Abstract
Common fragile sites (CFSs) are seen as chromosomal gaps and breaks brought about by inhibition of replication, and it is thought that they cluster with tumor breakpoints. This study presents a comprehensive analysis using conventional and molecular cytogenetic mapping of CFSs and their expression frequencies in two mouse strains, BALB/c and C57BL/6, and in human probands. Here we show that induced mouse CFSs relate to sites of spontaneous gaps and breaks and that CFS expression levels in chromosome bands are conserved between the two mouse strains and between syntenic mouse and human DNA segments. Furthermore, four additional mouse CFSs were found to be homologous to human CFSs on the molecular cytogenetic level (Fra2D-FRA2G, Fra4C2-FRA9E, Fra6A3.1-FRA7G, and Fra6B1-FRA7H), increasing the number of such CFSs already described in the literature to eight. Contrary to previous reports, DNA helix flexibility is not increased in the 15 human and eight mouse CFSs molecularly defined so far, compared to large nonfragile control regions. Our findings suggest that the mechanisms that provoke instability at CFSs are evolutionarily conserved. The role that large transcriptionally active genes may play in CFS expression is discussed.
Common fragile sites (CFSs) are chromosomal loci that become visible (expressed) as chromatid gaps or chromosome breaks in a cell-type-dependent manner (Elder and Robinson 1989; Murano et al. 1989a) after culturing cells under replicative stress. CFS expression has been observed in various mammalian species (Arlt et al. 2003). Cytogenetic CFS mapping studies on murine lymphocytes revealed 19 CFSs defined as chromosome bands with significantly elevated gap/break frequencies that expressed the lesion homozygously as well (Elder and Robinson 1989). Comparative analysis in lymphocyte chromosomes of human, three great apes (chimpanzee, gorilla, orangutan), and a simian monkey species (Macaca fascicularis) has demonstrated that a high percentage of human CFSs were found in the equivalent location on chromosomes of apes/monkeys, although with sometimes significantly different expression frequencies (Yunis and Soreng 1984; Smeets and van de Klundert 1990; Ruiz-Herrera et al. 2002). A possible coincidence of CFSs with evolutionarily rearranged sites was also described (Smeets and van de Klundert 1990; Ruiz-Herrera et al. 2002). However, considering only the most breakage-prone sites of each primate, very few appeared to correspond to a synteny break (SB). To summarize, cytogenetic studies gave varying results and could not clarify to what degree CFSs were conserved over species frontiers or if they colocalize with evolutionary rearrangements. Until now only four mouse CFSs have been molecularly defined. These are Fra6C1, which corresponds to the human CFS FRA4F (Rozier et al. 2004); Fra8E1, the homolog of human FRA16D (Krummel et al. 2002); Fra12C1 in the homologous region of FRA7K (Helmrich et al. 2006); and Fra14A2 in the FRA3B corresponding region (Glover et al. 1998). DNA sequence comparison detected highly conserved regions between mouse and human homologous CFSs (Shiraishi et al. 2001; Krummel et al. 2002). It is therefore possible that despite their instability, CFSs represent (at least over some time) evolutionarily stable regions.
But what is the molecular mechanism that forces breakage at specific sites? The amount and composition of repetitive elements within molecularly mapped CFSs were shown to be similar to overall distributions found in mouse and human genomes (Mishmar et al. 1998; Ried et al. 2000; Shiraishi et al. 2001; Krummel et al. 2002; Morelli et al. 2002; Limongi et al. 2003; Matsuyama et al. 2003). Several publications present evidence for the existence of areas with elevated DNA helix flexibility (potential local conformational variations in the molecule) within the CFS regions. Islands with high flexibility were found in all of the currently analyzed nine human and three mouse CFS sequences (Mishmar et al. 1998; Ried et al. 2000; Krummel et al. 2002; Morelli et al. 2002; Limongi et al. 2003; Matsuyama et al. 2003; Zlotorynski et al. 2003; Rozier et al. 2004). Unfortunately, an appropriate number of analyzed stringently selected control regions was always missing. The initial study summed up the results for 14 very small regions consisting of 1.1 Mb total length (Mishmar et al. 1998). In a recent paper, 17.2 Mb of control sequences were analyzed; however, these sequences were also combined from smaller regions, and therefore their individual helix flexibility values were not shown (Zlotorynski et al. 2003).
All four molecularly defined mouse CFSs and their corresponding human CFSs localize to genes that span >750 kb of genomic sequence: Grid2/GRID2 (Fra6C1/FRA4F), Wwox/WWOX (Fra8E1/FRA16D), Immp2l/IMMP2L (Fra12C1/FRA7K), and Fhit/ FHIT (Fra14A2/FRA3B). Moreover, three human CFSs span the sequences of large genes: FRA6E (PARK2) (Denison et al. 2003), FRA7H (EXOC4, formerly known as SEC8) (Mishmar et al. 1998), and FRA7I (CNTNAP2) (Ciullo et al. 2002). Therefore, features present in large genes were suggested to be causally related to CFS formation (Rozier et al. 2004).
In this study, we scored mouse CFSs, defined as chromosomal regions that express gaps, double-strand breaks, or exchange figures under induced replicative stress, and then used statistics to classify the different chromosomal bands as having low, middle,or high CFS expression. We mappedspontaneous gaps/breaks as well as induced CFSs in the genomes of twomouse strains (BALB/c and C57BL/6)and then compared the expression ofCFSs between the mouse and the humangenomes. Furthermore, we characterizedat the molecular level several mouseCFSs that showed synteny to humanCFSs. We also examined the DNA helixflexibility for 23 mapped human andmouse CFSs and large control regions,and searched for tissue-specific gene expression of large genes that reside withinCFSs and control regions.
Results
Cytogenetic mapping of mouse and human CFSs
We performed a comprehensive cytogenetic analysis of CFSs in lymphocytes of the two mouse strains BALB/c and C57BL/6 in comparison to human lymphocytes. In almost all of the mouse and human (K. Stout-Weider, unpubl.) chromosome bands observed at the 350–400 band level, we detected aphidicolin (APC)-induced lesions with different frequencies.
CFS expression levels are conserved in mouse strains BALB/c and C57BL/6
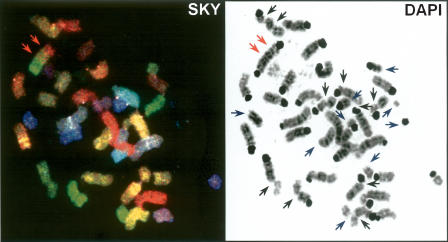
Spectral karyotyping (SKY) was applied in order to accomplish a reliable classification of the mouse chromosomes; chromosomal bands were identified with DAPI banding.
All spontaneously occurring or APC-induced chromatid gaps, deletions, and translocations were mapped along with their frequencies to their respective chromosomal band location (Fig. 1).
Figure 1.
Example of mouse CFS mapping. The lesions appearing as chromatid gaps (black arrows), chromosome breaks/deletions (blue arrows), or translocations (red arrows) were mapped to their cytogenetic locations according to the SKY classification (left) and the DAPI banding pattern (right). The figure shows an especially heavily rearranged metaphase with all three kinds of lesions.
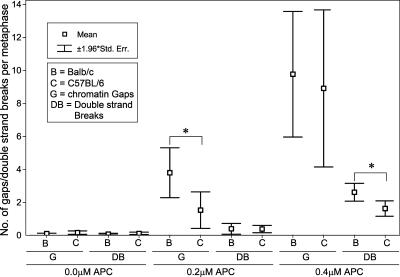
Chromatid gaps (G) and double- strand breaks (DB) including deletion breakpoints and exchange figures occurred at low frequencies of 0.2 ± 0.1 (BALB/c) and 0.3 ± 0.2 (C57BL/6) (events per metaphase, mean values ±1.96 SE) in control cells not exposed to APC. CFS expression showed a clear tendency to increase, with ascending APC concentrations, to higher levels in BALB/c as compared to C57BL/6 mice (BALB/c, G: 3.8 ± 1.7 at 0.2 μM APC, DB: 2.6 ± 0.6 at 0.4 μM APC; C57BL/6, G: 1.5 ± 1.3 at 0.2 μM APC, DB: 1.6 ± 0.5 at 0.4 μM APC; P < 0.05) (Fig. 2).
