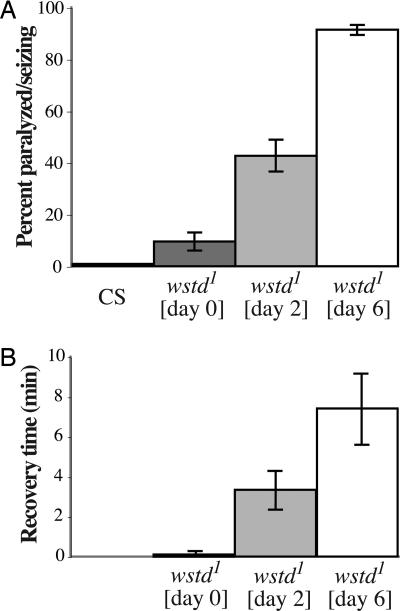
Paralytic behavior of wstd1 homozygotes at 38°C varied as a function of age (Fig. 1A). Within the first day after eclosion, only ≈10% of mutant adults were paralyzed after a 2-min exposure to 38°C. This fraction increased to >90% for 6-day-old flies (Fig. 1A). The time required for 100% of the flies to become paralyzed decreased from >10 min for 1-day-old wstd1 flies to 4 min for 7-day-old flies to ≈1 min for 14-day-old flies (data not shown).
Fig. 1.
ts paralysis of wstd1 flies becomes more severe with age. (A) Flies collected shortly after eclosion were aged for the indicated times at 22°C and aspirated into preheated vials at 38°C. The 0-day flies were 4–8 h posteclosion. Control Canton-S flies were 5–10 days old. The percentage of flies paralyzed or seizing on the bottom of the vial after a 2-min exposure was recorded. Results are from five independent samples of 15 flies each. Values shown are mean ± SEM. (B) Time required for 50% of flies of the indicated ages to recover the ability to stand at 22°C after a 10-min exposure to 38°C. Canton-S is not shown because they are not paralyzed at 38°C. Results are from five independent samples of 15 flies each. Values shown are mean ± SEM.
Kinetics of recovery from paralysis show the converse age dependence (Fig. 1B). After a 10-min exposure to 38°C, 50% of flies that were <1 day posteclosion recover the ability to stand almost immediately upon return to room temperature. This time increased to >3.3 min for 2-day-old flies and 7.4 min for 6-day-old flies (Fig. 1B). After 3 weeks of age, wstd1 flies required upwards of 30 min to recover from paralysis.
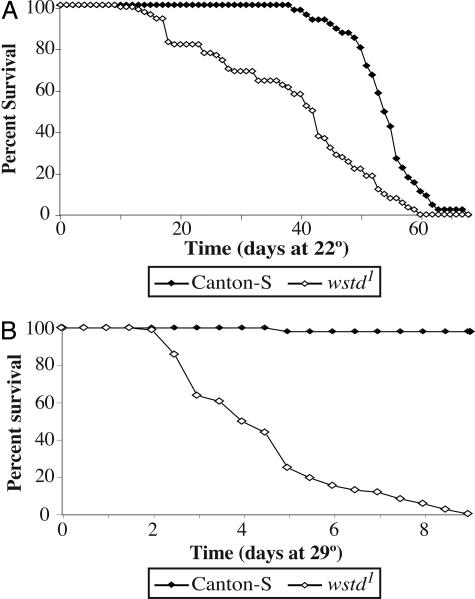
Lifespan measurements revealed that wstd1 mutants are short-lived compared with WT controls and that this difference is accentuated at elevated temperatures (Fig. 2). At 22°C, wstd1 flies reach the midpoint of their survival curve (T50) in 41.4 ± 2.8 days, compared with 54 ± 0.88 days for controls (P < 0.001). At 29°C, the lifespan of wstd1 flies is dramatically reduced with a T50 of only 4.08 ± 0.2 days, ≈10 times less than for control flies (38 ± 4) at this temperature (P < 0.001). We infer that wstd1 mutants have some underlying physiological impairment that results in reduced lifespan, even at temperatures below the threshold for causing paralysis.
Fig. 2.
wstd mutations reduce lifespan. (A) Flies were raised and aged at 22°C. (B) Animals were reared at 22°C, collected upon eclosion, and aged at 29°C. Lifespan of mutants is severely reduced at 29°C compared with controls. For wstd1, each point represents the mean from six independent samples, each containing 15 males. For Canton-S, each point represents the mean from three independent samples each containing 15 males.
Identification of Additional Mutant Alleles of wstd.
