Abstract
The emerging sequence of the heterochromatic portion of the Drosophila melanogaster genome, with the most recent update of euchromatic sequence, gives the first genome-wide view of the chromosomal distribution of the telomeric retrotransposons, HeT-A, TART, and Tahre. As expected, these elements are entirely excluded from euchromatin, although sequence fragments of HeT-A and TART 3 untranslated regions are found in nontelomeric heterochromatin on the Y chromosome. The proximal ends of HeT-A/TART arrays appear to be a transition zone because only here do other transposable elements mix in the array. The sharp distinction between the distribution of telomeric elements and that of other transposable elements suggests that chromatin structure is important in telomere element localization. Measurements reported here show (1) D. melanogaster telomeres are very long, in the size range reported for inbred mouse strains (averaging 46 kb per chromosome end in Drosophila stock 2057). As in organisms with telomerase, their length varies depending on genotype. There is also slight under-replication in polytene nuclei. (2) Surprisingly, the relationship between the number of HeT-A and TART elements is not stochastic but is strongly correlated across stocks, supporting the idea that the two elements are interdependent. Although currently assembled portions of the HeT-A/TART arrays are from the most-proximal part of long arrays, ~61% of the total HeT-A sequence in these regions consists of intact, potentially active elements with little evidence of sequence decay, making it likely that the content of the telomere arrays turns over more extensively than has been thought.
Telomeres of the genus Drosophila, like those of other eukaryotes, are made up of tandem arrays of nucleotide repeats copied from an RNA template. The distinctive feature of Drosophila telomeres lies in the sequence repeats themselves. In all other studied organisms, telomere repeats are very short simple sequences, which do not code for proteins. In contrast, the Drosophila telomere repeats are two retrotransposable elements, HeT-A and TART (see Pardue and DeBaryshe 2003). (In Drosophila melanogaster there are also a few copies of Tahre, an element combining sequences of HeT-A and TART [Abad et al. 2004b]. Tahre has not yet been reported in other Drosophila species.) The Drosophila telomere elements are 2 or 3 orders of magnitude larger than telomere repeats in other organisms, and they encode proteins used for their retrotransposition.
HeT-A, TART, and Tahre are non–long terminal repeat (non- LTR) retrotransposons, and the proteins they encode are closely related to the proteins encoded by a group of non-LTR retrotransposons that are abundant in Drosophila genomes. However, HeTA, TART, and Tahre are distinguished from these other retrotransposons in two important ways:
First, HeT-A, TART, and Tahre transpose specifically to chromosome ends, apparently identifying ends by some feature other than DNA sequence. In situ hybridization experiments have not detected these elements in euchromatic regions except when they have bound to the end of a chromosome that has broken in euchromatin (Traverse and Pardue 1988; Biessmann et al. 1990b). Other Drosophila retrotransposons transpose to many sites in euchromatin but have not been found in clones derived from telo- mere HeT-A/TART arrays.
Second, while most retrotransposons contain very little DNA that does not code for proteins, HeT-A, TART, and Tahre have 3 untranslated regions (3 UTRs) that make up nearly half their sequence. As is typical of UTRs, the sequence of these regions evolves rapidly. Nonetheless, HeT-A and TART arrays maintain the strand composition bias seen on other telomeres—the sense strand is always A+C-rich (Danilevskaya et al. 1998), like telomerase template sequences (Henderson 1995). In addition, HeT-A 3 UTRs have a pattern of irregularly spaced A-rich regions in every Drosophila species studied (Danilevskaya et al. 1998; Casacuberta and Pardue 2003).
This combination of unusual, but well-defined, chromosomal distribution and unusual sequence organization suggests that these features are related. The possibility of such a relationship is further emphasized by the fact that fragments of their 3 UTR sequence have been found not only in telomeric heterochromatin but also in other heterochromatic regions of the genome (Danilevskaya et al. 1991, 1993; Losada et al. 1999; Casacuberta and Pardue 2002). Such an association could result if these unusual sequences are positively selected for survival in heterochromatin, are deleterious if transposed into euchromatin, or both. The implications of this apparent relationship between chromosomal distribution and HeT-A/TART sequences can be investigated effectively only now as we begin to have a better idea of exactly how well our current limited information reflects the true distribution of these sequences in the genome.
The D. melanogaster euchromatic genome has been completely sequenced (Adams et al. 2000; Celniker et al. 2002), but the heterochromatic portion presents challenges for sequencing that are only slowly being overcome. Therefore, most available HeT-A and TART sequences have come from small fragments, either subcloned or amplified by PCR. There is little evidence for the exact genomic site from which these fragments originated; most of what we do know has been deduced from in situ hybridization to polytene chromosomes. In these giant chromosomes, in situ localization of cloned sequences is complicated by cross- hybridization to the many copies of HeT-A and TART and, at least sometimes, by under-replication of some heterochromatic sequences. For example, neither Y chromosomes nor pericentric satellite sequences are detectably polytenized (Gall et al. 1971). The replication status of other heterochromatic sequences is less well determined. If HeT-A and TART sequences are part of the set that is greatly under-replicated in salivary glands, then polytene chromosomes might not give a complete picture of the localization of these sequences.
The Drosophila Heterochromatin Genome Project is extending sequence of the D. melanogaster genome (stock 2057) into heterochromatic regions (Hoskins et al. 2002). Specifically, there is now assembled sequence extending into the telomere on the right end of chromosome 4 (4R) and the left end of the X chromosome (XL). These assemblies give the first detailed view of telomere structure in D. melanogaster. The following analysis of these sequences confirms that the chromosomal distribution of HeT-A and TART differs sharply from that of other retrotransposons in ways that are consistent with their roles at the telo- mere. They also provide new insight into the production and turnover of telomere arrays.
The results of this sequence analysis, in conjunction with quantitative hybridization measurements, allowed us to determine the magnitude and range of variation of telomere sequence in flies of different genotypes and in established cell lines. Previous molecular characterization of eukaryotic telomeres produced the unexpected finding that average lengths of telomerase- maintained telomeres fluctuate around equilibrium values specified by the genotype, tissue, and environment. Our measurements of Drosophila telomeres suggest that, despite their unusual retrotransposon mechanism, these telomeres have a similarly dynamic behavior.
Results
HeT-A, TART, and Tahre sequences are not found in euchromatic regions
BLAST searches of the finished euchromatic sequence in Release4.2 of the genome confirm in situ hybridization studies showing no HeT-A, TART, or Tahre sequences in euchromatin. There are no sequences with significant similarity (E < 0.002) to HeT-A or Tahre sequences. TART elements have 90 bp in the pol coding region with significant similarity to the pol region of BS, a non- LTR element found in several euchromatic sites. Euchromatin contains a few other sequences with similarity to TART 3 UTRs, but they are so short and scattered that it is likely that they are incidental rather than evidence of TART insertions (see Supplemental material, Section 1). All of these similarities localize to the 3 UTR sequences of TART.
These studies show that HeT-A, TART, and Tahre are strongly excluded from euchromatin. The exclusion of the telomeric elements from a major part of the genome is striking because the euchromatic regions contain elements from at least 76 families of other retrotransposons (Kaminker et al. 2002), which shows that retroelements can be accommodated in many sites. One possible mechanism for this exclusion is that HeT-A and TART RNA are specifically targeted by their Gag proteins to chromosome ends for reverse transcription (Rashkova et al. 2002, 2003). In addition, although we know nothing yet about how the 5 end of the reverse transcript is processed, it seems likely that HeT-A and TART have a special mechanism for forming free distal ends that helps preclude insertion at internal sites in the chromosome.
In contrast to their presence in euchromatin, other transposable elements are excluded from telomeric HeT-A/TART arrays except for a short transition region at the proximal edge of the array (see below).
Assembled sequence now extends into the HeT-A/TART arrays on the right end of chromosome 4 (4R) and the left endof chromosome X (XL)
Overall sequence organization
To date, in the currently assembled sequence, HeT-A/TART arrays form 75,943 bp on the end of 4R and 19,199 bp on XL (the structure of these arrays is depicted graphically in Fig. 1 and Supplemental Fig. 1). Restriction mapping of BACs used for the genome sequence shows that the terminal HeT-A/TART array on chromosome XL contains ~147 kb of sequence (Abad et al. 2004a). Thus the 19-kb XL HeT-A sequence reported here lies deep within the array. However, for 4R, the 76-kb sequence may include most, if not all, of the 4R telomere because the BAC used for the sequence is the longest BAC found for this telomere.