Figure 2.
Frequencies of chromatin gaps (G) and double-strand breaks (DB) at different APC concentrations in (B) BALB/c and (C) C57BL/6 mouse splenocytes. The y-axis shows the mean numbers of G and DB per metaphase; for each strain and APC concentration, means ± standard errors were calculated from counts in 125 metaphases (five mice). Statistically significant differences (Mann- Whitney U-test, P < 0.05) between strains are indicated with asterisks (*). The numbers of induced G and DB are slightly higher in BALB/c compared to C57BL/6 cells.
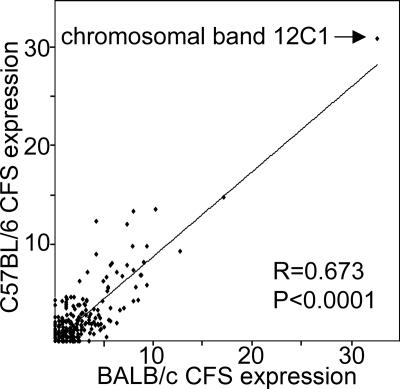
Correlation analysis of CFS frequencies of BALB/c and C57BL/6 mice at 0.4 μM APC showed a significant relation of medium strength (R = 0.67, P < 0.0001) (Fig. 3), that is, a conservation of the band-specific CFS expression levels in the two strains.
Figure 3.
Relation of CFS expression levels (including G and DB at0.4 μM APC, shown as percent of analyzed chromosomes) in 392 cyto- genetic bands of BALB/c and C57BL/6 mice. The line of best fit is shown. R is the Spearman test correlation coefficient. CFS expression on the chromosome band level is conserved in the two mouse strains.
Spontaneous gaps and breaks in mouse occur preferentially at chromosome bands with elevated APC-induced CFS expression
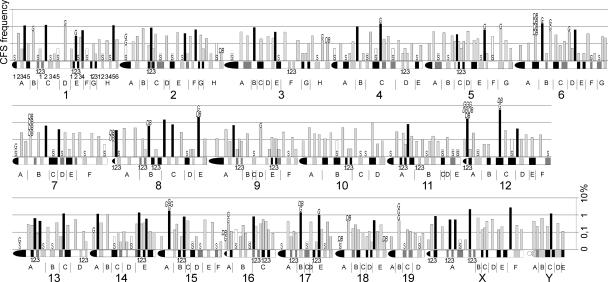
A detailed overview of the genome-wide CFS expression frequencies in mouse lymphocytes is shown in Figure 4. We were interested to see whether the rare spontaneous gaps and double- strand breaks occurred preferentially in bands with high APC- induced CFS expression. Therefore, we performed a mathematical calculation to divide the total of 392 chromosome bands into groups of low, medium, and high levels of APC-induced CFS expression using a nonrandom test (see Methods and Fig. 4).
Figure 4.
Log scale CFS expression levels in 392 mouse chromosome bands. The mouse chromosome ideogram (D. Adler, Department of Pathology at the University of Washington, supplemented according to http://www.ensembl.org/Mus_musculus/index.html) is displayed horizontally. All chromosome bands (A–H), sub-bands (1–6), and sub-sub-bands (1–3) are labeled for chromosome 1; sub-bands of the remaining chromosomes are, for the sake of clarity, not labeled and can be identified according to their position within chromosome bands and in relation to the indicated sub-sub-bands. CFS expression data were obtained in BALB/c and C57BL/6 splenocytes treated with 0.2 μM and 0.4 μM APC and are shown as the percent of analyzed chromosomes. Stable chromosome bands (S, white bars) with low APC sensitivity and bands with medium (gray) and high (black) CFS expression were identified with a nonrandom test (see Methods). Each spontaneous gap (G) and double-strand break (DB) found in untreated control cells are indicated.
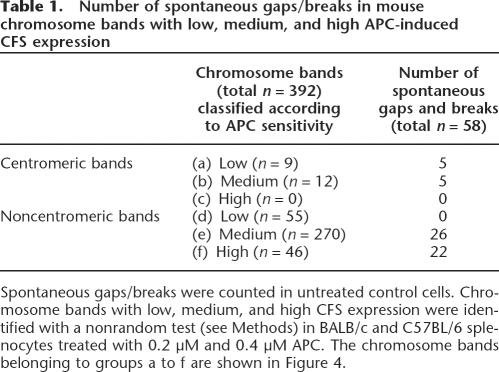
When comparing centromere and non-centromere chromo- some bands, we found that CFS expression increased after exposure to APC in non-centromeric chromosome bands (paired Wilcoxon test, P < 0.05) (data not shown), but not in centromeres (P = 0.07). Whereas spontaneous fragility in centromeric bands was relatively high (10 gaps/breaks in 21 centromeres), none of the 21 bands showed a high APC-induced fragility. On the other hand, all of the 48 spontaneous gaps/breaks that occurred in non-centromeric bands were found within the group of 316 bands with elevated CFS expression, and none were found within the group of 55 stable bands (Table 1; Fig. 4). Thus spontaneous non-centromeric gaps/breaks lie preferentially at sites with elevated APC-induced CFS expression (χ2 test, P < 0.0001).
Table 1.
Number of spontaneous gaps/breaks in mouse chromosome bands with low, medium, and high APC-induced CFS expression
CFS expression levels are conserved in syntenic regions of mouse and human chromosomes
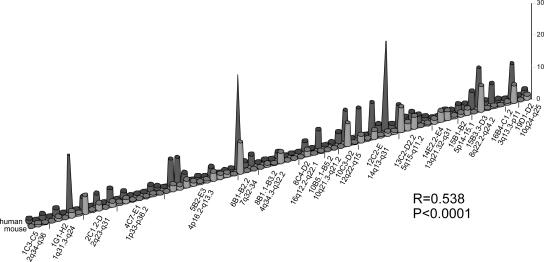
In order to analyze if CFS expression was conserved between mouse and human, as it was conserved between BALB/c and C57BL/6 strains (Fig. 3), we searched the Ensembl Genome Browser for large syntenic regions encompassing at least two chromosome bands. We detected 17 such conserved segments comprising a total of 77 bands (~500 Mb of mouse and human DNA, respectively). The median CFS expression frequencies of each of these 77 bands were subjected to a pairwise comparison between human and mouse. The Spearman test gave a correlation coefficient of R = 0.538 (P < 0.0001), showing a medium- strength correlation of CFS expression frequencies in homologous mouse and human bands (see also Fig. 5). This finding supports the view that CFS expression levels in syntenic regions are conserved during evolution.
Figure 5.
CFS expression levels in chromosome bands of 17 large (two or more cytogenetic bands) syntenic regions of the human and mouse (BALB/c and C57BL/6) genomes. x-axis: mouse and human chromosome bands; y-axis: frequency of CFS expression in percentages.
Variation of CFS expression in humans is not higher than that in inbred mice
To estimate variability in CFS expression between different individuals of the same species, the coefficient of variation (CV) was calculated for each chromosome band using CFS expression frequency data of five BALB/c and five C57BL/6 mice and two human groups with five persons each. CVs of all bands correlated between both human groups (R = 0.691) and between BALB/c and C57BL/6 (R = 0.410; P-values < 0.0001). Medians of the CV values including either all bands, all but stable bands, or only the bands expressing the highest level of CFSs (identified with the nonrandom test) were compared between the four groups. Each comparison showed lower median CV values for the groups of human probands as compared to the data obtained in mice (median CVs for all bands in human: 109 and 95; in BALB/c: 139; in C57BL/6: 158; for all but stable bands in human: 82 and 69; in BALB/c: 133; in C57BL/6: 153; for only high level CFS-expressing bands in human: 47 and 40; in BALB/c: 80; in C57BL/6: 102).
Molecular characterization of mouse and human CFSs
Four mouse CFSs syntenic to human CFSs were mapped at the DNA sequence level. Those newly mapped CFSs together with all other known molecularly identified CFSs were then analyzed for DNA helix flexibility, frequency of synteny breaks (SB), and presence of large genes, as well as gene expression levels of such genes. Altogether 74 Mb of DNA in human and mouse CFSs and 243 Mb of DNA in nonfragile control regions were analyzed.