Because we had just one mutant allele of wstd in our possession and wanted to be certain of obtaining a null allele, we conducted a failure of complementation screen for new alleles after γ-ray mutagenesis. Treated males were mated with homozygous wstd1 females, and the F1 progeny were screened for paralysis at 38°C. We recovered one lethal allele (wstd2) that failed to complement all wstd1 phenotypes, including ts paralysis, shortened lifespan, and neurodegeneration. We identified two additional alleles (wstd3 and wstd4) by testing a previously generated set of ethylmethane sulfonate-induced lethal alleles in the vicinity of wstd for failure to complement (27). The three new alleles are lethal in all heterozygous combinations with each other and when heterozygous for Df(3R)Exel6214, a deletion that completely uncovers the wstd gene. Flies heterozygous for wstd1 and any of the lethal alleles exhibit more extreme mutant phenotypes than wstd1 homozygotes and are comparable in severity to flies heterozygous for wstd1 and Df(3R)Exel6214. These results indicate that the original wstd1 mutation is a hypomorphic allele and that the lethal mutations represent the null phenotype. In addition, the failure of three different lethal alleles to complement all phenotypes associated with wstd strongly implies that all phenotypes result from mutations of the same single gene.
wstd Causes Progressive, Age-Dependent Neurodegeneration.
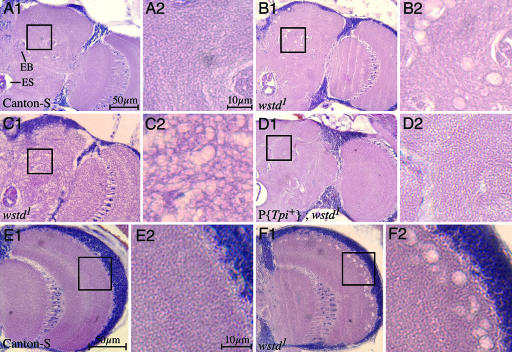
Histological examination of frontal head sections of wstd1 homozygotes incubated for 1–4 days at 29°C demonstrated the presence of characteristic neurodegenerative vacuolar lesions in the neuropil of the central brain that become more pronounced and extensive with age (Fig. 3). Immediately after shifting to 29°C, brains of wstd1 flies are indistinguishable from normal (data not shown). After 1 day at 29°C neuropathological lesions, evident as small, scattered holes in the neuropil of the central brain, begin to appear and increase in number over the next several days (Fig. 3 B1, B2, C1, C2, F1, and F2). Although lesions could be found throughout the brain in different individuals, they occurred most often in the neuropil lateral to the ellipsoid body and in the outer margin of the optic lobe (Fig. 3 B1, B2, F1, and F2). Even single lesions of this type were rare in WT flies (Fig. 3 A1 and A2). The range of severity in the appearance of lesions in wstd flies varied. A small percentage of mutant brains exhibited a severe spongiform appearance (Fig. 3 C1 and C2), whereas others had only sporadic lesions. Among >50 wstd brains examined, at least 75% could readily be distinguished from WT controls by their neuropathological appearance. Overall, flies heterozygous for wstd1 and any of the null alleles exhibited more extreme neurodegeneration; only ≈10% of these brains overlapped control brains in appearance.
Fig. 3.
wstd causes neurodegeneration. Frontal sections at approximately midbrain from adults of the indicated genotypes. (A1, B1, C1, and D1) Half of the central brain extending into the optic lobe. The esophagus (ES) is visible as the large hole at the bottom left in each panel. The ellipsoid body (EB) is also indicated. (A2, B2, C2, and D2) Magnified views of central brain regions corresponding to the respective boxed areas. (E1 and F1) Optic lobes with magnified views in E2 and F2, respectively. (A1 and A2) Canton-S. (B1 and B2) wstd1. Note vacuolar-like lesions. (C1 and C2) Example of extreme manifestation of spongiform phenotype observed in a small percentage of wstd1 flies. (D1 and D2) wstd1 carrying a WT transgene for Tpi. Among 10 brains of this genotype examined, no vacuolar lesions were observed and integrity of the brain tissue was indistinguishable from WT flies. (E1 and E2) Canton-S. (F1 and F2) wstd1. Note vacuolization along peripheral neuropil. Flies were aged for 1 day (C1, C2, F1, and F2) to 4 days (A1, A2, B1, B2, D1, D2, E1, and E2) at 22°C and then shifted to 29°C for 3 days before sectioning. (Scale bars: A1, B1, C1, D1, E1, and F1, 50 μm; A2, B2, C2, D2, E2, and F2, 10 μm.)
Molecular Identification of wstd.