Figure 1.
Elements in the HeT-A/TART array in the telomere of 4R. (A) Overview of elements in assembled sequence. Not drawn to scale. End of chromosome is on left. The right end begins with the most distal gene, CAPS, 425 bp from most proximal TART. All HeT-A and TART elements are in the same orientation, and except for those in the transition zone, all truncated elements have lost sequence from the 5 end. (For details on size and sequence content of particular elements, see B.) (B) Elements from the HeT-A/TART array in A. Each bar represents an element in the array drawn to scale with the total number of nucleotides indicated at the 5 end. The most distal element in the assembled array is on top, and the most proximal telomeric element is at the bottom. Because elements are variously truncated from the 5 end, they are aligned from the 3 end (at the right axis). In the array the 3 -most sequence of each element is actually connected to the 5 end of the element beneath it, but elements are shown separately for clarity. Open coding regions are light blue; complete coding regions are marked ORF. 3 ends marked A have an intact 3 end and oligo(A). HeT-A oligo(A) tails in this study range from 3 to 26 bp, averaging 8 bp; TART tails are longer, from 14 to 23 bp with an average of 18. Red “A” indicates elements thought to be “tags” (see text). Nontelomeric elements in the array (purple) are indicated by name on the right axis. Element marked “T” has an intact 3 end but has oligo(T) rather than oligo(A). Asterisk (*) indicates an element missing final GTT and oligo(A). Elements not marked “A” are 3 -truncated and shown displaced from the right axis to show their alignment with complete elements; in some cases there are enough missing 3 nucleotides to be seen as white space between the element and the right axis of the graph.
Each HeT-A and TART in these two arrays, whether complete or partial, is orientated with its 3 end toward the center of the chromosome. Both the XL and the 4R arrays contain mixtures of complete and partial elements (for details, see Supplemental Table 1). Because partial elements are truncated from the 5 end, ORFs, which all reside near the 5 end, are lost preferentially, and telomere regions are enriched in the 3 UTR sequences that make up an extraordinarily large part of the sequence of these telomere elements (Pardue and DeBaryshe 2003). For HeT-A there is also a special class of elements with <62 bp of the 3 -most sequence of the element plus an oligo(A). We call these “tags,” and, as discussed below, the unusual HeT-A promoter produces them. Such drastically truncated TART tags are not found, nor are they expected, because TART has a more conventional promoter.
The array on 4R begins only 425 bp distal to the first identified gene (CAPS), whereas the array on XL is separated from the most distal identified gene, Sp71, by a 52.5-kb subtelomeric region containing many transposable elements, including INE-1, Cr1a, and roo but neither HeT-A nor TART. In other D. melanogaster stocks, the sequence immediately interior to HeT-A and TART on the XL telomere is occupied by the repeats of Telomere Associated Sequences (TAS) (Karpen and Spradling 1992; Walter et al. 1995). However, in the stock sequenced by the Genome Project (hereafter referred to as 2057) the XL telomere lacks TAS (Abad et al. 2004a). Instead, in this 52.5-kb subtelomeric region, we identify only two small fragments of X TAS (49 bp and 147 bp).
HeT-A and TART elements are almost completely separated from the other sequences in the genome
On XL the array of HeT-A elements contains only one other sequence, 246 bp of novel sequence immediately distal to the most proximal (74 bp) HeT-A element. On 4R the 75.9-kb HeT-A/TART array has other transposable elements but only in the proximal 5.4 kb. This 5.4 kb appears to be a transition zone with fragments of HeT-A and TART mixed with fragments of nontelomeric elements 1360, Cr1a, and ninja. These HeT-A and TART elements are broken up in ways not seen in the rest of the array. (One HeT-A has been interrupted by insertion of 1360; also, with one minor exception discussed below, all HeT-As and TARTs without intact 3 ends are in this region.)
There is no obvious reason why other non-LTR elements, which are relatively abundant in euchomatin, should not transpose into telomere arrays. In fact, one might suppose that telomere arrays would be safe landing sites because they contain no known vital genes to be disrupted. However, previous studies of HeT-A and TART have not reported these other elements in Drosophila telomeres (see Pardue and DeBaryshe 2003). The 4R HeT-A/TART array is the first to show any mingling of other transposable elements, and that only in a narrow region adjacent to the interior of the chromosome. This mingling of nontelomeric with telomeric elements shows that their separation in other regions is not due to incompatibility of their DNA sequences. Possibly the segregation is dictated by chromatin structure; telo- mere arrays may have a chromatin structure that prevents insertion by other elements, a structure that becomes less regular near the proximal edge, permitting other elements to invade.
Much of the assembled sequences consist of complete HeT-A elements
Complete elements constitute 61.4% of the 61,467 bp of HeT-A DNA in the assembled sequences (~22% of the total HeT-A DNA measured by Southern hybridizations reported below). There are four (and possibly five) complete HeT-A elements distributed throughout the 4R array and one in the XL array (Fig. 1; Supplemental Fig. 1). We presume these four elements are competent because they contain what appears to be a complete set of the necessary sequences. The possibly competent fifth element in the 4R array, the second from the distal end, lacks the terminal GTT and oligo(A); however, RNA polymerase might continue into the truncated 3 UTR immediately downstream, yielding an RNA transposition intermediate with the terminal sequence necessary for reverse transcription onto the chromosome. If so, this illustrates one possible mechanism for forming length variants of HeT-A.
Not unexpectedly, these competent HeT-A elements differ by both nucleotide changes and insertions/deletions (indels). Significant sequence differences are typical of previously cloned HeT-A elements from various (unknown) chromosomal sites (Biessmann et al. 1992; Pardue et al. 1996; Danilevskaya et al. 1998); the 4R sequence shows similar heterogeneity in an array of elements on a single chromosome end (see Supplemental Tables 2 and 3).
Indels in the 5 and 3 UTRs might not be expected to have much effect on function because these regions are noncoding and have rather repetitive sequences. Surprisingly, there are also a number of indels in the coding sequences, yet none of these cause premature termination of translation. The longest of the encoded proteins is 30 amino acids longer than its shortest homolog, with most of the length difference due to a large indel near the N-terminal end, in a region previously identified as length polymorphic (Pardue et al. 1996). Furthermore, none of the sequence differences in the ORFs of these complete elements appear to affect the amino acid segments we have identified as important for telomere localization of these Gag proteins (Rashkova et al. 2003).
HeT-A tags
These numerous tiny 3 ends consist of <62 bp plus a terminal oligo(A). Rather than resulting from vigorous erosion of full- length elements, most of them are a product of the unusual HeT-A promoter (Danilevskaya et al. 1997). This promoter is located at the 3 end of each element and drives not its own transcription, but transcription of the HeT-A element immediately downstream, when such an element exists. HeT-A transcripts start either 31 or 62 nucleotides inside the element that contains the promoter, and thus the 5 end of each new HeT-A RNA transcript contains a tiny piece of the 3 end of the upstream neighbor, the “tag.” These tags form the extreme 5 end of the chromosome when an element is newly transposed. Their length is variable, which could result either from incomplete replication at transposition or from erosion while they form the extreme end of the chromosome.
As expected, eight of the 11 tags in the 4R and XL HeT-A/ TART arrays are immediately 5 of complete elements, either singly or in multiples (Fig. 1; Supplemental Fig. 1). In 4R, three are in tandem before the 5840-bp element (ignoring the 12-bp novel sequence), two are before the 5848-bp element, and one lies directly before the 6012-bp element. In XL a tandem pair precedes the 6006-bp element. We assume these tandem tags originate from multiple transpositions because an element should acquire an additional tag at each transposition. The other three tags are puzzling because none is directly 5 of a complete element; instead they are 5 of truncated HeT-A elements and should have been lost during truncation. We can only speculate on the origin of these last three tags.