Four CFSs are shown to be homologous between human and mouse at the molecular level
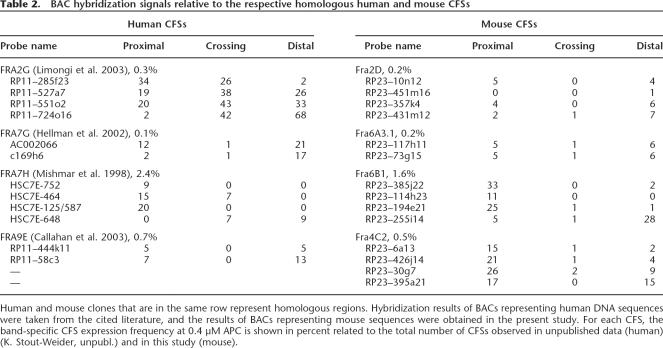
Multicolor FISH experiments with mouse BACs syntenic to human BACs located within the regions of CFSs FRA2G, FRA7G, FRA7H, and FRA9E allowed for the identification of the corresponding mouse CFSs (Table 2). Each of the four human CFSs has its corresponding CFS in the mouse genome spanning the exact homologous sequences. Together with the four already characterized CFSs—FRA3B/Fra14A2 (Glover et al. 1998), FRA4F/Fra6C1 (Rozier et al. 2004), FRA7K/Fra12C1 (Helmrich et al. 2006), and FRA16D/Fra8E1 (Krummel et al. 2002)—eight CFSs are now described to be conserved at the molecular level. The CFS expression frequencies (representing percentages of gaps/breaks/ exchange figures among all analyzed human and mouse chromosomes, respectively) of the syntenic chromosome bands containing the molecularly defined homologous human and mouse CFSs were 0.3%/0.2% for FRA2G/Fra2D, respectively,3.3%/1.1% for FRA3B/Fra14A2, 0.8%/0.4% for FRA4F/Fra6C1,2.0%/5.4% for FRA7K/Fra12C1, 0.1%/0.2% for FRA7G/Fra6A3.1,2.4%/1.6% for FRA7H/Fra6B1, 0.7%/0.5% for FRA9E/Fra4C2, and3.1%/1.8% for FRA16D/Fra8E1 (based on our human and mouse mapping studies). Correlation analysis revealed a significant association between the frequencies of CFS expression of these eight homologous CFSs (R = 0.762, P = 0.028).
Table 2.
BAC hybridization signals relative to the respective homologous human and mouse CFSs
Sites of evolutionary DNA rearrangements between the mouse and human genomes do not cluster in the 23 known CFS sequences
Since CFSs appear to represent genomic sites that are prone to DNA breakage, we opted to search in the human and mouse genomes for a possible coincidence of CFSs with regions that were rearranged during chromosome evolution. Out of the 15 human and eight mouse molecularly mapped CFSs, only two human CFSs and one mouse CFS contained a synteny break (SB) between the human and mouse genomes (the positions of SBs were defined based on the Ensembl Genome Browser) (see Supplemental Table S1). The average frequency of synteny breaks was lower in the CFS regions (one SB every 22.9 Mb in human and every 28.4 Mb in mouse) than in the control regions (one SB every 11.4 Mb in human and every 18.4 Mb in mouse). The control regions comprised 10 human and 10 mouse chromosome bands with the lowest CFS expression levels based on the 10 lowest Pd-values in the nonrandom test. Also, a part of human band 7q31 centromeric to FRA7K was included, where FISH experiments on APC-treated metaphase chromosomes detected no CFS expression (Helmrich et al. 2006). These data indicate that synteny breaks between the human and the mouse genome apparently do not cluster in CFSs.
DNA helix flexibility is not increased in mouse and human CFSs
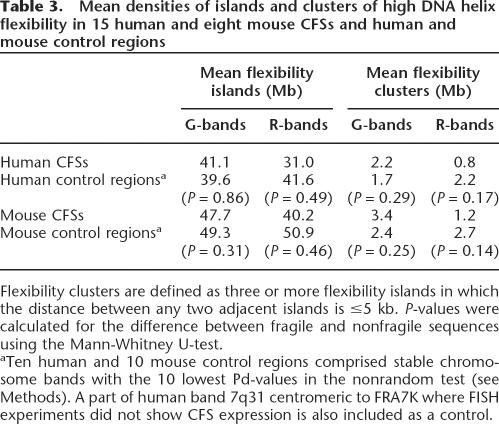
Using the software package TwistFlex developed by B. Kerem, we analyzed the helix flexibility of 74.2 Mb of DNA covering the 15 human and eight mouse precisely mapped CFSs and 243.1 Mb of DNA in control regions (defined by the nonrandom test). For every CFS and control region, the number of flexibility islands and their clusters per megabase were calculated (Supplemental Table S1). Since average numbers of flexibility islands in G-bands are higher than in R-bands owing to their higher AT content, we analyzed G- and R-bands separately.
Our analysis did not reveal significantly higher numbers of islands or clusters with increased helix flexibility in CFS regions compared to nonfragile regions (P ≥ 0.14) (Table 3). Moreover, neither in human nor in mouse did the number of flexibility islands relate to the expression frequency of the distinct CFSs (Spearman test; human: G-band CFSs, R = −0.476, P = 0.233; R- band CFSs, R = 0.071, P = 0.879; mouse: G-band CFSs, R = 0.500, P = 0.667; R-band CFSs, R = 0.100, P = 0.873). The same was found comparing the number of flexibility clusters to the CFS frequencies.
Table 3.
Mean densities of islands and clusters of high DNA helix flexibility in 15 human and eight mouse CFSs and human and mouse control regions
Comparing eight pairs of syntenic CFSs between human and mouse, the numbers of islands with increased flexibility were found to be conserved (Spearman R = 0.786, P = 0.021). However, the number of high flexibility clusters did not correlate (R = 0.265, P = 0.526).
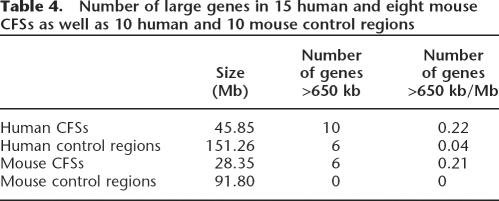
CFSs preferentially colocalize with large active genes
Nine of the 15 human and six of the eight mouse molecularly characterized CFSs include the coding regions of large genes (see Table 4 and Supplemental Table S1). Within 45.85 Mb of human CFS sequence, 10 genes >650 kb were found that map preferentially in the regions of the most frequently expressed CFSs. In contrast, among the 151.26 Mb of control sequence, only six large genes were found. In mouse, six large genes map within the28.35 Mb of known CFS sequence compared to none within 91.80 Mb of control sequence. Thus, large genes are found preferentially within CFS regions.
Table 4.
Number of large genes in 15 human and eight mouse CFSs as well as 10 human and 10 mouse control regions
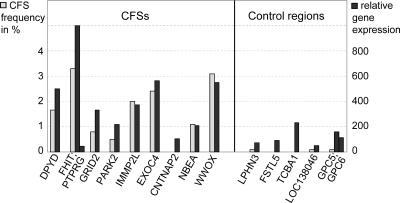
We determined the transcriptional activity of these large genes for human lymphocytes using RNA expression data published in the GNF SymAtlas (Fig. 6). Expression values >200 are common for genes at sites with frequent CFS expression. For example, the expression value of FHIT, which lies within the most frequent human lymphocyte CFS, FRA3B, is the highest among the analyzed large genes (Fig. 6; Supplemental Table S1). CNTNAP2, the large gene within FRA7I, is the only example showing an expression value close to the background level of 100. In agreement with the low CNTNAP2 expression, FRA7I is not detectable in lymphocytes. For all large genes in control regions except one (TCBA1 in 6q22), expression values below 200 were found. Moreover, we found a strong positive correlation between the expression values of large genes and the frequency of CFS formation at these sites (R = 0.857, P = 0.014).
Figure 6.
Expression levels of large human genes (≥650 kb) in CFSs and stable control regions. Shown on the x-axis are the large genes situated in nine of 15 CFSs and in five of 11 control regions. Gray bars indicate the frequency of CFS expression (left y-axis); black bars show the extent of gene expression (right y-axis, based on RNA expression array data from http://symatlas.gnf.org/SymAtlas/).
