Abstract
The Arabidopsis genome contains a highly complex and abundant population of small RNAs, and many of the endogenous siRNAs are dependent on RNA-Dependent RNA Polymerase 2 (RDR2) for their biogenesis. By analyzing an rdr2 loss-of-function mutant using two different parallel sequencing technologies, MPSS and 454, we characterized the complement of miRNAs expressed in Arabidopsis inflorescence to considerable depth. Nearly all known miRNAs were enriched in this mutant and we identified 13 new miRNAs, all of which were relatively low abundance and constitute new families. Trans-acting siRNAs (ta-siRNAs) were even more highly enriched. Computational and gel blot analyses suggested that the minimal number of miRNAs in Arabidopsis is ∼155. The size profile of small RNAs in rdr2 reflected enrichment of 21-nt miRNAs and other classes of siRNAs like ta-siRNAs, and a significant reduction in 24-nt heterochromatic siRNAs. Other classes of small RNAs were found to be RDR2-independent, particularly those derived from long inverted repeats and a subset of tandem repeats. The small RNA populations in other Arabidopsis small RNA biogenesis mutants were also examined; a dcl2/3/4 triple mutant showed a similar pattern to rdr2, whereas dcl1–7 and rdr6 showed reductions in miRNAs and ta-siRNAs consistent with their activities in the biogenesis of these types of small RNAs. Deep sequencing of mutants provides a genetic approach for the dissection and characterization of diverse small RNA populations and the identification of low abundance miRNAs.
Nearly all eukaryotes produce small RNAs that function in gene regulation and/or chromatin structure. The small RNA population in plants may be among the most complex because, in addition to producing microRNAs (miRNAs) that play critical roles in various developmental, stress, and signaling responses (for reviews, see Chen 2005; Zhang et al. 2006), plants also produce a complex set of small interfering RNAs (siRNAs) (for review, see Vaucheret 2006). Among the ∼77,000 different small RNAs that have been sequenced from Arabidopsis, it is likely that miRNAs account for <10%, so the non-redundant set of siRNAs must number >70,000 (Lu et al. 2005). Most of these siRNAs match to repeated sequences such as transposons and retrotransposons. Thus, in cereals and other plant species with larger genomes and correspondingly higher contents of repeated DNA, the complexity of siRNAs is expected to be far greater.
The identification and quantification of small RNAs using deep sequencing methods was first accomplished in Arabidopsis (Lu et al. 2005). This revealed, for the first time, the tremendous complexity of plant small RNAs. Specifically, nearly 2.2 million small RNAs were sequenced from inflorescence and seedlings, yielding >56,000 and 24,000 genome-matching distinct sequences from these tissues, respectively (Lu et al. 2005). Approximately 4300 of these sequences were observed in both libraries. This advance was accomplished using a technology called Massively Parallel Signature Sequencing (MPSS), an unconventional sequencing method that proceeds by a series of cycles involving adapter ligation, hybridization, and enzymatic digestion. The application of MPSS to small RNA sequencing exceeded by more than an order of magnitude the number of small RNAs discovered by all prior conventional sequencing approaches (Llave et al. 2002a; Reinhart et al. 2002; Sunkar and Zhu 2004; Gustafson et al. 2005; Meyers et al. 2006). However, the MPSS analysis was not saturating and limited to two tissue samples, so the full extent of the small RNA population remains to be discovered.
The large number of repeat-associated siRNAs makes it a major challenge to identify low-abundance miRNAs and unusual minor types of siRNAs. Prediction programs have been extremely useful to identify miRNA candidates, yet the output of these programs must be heavily filtered for removal of false positives and this can lead to the loss of bona fide miRNAs (Jones-Rhoades and Bartel 2004; Jones-Rhoades et al. 2006). And although the number of sequenced plant genomes is growing, comparative genomics may also miss important weakly or nonconserved miRNAs. Prediction algorithms also remain to be developed for a particularly interesting siRNA type, the trans-acting siRNAs (ta-siRNAs), as well as for natural antisense siRNAs, a type thus far observed only under stress conditions (Borsani et al. 2005). Like other siRNAs, the ta-siRNAs require an RNA-dependent RNA polymerase (RdRp) for their synthesis (Peragine et al. 2004; Vazquez et al. 2004; Allen et al. 2005; Yoshikawa et al. 2005). According to current models, production of ta-siRNAs is initiated by miRNA-mediated cleavage of a precursor transcript. The RNA is then reverse-transcribed by RDR6 to form dsRNA, and this dsRNA is processed into 21-nt phased duplexes by a Dicer enzyme called DCL4 (Gasciolli et al. 2005; Xie et al. 2005b; Yoshikawa et al. 2005). In the absence of DCL4, DCL2 and DCL3 may also contribute to ta-siRNA production (Gasciolli et al. 2005; Xie et al. 2005b; Henderson et al. 2006). Some of the 21-nt species mediate the cleavage of specific mRNAs in trans, similar to miRNAs, hence the name ta-siRNAs. Because ta-siRNA production requires an initial miRNA cleavage reaction, the Dicer required for miRNA production, DCL1, is also required for ta-siRNA production (Peragine et al. 2004; Vazquez et al. 2004; Yoshikawa et al. 2005). Neither miRNAs nor ta-siRNAs require RDR2 for their biosynthesis. Yet, RDR2 as well as DCL3 are critical for synthesis of the majority of repeat-associated siRNAs, often referred to as heterochromatic siRNAs (Xie et al. 2004). Interestingly, insect and mammal genomes do not appear to encode any RdRp genes (Wassenegger and Krczal 2006).