A cluster of TARTs on 4R
The array on 4R contains sequence from all three subfamilies of TART, but the largest contribution is from a cluster containing two complete and three partial copies of TART A. This is a significant fraction of all TART elements thought to be in this stock. However, it is known that TART elements are not evenly distributed on telomeres. In situ hybridization to polytene chromosomes from seven stocks showed no stock had detectable TART sequence on all telomeres; indeed, each stock had a different set of labeled telomeres (Levis et al. 1993). Clustering of HeT-A elements should be positively selected because any individual HeT-A is efficiently transcribed only if its upstream neighbor is also a HeT-A (Danilevskaya et al. 1997); as a consequence, TART elements should also cluster by default.
Surprisingly all the TART A elements in this cluster have >99.9% identical sequences in the regions where they overlap with each other (several are truncated) and with a complete TART A element (AJ566116) sequenced by Abad et al. (2004a) from the X chromosome of this same Canton S–derived stock. Furthermore, this sequence is 99.3% identical to a TART A from the distantly related Oregon R stock (AY561850). Sequence similarity within this TART subfamily is in sharp contrast to the strong sequence divergence seen between subfamilies; the three TART subfamilies have only 68%–70% nucleotide identity with indels and nucleotide changes in both coding and untranslated regions (Supplemental Tables 4, 5).
The sequences of these TART A elements provide strong support for the conclusion that the Perfect Non-Terminal Repeats (PNTRs) found at the 5 end and near the 3 end of TART elements are evolving together, somehow achieving the same sequence changes at each end of the element (Danilevskaya et al. 1999). In the PNTR regions, these Canton S elements differ from the Oregon R element by four indels of 1–35 bp and three nucleotide changes. All four indels and two of the three nucleotide changes are found in both 5 and 3 PNTRs. This concerted evolution of the 5 and 3 UTRs suggests that at some point in the life cycle there is recombination or gene conversion between the two PNTRs. Such a process may explain the large differences in length between TART 5 UTRs. The apparently complete elements on 4R have 5 UTRs of 717 and 670 bp. The one sequenced by Abad et al. (2004a) on X has a 5 UTR of 3934 bp. For each of these elements the entire length of the 5 UTR is identical to sequence within the 3 UTR. Thus much of the longer 5 UTRs may simply be a by-product of an interaction that occurs for other reasons, and the extended sequence may not be necessary for subsequent transposition. We tentatively consider the three apparently intact TART A elements to be potentially active.
Telomeric fragments in nontelomeric regions of the heterochromatic Y chromosome
The Y is the only completely heterochromatic Drosophila chromosome. The many repeated sequences on the Y make sequence assembly very difficult; however, seven sequence scaffolds of Y chromosome genes have been assembled (Carvalho et al. 2001). In these recent extensions of the Y sequence, we find HeT-A 3 UTR sequences 5 of CG40441, CG40442, and CG49448 (Ppl-Y2). TART 3 UTR sequences are associated with CG17866 (Kl-2). Within each scaffold, all HeT-A/TART sequences are in the same orientation, whether in tandem or separated by other elements.
These nontelomeric HeT-A/TART sequences in the Y heterochromatin differ from those in telomeric arrays in two important ways: First, they are inserted into the chromosome, rather than onto an end. Second, the fragments contain only 3 UTR sequence, yet some elements lack the extreme 3 sequences that appear to be required for transposition. These differences suggest that these nontelomeric HeT-A/TART elements did not transpose into the Y in the way elements transpose to the telomere.
In these Y chromosomal regions, HeT-A and TART 3 fragments are associated with other elements (Cr1a, Idefix, Ine-1, X- element, Gypsy, and Stalker), in a way reminiscent of the organization of the transition zone on 4R, suggesting that a segment of such a region could have been moved into the Y in a yet undefined way. HeT-A and TART 3 UTR sequences are a significant fraction of the very small amount of nontelomeric heterochromatic sequence now assembled, whereas essentially none of this sequence is found in euchromatin, striking evidence for our belief that HeT-A and TART sequences cannot enter euchromatin or are detrimental once entered, or both.
Much of the Y chromosome sequence is not yet assembled. We expect more HeT-A and TART fragments to be found because, in addition to the scattered fragments reported here, the Y has at least two large regions of nontelomeric tandem repeats containing HeT-A and TART fragments not yet included in the assembled scaffolds (Danilevskaya et al. 1991, 1993; Losada et al. 1999).
Estimating the number of HeT-A/TART elements in telomere arrays
Measurement of HeT-A/TART sequences on telomeres presents the usual problems of measuring complex sequence repeats. In addition, there is the added complication, described above, of the HeT-A and TART 3 UTR fragments present in Y chromosome heterochromatin.
Nevertheless, it is important to have estimates of the total length of telomere transposon sequence and its variation between stocks. We have made such estimates using quantitative Southern hybridization (Fig. 2). All experiments were done entirely with DNA from female flies to avoid sequences on the Y. The blots were probed with HeT-A and TART open reading frame (ORF) sequences, which are the most conserved part of the elements and which also avoid hybridization with 3 UTR sequences in nontelomere regions (for discussion of probe choice, see Methods). DNA was extracted from adult heads because they are a relatively homogeneous source of diploid cells. We obtain similar results from female larval brains (data not shown).
Figure 2.
An example of Southern blots used to measure genome content of telomere elements. Samples of DNA from the Oregon R and 2057 stocks probed with HeT-A ORF (lanes on left) and probed with TART ORF (lanes on right) were cut with BamHI (lane B), EcoRI (lane E), HindIII (lane H), or XhoI (lane X). Filters were first probed with sequence for rp49 as a loading control, and the relevant region of each filter is shown below the HeT-A/TART blots. Comparison of the restriction fragments shows that the two stocks differ significantly in the number and linear arrangement of different sequence variants of the telomere elements.
Because the ORF sequences used for probes are near the 5 (distal) end of the element, which is most likely to be truncated, these probes allow an approximate count of full-length elements. More precisely, these hybridization experiments measure the total amount of ORF DNA in the sample (both complete and partial ORFs), which we report in units of full-length ORF (“ORF equivalents”). In conjunction with the fragment distribution measured in the 4R and XL telomere arrays (see Discussion), these data can be used to estimate the number of complete and partial HeT-A or TART elements in the genome. (As discussed later ~80% of the total HeT-A ORF and ~90% of TART ORF is found in complete elements). Tahre is subsumed in the total because of cross- hybridization to HeT-A sequences.
In Figure 3 we show the measured number of HeT-A and TART ORF equivalents in the genome of flies from 2057 (the inbred Canton S stock sequenced by the Genome project), from Oregon R (a commonly used wild-type stock), and from the S2 cell line, derived from Oregon R embryos >30 yr ago (Schneider 1972). By correcting for sequence in incomplete ORFs (see Discussion), we can calculate the number of complete elements in 2057 (29 HeT-A and 7 TART) and, by inference in Oregon R (34 HeT-A and 11 TART). The difference between these stocks is statistically significant for HeT-A at the 68% confidence level and for TART at the 95% confidence level. We refer to these measurements on these two most used wild-type stocks and our cultured S2 cell line as our “gold standard” measurements because they form the basis for estimates on other genomes. Figure 4 and Table 1 summarize our results for all stocks and tissues reported here.
Figure 3.
HeT-A and TART ORF equivalents in D. melanogaster. Numbers of full-length ORF equivalents were calculated from measurements of Southern blots (see Fig. 2). Data were corrected for DNA loading by first hybridizing with a probe for a single copy gene (rp49). The relative activity of the two probes (rp49 and ORF) was determined by measurement of a dose curve for each probe on the same filter. Fly DNA was prepared from adult female heads. For each DNA sample, the first four bars show the results of separate experiments, each of which averages several measurements (light gray indicates HeT-A; dark gray, TART). The final two bars (rising stripes, HeT-A; falling stripes, TART) show the results of averaging all data for that DNA. Error bars indicate the standard deviation of these averages (defined as the standard error of the underlying population).
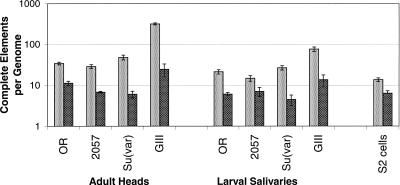
Figure 4.
Estimated number of complete HeT-A and TART elements in D. melanogaster. Two features illustrated by this graph are the under-replication in salivary glands over a data range so wide that the vertical axis has been chosen to be logarithmic, and also the near proportionality over such a wide range in the relationship between the number of HeT-A and TART elements across different stocks and tissues (correlation coefficient, r = 0.88–0.96, P = 0.07–0.0001, depending on tissue type) (see Supplemental material, Section 3.) Less obvious, but statistically significant, is the tracking of elements in heads vs. that same element in salivary glands (r = 0.95–0.99, P = 0.05 to <0.0001). Light gray indicates HeT-A; dark gray, TART.