Discussion
In this work we present a whole-genome mapping study for CFSs in lymphocytes of mouse strains BALB/c and C57BL/6. Both mouse strains expressed a similar number of CFSs in control cells that were not subjected to APC treatment during cell culture. We show here that these spontaneous aberrations occur preferentially in the same non-centromeric bands that also show elevated levels of induced CFS expression, which means that APC increases the expression of natural fragile sites and does not accumulate artifacts. Interestingly, centromeric bands showed slightly higher levels of spontaneous gaps and breaks than noncentromeric bands, but the expression of centromeric CFSs did not increase after APC treatment. Thissuggests that gaps/breaks in centromeresoccur via separate mechanisms. Following APC induction, we found an elevated overall CFS expression in BALB/ccompared to C57BL/6 mice. This mightbear a relation to the high post-irradiation breast cancer risk of BALB/canimals (Storer et al. 1988) that is due tostrain-specific polymorphisms in thecoding region of Prkdc, causing chromatid-type aberrations (Ponnaiya et al.1997; Yu et al. 2001). The Prkdc gene encodes the DNA-dependent protein kinase catalytic subunit, which is knownto be involved in DNA double-strandedbreak repair and post-IR signal transduction (Yu et al. 2001). Furthermore, defects in different repair mechanismswere shown to increase the expression of CFSs (Turner et al. 2002), so that the Prkdc deficiency may contribute to the observed high number of CFSs induced by APC in BALB/c mice.
As shown in Figure 2, differences between CFS levels in BALB/c and C57BL/6 mice reached statistical significance for the number of gaps at 0.2 μM APC but not at 0.4 μM APC and for the number of DBs at 0.4 μM APC but not at 0.2 μM APC. Mechanistically, this could mean that the gaps seen at lower dose are converted to DBs at higher dose. Despite different sensitivities to APC between BALB/c and C57BL/6, we showed a high degree of conservation of CFS expression frequencies per chromosome band. A strain-overlapping conservation was furthermore affirmed in a comparison of our data to published results from APC-treated lymphocytes of ICR mice (Elder and Robinson 1989). Out of the 19 bands cited, 14 belong to the 23 most frequently observed CFSs in our study at comparable conditions of induction. In addition to the conservation of CFS expression levels among different mouse strains, we found the same for syntenic regions of the mouse and human genomes. Such conservation was already described for the most frequent human CFSs compared to the genomes of different great apes (Yunis and Soreng 1984; Smeets and van de Klundert 1990). Our data shown in Figure 5 support the conclusion that a conservation is also true for less frequent CFSs. Therefore, since every chromosome segment apparently has its specific tendency to form a CFS that is conserved during evolution, we suggest basing the fragility classification on the whole continuum of CFS expression levels (e.g., low, medium, and high) rather than on division into fragile or nonfragile DNA regions.
On a molecular basis, we showed four CFSs to be conserved between the human and mouse genomes (FRA2G/Fra2D, FRA7G/ Fra6A3.1, FRA7H/Fra6B1, and FRA9E/Fra4C2). Thus, together with FRA3B, FRA4F, FRA7K, and FRA16D, which correspond to mouse Fra14A2 (Glover et al. 1998), Fra6C1 (Rozier et al. 2004), Fra12C1 (Helmrich et al. 2006), and Fra8E1 (Krummel et al. 2002), respectively, eight CFSs are now found to be conserved on the DNA sequence level. Their mouse versus human expression frequencies correlate significantly.
To answer the question whether SBs might have occurred as a consequence of breakage at CFSs, we looked for SBs between mouse and human genomes in mapped fragile sequences and in low-fragility areas and found CFSs without SBs as well as control regions with SBs. The comparison of SB frequencies between CFSs and control regions revealed no elevation in CFSs. Still, the problem remains that to date we only have 15 CFS sequences to include in the analysis, representing only a small population of the total number of CFSs. Clearly, the analysis needs to be extended in the future when additional CFSs are precisely mapped on the DNA sequence level. Yet our results include some of the most frequently expressed CFSs (as FRA3B, 16D, XB, 7H, 7K, 13A), adding weight to the observation that SBs do not cluster into this group of 15 CFSs. To explain the observed result, one can speculate that mechanisms different from CFS expression are involved in the formation of evolutionary chromosomal rearrangements, for example, random breakage as proposed by Nadeau and Taylor (1984). However, it is also possible that synteny breaks do occur in CFSs but as a result of the breakage reduce or lose the ability to appear as a CFS in the split regions.
The characterization of 15 human and eight mouse CFS- expressing sequences and large nonfragile regions provides a powerful tool to search for further molecular features of CFSs. To date, the cellular mechanisms leading to the formation of CFSs are still largely unknown. Several recent studies showed that areas with elevated DNA helix flexibility lie within CFS sequences (Mishmar et al. 1998; Zlotorynski et al. 2003; Rozier et al. 2004). However, these studies lack a sufficient number of nonfragile control regions. We used stringent criteria to choose control regions (243 Mb) and showed that human and mouse CFS regions do not contain an elevated number of highly flexible structures in comparison to large low-fragility control regions. It has to be noted that the mouse DNA sequence still shows gaps that might lead to false results, even though the number of these gaps is similar for CFS and control regions. Thus, our comprehensive analysis demonstrated that, in contrast to the current view in the literature, elevated DNA helix flexibility is obviously not related to the formation of CFSs. Also, because of the following observations, it is rather improbable that the DNA sequence per se accounts for the expression of CFSs. Firstly, the interindividual CFS expression frequency variation (expressed as median CV values of all chromosome bands) in humans is not higher than among inbred mouse strains that are characterized by genomic sequence identities of almost 100%. Secondly, CFSs were found to be expressed in a tissue-dependent manner (Elder and Robinson 1989; Murano et al. 1989b; Simonic and Gericke 1996). Here we showed that human CFSs differ from nonfragile regions in that the former frequently house transcriptionally active genes that span >650 kb of genomic sequence. Large genes appear also very frequently in the mouse CFS regions and were absent in control areas. Owing to the lack of gene expression data for mouse tissues, we are not able to conclude on the transcriptional activity of these genes in mouse lymphocytes.
The large genes located within CFSs (see Supplemental Table S1) encode very different proteins varying from neurotransmitter receptors (GRID2), mediators of cell–cell interactions in the nervous system (CNTNAP2 and EXOC4), enzymes in uracil and thymidine catabolism (DPYD), or mediators of protein kinase A sub- cellular location (NBEA) to proteins involved in protein degradation (PARK2) and cell cycle control (FHIT). Some of the proteins are thought to act as tumor suppressors (FHIT, WWOX, PTPRG). However, all these genes are similar in that they are expressed in the tissue in which the CFSs are inducible. This clearly separates the large genes of the CFS regions from those in control areas.
The concept of an association of the localization of CFSs with active gene regions is furthermore supported by the expression of a CFS at Xp22.3 (a region escaping X-inactivation) in both homologous chromosomes in females, whereas the CFS at Xq22.1 is expressed only in the transcriptionally active X (Austin et al. 1992).
It is known that the DNA polymerase inhibitor APC acts during S phase by reducing or stopping DNA synthesis (Pedrali-Noy et al. 1981). Furthermore, CFSs have always been studied in mitotic chromosomes, which means in cells that have been exposed to APC in S phase and (erroneously) have passed the G2/M checkpoint. The function of proteins that are involved in checkpoint control such as ATR or BRCA1 was described to be essential for the integrity of the CFS regions (Casper et al. 2002; Turner et al. 2002; Arlt et al. 2004). Therefore, we conclude that stalled replication forks at large transcriptionally active sites coupled with escape of checkpoint control may contribute to CFS expression and the induction of DNA double-strand breaks. However, this concept needs further experimental validation. To date, we can only speculate what molecular mechanisms might link CFSs to regions of large active genes. A direct interaction with the transcription machinery, which has also been found to be active during (mammalian) S phase (Jensen et al. 1993), might slow down the replicating polymerase, rendering the sites of interaction favored for replication stagnation. The probability of such interaction may increase with the size of the actively transcribed gene. Evidence for delayed replication (in late-replicating DNA) has been shown for CFSs FRA3B and FRA16D (Wang et al. 1999; Palakodeti et al. 2004). Furthermore, recent genome-wide studies addressing the issue of interplay between nuclear RNA and DNA synthesis showed a strong link between transcription and replication timing (Schubeler et al. 2002; Woodfine et al. 2004). Therefore, CFSs may represent regions where the temporal coordination of the two processes is especially critical and sensitive to problems.