Although the endogenous small RNAs of plants are particularly complex, the availability of mutants in small RNA biogenesis pathways of Arabidopsis often makes categorizing small RNAs a straightforward task (for review, see Vaucheret 2006). Typically, an analysis of DCL and RdRp dependencies is performed by preparing RNA gel blots from various mutants and probing for individual small RNAs (Xie et al. 2004). In this report, we sought to take an alternative approach, namely to deeply sequence the Arabidopsis small RNA population in several of these small RNA biosynthetic mutants. To this end, we sequenced >900,000 small RNAs from the inflorescence of the rdr2 mutant, a genotype that is depleted for most repeat-associated, heterochromatic small RNAs. We found the small RNA population from this mutant to be highly enriched for miRNAs, ta-siRNAs, and other interesting RDR2-independent siRNAs. By combining deep sequencing, along with computational and gel blot analyses, we were able to identify 13 new miRNAs as well as estimate the minimal number of different miRNA loci in the Arabidopsis inflorescence. We also applied 454 sequencing, another parallel sequencing technology (Margulies et al. 2005), to compare small RNA populations of rdr2 with those of several other mutants of Arabidopsis, including dcl1, rdr6, and a dcl2/3/4 triple mutant. Taken together, our data indicate that parallel sequencing of different small RNA populations using key mutants is a genetic approach that will greatly contribute to our ability to characterize the small RNAs from plants and other organisms.
Results
Sequencing of Arabidopsis rdr2 by MPSS and 454
Previous reports have indicated that rdr2 mutants show a dramatic reduction in endogenous siRNAs and a corresponding increase in miRNAs (Xie et al. 2004). We reasoned that deep sequencing in this mutant would reveal the full complement of miRNAs in Arabidopsis. We applied two methods for the high-throughout sequencing of small RNAs (for review, see Meyers et al. 2006), including the approach we have described based on MPSS (Lu et al. 2005), and a similar approach using the 454 technology (Margulies et al. 2005). MPSS provides extraordinary depth, sequencing a half million or more molecules per library, while 454 has longer reads and thereby provides information about length. Both methods provide quantitative data based on the frequency of the molecules that were sequenced. The small RNA molecules were isolated by size fractionation, sequentially ligated to RNA adapters at the 5′ and 3′ ends, and used to make cDNA template for sequencing. We generated the libraries using mixed stage inflorescences, which are known to be a rich source of small RNAs (Lu et al. 2005). MPSS produced 915,856 17-nt signatures from rdr2 (Table 1), which is comparable with the 721,044 signatures previously obtained for wild-type Arabidopsis inflorescence (Lu et al. 2005). However, the rdr2 complexity was reduced by more than 80% compared with wild type in terms of sequence diversity (9066 different genome-matched sequences in rdr2 compared with 56,920 in wild type). This dramatic difference was despite the larger total number of sequencing reads. Similarly, the 454 sequencing data demonstrated a reduced complexity for rdr2 small RNAs. Using 454, we sequenced 11,631 small RNAs from wild-type inflorescence (5713 distinct, genome-matching) and 7134 from rdr2 (686 distinct, genome-matching). The rdr2 diversity was <13% that of wild type, although in the case of the 454 data, fewer small RNAs were sequenced than with MPSS. The MPSS and 454 data correlated much better for the rdr2 mutant than the wild type, probably because the reduced complexity of rdr2 allowed a more saturating level of sampling for even low levels of sequences (Supplemental Fig. S1).
Table 1.
Summary statistics of MPSS and 454 libraries of rdr2 and wild-type inflorescence
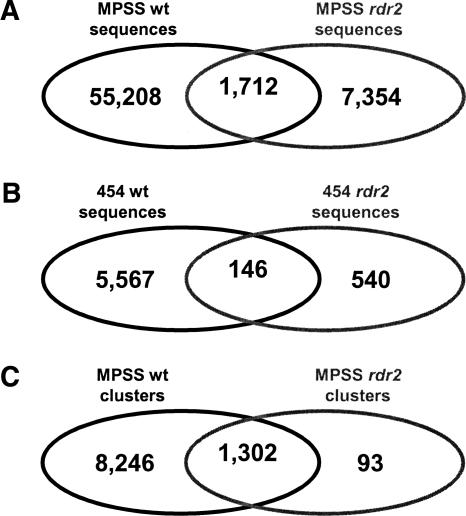
Because rdr2 is known to lack many heterochromatic siRNAs (Xie et al. 2004), we compared wild-type and rdr2 sequences to determine if the small RNAs remaining in rdr2 are primarily a subset of those in wild type. As measured by both MPSS and 454, ∼20% of the rdr2 small RNAs were also observed in the wild-type library (Fig. 1A,B). We hypothesized that this low level of similarity was largely the result of different siRNAs that represent the same regions. Therefore, we next determined if the genomic loci generating small RNAs in rdr2 were the same as wild type. To do this, we clustered the small RNAs in both rdr2 and wild type using our proximity-based algorithm (Lu et al. 2005) and compared clusters across the two libraries for the MPSS data. This analysis demonstrated that nearly all of the clusters (93%) containing at least three small RNAs that were detected in rdr2 were also detected in the wild-type inflorescence (Fig. 1C). Therefore, most of the small RNA-producing loci in rdr2 are also producing small RNAs in wild-type inflorescences. Most of the rdr2-only clusters were low-abundance sequences that may not have been detected in wild type due to the complexity of wild-type small RNAs and an unsaturated sample size.
Figure 1.