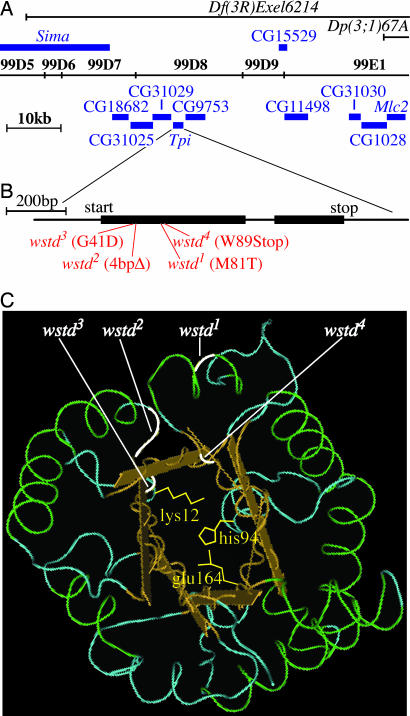
By recombination, wstd mapped to 3–100.0 ± 3, to the right of the marker Dr, which is located at 99B2 on the cytological map. To refine the cytological location of wstd, we performed complementation tests with deletions that spanned the distal tip of the chromosome. Df(3R)Exel6214, deleted for 99D5-99E2, uncovered the ts-paralytic phenotype of wstd1. Dp(3;1)67A, an insertional duplication of 99D9-E1;100 into the X chromosome, did not complement wstd1, indicating that the duplication does not include wstd+. Because the left endpoint of Dp(3;1)67A includes the myosin light chain-2 (Mlc2) gene (27), wstd is located to the left of this gene (Fig. 4). These results placed wstd in the interval, 99D5;99D9-E1 (Fig. 4). The chromosome segment that includes wstd encompasses ≈50 kb on the molecular map and contains 10 genes, most of unknown function. We initiated sequence analysis of these genes and discovered coding changes in all four wstd alleles in the gene that encodes Tpi, an essential glycolytic enzyme that is highly conserved from bacteria to humans. Two wstd alleles introduce premature stop codons and two produce single amino acid changes. (Fig. 4). The presence of mutational changes in Tpi in four different wstd alleles strongly indicates that this enzyme is the wstd gene product.
Fig. 4.
wstd mutations reside within the gene encoding Tpi. (A) Complementation tests indicated that Df(3R)Exel6214 uncovers wstd mutations, whereas Dp(3;1)67A does not contain wstd+. The location of the relevant breakpoints of these chromosomes on the cytological and molecular map of the region is shown. All known transcription units within the defined interval are indicated. Sequence analysis revealed that four different wstd alleles contained mutations in the Tpi coding sequence. (B) Expanded view of the Tpi transcription unit showing the location of the mutations in the four wstd mutant alleles examined here. wstd1 causes a Met-to-Thr substitution at amino acid position 81. wstd2 contains a 4-bp deletion that introduces a frameshift beginning in codon 43 and generates a premature stop at codon 47. wstd3 causes a Gly-to-Asp substitution at amino acid position 41. wstd4 introduces a premature stop codon at amino acid position 89. (C) Location of the mutations on the 3D structure of the Tpi protein. The three amino acid residues (Lys-12, Glu-64, and His-94) that directly participate in the enzymatic reaction are indicated.
To verify that Tpi is the affected gene, we performed germ-line transformation rescue experiments with a genomic clone containing the entire WT wstd gene expressed from its endogenous promoter (P{Tpi+}). Flies homozygous for wstd1 and carrying one copy of the rescue construct were indistinguishable from WT flies with respect to locomotor behavior (0% paralysis after 20 min at 38°C), lifespan (100% survival after 21 days at 29°C), and neuronal survival (Fig. 3 D1 and D2). In addition, one copy of P{Tpi+} rescued the lethal phenotype of wstd2, wstd3, and wstd4. Thus, P{Tpi+} fully rescues all phenotypes associated with wstd mutations and establishes that all wstd phenotypes arise from mutations of Tpi.
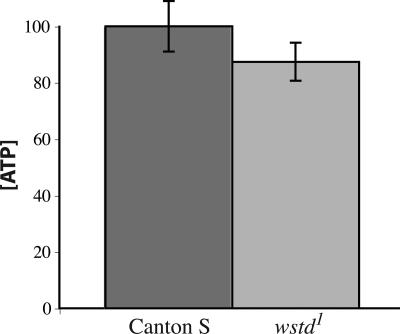
wstd Does Not Cause Significant Reduction in ATP Levels.
Mutations of Tpi in humans also result in neurodegenerative disease (20, 21). However, the mechanism underlying the pathology of Tpi deficiency remains unclear. Because Tpi is an essential glycolytic enzyme and we have previously found that metabolic disruptions are associated with neurodegeneration in other Drosophila mutants, it seemed possible that a metabolic impairment resulting in reduced ATP levels could underlie the wstd mutant phenotypes. To test this possibility, we performed quantitative ATP assays. Although ATP levels are slightly lower in wstd homozygotes compared with WT controls, this difference was not significant (P > 0.37; Fig. 5). This result is consistent with human studies, which found no decrease in ATP levels in TPI-deficient patients (21, 28). Thus, in flies, as in humans, a deficit in ATP is not the apparent cause of the neuropathological phenotypes associated with loss of Tpi activity. Instead, as described in Discussion, we favor the model that neuropathogenesis in flies and people deficient for Tpi activity results from accumulation of toxic metabolites upstream of the enzymatic block.