Methods
Metaphase chromosome preparation and induction of CFSs
Mouse splenocytes from five BALB/c and five C57BL/6 mice (BALB/c: three female and two male; C57BL/6: two female and three male) were cultured for 48 h in RPMI 1640 medium plus 20% FCS and 0.0006% β-mercaptoethanol. T-lymphocytes were stimulated using 6 ng/μL Concanavalin A, B-lymphocytes by 2.5 ng/μL lipopolysaccharide. CFSs were induced by adding aphidicolin (APC) at concentrations of 0.0 μM, 0.2 μM, or 0.4 μM to the cell culture medium 24 h prior to the harvest. Human T- lymphocytes were obtained from peripheral blood of 10 unrelated volunteers and cultured for 72 h in RPMI medium 1640, 20% FBS, and 3 μg/mL phytohemaglutinine (Biochrom) supplemented with 0.4 μM APC for 24 h.
Scoring of CFSs
CFSs, defined as lesions that appeared as gaps, double-strand breaks, or exchange figures, were mapped by analyzing the DAPI images of 25 complete metaphase spreads of each mouse and for three different APC concentrations (750 metaphases in total). To ensure the proper identification of the mouse chromosomes, Spectral Karyotyping was performed as previously described (Schrock et al. 1996). Each lesion observed was taken into consideration to define the CFS expression frequencies of individual chromosomal bands (with a resolution of ~5–20 Mb). A subsequent statistical calculation was done to distinguish between bands with high, medium, or low CFS levels (see below).
Preparation of BACs and their use for fluorescence in situ hybridization (FISH)
BACs were obtained from the library RP23 (see http://genomics. roswellpark.org/). DNA was prepared following standard protocols. BAC DNA was labeled via nick translation with Biotin-16- dUTP, DEAC-dUTP, Digoxygenin-dUTP, or TAMRA-dUTP. Hybridization and detection procedures were performed as described (Schrock et al. 1996).
Statistical analysis
The nonrandom test (Dahm et al. 2002) was applied to identify chromosome regions with extremely low, medium, and high CFS expression frequencies. For this, the CFS mapping data from 0.2 μM and 0.4 μM APC-treated lymphocytes of mouse strains BALB/c and C57BL/6 were combined. For each of the 392 mouse chromosome bands, the number of observed APC-induced CFSs (a), the number of analyzed chromosomes (b), and the size of the band in megabases (obtained from http://www.ensembl.org) (c) were regarded. Because b was slightly varying for different chromosomes and close to 1000, a was normalized to d: d = (1000/ b) × a. The expected number of CFSs for a specific band (e) was calculated taking into account the size of chromosomal bands: e = (sum of d-values) × [c/(sum of all c-values)]. Using Microsoft Excel and applying the binomial distribution, P-values were calculated for increased CFS expression: Pi = 1-BINOMVERT(a,b,e/ 1000,TRUE) + BINOMVERT(a,b,e/1000,FALSE), and for decreased CFS expression: Pd = BINOMVERT(a,b,e/1000,TRUE). Bands were considered to show extremely low or high CFS expression frequencies if their Pd or Pi values, respectively, were below the Bonferroni bound of 1.28 × 10−4 (0.05/392). All other bands were classified as medium fragile.
Furthermore, standard statistical tests such as the Mann- Whitney U-test, the paired Wilcoxon test, and the Spearman test were applied using the software package Statistica for Windows Version 5.5 (StatSoft Inc.). Coefficient of variation (CV) values were calculated as 100 × SD/(mean value).
Use of databases
Positions of BACs and genes as well as DNA sequence data of CFSs and control regions were taken from http://genome.ucsc.edu, version May 2004. Human/mouse syntenic segments were defined by use of the Ensembl Genome Browser (http://www.ensembl.org, July 2004 version).
Mouse sequences were allocated to their chromosome location based on gene mapping data as indicated in the NCBILocusLink. The exact boundaries of mouse G- and R-bands were determined according to gene and CpG-island densities as shown in the UCSC Genome Browser.
Evolutionary breakpoints were identified by analyzing data from http://www.ensembl.org (version July 2004). Human gene expression data were obtained from the GNF SymAtlas (http:// symatlas.gnf.org/SymAtlas/, Version 1.2.4) (Su et al. 2002). Because the data were based on hybridization of fixed amounts of overall mRNA, values of different genes within one tissue can be compared directly. Values were obtained from Pb-CD4 + Tcells and Pb-CD8 + Tcells (T-lymphocytes). For genes with more than one given RNA expression result, mean values were calculated. Gene expression values were compared to the CFS frequencies using the Spearman correlation test; at regions including two large genes, only the gene with the higher expression value was taken into account (FHIT but not PTPRG, and GPC5 but not GPC6). Mouse gene expression analysis data are not included because of the lack of large genes in mouse control regions.
Analysis of DNA helix flexibility
To evaluate DNA flexibility, we used a measure of the potential local variations in the DNA structure, expressed as fluctuations in the twist angle (Sarai et al. 1989). This measure provides average twist angle fluctuations for each of the possible dinucleotides and thus enables the evaluation of the flexibility of a DNA sequence by summation of these values. To carry out the analysis, TwistFlex was used as provided by B. Kerem at http://www.jail.cs.huji.ac.il/~netab/index.html. The analysis was performed according to B. Kerem and coworkers (Zlotorynski et al. 2003) in windows of 100 bp with a shift increment of 1 bp. Dinucleotide values were summed along the window and averaged by the window length. Windows with values >13.7° were considered as flexibility peaks. Since the window length is 100 bp, flexibility peaks that were <100 bp apart were considered as one flexibility peak. Flexibility peaks as well as clusters of flexibility peaks (defined as at least three flexibility peaks in which the distance between any two adjacent peaks is ≤5 kb) were calculated. As a control, we recalculated the helix flexibility for published DNA sequences submitted to flexibility analysis (Zlotorynski et al. 2003) and obtained identical results.
ACKNOWLEDGMENTS
We regret to announce that Klaus Hermann died in November 2005. This work was funded by the German Federal Ministry of Education and Research (BioFuture Award 0811375 to E.S.).
Footnotes
Supplemental material is available online at www.genome.org.
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5335506.