Table 1.
HeT-A and TART in D. melanogaster
The cultured S2 cells have 14 HeT-A and 6 TART elements. Although they have fewer total elements than any of the fly stocks, S2 cells have maintained significant numbers of telomere elements over the >30 yr the line has been in culture. Similar numbers of elements were found in Schneider 3 cells and in a sample of S2 cells that has been frozen for the past six years (data not shown). These results are reminiscent of mammalian immortal cell lines, which tend to have shorter telomere arrays than do primary cultures (Counter et al. 1994; Harley et al. 1994; Tommerup et al. 1994).
D. melanogaster telomere length is influenced by genotype
Because of the difficulties in measuring telomere length in Drosophila, length variability has not received the attention given telomeres in other organisms. However, mutations in two genes, Tel-1 and Su(var)205, have been reported to cause telomere lengthening (Savitsky et al. 2002; Siriaco et al. 2002). Su(var)205 encodes the chromatin protein HP1; the product of Tel-1 is not known. We have compared telomere length in stocks carrying these mutations with our Oregon R stock because our Tel-1 mutant (in the GIII stock) is in an Oregon R background (Siriaco et al. 2002). We find that HeT-A elements in GIII heads are approximately nine times more abundant than in Oregon R heads, while TART elements are approximately twice as abundant. For our allele, Su(var)2054, HeT-A is slightly more abundant and TART slightly less than in Oregon R, both differences are statistically significant at the 95% confidence level (see Supplemental material).
We note that all of the D. melanogaster stocks we have analyzed have both HeT-A and TART elements. TART is always less abundant than HeT-A. An unexpected feature of Figure 4 is the nearly proportional relationship, over a hundredfold range, between the relative amounts of HeT-A and TART across stock lines and tissue type. (Correlation analysis of the nine experiments illustrated in Figure 4 implies that the relative number of HeT-A and TART elements is linearly correlated across genomes at >99% confidence level. [For details, see Supplemental material, Sections 3 and 4.]) Although we believe that HeT-A and TART depend on each other during transposition (Rashkova et al. 2002), there was no a priori reason to believe that their relative abundances would be so strongly correlated.
Telomere arrays are under-replicated in polytene cells
In his early studies of Drosophila salivary gland polytene chromosomes, Heitz (1934) suggested that heterochromatic sequences were under-replicated in these nuclei, and Gall et al. (1971) showed that the highly repeated satellite DNA sequences were under-replicated there. In the four stocks studied here, we find that telomere arrays, which are also heterochromatic, evidence similar tissue-dependent replicative control with the length of HeT-A and TART coding sequence in larval salivary glands being shorter than in adult heads, our standard diploid tissue (Fig. 4).
pecifically, we find that salivary gland HeT-A is under- replicated at the 95% confidence level in the Oregon R, 2057, Su(var)2054, and GIII stocks. For TART, pairwise comparison finds under-replication at the 95% confidencelevel for Oregon R and at the 68% confidence level for Su(var)2054, and GIII. For TART in 2057 our data do not show evidence of salivary gland under- replication. Figure 5 shows the replication ratio (the ratio of each element in salivary glands to the same element in heads). When averaged over all stocks HeT-A is significantly less under- replicated than TART. For details, see Supplemental material, Section 3.
Figure 5.
Salivary under-replication in female larvae. The ratio of the number of elements in salivary glands to the number in heads is shown for each stock and for an average over all stocks. Error bars indicate the standard deviations in these averages (see Fig. 3 legend). All are under- replicated except, perhaps, TART in 2057. Light bars are HeT-A; dark bars, TART.
HeT-A/TART arrays are easily detectable by in situ hybridization on salivary gland chromosomes, showing that these sequences, like the rest of the chromosome, undergo multiple rounds of replication to make the polytene chromosomes. However, there is reason to believe that new transpositions do not occur in polytene nuclei because HeT-A RNA is not found in these cells (George and Pardue 2003), nor does HeT-A Gag enter polytene nuclei (Pardue et al. 2005). Nevertheless, our measurements cannot be explained by a simple failure to replace RNA primers of DNA synthesis (measurements on broken Drosophila chromosome ends average 2 nt per round of replication and are consistent with that mechanism for end shortening) (Levis 1989; Biessmann et al. 1990a). Polytene chromosomes usually undergo a total of ≤10 rounds of replication and would be almost undetectably shorter after replication at this loss rate Thus, the decreased amount of telomeric DNA we detect in polytenes (Fig.4) must be due to something more than failure to replace these primers.
Discussion
Intact and apparently functional HeT-A elements are found near the proximal end of the telomere arrays
A surprising finding of this study has been the number of apparently functional HeT-A elements deep within the telomere arrays. If addition of telomere repeats serves only to replace eroded sequence on the chromosome end, one would expect sequences deep inside the arrays to decay because, once added to the end, there should be little constraint to maintain function if their only function is to buffer a chromosome end. Instead, the full-length sequences here have maintained ORFs and other regions needed for function. The existence of functional elements in proximal regions of these long telomere arrays suggests that these interior sequences may be renewed more frequently than has been thought and that turnover in these arrays does not simply replace terminal sequence lost in DNA replication. A likely possibility is that telomeres sometimes undergo drastic shortening, perhaps by a mechanism such as that proposed for yeast telomere rapid deletion (Lustig 2003) or mammalian t-loop homologous recombination (Wang et al. 2004). This shortening would then be followed by rapid multiple transpositions to restore the telomere to an appropriate length. Such a drastic shortening might also explain the loss of most of the TASs on the 2057 XL telomere.
More HeT-A and TART elements than expected have intact 5 ends
Non-LTR elements are frequently 5'-truncated, presumably because reverse transcription, which begins at the 3' end, is incomplete. In their analysis of sequence from the euchromatic parts of the D. melanogaster genome, Kaminker et al. (2002) found that 79% of the non-LTR retroelements identified were partial elements. We expected that HeT-A and TART would be as likely to undergo incomplete reverse transcription as other non-LTR elements and, in addition, to suffer perhaps significant erosion during the time when each element forms the end of a telomere.
The data do not support the expectation that significantly more telomere elements would be truncated; 70% (14 of 20) of our HeT-A and 71% (5 of 7) of our TART elements are truncated, slightly less than the 79% seen for elements not subject to end erosion. For this calculation, we omit the tiny “tags,” which we believe are byproducts of the unusual HeT-A promoter. These tags all have <50 bp of homology and therefore would also have been omitted in the calculation of Kaminker et al. (2002).
This observation that a significant fraction of HeT-A elements in the array shows little, if any, terminal erosion suggests that ends are protected from degradation or that transpositions frequently occur in rapid succession before erosional loss. These possibilities are not mutually exclusive. Protection could be provided by terminal structures like the t-loops seen on chromosomes in other organisms (Griffith et al. 1999); however, it is not yet known whether Drosophila telomeres have such structures.
D. melanogaster has very long telomere arrays
Quantitative Southern hybridization analyses give us a reasonably accurate measurement of the number of HeT-A and TART ORF equivalents in the female genomes of several stocks and, with the sequence analysis reported above, provides a basis for estimating the total length of HeT-A and TART sequence in telomeres.
That estimate has several uncertainties. Apparently intact elements can differ by indels that add up to several hundred base pairs; the 5 end of TART presents technical problems because of its PNTRs (see Pardue and DeBaryshe 2003) and may not be completely defined; also, telomere arrays have severely 5 truncated elements (without any ORF) not detected by our Southern hybridizations. By using our data from the assembled sequence on XL and 4R, we can correct for truncated elements (in doing so we remove from consideration the two elements truncated by cloning, distal TART on 4R and distal HeT-A on XL). Although the most distal element in the 4R sequence is a 5 truncated TART, which could be the true end of the chromosome, we treat it like the most distal, 5 truncated, HeT-A in XL, which is clearly truncated by cloning, and, in order not to bias estimation of the results, exclude both. From these measurements we determine that
#9679; The average length of the complete HeT-A element is 5893 169 bp (SD) (based on the seven elements listed in Supplemental Table 2).