Small RNAs or clusters common to wild type and rdr2. Venn diagrams representing genome-matched rdr2 454 and MPSS sequences from Table 1. (A) A comparison of distinct signatures in the MPSS libraries indicates 19% of rdr2 sequences were also found in wild type. (B) A comparison of distinct signatures in the 454 libraries indicates 21% of rdr2 sequences were also found in wild type. (C) A comparison of genomic clusters of MPSS signatures indicates 93% of small RNA clusters represented in rdr2 were also found in wild type. For this analysis, clusters contained at least three small RNAs across both libraries; this cutoff was chosen arbitrarily to remove clusters with only one or two small RNAs that could be background. Most of the rdr2-only clusters are low-abundance miRNAs or other “real” sequences that were not detected due to depth of coverage in the wild-type library.
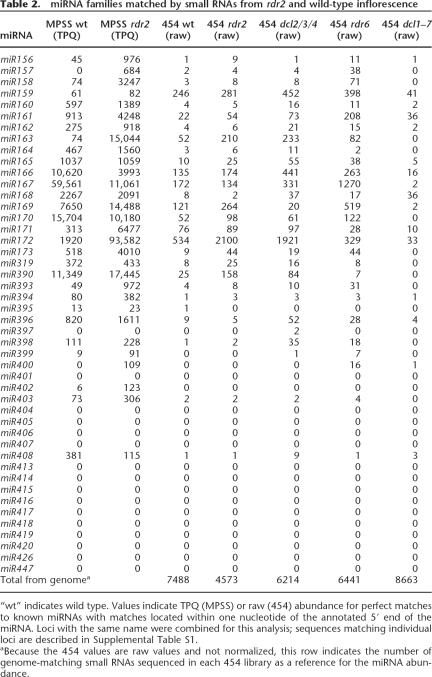
We next examined the population of miRNAs in the rdr2 mutant compared with wild type. The most obvious trend was the expected enrichment of nearly all miRNAs in rdr2 compared with the wild type (Table 2; Supplemental Table S1). The overall enrichment of miRNAs in rdr2 was 1.8-fold, based on the proportion of small RNAs represented by known miRNAs (Table 3), a level similar to the 2.2-fold enrichment reported for a low level of sequencing (Xie et al. 2005a). Eight miRNAs were enriched more than fivefold in rdr2, including miR158, miR163, miR171, miR172, miR173, miR393, miR399, and miR402 (Table 2). The most abundant miRNA in rdr2 was miR172. This miRNA was also the most abundant in a dcl2/3/4 triple mutant (Henderson et al. 2006), which, as discussed below, has a small RNA profile similar to rdr2. Both of these mutants lack many common siRNAs, and perhaps this indirectly and positively impacts miR172 abundance. At the other extreme, miR167 had a lower abundance in rdr2 than wild type, that was most prominent in the MPSS data. Across the remaining miRNAs, relatively few qualitative differences were observed in terms of miRNAs that were present or absent (Table 2; Supplemental Table S1). For example, the MPSS data showed that only two known miRNA families were present in rdr2 that had not been detected in wild-type inflorescence (miR157, miR400), while only miR395 was observed in wild-type but not the rdr2 454 library (and this may be due to the low sampling depth of the 454 data). Fifteen known miRNAs were never observed in either wild-type or rdr2 libraries (Table 2; Supplemental Table S1); this could indicate that these miRNAs are not expressed in the tissues or conditions that we sampled, some of these are not bona fide miRNAs as previously suggested (Xie et al. 2005a), or biases in cloning and/or sequencing steps led to their absence.
Table 2.
miRNA families matched by small RNAs from rdr2 and wild-type inflorescence
Table 3.
Small RNAs from MPSS libraries matching different types of repeats
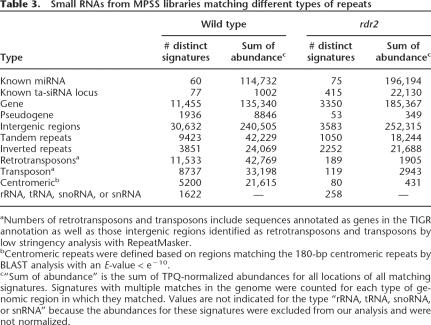
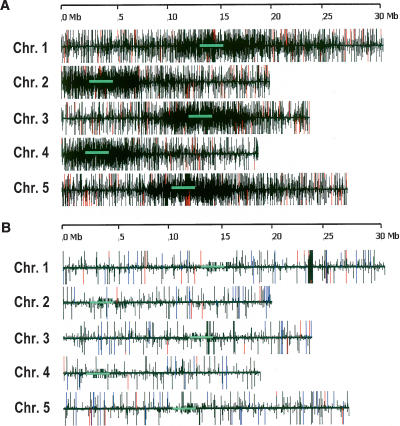
The rdr2 small RNAs showed a much more limited distribution on the Arabidopsis chromosomes compared with wild type, due to their reduced complexity. The small RNAs from the rdr2 mutant did not show a pericentromeric concentration, which is a noticeable contrast with wild-type small RNAs (Fig. 2A,B); this is consistent with a loss of heterochromatic siRNAs in rdr2 (Lippman and Martienssen 2004; Xie et al. 2004). However, there were many more loci matching small RNAs in rdr2 than are represented by the 117 known miRNA loci (Fig. 2B). This could indicate that many miRNAs, ta-siRNAs, or other RDR2-independent small RNAs have yet to be described. As a first step to determine the nature of these RDR2-independent small RNAs, we examined the relationship between rdr2 small RNAs and different genomic regions. Compared with wild type, small RNAs were reduced in rdr2 in each class of genomic sequence that we investigated (Table 3; Supplemental Fig. S2). Based on the normalized abundances, there was a proportionally greater reduction in small RNAs associated with pseudogenes, transposons, and retrotransposons, compared with genes and unclassified intergenic regions, consistent with a loss of heterochromatic siRNAs (Table 3; Fig. 2B; Supplemental Fig. S2). Small RNAs in the intergenic regions potentially represent unannotated miRNAs or siRNAs from unannotated repeats such as tandem or inverted genomic repeats (Martienssen 2003; Lippman and Martienssen 2004; Lu et al. 2005). Inverted repeats showed one of the lowest reductions in small RNAs in rdr2, while small RNAs from tandem repeats were fewer but still well-represented; these two classes of repeats are discussed in more detail below. Because miRNAs are one of the most well-known and interesting classes of small RNAs, and because they do not require RDR2, we focused first on the identification of new miRNAs from the rdr2 sequence data.