Fig. 5.
ATP levels in wstd1 homozygotes were not significantly reduced. Duplicate measurements were made on 14 independent samples of each genotype. All flies were aged 4–7 days at 22°C. Values shown (mean ± SEM) are corrected for protein content and normalized as a percentage of control samples. Although the values for wstd1 tended to be slightly lower than controls, this difference was not significant (P > 0.37, Whitney–Mann test).
Discussion
Mutations of Tpi Cause Neurological Disease in Flies and Humans.
In this study, we have identified mutations in the Drosophila gene that encodes the glycolytic enzyme, Tpi, on the basis of a forward genetic screen for neurodegeneration mutants. Mutations of the corresponding gene in humans cause the disorder, Tpi deficiency, which is associated with phenotypic manifestations similar to those seen in Drosophila, including hemolytic anemia, progressive neurologic dysfunction, neurodegeneration, and early death (20, 21). Neurological symptoms first appear at 6 months to 2 years of age and involve lower motor neurons combined with dystonic and dyskinetic features of central origin. Death generally occurs by 6 years of age (21, 28). Although TPI deficiency was first described in 1965, the underlying pathophysiology has remained elusive. The additional information derived from analysis of the Drosophila mutants offers insights into the underlying disease mechanism.
Several lines of evidence argue against the most obvious possibility, that the neurological phenotypes result from a metabolic impairment and a consequent deficit of ATP. First, quantitative assays both in humans and flies indicate there is no decrement in ATP levels when Tpi activity is impaired (21, 28). Second, mutations affecting other glycolytic enzymes do not cause comparable neurological phenotypes. Mutations have been identified in humans that perturb most steps in glycolysis (28). All of these mutations are associated with hemolytic anemia but, with the exception of phosphoglycerate kinase (PGK), none of the other enzymopathies cause neurological impairment even though several of them do cause significant reductions in ATP levels (28). Thus, the mechanism by which Tpi deficiency leads to neurodegeneration appears to be distinct and not just a direct consequence of impaired glycolysis. We discuss later a possible mechanistic link between TPI deficiency and PGK deficiency.
Studies of several human TPI patients have led to the proposal that neurodegeneration is associated with misfolding and aberrant or inappropriate protein–protein associations of the mutant enzyme (29–31). Although we cannot rule out this possibility in humans, it does not appear to be a key factor in Drosophila. The original wstd allele behaves as a fully recessive, hypomorphic mutation whose associated phenotypes are more extreme in flies heterozygous for wstd and a lethal null allele or a deletion of the gene than in wstd homozygotes. In addition, expression of a transgene containing a WT copy of the gene fully rescues all wstd phenotypes. Thus, we see no indication of dominant neomorphic or other gain-of-function phenotypes as would be expected if the mutant protein engages in aberrant or inappropriate associations with other cellular targets.
A Proposed Disease Mechanism for Tpi Deficiency.
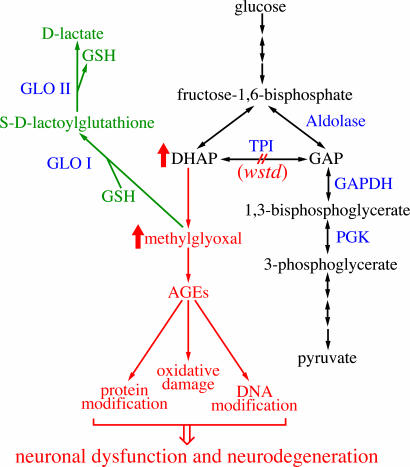
What is the basis of neurodegeneration in Tpi-deficient flies and humans if not a deficit in metabolism or a result of aberrant protein–protein associations? We favor the following model (Fig. 6), which is based on the available information for Tpi mutations in flies and humans together with the growing body of data about methylglyoxal and its biological effects (22–26). In glycolysis, fructose bisphosphate is broken down into GAP and DHAP. The glycolytic pathway can further metabolize only GAP. Tpi interconverts DHAP and GAP, enabling DHAP to be metabolically used (Fig. 6). Loss of Tpi activity should therefore result in excess accumulation of DHAP. Indeed, the most apparent biochemical alteration reported in TPI-deficient patients is the striking (>20-fold) increase in cellular concentration of DHAP (21, 28). Although there is no indication that DHAP itself is toxic, it decomposes nonenzymatically to methylglyoxal (23, 25). Methylglyoxal is a highly reactive α-oxoaldehyde that can modify both proteins and DNA to form AGEs (22–25). The deleterious consequences of accumulated of glycation adducts include protein inactivation, denaturation, and crosslinking; altered gene expression; increased oxidative stress; DNA damage; and apoptosis. In humans, methylglyoxal and AGEs have been linked with a variety of conditions, including diabetes, uremia, cancer, and aging. Most important with respect to the phenotypes under consideration here, methylglyoxal is toxic to neurons (24, 32, 33).