#9679; ~61% of the total HeT-A sequence in the 2057 stock is in complete elements.
#9679; ~80% of the total HeT-A coding sequence in 2057 is in complete coding sequences.
#9679; The consensus length of the TART element of 11,734 bp (sub- families A, B, and C) (data not shown) is consistent with our new data.
#9679; ~89% of the total TART sequence in 2057 is in complete elements.
#9679; ~100% of the total TART coding sequence in 2057 is in complete coding sequences.
Using these numbers, we calculate from our hybridization results that the 2057 genome contains polarized HeT-A/TART arrays with ~29 complete HeT-A elements and approximately seven complete TART elements. Correcting for partial elements, we calculate ~365 kb of total HeT-A and TART sequence on eight telomeres, an average of ~45.6 kb of HeT-A and TART sequence per telomere. Perforce, we use the same correction factors for estimates of other genomes.
We can compare this estimate to the one other measurement done for D. melanogaster telomere arrays. Abad et al. (2004a) have selected clones with telomere sequences from BAC libraries of 2057 DNA, measured them by restriction mapping and published results for six of the eight chromosome ends. Length ranged from 147 kb on XL to 0 kb on 3L, averaging 50 kb for each of the six telomeres in the BACs that they analyzed. Both Southern hybridization and BAC mapping have uncertainties, but the uncertainties differ with technique. For example, a major uncertainty of BAC mapping is whether the library contains all of the telomere sequences. Nevertheless, the two techniques yield a satisfyingly similar number for the average amount of DNA in the HeT-A/TART array on each telomere.
Although most eukaryotes have very similar telomere sequences, multicellular eukaryotes have much longer telomere arrays than do unicellular eukaryotes. Among the longest studied telomeres are those of inbred strains of laboratory mice. These telomeres range from 30–150 kb (Kipling and Cooke 1990), approximately the length of D. melanogaster HeT-A/TART arrays. In contrast, wild-derived inbred mouse strains have telomeres in the 4–15 kb range (Hemann and Greider 2000), approximately the size of human telomeres.
It is interesting that mice and flies, the two organisms known to have unusually long telomeres, are also unusual because they have been kept in small isolated laboratory populations for many years, suggesting that something about the population structure or relatively luxurious laboratory conditions may affect telomere length. It will be interesting to see whether wild- derived D. melanogaster have shorter telomeres, like wild-derived mice.
D. melanogaster telomere length is influenced by genotype
Studies of several organisms have shown that, although telomere length varies, these variations are held within a relatively narrow range and the center of this range can be changed by external conditions or by changes in genotype (see Greider 1996; Smogorzewska and de Lange 2004). For example, a recent study identified ~150 nonessential genes in Saccharomyces cerevisiae that changed the average around which telomere length fluctuated (Askree et al. 2004). Loss of some of these genes led to longer telomeres; conversely, loss of other genes led to shorter telomeres. These studies show that addition and loss of telomere sequence is under complex control.
The retrotransposon telomeres of Drosophila, similar to those maintained by telomerase, have genetically modulated length control. Savitsky et al. (2002) reported that three stocks carrying different mutant alleles of Su(var)205 had high levels of telomeric DNA. However, stocks from a different laboratory but carrying the same alleles had lower amounts of telomeric DNA. The Su(var)2054 stock we studied is from that second laboratory, and like Savitsky et al., we found a lesser amount of telomeric DNA than reported for the other Su(var)205 stocks. Comparison of these two sets of mutant stocks suggests that different genetic backgrounds can modify the effect of the Su(var)205 mutation on telomere length.**
Tel-1 mutant flies have significantly more telomeric DNA than the other stocks, and the amounts are influenced by genetic background, as seen by comparing the Gaiano and GIII strains (Gaiano is the source of the Tel-1 mutation, which was moved into Oregon R to make GIII) (Siriaco et al. (2002). Our measurements on GIII show a larger increase in HeT-A (approximately nine times) than reported by Siriaco et al. (four times). The difference in these results could be due either to differences in methodology or to differences in the flies, or both. Siriaco et al. used GIII flies 130 generations after Tel-1 had been moved into an Oregon R background. We studied this stock several years later, and telomere lengths may have changed. Nevertheless, both studies show that the Tel-1 mutation causes significant lengthening of telomeres.
Analysis of the assembled sequence suggested that Drosophila telomeres occasionally undergo large deletions of the type reported in yeast and humans (Lustig 2003; Wang et al. 2004). On the other hand, our DNA measurements show that stocks and cell lines maintain relatively constant equilibrium telomere lengths, under some genetic controls, so deleted material must be rapidly replaced (while maintaining significant correlation of the numbers of HeT-A and TART elements).
Methods
Fly stocks and cell lines
Oregon R, a wild-type stock maintained in our laboratory for many years; 2057, an isogenic y1, cn1, bw,1sp1 Canton S stock obtained from A. Villasante (Centro Servero Ochoa); GIII, Tel-1 mutant in Oregon R background made by J. Mason (NIEHS) (Siriaco et al. 2002); Su(var)2054/CyO, y+, obtained from J. Eissenberg (St. Louis University). The Schneider S2 and S3 cell lines (Schneider 1972) have been in our laboratory for >30 and 3 yr, respectively.
Sequence analyses
Release 4.2 of the Drosophila Genome was analyzed by BLAST through FlyBase (http://flybase.bio.indiana.edu), using the default parameters and accepting all similarities below E = 0.002. Queries were sequences of the elements we use as type sequences: HeT-A, U06920 (bp 1015–7097); TART A, AY561850; TART B, U14101; TART C, AY600955; Tahre, AJ542581 (bp 81–10543). The same queries were used to search the FlyBase “euchromatin and heterochromatin scaffolds,” accepting only sequence from scaffolds assigned to specific chromosome arms. The 4R telomere sequence was assembled from AC010841. Y chromosome scaffolds were AABU01002777–783. X chromosome sequences are from CP000372. Regions of BLAST similarity were ordered, and sequences between these regions were analyzed by multiple alignments with other copies of HeT-A and TART and by dot matrix analyses. Sequences not identified by these analyses were blasted against the whole genome. Taken together, these analyses give the number of full-length and partial elements on the HeT-A/TART arrays we analyzed and also the number of nucleotides in each. Insofar as these distributions are typical of all D. melanogaster chromosomes, they can be used to estimate the total number of full-length and partial elements in the fly genome.
The telomeric sequence assemblies (4th and X) have been validated by comparison to restriction fingerprint digests (BACN05O16, CH221–48I20, respectively) using five enzymes: ApaLI, BamHI, EcoRI, HindIII, and XhoI (collaboration between the Berkeley Drosophila Genome Project and BC Cancer Agency Genome Sciences Center and BC Cancer Agency, Vancouver, Canada [in prep.]).
Probes for Southern hybridization
Hybridization of sequences as complex as HeT-A and TART risks undercounting if the probe does not hybridize equally with all the different variant sequences and overcounting if the probe hybridizes with background sequences. The probes used were chosen because there is a window of hybridization stringency that should allow efficient detection of known variants while, with the possible exception noted below, minimizing hybridization to sequences that are not HeT-A or TART. Although HeT-A elements can have as little as 67% nucleotide identity, ORFs are more conserved, and we use only these as probes. The ORF used (element 23Zn-1) has ~82% nucleotide identity with all but two other known elements (it has ~80% identity with one of these two and ~97% with the other) (Supplemental Table 3). Multiple alignments of these coding regions show a remarkably scattered distribution of nucleotide changes and of small (1 and 2 codon) deletions. This scattering ensures that the identities between any two elements are evenly distributed. There is a polymorphic region where ORFs can differ by as many as 93 nt, but 23Zn-1 lacks all 93 nt so it will not preferentially hybridize in that region. For TART we use ORF 2 as a probe because it has higher identities (≥88%) and lacks the very large polymorphic region in ORF1. We have searched the genome sequence with the exact sequences of the two probes used, and the only detectable possible cross reacting sequence is 69 nt in ORF2 of the BS element, which has 76% identity with the TART probe. BS is a minor element; there are eight copies of this sequence in the sequenced genome so even if it cross-reacts, it contributes little to the total.
These probes allow direct comparison of genomes on the basis of their content of ORF coding sequence. Information from the sequence assembly can then be used to derive an estimation of complete and partial elements from these ORF equivalents.