Figure 2.
Chromosomal distribution of small RNAs from wild-type and rdr2 inflorescence. Inflorescence small RNAs matched to the Arabidopsis chromosomes as measured by MPSS. The height of the vertical lines indicates the abundance of the small RNA. Maximum height of black bar, >25. The red bars indicate signatures with >125 transcripts per quarter million (TPQ) and highest black bars indicate signatures with >25 and ≤125 TPQ; other black bars indicate signature with ≤25 TPQ. Horizontal green bars indicate the approximate location of the centromere. (A) Wild-type inflorescence (adapted from Lu et al. 2005). (B) rdr2 inflorescence; the abundance of known miRNAs is indicated by blue lines.
Experimental validation of novel miRNAs
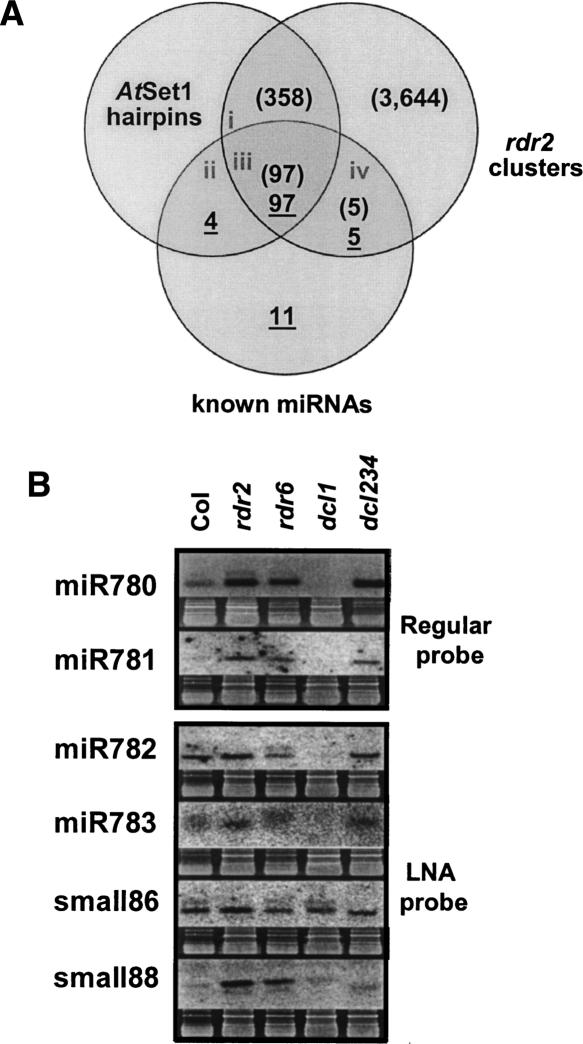
As a first step toward the identification of novel miRNAs, rdr2 MPSS sequences were compared with previously identified wild-type small RNAs in a five-way Venn diagram (Fig. 3). Among those small RNAs that are present in both libraries, the sequences were chosen for further analysis from boxes 3–6 and 9–12; these sequences matched genomic regions that can form hairpin structures and they passed the sparse cluster filter typical of miRNAs (Lu et al. 2005). Eliminating known miRNA genes (101 sequences) and transposons (eight sequences) resulted in a set of 54 small RNA sequences and a total of 31 candidate genomic loci. Because most of the novel candidate miRNAs were sequenced by MPSS multiple times and all were independently detected in two different samples (rdr2 and wild type), they may represent good candidates for novel Arabidopsis miRNAs that are expressed at low levels, may not be conserved between plant species, and have not been described as miRNAs by previous approaches or experiments.
Figure 3.
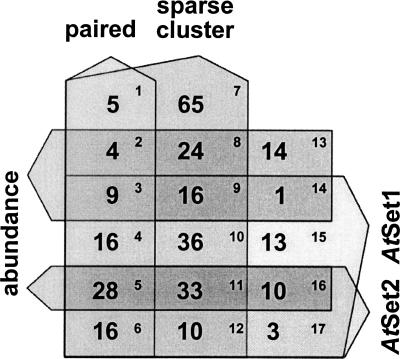
Use of rdr2 sequences to select miRNA candidates from previously identified wild-type small RNAs. Five-way Venn diagram of selection criteria for miRNAs. The number of distinct rdr2 MPSS signatures matching the criteria is indicated in each box numbered in upper right; only rdr2 signatures also found in the wild-type library are represented. The figure excludes 13,153 distinct signatures that did not pass any of the criteria (of which 1583 were found in both rdr2 and wild-type inflorescence libraries) and 54 matching to the criteria in the Venn which were present in rdr2 but not wild-type inflorescence (these 54 are included in Supplemental Fig. S2). The paired, sparse, abundance filters, and AtSet1 and AtSet2 filters are described elsewhere (Jones-Rhoades and Bartel 2004; Lu et al. 2005) but represent potential hairpin structures typical of miRNA precursors and conservation of those structures in rice, respectively.