Fig. 6.
A model of how mutations in wstd lead to neuronal dysfunction and neurodegeneration. Relevant enzymatic steps in glycolysis are shown (black). Double-headed arrows represent reactions that can flow in either direction. Loss of TPI activity (red) causes accumulation of excess DHAP, which converts to methylglyoxal nonenzymatically. Elevated methylglyoxal levels result in production of AGEs that impair neuronal function and survival. Methylglyoxal is normally detoxified by the glyoxalase pathway (green). Glyoxalase I (GloI) and glyoxalase II (GloII) use glutathione (GSH) as a cofactor to convert methylglyoxal to d-lactate.
Normally the glyoxalase pathway protects cells from excess methylglyoxal (34). Glyoxalase I and II detoxify methylglyoxal by converting it to lactic acid, using catalytic amounts of glutathione as a cofactor. However, under conditions of oxidative stress, glutathione levels are reduced, resulting in elevated levels of methylglyoxal. This effect can be further magnified in a positive feedback loop because AGEs themselves contribute to the production of reactive oxygen species (26).
On the basis of these considerations, we suggest that the defects in the function and survival of Tpi-deficient neurons result from the toxic effects of elevated methylglyoxal levels. In the case of wstd mutants, we expect that reduction in Tpi activity causes methylglyoxal and AGEs to be elevated even at permissive temperatures. We propose that the affected targets include one or more proteins essential for neural activity and that AGE modifications compromise signaling capabilities of mutant neurons. As with other ts-paralytic mutants that directly affect membrane excitability and synaptic transmission, under nonstressful conditions neurons in wstd mutants function sufficiently well that behavior is overtly normal. However, the weakened neurons are not able to withstand the greater physiological demands of elevated temperature, resulting in paralysis of wstd flies at 38°C. Chronic exposure to the intermediate temperature of 29°C results in accelerated neuronal death and shortened lifespan.
A similar mechanism may explain, in part, the neurological phenotypes associated with PGK deficiency. Although generally less severe than TPI deficiency, PGK deficiency results in exercise intolerance, seizures, mental retardation, and progressive extrapyramidal tract disease (28). Interestingly, PGK mutations in Drosophila were also identified as ts-paralytic mutants (35). It is not yet known whether these flies exhibit neurodegeneration, although they have a very reduced lifespan. The basis of the neurological phenotypes of PGK-deficient flies and humans is not known. Because PGK acts two steps downstream from Tpi (Fig. 6), a block in PGK could be predicted to cause an accumulation of GAP and DHAP, whose conversion to methylglyoxal could contribute, in part, to the disease manifestations.
Methyglyoxal and AGEs May Be Linked with Other Neurological Disorders.
From the proposed model for TPI deficiency and PGK deficiency, one might also expect that mutations of GAPDH, which acts immediately after TPI (Fig. 6), would also cause neurological disease. Because appropriate mutations are lacking, this possibility remains untested. Nonetheless, it is notable that the polyglutamine tracts of Huntingtin and other polyglutamine disease proteins have been found to bind specifically to GAPDH and inhibit its enzymatic activity (36, 37). As suggested for PGK deficiency, this block in enzymatic activity could lead to elevated production of methylglyoxal and AGEs in polyglutamine diseases (Fig. 6).
Discovery that exposure to 1-methyl-4phenyl-1,2,3,6-tetrahydropridine (MPTP) could cause Parkinson's disease provided the prototypical example of how environmental toxins could be linked to sporadic occurrence of this disease (reviewed in ref. 38). Furthermore, it was proposed that even in the absence of exogenous toxins, endogenous toxins derived from metabolism could have similar consequences. One example is the natural β-carbolines (BC), endogenous neurotoxins formed from tryptophan and related compounds, which closely resemble MPTP in structure (39). Injection of BC into the substantia nigra in animal models cause neurodegeneration. However, the mechanism of BC (and MPTP) toxicity remains unknown. Recent studies demonstrate that BC binds with very high affinity to Tpi and is an extremely potent inhibitor of its enzymatic activity (39). Thus, decreased Tpi activity, and a resultant increase in AGEs, could be an important factor contributing to Parkinson's disease.
Methylglyoxal and AGEs have also been linked with Alzheimer's disease. AGE residues accumulate in the characteristic extracellular deposits of amyloid peptide and intracellular deposits of tau, contributing to the insolubility and protease resistance of these deposits (24, 40). Moreover, microarray analysis of a mouse model of Alzheimer's disease identified the gene for the detoxifying enzyme, glyoxalase I, as the only one whose expression was significantly up-regulated (41).
Methylglyoxal and AGEs may even play a role in autism. Recent proteomic studies identified a single nucleotide polymorphism in glyoxalase I as an autism susceptibility factor (42). In addition, biochemical measurements showed a significant decrement in glyoxalase I activity and an accumulation of AGEs in autism brains (42).