Southern hybridization analyses
All DNA was prepared from females to eliminate the nontelomeric HeT-A/TART-related sequences found in the Y (Danilevskaya et al. 1991, 1993; Losada et al. 1999; this study). Three2.5-μg aliquots per sample were run side-by-side on an 0.7% aga- rose gel, each having been digested with a different restriction enzyme to provide simultaneous, multiple, independent measurements for statistical analysis while minimizing bias in the size and number of fragments produced by any one enzyme. DNA was transferred to Hybond-N (Amersham Biosciences) filters and hybridized first with a probe for the single copy gene, rp49 (now named RpL32, bp 241–880 of X00848) to correct for variations in the amount of DNA loaded in each lane. After the measurements described below, filters were stripped (or for some experiments, a probe was allowed to decay) and probed with sequence from the ORFs of either HeT-A (U06920, bp 1746–4511) or TART (U02279, bp 439–2683).
Hybridization was in 4 SET (1 SET is 0.15 M NaCl, 2 mM EDTA, 0.03 M Tris-Cl at pH 7.0), 2 Denhart's solution, 0.5% SDS, and 100 μg salmon sperm DNA per milliliter at 65°C. Washes were 1 SSC (0.15 M NaCl, 0.015 M Na citrate at pH 7.0) and 0.5% SDS at 65°C. Hybridized 32P-labeled probes were detected by scanning the filters with Molecular Dynamics PhosphorImagers, and the data were reduced to usable digital form by Molecular Dynamics ImageQuant software. In reducing the data, background subtraction, utilizing measuremen t of interlane exposure adjacent to each lane of DNA, was necessary, primarily to avoid overestimation of the rp49-probed DNA measurements and, hence, underestimation of the quantity of HeT-A and TART DNA present.
Gels used to determine the “gold standard” absolute number of ORF equivalents per genome (in S2 cells and Oregon R and 2057 Female Adult Heads) also contained three lanes each of the rp49 and retrotransposon probes, which were used for determining the relative activity of each probe. Actually, as discussed above and in detail in Supplemental material, Section 4, we report the number of ORF equivalents per genome in each stock,i.e., the total length of ORF sequence, both full length and partial, measured in units of full-length ORF sequence.
For this set of measurements, two independent experiments were performed. This procedure gave multiple independent measurements of each stock for each probe. Data processing was performed using an Excel spreadsheet developed for the purpose.
For other samples, three lanes of Oregon R Female Adult Heads were run on each gel as a control, and the number of genome equivalents per stock was determined relative to OregonR. The actual number of ORF equivalents was then determined by multiplying by the reference value determined for Oregon R heads in the more extensive measurements described above.
Data from the six probe lanes were also analyzed to demonstrate that our experimental techniques (filter preparation, digitizing, and analysis) are linear over the full range of DNA deposited in the gel. Measurements were linear within a few percent over all source strengths (measured in counts per pixel) (data not shown).
For details of the data analysis and statistical controls used, see Supplemental material, Section 4.
ACKNOWLEDGMENTS
We thank J. Eissenberg for the Su(var)2054 stock, J. Mason for the GIII stock, and A. Villasante for the 2057 stock. We are grateful to R.A. Hoskins, C.D. Smith, J. Carlson, and A.B. de Carvalho for helpful advice. We thank E. Casacuberta, J. Collett, R. Dudley, and K. Lowenhaupt for stimulating discussions. This work has been supported by National Institutes of Health Grant GM50315 to M.L.P. The sequencing portion of this work was supported by NIH grant P50-HG00750 (G.M.R.) and carried out under U.S. Department of Energy contract DE-AC0376SF00098.
Footnotes
Supplemental material is available online at http://www.genome.org.
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5348806.
REFERENCES
- Abad J.P., De Pablos B., Osoegawa K., De Jong P.J., Martin-Gallardo A., Villasante A., De Pablos B., Osoegawa K., De Jong P.J., Martin-Gallardo A., Villasante A., Osoegawa K., De Jong P.J., Martin-Gallardo A., Villasante A., De Jong P.J., Martin-Gallardo A., Villasante A., Martin-Gallardo A., Villasante A., Villasante A. Genomic analysis of Drosophila melanogaster telomeres: Full-length copies of HeT-A and TART elements at telomeres. Mol. Biol. Evol. 2004a;21:1613–1619. doi: 10.1093/molbev/msh174. [DOI] [PubMed] [Google Scholar]
- Abad J.P., De Pablos B., Osoegawa K., De Jong P.J., Martin-Gallardo A., Villasante A., De Pablos B., Osoegawa K., De Jong P.J., Martin-Gallardo A., Villasante A., Osoegawa K., De Jong P.J., Martin-Gallardo A., Villasante A., De Jong P.J., Martin-Gallardo A., Villasante A., Martin-Gallardo A., Villasante A., Villasante A. TAHRE, a novel telomeric retrotransposon from Drosophila melanogaster, reveals the origin of Drosophila telomeres. Mol. Biol. Evol. 2004b;21:1620–1624. doi: 10.1093/molbev/msh180. [DOI] [PubMed] [Google Scholar]
- Adams M.D., Celniker S.E., Holt R.A., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Celniker S.E., Holt R.A., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Holt R.A., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Li P.W., Hoskins R.A., Galle R.F., Hoskins R.A., Galle R.F., Galle R.F., et al. The genome sequence of Drosophila melanogaster . Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Askree A.H., Yehuda T., Smolikov S., Gurevich R., Hawk J., Coker C., Krauskopf A., Kupiec M., McEachern M.J., Yehuda T., Smolikov S., Gurevich R., Hawk J., Coker C., Krauskopf A., Kupiec M., McEachern M.J., Smolikov S., Gurevich R., Hawk J., Coker C., Krauskopf A., Kupiec M., McEachern M.J., Gurevich R., Hawk J., Coker C., Krauskopf A., Kupiec M., McEachern M.J., Hawk J., Coker C., Krauskopf A., Kupiec M., McEachern M.J., Coker C., Krauskopf A., Kupiec M., McEachern M.J., Krauskopf A., Kupiec M., McEachern M.J., Kupiec M., McEachern M.J., McEachern M.J. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc. Natl. Acad. Sci. 2004;101:8658–8663. doi: 10.1073/pnas.0401263101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biessmann H., Carter S.B., Mason J.M., Carter S.B., Mason J.M., Mason J.M. Chromosome ends in Drosophila without telomeric DNA sequences. Proc. Natl. Acad. Sci. 1990a;87:1758–1761. doi: 10.1073/pnas.87.5.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biessmann H., Mason J.M., Ferry K., d'Hulst M., Valgeirsdottir K., Traverse K.L., Pardue M.L., Mason J.M., Ferry K., d'Hulst M., Valgeirsdottir K., Traverse K.L., Pardue M.L., Ferry K., d'Hulst M., Valgeirsdottir K., Traverse K.L., Pardue M.L., d'Hulst M., Valgeirsdottir K., Traverse K.L., Pardue M.L., Valgeirsdottir K., Traverse K.L., Pardue M.L., Traverse K.L., Pardue M.L., Pardue M.L. Addition of telomere-associated HeT DNA sequences “heals” broken chromosome ends in Drosophila . Cell. 1990b;61:663–673. doi: 10.1016/0092-8674(90)90478-w. [DOI] [PubMed] [Google Scholar]