As a complementary experimental approach to validate candidate miRNAs, the expression of candidate miRNAs in different genetic backgrounds was evaluated by RNA gel blot analysis of low molecular weight RNA isolated from inflorescence tissues. Canonical miRNAs generally require DCL1 (not DCL2, 3 or 4), but not RDR2 or RDR6, while 21-nt siRNAs from ta-siRNA loci require DCL1, DCL4, and RDR6 but not RDR2. Arabidopsis mutants with defects in Dicer and RdRp genes, therefore, are important tools to distinguish among different classes of small RNAs (Xie et al. 2004). Of the 31 candidate hairpin-forming genomic loci from the Venn diagram, we conducted RNA gel blot analysis of 13 from boxes containing small RNA signatures with an MPSS abundance of ≥40 TPQ, including three small RNAs that we previously predicted to be miRNAs (Lu et al. 2005). Bands within the size range of 21–24 nt expected for mature miRNAs were observed for 12 of 13 candidates that we tested and, of these, nine small RNAs had genetic requirements similar to those of typical, known miRNAs (Fig. 4; Supplemental Table S2A); our blots indicated the small RNAs are present in inflorescence tissue of wild type, rdr2, rdr6, and a dcl2/3/4 triple mutant (Henderson et al. 2006), but are absent in dcl1–7. Furthermore, these nine small RNAs can form stable foldback structures with the flanking genomic sequence, which is typical of a miRNA precursor, and contain the sequenced small RNA within one arm of the hairpin (Supplemental Fig. S3). Like the majority of known miRNAs, the first 5′ nucleotide of these new miRNAs was predominantly a uracil residue (Zhang et al. 2006). Based on the mutant analysis and folding, these are new miRNAs. We focused on boxes 3, 9, and 10 (Fig. 3) to identify new miRNAs because sequences in these boxes lacked a match in AtSet2, indicating that the Arabidopsis hairpin sequences were not well conserved with rice. Thus, it is not surprising that among these nine new Arabidopsis miRNAs, five do not have identifiable homologs in rice or Medicago truncatula based on sequence rather than hairpin comparisons. Like other nonconserved miRNAs, such as miR161 and miR163, these five miRNAs are represented by single loci rather than multigene families (Allen et al. 2004; Jones-Rhoades et al. 2006).
Figure 4.
Novel miRNAs identified from Venn analysis of rdr2 sequences. Small RNAs were selected for validation by RNA gel blots, as described in the text. Low molecular weight RNA isolated from inflorescence tissues was probed with labeled oligonucleotides. The lanes in the blots include the following samples: wild type, rdr2, rdr6, dcl1–7, and dcl2/3/4. The normalized abundance level from the MPSS data for rdr2 and wild type is listed to the right of the identifier for each small RNA. The reason for the apparent increases in abundance in rdr2 vs. wild type in the blots is not clear; approximately equal amounts of RNA were loaded. It does not appear to be due to RDR2-dependent small RNAs in the 5′ flanking regions, although for miR775, the most extreme case, there is an overlapping small RNA that is largely RDR2-dependent that might interfere with miR775 production in wild type.
Plant miRNAs function in the regulation of gene expression either by inducing cleavage of their mRNA targets or by translational repression. Therefore, to characterize the function of the new miRNAs we have identified, we predicted the regulatory targets using an algorithm similar to the one described by Jones-Rhoades and Bartel (2004). In general, cleavage is predominant and can be experimentally assessed using a modified 5′-RACE approach to validate these mRNA targets (Llave et al. 2002b; Reinhart et al. 2002; Kasschau et al. 2003; Jones-Rhoades and Bartel 2004). Targets were predicted with a penalty score of 2.5 or better for seven of the nine new miRNAs (Supplemental Table S3A), using the 21-nt sequence derived from the 17-nt MPSS tag plus four adjacent nucleotides from the matching genomic location. The new Arabidopsis miRNA genes we have identified are expressed at relatively low abundances as demonstrated by our MPSS data and RNA gel blots (Fig. 4), and most of them were also absent or marginally represented in other small RNA libraries sequenced by traditional methods (Gustafson et al. 2005). Consequently, mapping of cleavage products generated from these new miRNAs may be challenging due to the low and/or differential expression of the predicted target mRNAs.
Nevertheless, we verified three new miRNA targets among which two have a predicted role in plant defense responses. Two transcripts encoding the CC-NBS-LRR class of putative disease resistance proteins (At5g43740 and At1g51480) were experimentally validated as in vivo targets of miR772 (Supplemental Fig. S4A). The predicted target site for miR772 is the region encoding the P-loop domain which is highly conserved in this class of CCNBS-LRR disease resistance proteins (Meyers et al. 2003). Because of this conservation, miR772 is predicted to target at least 10 more relatives of this gene family (Supplemental Table S3A); the targeting of multiple members of a gene family by a miRNA has previously been reported for several known miRNAs (Llave et al. 2002b; Kasschau et al. 2003; Jones-Rhoades and Bartel 2004). Interestingly, two additional cleavage sites in At1g51480 were mapped, one 31 nt upstream and the other 16 nt downstream of the expected miR772 cleavage site (data not shown). This may result from the activities of other small RNAs that have not yet been identified. MiR773, miR774, and miR778 were also predicted to target several members of a gene family; for instance, miR774 is predicted to target transcripts for two genes that encode F-box proteins (Supplemental Fig. S4B; Supplemental Table S3A). Notably, several other F-box mRNAs are known targets of miRNA394 and 396 (Jones-Rhoades and Bartel 2004), and target validation assays indicated that the mRNA for another member of this extended gene family (At3g19890) is being cleaved by miR774 (Supplemental Fig. S4B). Although multiple attempts failed to confirm miR778 and miR773-mediated cleavage, the cleavage products of the transcripts predicted to be targets of these miRNAs may be detected in the future, under different conditions that elevate their abundance, for example. These predicted targets include components associated with silencing: two putative SU(VAR)3–9 like histone methyltransferase (SUVH5 and SUVH6) transcripts that are potential targets of miR778 (Naumann et al. 2005) and members of the family of DNA (cytosine-5)-methyltransferases that are potentially targeted by miR773. Previous reports have described miRNA targets involved in silencing, including DCL1 and Argonaute1 (AGO1), targets of miR162 and miR168, respectively (Rhoades et al. 2002; Xie et al. 2003; Vaucheret et al. 2004).