REFERENCES
- Arlt M.F., Casper A.M., Glover T.W., Casper A.M., Glover T.W., Glover T.W. Common fragile sites. Cytogenet. Genome Res. 2003;100:92–100. doi: 10.1159/000072843. [DOI] [PubMed] [Google Scholar]
- Arlt M.F., Xu B., Durkin S.G., Casper A.M., Kastan M.B., Glover T.W., Xu B., Durkin S.G., Casper A.M., Kastan M.B., Glover T.W., Durkin S.G., Casper A.M., Kastan M.B., Glover T.W., Casper A.M., Kastan M.B., Glover T.W., Kastan M.B., Glover T.W., Glover T.W. BRCA1 is required for common-fragile-site stability via its G2/M checkpoint function. Mol. Cell. Biol. 2004;24:6701–6709. doi: 10.1128/MCB.24.15.6701-6709.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin M.J., Collins J.M., Corey L.A., Nance W.E., Neale M.C., Schieken R.M., Brown J.A., Collins J.M., Corey L.A., Nance W.E., Neale M.C., Schieken R.M., Brown J.A., Corey L.A., Nance W.E., Neale M.C., Schieken R.M., Brown J.A., Nance W.E., Neale M.C., Schieken R.M., Brown J.A., Neale M.C., Schieken R.M., Brown J.A., Schieken R.M., Brown J.A., Brown J.A. Aphidicolin-inducible common fragile-site expression: Results from a population survey of twins. Am. J. Hum. Genet. 1992;50:76–83. [PMC free article] [PubMed] [Google Scholar]
- Callahan G., Denison S.R., Phillips L.A., Shridhar V., Smith D.I., Denison S.R., Phillips L.A., Shridhar V., Smith D.I., Phillips L.A., Shridhar V., Smith D.I., Shridhar V., Smith D.I., Smith D.I. Characterization of the common fragile site FRA9E and its potential role in ovarian cancer. Oncogene. 2003;22:590–601. doi: 10.1038/sj.onc.1206171. [DOI] [PubMed] [Google Scholar]
- Casper A.M., Nghiem P., Arlt M.F., Glover T.W., Nghiem P., Arlt M.F., Glover T.W., Arlt M.F., Glover T.W., Glover T.W. ATR regulates fragile site stability. Cell. 2002;111:779–789. doi: 10.1016/s0092-8674(02)01113-3. [DOI] [PubMed] [Google Scholar]
- Ciullo M., Debily M.A., Rozier L., Autiero M., Billault A., Mayau V., El Marhomy S., Guardiola J., Bernheim A., Coullin P., Debily M.A., Rozier L., Autiero M., Billault A., Mayau V., El Marhomy S., Guardiola J., Bernheim A., Coullin P., Rozier L., Autiero M., Billault A., Mayau V., El Marhomy S., Guardiola J., Bernheim A., Coullin P., Autiero M., Billault A., Mayau V., El Marhomy S., Guardiola J., Bernheim A., Coullin P., Billault A., Mayau V., El Marhomy S., Guardiola J., Bernheim A., Coullin P., Mayau V., El Marhomy S., Guardiola J., Bernheim A., Coullin P., El Marhomy S., Guardiola J., Bernheim A., Coullin P., Guardiola J., Bernheim A., Coullin P., Bernheim A., Coullin P., Coullin P., et al. Initiation of the breakage-fusion-bridge mechanism through common fragile site activation in human breast cancer cells:The model of PIP gene duplication from a break at FRA7I. Hum. Mol. Genet. 2002;11:2887–2894. doi: 10.1093/hmg/11.23.2887. [DOI] [PubMed] [Google Scholar]
- Dahm P.F., Olmsted A.W., Greenbaum I.F., Olmsted A.W., Greenbaum I.F., Greenbaum I.F. Probability models and the applicability of statistical procedures in the identification of chromosomal fragile sites. Biometrics. 2002;58:1028–1033. doi: 10.1111/j.0006-341x.2002.01028.x. [DOI] [PubMed] [Google Scholar]
- Denison S.R., Callahan G., Becker N.A., Phillips L.A., Smith D.I., Callahan G., Becker N.A., Phillips L.A., Smith D.I., Becker N.A., Phillips L.A., Smith D.I., Phillips L.A., Smith D.I., Smith D.I. Characterization of FRA6E and its potential role in autosomal recessive juvenile parkinsonism and ovarian cancer. Genes Chromosomes Cancer. 2003;38:40–52. doi: 10.1002/gcc.10236. [DOI] [PubMed] [Google Scholar]
- Elder F.F., Robinson T.J., Robinson T.J. Rodent common fragile sites: Are they conserved? Evidence from mouse and rat. Chromosoma. 1989;97:459–464. doi: 10.1007/BF00295030. [DOI] [PubMed] [Google Scholar]
- Glover T.W., Hoge A.W., Miller D.E., Ascara-Wilke J.E., Adam A.N., Dagenais S.L., Wilke C.M., Dierick H.A., Beer D.G., Hoge A.W., Miller D.E., Ascara-Wilke J.E., Adam A.N., Dagenais S.L., Wilke C.M., Dierick H.A., Beer D.G., Miller D.E., Ascara-Wilke J.E., Adam A.N., Dagenais S.L., Wilke C.M., Dierick H.A., Beer D.G., Ascara-Wilke J.E., Adam A.N., Dagenais S.L., Wilke C.M., Dierick H.A., Beer D.G., Adam A.N., Dagenais S.L., Wilke C.M., Dierick H.A., Beer D.G., Dagenais S.L., Wilke C.M., Dierick H.A., Beer D.G., Wilke C.M., Dierick H.A., Beer D.G., Dierick H.A., Beer D.G., Beer D.G. The murine Fhit gene is highly similar to its human orthologue and maps to a common fragile site region. Cancer Res. 1998;58:3409–3414. [PubMed] [Google Scholar]
- Hellman A., Zlotorynski E., Scherer S.W., Cheung J., Vincent J.B., Smith D.I., Trakhtenbrot L., Kerem B., Zlotorynski E., Scherer S.W., Cheung J., Vincent J.B., Smith D.I., Trakhtenbrot L., Kerem B., Scherer S.W., Cheung J., Vincent J.B., Smith D.I., Trakhtenbrot L., Kerem B., Cheung J., Vincent J.B., Smith D.I., Trakhtenbrot L., Kerem B., Vincent J.B., Smith D.I., Trakhtenbrot L., Kerem B., Smith D.I., Trakhtenbrot L., Kerem B., Trakhtenbrot L., Kerem B., Kerem B. A role for common fragile site induction in amplification of human oncogenes. Cancer Cell. 2002;1:89–97. doi: 10.1016/s1535-6108(02)00017-x. [DOI] [PubMed] [Google Scholar]
- Helmrich A., Stout-Weider K., Matthaei A., Hermann K., Heiden T., Schrock E., Stout-Weider K., Matthaei A., Hermann K., Heiden T., Schrock E., Matthaei A., Hermann K., Heiden T., Schrock E., Hermann K., Heiden T., Schrock E., Heiden T., Schrock E., Schrock E. Identification of the human/mouse syntenic common fragile site FRA7K/Fra12C1—Relation of FRA7K and other human common fragile sites on chromosome 7 to evolutionary breakpoints. Int. J. Cancer. 2006 doi: 10.1002/ijc.22049. (in press) [DOI] [PubMed] [Google Scholar]
- Jensen P.O., Larsen J., Christiansen J., Larsen J.K., Larsen J., Christiansen J., Larsen J.K., Christiansen J., Larsen J.K., Larsen J.K. Flow cytometric measurement of RNA synthesis using bromouridine labeling and bromodeoxyuridine antibodies. Cytometry. 1993;14:455–458. doi: 10.1002/cyto.990140416. [DOI] [PubMed] [Google Scholar]
- Krummel K.A., Denison S.R., Calhoun E., Phillips L.A., Smith D.I., Denison S.R., Calhoun E., Phillips L.A., Smith D.I., Calhoun E., Phillips L.A., Smith D.I., Phillips L.A., Smith D.I., Smith D.I. The common fragile site FRA16D and its associated gene WWOX are highly conserved in the mouse at Fra8E1. Genes Chromosomes Cancer. 2002;34:154–167. doi: 10.1002/gcc.10047. [DOI] [PubMed] [Google Scholar]
- Limongi M.Z., Pelliccia F., Rocchi A., Pelliccia F., Rocchi A., Rocchi A. Characterization of the human common fragile site FRA2G. Genomics. 2003;81:93–97. doi: 10.1016/s0888-7543(03)00007-7. [DOI] [PubMed] [Google Scholar]