- Biessmann H., Champion L.E., O'Hair M., Ikenaga K., Kasravi B., Mason J.M., Champion L.E., O'Hair M., Ikenaga K., Kasravi B., Mason J.M., O'Hair M., Ikenaga K., Kasravi B., Mason J.M., Ikenaga K., Kasravi B., Mason J.M., Kasravi B., Mason J.M., Mason J.M. Frequent transpositions of Drosophila melanogaster HeT-A elements to receding chromosome ends. EMBO J. 1992;11:4459–4469. doi: 10.1002/j.1460-2075.1992.tb05547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho A.B., Dobo B.A., Vibranovski M.D., Clark A.G., Dobo B.A., Vibranovski M.D., Clark A.G., Vibranovski M.D., Clark A.G., Clark A.G. Identification of five new genes on the Y chromosome of Drosophila melanogaster . Proc. Natl. Acad. Sci. 2001;98:13225–13230. doi: 10.1073/pnas.231484998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casacuberta E., Pardue M.-L., Pardue M.-L. Coevolution of the telomeric retrotransposons across Drosophila species. Genetics. 2002;161:1113–1124. doi: 10.1093/genetics/161.3.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casacuberta E., Pardue M.-L., Pardue M.-L. HeT-A elements in D. virilis:Retrotransposon telomeres are conserved across the Drosophila genus . Proc. Natl. Acad. Sci. 2003;100:14091–14096. doi: 10.1073/pnas.1936193100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker S.E., Wheeler D.L., Kronmiller B., Carlson J., Halpern A., Patel S., Adams M., Champe M., Dugan S.P., Frise E., Wheeler D.L., Kronmiller B., Carlson J., Halpern A., Patel S., Adams M., Champe M., Dugan S.P., Frise E., Kronmiller B., Carlson J., Halpern A., Patel S., Adams M., Champe M., Dugan S.P., Frise E., Carlson J., Halpern A., Patel S., Adams M., Champe M., Dugan S.P., Frise E., Halpern A., Patel S., Adams M., Champe M., Dugan S.P., Frise E., Patel S., Adams M., Champe M., Dugan S.P., Frise E., Adams M., Champe M., Dugan S.P., Frise E., Champe M., Dugan S.P., Frise E., Dugan S.P., Frise E., Frise E., et al. Finishing a whole genome shotgun: Release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol. 2002;3:research0079. doi: 10.1186/gb-2002-3-12-research0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Counter C.M., Botelho F.M., Wang P., Harley C.B., Bacchetti S., Botelho F.M., Wang P., Harley C.B., Bacchetti S., Wang P., Harley C.B., Bacchetti S., Harley C.B., Bacchetti S., Bacchetti S. Stabilization of short telomeres and telomerase activity accompany immortalization of Epstein-Barr virus-transformed human B lymphocytes. J. Virol. 1994;68:3410–3414. doi: 10.1128/jvi.68.5.3410-3414.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilevskaya O.N., Kurenova E.V., Pavlova M.N., Bebehov D.V., Link A.J., Koga A., Velek V., Hartl D.L., Kurenova E.V., Pavlova M.N., Bebehov D.V., Link A.J., Koga A., Velek V., Hartl D.L., Pavlova M.N., Bebehov D.V., Link A.J., Koga A., Velek V., Hartl D.L., Bebehov D.V., Link A.J., Koga A., Velek V., Hartl D.L., Link A.J., Koga A., Velek V., Hartl D.L., Koga A., Velek V., Hartl D.L., Velek V., Hartl D.L., Hartl D.L. HeT-A family DNA sequences in the Y chromosome of Drosophila melanogaster share homology with X-linked Stellate genes. Chromosoma. 1991;100:118–124. doi: 10.1007/BF00418245. [DOI] [PubMed] [Google Scholar]
- Danilevskaya O., Lofsky A., Kurenova E.V., Pardue M.-L., Lofsky A., Kurenova E.V., Pardue M.-L., Kurenova E.V., Pardue M.-L., Pardue M.-L. The Y chromosome of Drosophila melanogaster contains a distinctive subclass of HeT-A-related repeats. Genetics. 1993;134:531–543. doi: 10.1093/genetics/134.2.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilevskaya O.N., Arkhipova I.R., Traverse K.L., Pardue M.-L., Arkhipova I.R., Traverse K.L., Pardue M.-L., Traverse K.L., Pardue M.-L., Pardue M.-L. Promoting in tandem: The promoter for telomere transposon HeT-A and implications for the evolution of retroviral LTRs. Cell. 1997;86:647–655. doi: 10.1016/s0092-8674(00)81907-8. [DOI] [PubMed] [Google Scholar]
- Danilevskaya O.N., Lowenhaupt K., Pardue M.L., Lowenhaupt K., Pardue M.L., Pardue M.L. Conserved subfamilies of the Drosophila HeT-A telomere-specific retrotransposon. Genetics. 1998;148:233–242. doi: 10.1093/genetics/148.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilevskaya O.N., Traverse K.L., Hogan N.C., DeBaryshe P.G., Pardue M.L., Traverse K.L., Hogan N.C., DeBaryshe P.G., Pardue M.L., Hogan N.C., DeBaryshe P.G., Pardue M.L., DeBaryshe P.G., Pardue M.L., Pardue M.L. The two Drosophila telomeric transposable elements have very different patterns of transcription. Mol. Cell. Biol. 1999;19:873–881. doi: 10.1128/mcb.19.1.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall J.G., Cohen E.H., Polan M.L., Cohen E.H., Polan M.L., Polan M.L. Repetitive DNA sequences in Drosophila chromosomes. Chromosoma. 1971;33:319–344. doi: 10.1007/BF00284948. [DOI] [PubMed] [Google Scholar]
- George J.A., Pardue M.-L., Pardue M.-L. The promoter of the heterochromatic Drosophila telomeric retrotransposon, HeT-A, is active when moved into euchromatic locations. Genetics. 2003;163:625–635. doi: 10.1093/genetics/163.2.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider C.W. Telomere length regulation. Annu. Rev. Biochem. 1996;65:337–365. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- Griffith J., Comeau L., Rosenfield S., Stansel R., Bianchi A., Moss H., de Lange T., Comeau L., Rosenfield S., Stansel R., Bianchi A., Moss H., de Lange T., Rosenfield S., Stansel R., Bianchi A., Moss H., de Lange T., Stansel R., Bianchi A., Moss H., de Lange T., Bianchi A., Moss H., de Lange T., Moss H., de Lange T., de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- Harley C.B., Kim N.W., Prowse K.R., Weinrich S.L., Hirsch K.S., West M.D., Bacchetti S., Hirte H.W., Counter C.M., Greider C.W., Kim N.W., Prowse K.R., Weinrich S.L., Hirsch K.S., West M.D., Bacchetti S., Hirte H.W., Counter C.M., Greider C.W., Prowse K.R., Weinrich S.L., Hirsch K.S., West M.D., Bacchetti S., Hirte H.W., Counter C.M., Greider C.W., Weinrich S.L., Hirsch K.S., West M.D., Bacchetti S., Hirte H.W., Counter C.M., Greider C.W., Hirsch K.S., West M.D., Bacchetti S., Hirte H.W., Counter C.M., Greider C.W., West M.D., Bacchetti S., Hirte H.W., Counter C.M., Greider C.W., Bacchetti S., Hirte H.W., Counter C.M., Greider C.W., Hirte H.W., Counter C.M., Greider C.W., Counter C.M., Greider C.W., Greider C.W., et al. Telomerase, cell immortality, and cancer. Cold Spring Harb. Symp. Quant. Biol. 1994;59:307–315. doi: 10.1101/sqb.1994.059.01.035. [DOI] [PubMed] [Google Scholar]
- Heitz E. Uber α- und β-Heterochromatin sowie Konstanz und Bau der Chromomeren bei Drosophila . Biol. Zent. Bl. 1934;54:588–609. [Google Scholar]
- Hemann M.T., Greider C.W., Greider C.W. Wild-derived inbred mouse strains have short telomeres. Nucleic Acids Res. 2000;28:4474–4478. doi: 10.1093/nar/28.22.4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson E. Telomere DNA structure. In: Blackburn E.H., Greider C.W., Greider C.W., editors. Telomeres. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1995. pp. 11–34. [Google Scholar]