Estimation of the number of Arabidopsis miRNAs in Arabidopsis
Given the depth of MPSS coverage and relatively low complexity of rdr2 small RNA sequences, one could reasonably assume that the majority of inflorescence-expressed Arabidopsis miRNAs have been detected in the rdr2 library (since inflorescence is the richest source of small RNAs among the tissues we have investigated). Using the data from our rdr2 library, we sought to estimate the total number of miRNAs in Arabidopsis. Since we did not want to bias our analysis by including only the best candidates, we reassessed the small RNA sequence data using less stringent criteria. In particular, we allowed into the analysis miRNA candidates only found in the rdr2 library instead of both libraries (Supplemental Fig. S5) and lowered the acceptable abundance value from 40 TPQ to 10 TPQ. Using this approach, we identified a group of 78 miRNA candidates from the 358 rdr2 clusters that match to hairpin-like folds and have signatures expressed at levels ≥10 TPQ (Fig. 5A). By performing conventional RNA gel blots for 10 candidates that were randomly chosen from the set of 78 and had not already been analyzed during our first round of miRNA validation, we were able to identify an additional two candidate miRNAs (Fig. 5B). Because the RNA gel blot assay for the detection of miRNAs is not particularly sensitive, the expression level of the other eight candidate miRNAs may have fallen below the detection limit of our assay conditions. To increase the sensitivity of detection, we used LNA-modified oligo probes (Valoczi et al. 2004) to test these eight miRNA candidates that were previously undetectable by RNA gel blot analysis. Of these, four candidates were positive by LNA oligo assay, but the expression of two of the four was not reduced in dcl1–7 (Fig. 5B). Overall, a total of four of the eight candidates had canonical miRNA expression patterns as detected by either conventional (miR780, miR781) or LNA probes (miR782, miR783) (Fig. 5B). According to miRNA annotation criteria (Ambros et al. 2003), the four of 10 candidates assayed by RNA gel blot hybridization can be considered as bona fide miRNAs (Supplemental Fig. S6); this brings the total number of new miRNAs that we identified to 13. Therefore, these gel blot analyses of the randomly selected small RNAs produced an overall success rate of 40%. Extrapolating from this rate, we would anticipate finding ∼31 new miRNAs (from the calculation of 78 × 40%) in the group of 78 miRNA candidates described above, if we had done a comprehensive and exhaustive analysis of inflorescence-expressed small RNAs.
Figure 5.
Estimating the minimal Arabidopsis miRNA population. (A) A Venn diagram to identify the number of miRNAs that may exist in Arabidopsis. Numbers in each section indicate sequences that satisfy each criterion, include AtSet1 hairpin folds (Jones-Rhoades and Bartel 2004), known miRNAs (underlined numbers), and genomic clusters of rdr2 MPSS signatures (numbers in brackets). AtSet1 and rdr2 clusters were considered matching if the start or end of either overlapped. Lowercase Roman numerals indicate sections from which sequences were selected in B. (B) Gel blots to evaluate the fraction of bona fide miRNAs in selected sectors. Small RNAs identified by rdr2 signatures in section (i) in A were randomly selected and evaluated by RNA gel blots to determine their presence/absence in the dcl1–7, dcl2/3/4, and rdr6 mutants. LNA probes were used for miR782, miR783, small86, and small88 (see Methods).
To estimate the total number of Arabidopsis miRNAs, two adjustment factors were taken into account for the calculations. Of the 101 known miRNA loci present in the genome-wide folding data of AtSet1, 97 were detected by MPSS in the inflorescence libraries. Based on this result, we determined the MPSS detection rate for inflorescence of 0.96 (97 of 101 known miRNAs in AtSet1 detected by MPSS). We also determined the AtSet1 detection rate of 0.86 (101 of 117 known miRNAs in AtSet1). Accounting for these two rates, we estimate that there are at least 155 miRNA loci in the Arabidopsis genome (calculated from ([97 + 31]/0.96)/0.86, and with the caveat that these sample sizes are too small to draw definitive conclusions). In fact, this is probably an underestimate, if the failure to detect miRNA candidates by our assays was due to very low abundance, restricted cell-type expression, or induction only in response to specific environmental cues (and assuming all annotated miRNAs are real). In addition to the known and new miRNAs, the rdr2 data demonstrated the presence of numerous RDR2-independent small RNAs, some of which may yet be characterized as new miRNAs.