Although a direct causal connection between methylglyoxal and AGEs in the pathogenesis of any of these neurological disorders remains to be proven, as it has been for diabetes (43), the number of potential links is remarkable and illustrates the broad potential for AGEs to be a significant factor in neurological disease. From this perspective, Tpi can be considered not just a glycolytic enzyme but also a component of a protective pathway that limits the potentially deleterious accumulation of DHAP, a toxigenic compound. The mutations in Drosophila Tpi that we have isolated and characterized here should therefore not only provide an excellent experimental system for elucidating the biological mechanisms responsible for the syndrome of human TPI deficiency but could also lead to insights about other human neurological disorders.
Materials and Methods
Fly Strains.
Flies were cultured on cornmeal-molasses agar medium at room temperature (≈22°C) unless otherwise noted. The wstd1 mutation was induced by mutagenesis with ethylmethane sulfonate and is part of our collection of ts-paralytic mutants. The wstd2, wstd3, and wstd4 alleles identified here are recessive lethals. The lethal phase for all three of these alleles was between the first and second larval instars. WT controls were Canton-S. Strains used in the cytological mapping of wstd were obtained from the Bloomington Stock Center (Bloomington, IN).
Lifespan Analysis.
Flies were raised to adulthood at 22°C. Cohorts of 15 newly eclosed males were placed in single vials and incubated at 22°C or 29°C. Flies incubated at 29°C were transferred daily into fresh vials, whereas flies incubated at 22°C were transferred to fresh vials every other day. The number of surviving flies in each vial was recorded daily. Each set of 15 flies was treated as an independent sample. Survival curves were plotted as the mean percentage of surviving flies at each time point. T50 is defined as the time in days at which 50% of the sample still survives. Statistical comparisons were done by using Welch's t test.
Behavioral Tests.
Newly eclosed flies were collected under CO2, placed in vials, and aged at 22°C until testing. Groups of 15 flies were aspirated into preheated vials in a water bath at 38° and observed for 10 min. The number of flies paralyzed or seizing was recorded at timed intervals. After 10 min the flies were aspirated into fresh vials at 22°C. The number of flies that regained the ability to stand was recorded at timed intervals until all flies were standing.
Histology.
Newly eclosed flies were aged for 1–4 days at 22°C before being shifted to 29°C for 2–3 days. Heads were dissected and placed in fresh Carnoy's fixative for 24 h at 22°C, washed in 70% ethanol, and processed into paraffin by using standard histological procedures. Serial 6-μm frontal sections were taken, stained in hematoxylin and eosin, and examined under a light microscope. Neurodegeneration was indicated by the appearance of vacuolar lesions in the brain neuropil.
ATP Assays.
ATP content was determined by using a luciferase-based assay (Invitrogen, Carlsbad, CA) as described (44) with a Harta MicrolumiXS Microplate Luminometer with Lumiterm II acquisition software (Harta Instruments, Gaithersburg, MD). Eight adult females and eight adult males were used per genotype per sample. Duplicate measurements were made for each sample, and the values were corrected for protein content as determined by DC protein assays (Bio-Rad, Hercules, CA). Assays using ATP standards of known concentration were run in parallel to determine sensitivity and linear range of measurement. ATP concentrations of wstd samples were normalized as a percentage of WT control samples.
Molecular Analysis.
Genomic DNA was isolated from Canton-S and wstd1 adults. For wstd2, wstd3, and wstd4 genomic DNA was obtained from homozygous L1 larvae identified by using a GFP-tagged balancer chromosome. The genomic region that includes Tpi was amplified by PCR from each sample by using the primers 5′-CAACTTTGGACACCTTGTCG-3′and 5′-CCCCATCAGATGGTTGGAG-3′. The amplified PCR product was sequenced by using the above primers and the additional primer 5′-CGGGTGTGGCGGTCTGG-3′.
The genomic rescue construct was created by amplification of the Tpi region from Canton-S genomic DNA by using the primers, 5′-AAGCGGCCGCAGGATCACAAGGATGCGAAAGG-3′ and 5′-AAGCGGCCGCGTATCTAAGGCCACTTGGGTTCAAG-3′. These primers introduced NotI sites at the ends of the amplified fragment. The PCR product was digested with NotI and inserted into the NotI site of pCasper. Germ-line transformation with this construct via injection of embryos was carried out by standard methods as described (45).
Acknowledgments
We thank Tim Fergestad, Lingling Ho, Kate O'Connor-Giles, and Julie Simpson for advice, help, and comments on the manuscript and Allen Laughon, Francisco Pelegri, and Aki Ikeda for use of equipment. This work was supported by National Institutes of Health Grant R01-NS15390 and National Institutes of Health Genetics Training Grant 5T32GM07133. This is paper no. 3631 from the Laboratory of Genetics.