- Hoskins R.A., Smith C.D., Carlson J., Carvalho B.A., Halpern A., Kaminker J.S., Kennedy C., Mungall C.J., Sullivan B.A., Sutton G.G., Smith C.D., Carlson J., Carvalho B.A., Halpern A., Kaminker J.S., Kennedy C., Mungall C.J., Sullivan B.A., Sutton G.G., Carlson J., Carvalho B.A., Halpern A., Kaminker J.S., Kennedy C., Mungall C.J., Sullivan B.A., Sutton G.G., Carvalho B.A., Halpern A., Kaminker J.S., Kennedy C., Mungall C.J., Sullivan B.A., Sutton G.G., Halpern A., Kaminker J.S., Kennedy C., Mungall C.J., Sullivan B.A., Sutton G.G., Kaminker J.S., Kennedy C., Mungall C.J., Sullivan B.A., Sutton G.G., Kennedy C., Mungall C.J., Sullivan B.A., Sutton G.G., Mungall C.J., Sullivan B.A., Sutton G.G., Sullivan B.A., Sutton G.G., Sutton G.G., et al. Heterochromatic sequences in a Drosophila whole genome shotgun assembly. Genome Biol. 2002;3:search0085. doi: 10.1186/gb-2002-3-12-research0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminker J.S., Bergman C.M., Kronmiller B., Carlson J., Svirskas R., Patel S., Frise E., Wheeler D.A., Lewis S.E., Rubin G.M., Bergman C.M., Kronmiller B., Carlson J., Svirskas R., Patel S., Frise E., Wheeler D.A., Lewis S.E., Rubin G.M., Kronmiller B., Carlson J., Svirskas R., Patel S., Frise E., Wheeler D.A., Lewis S.E., Rubin G.M., Carlson J., Svirskas R., Patel S., Frise E., Wheeler D.A., Lewis S.E., Rubin G.M., Svirskas R., Patel S., Frise E., Wheeler D.A., Lewis S.E., Rubin G.M., Patel S., Frise E., Wheeler D.A., Lewis S.E., Rubin G.M., Frise E., Wheeler D.A., Lewis S.E., Rubin G.M., Wheeler D.A., Lewis S.E., Rubin G.M., Lewis S.E., Rubin G.M., Rubin G.M., et al. The transposable elements of the Drosophila melanogaster euchromatin: A genomics perspective. Genome Biol. 2002;3:research0084. doi: 10.1186/gb-2002-3-12-research0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen G.H., Spradling A.C., Spradling A.C. Analysis of subtelomeric heterochromatin in the Drosophila minichromosome Dp1187 by single P element insertional mutagenesis. Genetics. 1992;132:737–753. doi: 10.1093/genetics/132.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipling D., Cooke H.J., Cooke H.J. Hypervariable ultra-long telomeres in mice. Nature. 1990;374:400–402. doi: 10.1038/347400a0. [DOI] [PubMed] [Google Scholar]
- Levis R.W. Viable deletions of a telomere from a Drosophila chromosome. Cell. 1989;58:791–801. doi: 10.1016/0092-8674(89)90112-8. [DOI] [PubMed] [Google Scholar]
- Levis R.W., Ganesan R., Houtchens K., Tolar L.A., Sheen F.-M., Ganesan R., Houtchens K., Tolar L.A., Sheen F.-M., Houtchens K., Tolar L.A., Sheen F.-M., Tolar L.A., Sheen F.-M., Sheen F.-M. Transposons in place of telomeric repeats at a Drosophila telomere. Cell. 1993;75:1083–1093. doi: 10.1016/0092-8674(93)90318-k. [DOI] [PubMed] [Google Scholar]
- Losada A., Agudo M., Abad J.P., Villesante A., Agudo M., Abad J.P., Villesante A., Abad J.P., Villesante A., Villesante A. HeT-A telomere-specific retrotransposons in the centric heterochromatin of Drosophila melanogaster chromosome 3. Mol. Gen. Genet. 1999;262:618–622. doi: 10.1007/s004380051124. [DOI] [PubMed] [Google Scholar]
- Lustig A.J. Clues to catastrophic telomere loss in mammals from yeast telomere rapid deletion. Nat. Rev. Genet. 2003;4:916–923. doi: 10.1038/nrg1207. [DOI] [PubMed] [Google Scholar]
- Pardue M.-L., DeBaryshe P.G., DeBaryshe P.G. Retrotransposons provide an evolutionarily robust non-telomerase mechanism to maintain telomeres. Annu. Rev. Genet. 2003;37:485–511. doi: 10.1146/annurev.genet.38.072902.093115. [DOI] [PubMed] [Google Scholar]
- Pardue M.-L., Danilevskaya O.N., Lowenhaupt K., Wong J., Erby K., Danilevskaya O.N., Lowenhaupt K., Wong J., Erby K., Lowenhaupt K., Wong J., Erby K., Wong J., Erby K., Erby K. The “gag” coding region of the Drosophila telomeric retrotransposon, HeT-A, has an internal frame shift and a length polymorphic region. J. Mol. Evol. 1996;43:572–583. doi: 10.1007/BF02202105. [DOI] [PubMed] [Google Scholar]
- Pardue M.-L., Rashkova S., Casacuberta E., DeBaryshe P.G., George J.A., Traverse K.L., Rashkova S., Casacuberta E., DeBaryshe P.G., George J.A., Traverse K.L., Casacuberta E., DeBaryshe P.G., George J.A., Traverse K.L., DeBaryshe P.G., George J.A., Traverse K.L., George J.A., Traverse K.L., Traverse K.L. Two retrotransposons maintain telomeres in Drosophila . Chromosome Res. 2005;13:443–453. doi: 10.1007/s10577-005-0993-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashkova S., Karam S.E., Kellum R., Pardue M.-L., Karam S.E., Kellum R., Pardue M.-L., Kellum R., Pardue M.-L., Pardue M.-L. Gag proteins of the two Drosophila telomeric retrotransposons are targeted to chromosome ends. J. Cell Biol. 2002;159:397–402. doi: 10.1083/jcb.200205039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashkova S., Athanasiadis A., Pardue M.-L., Athanasiadis A., Pardue M.-L., Pardue M.-L. Intracellular targeting of Gag proteins of the Drosophila telomeric retrotransposons. J. Virol. 2003;77:6376–6384. doi: 10.1128/JVI.77.11.6376-6384.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitsky M., Kravchuk O., Melnikova L., Georgiev P., Kravchuk O., Melnikova L., Georgiev P., Melnikova L., Georgiev P., Georgiev P. Heterochromatin protein 1 is involved in control of telomere elongation in Drosophila melanogaster . Mol. Cell. Biol. 2002;22:3204–3218. doi: 10.1128/MCB.22.9.3204-3218.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider I. Cell lines derived from late embryonic stages of Drosophila melanogaster . J. Embryol. Exp. Morphol. 1972;27:353–365. [PubMed] [Google Scholar]
- Siriaco G.M., Cenci G., Haoudi A., Champion L.E., Zhou C., Gatti M., Mason J.M., Cenci G., Haoudi A., Champion L.E., Zhou C., Gatti M., Mason J.M., Haoudi A., Champion L.E., Zhou C., Gatti M., Mason J.M., Champion L.E., Zhou C., Gatti M., Mason J.M., Zhou C., Gatti M., Mason J.M., Gatti M., Mason J.M., Mason J.M. Telomere elongation (Tel), a new mutation in Drosophila melanogaster that produces long telomeres. Genetics. 2002;160:235–245. doi: 10.1093/genetics/160.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A., de Lange T., de Lange T. Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 2004;73:177–208. doi: 10.1146/annurev.biochem.73.071403.160049. [DOI] [PubMed] [Google Scholar]
- Tommerup H., Dousmanis A., de Lange T., Dousmanis A., de Lange T., de Lange T. Unusual chromatin in human telomeres. Mol. Cell. Biol. 1994;14:5777–5785. doi: 10.1128/mcb.14.9.5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traverse K.L., Pardue M.-L., Pardue M.-L. A spontaneously opened ring chromosome of Drosophila melanogaster has acquired He-T DNA sequences at both new telomeres. Proc. Natl. Acad. Sci. 1988;85:8116–8120. doi: 10.1073/pnas.85.21.8116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M.F., Jang C., Kasravi D., Donath J., Mechler B.M., Mason J.M., Biessmann H., Jang C., Kasravi D., Donath J., Mechler B.M., Mason J.M., Biessmann H., Kasravi D., Donath J., Mechler B.M., Mason J.M., Biessmann H., Donath J., Mechler B.M., Mason J.M., Biessmann H., Mechler B.M., Mason J.M., Biessmann H., Mason J.M., Biessmann H., Biessmann H. DNA organization and polymorphism of a wild-type Drosophila telomere region. Chromosoma. 1995;104:229–241. doi: 10.1007/BF00352254. [DOI] [PubMed] [Google Scholar]
- Wang R.C., Smogorzewska A., de Lange T., Smogorzewska A., de Lange T., de Lange T. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell. 2004;119:355–368. doi: 10.1016/j.cell.2004.10.011. [DOI] [PubMed] [Google Scholar]