Other RDR2-independent small RNAs in Arabidopsis
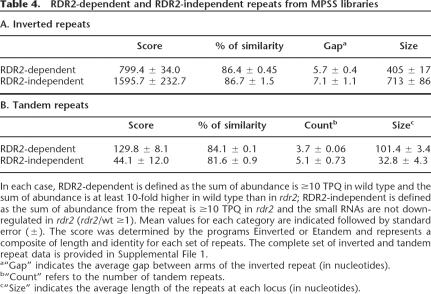
We and other groups have previously shown that a significant number of Arabidopsis endogenous siRNAs match to various kinds of repeats (Martienssen 2003; Lippman and Martienssen 2004; Lu et al. 2005). Xie et al. (2004) have shown the requirement of RDR2 and DCL3 for the biosynthesis of a subset of repeat-associated siRNAs. However, considering the presence of multiple RdRps in Arabidopsis and the diversity of repeats, it is unclear which populations of siRNAs generated from repeat sequences are dependent on RDR2 activity. We separately characterized the RDR2-dependent and RDR2-independent inverted and tandem repeats, as these repeats are known to be sources of small RNAs. The RDR2-dependent inverted repeat set, comprising a total of 461 genomic locations, was defined as those for which (1) the sum of abundance is ≥10 TPQ in wild type; (2) the sum of abundance is at least 10-fold higher in wild type than in rdr2. Similarly, we considered a repeat to be RDR2-independent only if the sum of abundance from the repeat is ≥10 TPQ and not down-regulated (rdr2/wt ≥1) in rdr2; we found 55 loci for this set (12% of the total). As shown in Table 4, the repeat score of the RDR2-independent set was significantly higher than that of the RDR2-dependent set (Mann-Whitney Test: P-value = 0.0048). One of the primary determinants of the score is the length of the repeat, suggesting that the RDR2 dependence of inverted repeats may be based on their length. This is consistent with a previous study suggesting that, for some inverted repeats, RDR2 may contribute to the formation or stability of a complex that contains active DCL3 (Xie et al. 2004). For genomic loci that contain long inverted duplications and can form extensive dsRNA structures (foldbacks), RDR2 is most likely dispensable for siRNA production (Table 4). One hypothesis is that one or more Dicers can efficiently process long dsRNA precursors even in the absence of RDR2. In agreement with this, closer examination of some RDR2-independent inverted repeats revealed that these loci usually showed complex patterns of siRNA accumulation with different size classes affected by different Dicer mutants (Supplemental Fig. S7).
Table 4.
RDR2-dependent and RDR2-independent repeats from MPSS libraries
We identified a potential foldback structure in the S-receptor kinase gene (SRK) as one of the most strongly expressed RDR2-independent siRNA-producing regions (Supplemental Fig. S8). The large number of sequenced small RNAs matching to this stem–loop suggests that it is a substrate for Dicer cleavage. The observation that small85 from this locus is still evident in the dcl1–7 and dcl2/3/4 mutants but not in a quadruple dcl1/2/3/4 mutant (data not shown) suggests the involvement of multiple Dicers (Supplemental Fig. S8). Functional copies of SRK and a gene called SCR are important for self-incompatibility in Brassica and Arabidopsis species (such as A. lyrata) (Nasrallah et al. 2002). Loss of this self-incompatibility system in Arabidopsis thaliana is one of the key factors that led to the selection of A. thaliana as a model system for plants (Redei 1975). Suggested explanations for this loss include the fragmented SCR gene or the alternatively spliced SRK transcripts that contain premature nonsense codons that are present in A. thaliana (Kusaba et al. 2001). Our data suggest that the SRK gene may be silenced by an inverted repeat, and these small RNAs may have played a previously unknown role in the loss of SRK function in A. thaliana.
Unlike inverted repeats from which dsRNA is readily generated simply by folding of a single RNA, tandem repeats should require an RdRp to form dsRNA structures. Indeed, tandem repeats show a higher overall dependence on RDR2 than inverted repeats (Table 4). Our RDR2-dependent tandem repeat set contained 3491 genomic locations whereas the RDR2-independent tandem repeat set contained only 82 loci (2% of the total). Interestingly, we also found that the average length of the tandem repeat unit in the RDR2-dependent set is significantly larger than that of the RDR2-independent set (Mann-Whitney Test: P-value = 0.0001). Therefore, high quality and long tandem repeats generally appear to require RDR2 to generate dsRNAs and sustain siRNA production. Other RdRps probably facilitate dsRNA production from these short tandem repeats because the Arabidopsis genome contains six RdRp homologs (Yu et al. 2003). One likely hypothesis is that different RdRps could function redundantly on tandem repeats.