- Matsuyama A., Shiraishi T., Trapasso F., Kuroki T., Alder H., Mori M., Huebner K., Croce C.M., Shiraishi T., Trapasso F., Kuroki T., Alder H., Mori M., Huebner K., Croce C.M., Trapasso F., Kuroki T., Alder H., Mori M., Huebner K., Croce C.M., Kuroki T., Alder H., Mori M., Huebner K., Croce C.M., Alder H., Mori M., Huebner K., Croce C.M., Mori M., Huebner K., Croce C.M., Huebner K., Croce C.M., Croce C.M. Fragile site orthologs FHIT/FRA3B and Fhit/Fra14A2: Evolutionarily conserved but highly recombinogenic. Proc. Natl. Acad. Sci. 2003;100:14988–14993. doi: 10.1073/pnas.2336256100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishmar D., Rahat A., Scherer S.W., Nyakatura G., Hinzmann B., Kohwi Y., Mandel-Gutfroind Y., Lee J.R., Drescher B., Sas D.E., Rahat A., Scherer S.W., Nyakatura G., Hinzmann B., Kohwi Y., Mandel-Gutfroind Y., Lee J.R., Drescher B., Sas D.E., Scherer S.W., Nyakatura G., Hinzmann B., Kohwi Y., Mandel-Gutfroind Y., Lee J.R., Drescher B., Sas D.E., Nyakatura G., Hinzmann B., Kohwi Y., Mandel-Gutfroind Y., Lee J.R., Drescher B., Sas D.E., Hinzmann B., Kohwi Y., Mandel-Gutfroind Y., Lee J.R., Drescher B., Sas D.E., Kohwi Y., Mandel-Gutfroind Y., Lee J.R., Drescher B., Sas D.E., Mandel-Gutfroind Y., Lee J.R., Drescher B., Sas D.E., Lee J.R., Drescher B., Sas D.E., Drescher B., Sas D.E., Sas D.E., et al. Molecular characterization of a common fragile site(FRA7H) on human chromosome 7 by the cloning of a simian virus 40 integration site. Proc. Natl. Acad. Sci. 1998;95:8141–8146. doi: 10.1073/pnas.95.14.8141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morelli C., Karayianni E., Magnanini C., Mungall A.J., Thorland E., Negrini M., Smith D.I., Barbanti-Brodano G., Karayianni E., Magnanini C., Mungall A.J., Thorland E., Negrini M., Smith D.I., Barbanti-Brodano G., Magnanini C., Mungall A.J., Thorland E., Negrini M., Smith D.I., Barbanti-Brodano G., Mungall A.J., Thorland E., Negrini M., Smith D.I., Barbanti-Brodano G., Thorland E., Negrini M., Smith D.I., Barbanti-Brodano G., Negrini M., Smith D.I., Barbanti-Brodano G., Smith D.I., Barbanti-Brodano G., Barbanti-Brodano G. Cloning and characterization of the common fragile site FRA6F harboring a replicative senescence gene and frequently deleted in human tumors. Oncogene. 2002;21:7266–7276. doi: 10.1038/sj.onc.1205573. [DOI] [PubMed] [Google Scholar]
- Murano I., Kuwano A., Kajii T., Kuwano A., Kajii T., Kajii T. Cell type-dependent difference in the distribution and frequency of aphidicolin-induced fragile sites: T and B lymphocytes and bone marrow cells. Hum. Genet. 1989a;84:71–74. doi: 10.1007/BF00210675. [DOI] [PubMed] [Google Scholar]
- Murano I., Kuwano A., Kajii T., Kuwano A., Kajii T., Kajii T. Fibroblast-specific common fragile sites induced by aphidicolin. Hum. Genet. 1989b;83:45–48. doi: 10.1007/BF00274145. [DOI] [PubMed] [Google Scholar]
- Nadeau J.H., Taylor B.A., Taylor B.A. Lengths of chromosomal segments conserved since divergence of man and mouse. Proc. Natl. Acad. Sci. 1984;81:814–818. doi: 10.1073/pnas.81.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palakodeti A., Han Y., Jiang Y., Le Beau M.M., Han Y., Jiang Y., Le Beau M.M., Jiang Y., Le Beau M.M., Le Beau M.M. The role of late/slow replication of the FRA16D in common fragile site induction. Genes Chromosomes Cancer. 2004;39:71–76. doi: 10.1002/gcc.10290. [DOI] [PubMed] [Google Scholar]
- Pedrali-Noy G., Kuenzle C.C., Focher F., Belvedere M., Spadari S., Kuenzle C.C., Focher F., Belvedere M., Spadari S., Focher F., Belvedere M., Spadari S., Belvedere M., Spadari S., Spadari S. An enzymatic method for microdetermination of aphidicolin: A promising anticancer drug. J. Biochem. Biophys. Methods. 1981;4:113–121. doi: 10.1016/0165-022x(81)90024-5. [DOI] [PubMed] [Google Scholar]
- Ponnaiya B., Cornforth M.N., Ullrich R.L., Cornforth M.N., Ullrich R.L., Ullrich R.L. Radiation-induced chromosomal instability in BALB/c and C57BL/6 mice: The difference is as clear as black and white. Radiat. Res. 1997;147:121–125. [PubMed] [Google Scholar]
- Ried K., Finnis M., Hobson L., Mangelsdorf M., Dayan S., Nancarrow J.K., Woollatt E., Kremmidiotis G., Gardner A., Venter D., Finnis M., Hobson L., Mangelsdorf M., Dayan S., Nancarrow J.K., Woollatt E., Kremmidiotis G., Gardner A., Venter D., Hobson L., Mangelsdorf M., Dayan S., Nancarrow J.K., Woollatt E., Kremmidiotis G., Gardner A., Venter D., Mangelsdorf M., Dayan S., Nancarrow J.K., Woollatt E., Kremmidiotis G., Gardner A., Venter D., Dayan S., Nancarrow J.K., Woollatt E., Kremmidiotis G., Gardner A., Venter D., Nancarrow J.K., Woollatt E., Kremmidiotis G., Gardner A., Venter D., Woollatt E., Kremmidiotis G., Gardner A., Venter D., Kremmidiotis G., Gardner A., Venter D., Gardner A., Venter D., Venter D., et al. Common chromosomal fragile site FRA16D sequence: Identification of the FOR gene spanning FRA16D and homozygous deletions and translocation breakpoints in cancer cells. Hum. Mol. Genet. 2000;9:1651–1663. doi: 10.1093/hmg/9.11.1651. [DOI] [PubMed] [Google Scholar]
- Rozier L., El-Achkar E., Apiou F., Debatisse M., El-Achkar E., Apiou F., Debatisse M., Apiou F., Debatisse M., Debatisse M. Characterization of a conserved aphidicolin-sensitive common fragile site at human 4q22 and mouse 6C1: Possible association with an inherited disease and cancer. Oncogene. 2004;23:6872–6880. doi: 10.1038/sj.onc.1207809. [DOI] [PubMed] [Google Scholar]
- Ruiz-Herrera A., Ponsa M., Garcia F., Egozcue J., Garcia M., Ponsa M., Garcia F., Egozcue J., Garcia M., Garcia F., Egozcue J., Garcia M., Egozcue J., Garcia M., Garcia M. Fragile sites in human and Macaca fascicularis chromosomes are breakpoints in chromosome evolution. Chromosome Res. 2002;10:33–44. doi: 10.1023/a:1014261909613. [DOI] [PubMed] [Google Scholar]
- Sarai A., Mazur J., Nussinov R., Jernigan R.L., Mazur J., Nussinov R., Jernigan R.L., Nussinov R., Jernigan R.L., Jernigan R.L. Sequence dependence of DNA conformational flexibility. Biochemistry. 1989;28:7842–7849. doi: 10.1021/bi00445a046. [DOI] [PubMed] [Google Scholar]
- Schrock E., du Manoir S., Veldman T., Schoell B., Wienberg J., Ferguson-Smith M.A., Ning Y., Ledbetter D.H., Bar-Am I., Soenksen D., du Manoir S., Veldman T., Schoell B., Wienberg J., Ferguson-Smith M.A., Ning Y., Ledbetter D.H., Bar-Am I., Soenksen D., Veldman T., Schoell B., Wienberg J., Ferguson-Smith M.A., Ning Y., Ledbetter D.H., Bar-Am I., Soenksen D., Schoell B., Wienberg J., Ferguson-Smith M.A., Ning Y., Ledbetter D.H., Bar-Am I., Soenksen D., Wienberg J., Ferguson-Smith M.A., Ning Y., Ledbetter D.H., Bar-Am I., Soenksen D., Ferguson-Smith M.A., Ning Y., Ledbetter D.H., Bar-Am I., Soenksen D., Ning Y., Ledbetter D.H., Bar-Am I., Soenksen D., Ledbetter D.H., Bar-Am I., Soenksen D., Bar-Am I., Soenksen D., Soenksen D., et al. Multicolor spectral karyotyping of human chromosomes. Science. 1996;273:494–497. doi: 10.1126/science.273.5274.494. [DOI] [PubMed] [Google Scholar]