Abbreviations
- ts
temperature-sensitive
- Tpi
triosephosphate isomerase
- PGK
phosphoglycerate kinase
- GAP
glyceraldehyde-3-phosphate
- DHAP
dihydroxyacetone phosphate
- AGE
advanced glycation end product.
Footnotes
The authors declare no conflict of interest.
References
- 1.Siddiqi O, Benzer S. Proc Natl Acad Sci USA. 1976;73:3253–3257. doi: 10.1073/pnas.73.9.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu CF, Ganetzky B, Jan LY, Jan YN, Benzer S. Proc Natl Acad Sci USA. 1978;75:4047–4051. doi: 10.1073/pnas.75.8.4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ganetzky B, Wu CF. Annu Rev Genet. 1986;20:13–44. doi: 10.1146/annurev.ge.20.120186.000305. [DOI] [PubMed] [Google Scholar]
- 4.Loughney K, Kreber R, Ganetzky B. Cell. 1989;58:1143–1154. doi: 10.1016/0092-8674(89)90512-6. [DOI] [PubMed] [Google Scholar]
- 5.Atkinson NS, Robertson GA, Ganetzky B. Science. 1991;253:551–555. doi: 10.1126/science.1857984. [DOI] [PubMed] [Google Scholar]
- 6.Titus SA, Warmke JW, Ganetzky B. J Neurosci. 1997;17:875–881. doi: 10.1523/JNEUROSCI.17-03-00875.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuan LL, Ganetzky B. Science. 1999;283:1343–1345. doi: 10.1126/science.283.5406.1343. [DOI] [PubMed] [Google Scholar]
- 8.Coyle IP, Koh YH, Lee WC, Slind J, Fergestad T, Littleton JT, Ganetzky B. Neuron. 2004;41:521–534. doi: 10.1016/s0896-6273(04)00016-9. [DOI] [PubMed] [Google Scholar]
- 9.Bilen J, Bonini NM. Annu Rev Genet. 2005;39:153–171. doi: 10.1146/annurev.genet.39.110304.095804. [DOI] [PubMed] [Google Scholar]
- 10.Buchanan RL, Benzer S. Neuron. 1993;10:839–850. doi: 10.1016/0896-6273(93)90200-b. [DOI] [PubMed] [Google Scholar]
- 11.Min KT, Benzer S. Curr Biol. 1997;7:885–888. doi: 10.1016/s0960-9822(06)00378-2. [DOI] [PubMed] [Google Scholar]
- 12.Min KT, Benzer S. Science. 1999;284:1985–1988. doi: 10.1126/science.284.5422.1985. [DOI] [PubMed] [Google Scholar]
- 13.Tschape JA, Hammerschmied C, Muhlig-Versen M, Athenstaedt K, Daum G, Kretzschmar D. EMBO J. 2002;21:6367–6376. doi: 10.1093/emboj/cdf636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kretzschmar D, Hasan G, Sharma S, Heisenberg M, Benzer S. J Neurosci. 1997;17:7425–7432. doi: 10.1523/JNEUROSCI.17-19-07425.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finley KD, Edeen PT, Cumming RC, Mardahl-Dumesnil MD, Taylor BJ, Rodriguez MH, Hwang CE, Benedetti M, McKeown M. J Neurosci. 2003;23:1254–1264. doi: 10.1523/JNEUROSCI.23-04-01254.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Botella JA, Ulschmid JK, Gruenewald C, Moehle C, Kretzschmar D, Becker K, Schneuwly S. Curr Biol. 2004;14:782–786. doi: 10.1016/j.cub.2004.04.036. [DOI] [PubMed] [Google Scholar]
- 17.Palladino MJ, Hadley TJ, Ganetzky B. Genetics. 2002;161:1197–1208. doi: 10.1093/genetics/161.3.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palladino MJ, Bower JE, Kreber R, Ganetzky B. J Neurosci. 2003;23:1276–1286. doi: 10.1523/JNEUROSCI.23-04-01276.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fergestad T, Ganetzky B, Palladino MJ. Genetics. 2006;172:1031–1042. doi: 10.1534/genetics.105.050625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schneider AS, Valentine WN, Hattori M, Heins HL., Jr N Engl J Med. 1965;272:229–235. doi: 10.1056/NEJM196502042720503. [DOI] [PubMed] [Google Scholar]
- 21.Schneider AS. Baillieres Best Pract Res Clin Haematol. 2000;13:119–140. doi: 10.1053/beha.2000.0061. [DOI] [PubMed] [Google Scholar]
- 22.Lo TW, Westwood ME, McLellan AC, Selwood T, Thornalley PJ. J Biol Chem. 1994;269:32299–32305. [PubMed] [Google Scholar]
- 23.Kalapos MP. Toxicol Lett. 1999;110:145–175. doi: 10.1016/s0378-4274(99)00160-5. [DOI] [PubMed] [Google Scholar]