- Schubeler D., Scalzo D., Kooperberg C., van Steensel B., Delrow J., Groudine M., Scalzo D., Kooperberg C., van Steensel B., Delrow J., Groudine M., Kooperberg C., van Steensel B., Delrow J., Groudine M., van Steensel B., Delrow J., Groudine M., Delrow J., Groudine M., Groudine M. Genome-wide DNA replication profile for Drosophila melanogaster:A link between transcription and replication timing. Nat. Genet. 2002;32:438–442. doi: 10.1038/ng1005. [DOI] [PubMed] [Google Scholar]
- Shiraishi T., Druck T., Mimori K., Flomenberg J., Berk L., Alder H., Miller W., Huebner K., Croce C.M., Druck T., Mimori K., Flomenberg J., Berk L., Alder H., Miller W., Huebner K., Croce C.M., Mimori K., Flomenberg J., Berk L., Alder H., Miller W., Huebner K., Croce C.M., Flomenberg J., Berk L., Alder H., Miller W., Huebner K., Croce C.M., Berk L., Alder H., Miller W., Huebner K., Croce C.M., Alder H., Miller W., Huebner K., Croce C.M., Miller W., Huebner K., Croce C.M., Huebner K., Croce C.M., Croce C.M. Sequence conservation at human and mouse orthologous common fragile regions, FRA3B/FHIT and Fra14A2/Fhit. Proc. Natl. Acad. Sci. 2001;98:5722–5727. doi: 10.1073/pnas.091095898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonic I., Gericke G.S., Gericke G.S. The enigma of common fragile sites. Hum. Genet. 1996;97:524–531. doi: 10.1007/BF02267080. [DOI] [PubMed] [Google Scholar]
- Smeets D.F., van de Klundert F.A., van de Klundert F.A. Common fragile sites in man and three closely related primate species. Cytogenet. Cell Genet. 1990;53:8–14. doi: 10.1159/000132885. [DOI] [PubMed] [Google Scholar]
- Storer J.B., Mitchell T.J., Fry R.J., Mitchell T.J., Fry R.J., Fry R.J. Extrapolation of the relative risk of radiogenic neoplasms across mouse strains and to man. Radiat. Res. 1988;114:331–353. [PubMed] [Google Scholar]
- Su A.I., Cooke M.P., Ching K.A., Hakak Y., Walker J.R., Wiltshire T., Orth A.P., Vega R.G., Sapinoso L.M., Moqrich A., Cooke M.P., Ching K.A., Hakak Y., Walker J.R., Wiltshire T., Orth A.P., Vega R.G., Sapinoso L.M., Moqrich A., Ching K.A., Hakak Y., Walker J.R., Wiltshire T., Orth A.P., Vega R.G., Sapinoso L.M., Moqrich A., Hakak Y., Walker J.R., Wiltshire T., Orth A.P., Vega R.G., Sapinoso L.M., Moqrich A., Walker J.R., Wiltshire T., Orth A.P., Vega R.G., Sapinoso L.M., Moqrich A., Wiltshire T., Orth A.P., Vega R.G., Sapinoso L.M., Moqrich A., Orth A.P., Vega R.G., Sapinoso L.M., Moqrich A., Vega R.G., Sapinoso L.M., Moqrich A., Sapinoso L.M., Moqrich A., Moqrich A., et al. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl. Acad. Sci. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner B.C., Ottey M., Zimonjic D.B., Potoczek M., Hauck W.W., Pequignot E., Keck-Waggoner C.L., Sevignani C., Aldaz C.M., McCue P.A., Ottey M., Zimonjic D.B., Potoczek M., Hauck W.W., Pequignot E., Keck-Waggoner C.L., Sevignani C., Aldaz C.M., McCue P.A., Zimonjic D.B., Potoczek M., Hauck W.W., Pequignot E., Keck-Waggoner C.L., Sevignani C., Aldaz C.M., McCue P.A., Potoczek M., Hauck W.W., Pequignot E., Keck-Waggoner C.L., Sevignani C., Aldaz C.M., McCue P.A., Hauck W.W., Pequignot E., Keck-Waggoner C.L., Sevignani C., Aldaz C.M., McCue P.A., Pequignot E., Keck-Waggoner C.L., Sevignani C., Aldaz C.M., McCue P.A., Keck-Waggoner C.L., Sevignani C., Aldaz C.M., McCue P.A., Sevignani C., Aldaz C.M., McCue P.A., Aldaz C.M., McCue P.A., McCue P.A., et al. The fragile histidine triad/common chromosome fragile site 3B locus and repair-deficient cancers. Cancer Res. 2002;62:4054–4060. [PubMed] [Google Scholar]
- Wang L., Darling J., Zhang J.S., Huang H., Liu W., Smith D.I., Darling J., Zhang J.S., Huang H., Liu W., Smith D.I., Zhang J.S., Huang H., Liu W., Smith D.I., Huang H., Liu W., Smith D.I., Liu W., Smith D.I., Smith D.I. Allele-specific late replication and fragility of the most active common fragile site, FRA3B. Hum. Mol. Genet. 1999;8:431–437. doi: 10.1093/hmg/8.3.431. [DOI] [PubMed] [Google Scholar]
- Woodfine K., Fiegler H., Beare D.M., Collins J.E., McCann O.T., Young B.D., Debernardi S., Mott R., Dunham I., Carter N.P., Fiegler H., Beare D.M., Collins J.E., McCann O.T., Young B.D., Debernardi S., Mott R., Dunham I., Carter N.P., Beare D.M., Collins J.E., McCann O.T., Young B.D., Debernardi S., Mott R., Dunham I., Carter N.P., Collins J.E., McCann O.T., Young B.D., Debernardi S., Mott R., Dunham I., Carter N.P., McCann O.T., Young B.D., Debernardi S., Mott R., Dunham I., Carter N.P., Young B.D., Debernardi S., Mott R., Dunham I., Carter N.P., Debernardi S., Mott R., Dunham I., Carter N.P., Mott R., Dunham I., Carter N.P., Dunham I., Carter N.P., Carter N.P. Replication timing of the human genome. Hum. Mol. Genet. 2004;13:191–202. doi: 10.1093/hmg/ddh016. [DOI] [PubMed] [Google Scholar]
- Yu Y., Okayasu R., Weil M.M., Silver A., McCarthy M., Zabriskie R., Long S., Cox R., Ullrich R.L., Okayasu R., Weil M.M., Silver A., McCarthy M., Zabriskie R., Long S., Cox R., Ullrich R.L., Weil M.M., Silver A., McCarthy M., Zabriskie R., Long S., Cox R., Ullrich R.L., Silver A., McCarthy M., Zabriskie R., Long S., Cox R., Ullrich R.L., McCarthy M., Zabriskie R., Long S., Cox R., Ullrich R.L., Zabriskie R., Long S., Cox R., Ullrich R.L., Long S., Cox R., Ullrich R.L., Cox R., Ullrich R.L., Ullrich R.L. Elevated breast cancer risk in irradiated BALB/c mice associates with unique functional polymorphism of the Prkdc (DNA-dependent protein kinase catalytic subunit) gene. Cancer Res. 2001;61:1820–1824. [PubMed] [Google Scholar]
- Yunis J.J., Soreng A.L., Soreng A.L. Constitutive fragile sites and cancer. Science. 1984;226:1199–1204. doi: 10.1126/science.6239375. [DOI] [PubMed] [Google Scholar]
- Zlotorynski E., Rahat A., Skaug J., Ben-Porat N., Ozeri E., Hershberg R., Levi A., Scherer S.W., Margalit H., Kerem B., Rahat A., Skaug J., Ben-Porat N., Ozeri E., Hershberg R., Levi A., Scherer S.W., Margalit H., Kerem B., Skaug J., Ben-Porat N., Ozeri E., Hershberg R., Levi A., Scherer S.W., Margalit H., Kerem B., Ben-Porat N., Ozeri E., Hershberg R., Levi A., Scherer S.W., Margalit H., Kerem B., Ozeri E., Hershberg R., Levi A., Scherer S.W., Margalit H., Kerem B., Hershberg R., Levi A., Scherer S.W., Margalit H., Kerem B., Levi A., Scherer S.W., Margalit H., Kerem B., Scherer S.W., Margalit H., Kerem B., Margalit H., Kerem B., Kerem B. Molecular basis for expression of common and rare fragile sites. Mol. Cell. Biol. 2003;23:7143–7151. doi: 10.1128/MCB.23.20.7143-7151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]