- 24.Ramasamy R, Vannucci SJ, Yan SS, Herold K, Yan SF, Schmidt AM. Glycobiology. 2005;15:16R–28R. doi: 10.1093/glycob/cwi053. [DOI] [PubMed] [Google Scholar]
- 25.Ramasamy R, Yan SF, Schmidt AM. Cell. 2006;124:258–260. doi: 10.1016/j.cell.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 26.Ahmed N, Battah S, Karachalias N, Babaei-Jadidi R, Horanyi M, Baroti K, Hollan S, Thornalley PJ. Biochim Biophys Acta. 2003;1639:121–132. doi: 10.1016/j.bbadis.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 27.Warmke JW, Kreuz AJ, Falkenthal S. Genetics. 1989;122:139–151. doi: 10.1093/genetics/122.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanaka KR, Zerez CR. Semin Hematol. 1990;27:165–185. [PubMed] [Google Scholar]
- 29.Orosz F, Wagner G, Liliom K, Kovacs J, Baroti K, Horanyi M, Farkas T, Hollan S, Ovadi J. Proc Natl Acad Sci USA. 2000;97:1026–1031. doi: 10.1073/pnas.97.3.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olah J, Orosz F, Keseru GM, Kovari Z, Kovacs J, Hollan S, Ovadi J. Biochem Soc Trans. 2002;30:30–38. doi: 10.1042/. [DOI] [PubMed] [Google Scholar]
- 31.Ovadi J, Orosz F, Hollan S. Mol Cell Biochem. 2004;256/257:83–93. doi: 10.1023/b:mcbi.0000009860.86969.72. [DOI] [PubMed] [Google Scholar]
- 32.Kikuchi S, Shinpo K, Moriwaka F, Makita Z, Miyata T, Tashiro K. J Neurosci Res. 1999;57:280–289. doi: 10.1002/(SICI)1097-4547(19990715)57:2<280::AID-JNR14>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 33.Kikuchi S, Shinpo K, Takeuchi M, Yamagishi S, Makita Z, Sasaki N, Tashiro K. Brain Res Brain Res Rev. 2003;41:306–323. doi: 10.1016/s0165-0173(02)00273-4. [DOI] [PubMed] [Google Scholar]
- 34.Thornalley PJ. Biochem Soc Trans. 2003;31:1343–1348. doi: 10.1042/bst0311343. [DOI] [PubMed] [Google Scholar]
- 35.Wang P, Saraswati S, Guan Z, Watkins CJ, Wurtman RJ, Littleton JT. J Neurosci. 2004;24:4518–4529. doi: 10.1523/JNEUROSCI.0542-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mazzola JL, Sirover MA. J Neurochem. 2001;76:442–449. doi: 10.1046/j.1471-4159.2001.00033.x. [DOI] [PubMed] [Google Scholar]
- 37.Sirover MA. Biochim Biophys Acta. 1999;1432:159–184. doi: 10.1016/s0167-4838(99)00119-3. [DOI] [PubMed] [Google Scholar]
- 38.Dauer W, Przedborski S. Neuron. 2003;39:889–909. doi: 10.1016/s0896-6273(03)00568-3. [DOI] [PubMed] [Google Scholar]
- 39.Bonnet R, Pavlovic S, Lehmann J, Rommelspacher H. Neuroscience. 2004;127:443–453. doi: 10.1016/j.neuroscience.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 40.Ahmed N, Ahmed U, Thornalley PJ, Hager K, Fleischer G, Munch G. J Neurochem. 2005;92:255–263. doi: 10.1111/j.1471-4159.2004.02864.x. [DOI] [PubMed] [Google Scholar]
- 41.Chen F, Wollmer MA, Hoerndli F, Munch G, Kuhla B, Rogaev EI, Tsolaki M, Papassotiropoulos A, Gotz J. Proc Natl Acad Sci USA. 2004;101:7687–7692. doi: 10.1073/pnas.0402338101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Junaid MA, Kowal D, Barua M, Pullarkat PS, Sklower Brooks S, Pullarkat RK. Am J Med Genet A. 2004;131:11–17. doi: 10.1002/ajmg.a.30349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yao D, Taguchi T, Matsumura T, Pestell R, Edelstein D, Giardino I, Suske G, Ahmed N, Thornalley PJ, Sarthy VP, et al. Cell. 2006;124:275–286. doi: 10.1016/j.cell.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 44.Fergestad T, Bostwick B, Ganetzky B. Genetics. 2006;173:1357–1364. doi: 10.1534/genetics.106.057463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kusano A, Staber C, Ganetzky B. Proc Natl Acad Sci USA. 2002;99:6866–6870. doi: 10.1073/pnas.102165099. [DOI] [PMC free article] [PubMed] [Google Scholar]