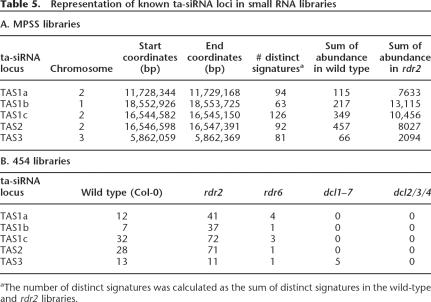
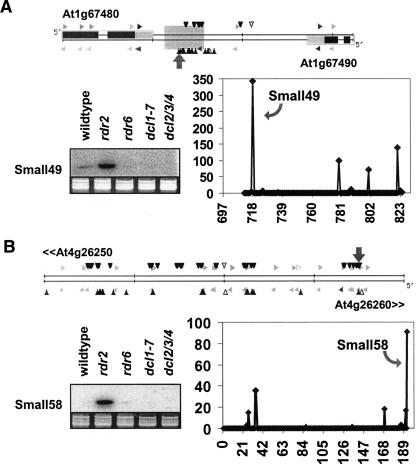
Known ta-siRNA loci were the most enriched small RNA sources in the rdr2 background. For the four previously characterized ta-siRNA loci, the sum of small RNA abundance was at least 20-fold higher in rdr2 than in wild type based on the MPSS data (Table 5A; Supplemental Fig. S9). This greatly exceeds the 1.8-fold for enrichment of total miRNA abundance mentioned earlier. Using known ta-siRNAs as reference, we developed a set of filters to enrich for new ta-siRNAs. Four filters were designed and applied to identify genomic locations representing potential tasiRNA loci: (1) The cluster contains at least 10 distinct signatures; (2) the sum of abun dance for the cluster is ≥100 TPQ; (3) the sum of abundance is at least 10-fold higher in rdr2 than in wild type; (4) the cluster does not match to known miRNAs, ta-siRNAs, transposons, retrotransposons, or centromere repeats. These filters generated 28 po tential ta-siRNA loci (Supplemental Table S4). Interestingly, among these, 14 loci (50% of the filter output) corresponded to different members of the PPR gene family, a group of genes known to be targeted by miRNAs, ta-siRNAs, and siRNAs (Allen et al. 2005). Seven of the 14 remaining candidate loci were further examined by RNA gel blot ting. We found two candidates (small49, small58) displaying typical ta-siRNA expres sion patterns (present in rdr2 but very low in rdr6, dcl1, and dcl2/3/4) (Supplemental Table S2B; Fig. 6). Furthermore, a clear 21-nt phased pattern was observed at the locus containing small49, consistent with Dicer activity (Fig. 6). With this low stringency filtering protocol that captures all known ta-siRNA loci, we found relatively few loci which had ta-siRNA characteristics. Therefore, we interpret these data as an indication that ta-siRNA genes are rare in the Arabidopsis genome. This result is consistent with the observation that mutations that block ta-siRNA production have a relatively weak phenotype (Peragine et al. 2004; Vazquez et al. 2004). However, it is also possible that other ta-siRNAs were expressed at very low levels or not at all under our sampling conditions.
Table 5.
Representation of known ta-siRNA loci in small RNA libraries
Figure 6.
RDR2-independent small RNAs from regions with ta-siRNA-like features. (A) The locus that includes small49 exhibits 21-nt phasing and accumulation characteristics in mutants similar to those of ta-siRNAs. Image of the MPSS Web viewer for the intergenic region that contains small49 in the position indicated by the large gray arrow (the shaded square covering both strands in this intergenic region indicates an inverted repeat); an RNA gel blot of small49; a plot of the y-axis indicating the small RNA abundance in the rdr2 mutant as measured by MPSS (in TPQ), and the x-axis indicating nucleotide position on Chr. 1, with the “697” indicating position 25,282,697. (B) The blot shows that small58 also has ta-siRNA-like accumulation features; images as described in A, with the 0 position in the x-axis of the plot indicating nucleotide 13,295,900 on Chr. 4. Due to limited space, only a portion of this intergenic region is shown, with the flanking genes indicated at the ends by their At identifiers.
Small RNA size distribution in rdr2 and the small RNA populations in other mutants
The enrichment of miRNAs and loss of heterochromatic siRNAs in rdr2 should correlate with a shift in the sizes of the small RNA population. Canonical miRNAs are 21 nt while canonical heterochromatic siRNAs are 24 nt. Because the MPSS sequence data are limited to 17 nt for small RNAs, we used the 454 sequence data to determine the size distribution of the small RNAs. As an additional comparison to wild-type and rdr2 inflorescences, we also sequenced small RNAs from the inflorescence of the Arabidopsis mutants rdr6 and dcl1–7, and compared these with data we recently obtained for dcl2/ 3/4 (Henderson et al. 2006). All of these mutants are altered in important genes for small RNA biogenesis. We assessed the size distribution based on both distinct sequences and total abundances (Fig. 7). Both rdr2 and the dcl2/3/4 triple mutant showed a similar pattern of 24-nt siRNA reduction and 21-nt miRNA enrichment (Fig. 7A,B). The increase in 21 mers in both mutants reflects an enrichment of miRNAs and is consistent with previous reports (Table 2; Fig. 7B,D; Xie et al. 2004). In contrast to miRNAs, 21-nt siRNAs from known ta-siRNA loci can be readily identified from rdr2 but were absent in dcl2/3/4 (Table 5B; Henderson et al. 2006), consistent with previous observations that DCL4 is required for ta-siRNA production (Gasciolli et al. 2005; Xie et al. 2005b; Yoshikawa et al. 2005; Henderson et al. 2006). Nevertheless, a strong correlation between the 454 data of rdr2 and dcl2/ 3/4/ was observed (R2 = 0.92 for all small RNAs present in both libraries; R2 = 0.95 for miRNAs; Supplemental Fig. S10). In contrast, the dcl1–7 mutant demonstrated a lower proportion of 21-nt small RNAs compared with wild type (Fig. 7B), and most of this difference can be attributed to a substantial reduction in known miRNAs (Table 2; Fig. 7B,D). This is consistent with the known reduction in the miRNA complement of dcl1–7 (Park et al. 2002). Both the wild type and rdr6 mutant have substantial peaks at both 21 and 24 nt, as expected. However, analysis of ta-siRNA abundance in the rdr6 mutants has revealed that indeed very few ta-siRNAs were detected in the absence of RDR6 (Table 5B).
Figure 7.
Small RNA size distribution in mutants evaluated with 454 sequencing. In each plot, gray indicates wild type, light blue is rdr2, green is dcl1–7, dark blue is rdr6, and red is the dcl2/3/4 triple mutant. (A) Number of distinct signatures vs. size. (B) Total abundance of sequences vs. size. (C) Number of distinct vs. size, with known miRNAs removed. (D) Total abundance vs. size, with known miRNAs removed.
Even the modest depth of the 454 sequencing was sufficient to identify differential effects of specific mutants on the accumulation on miRNA families. Although DCL1 appears to be the only Dicer protein responsible for miRNA biogenesis in Arabidopsis (Henderson et al. 2006; Jones-Rhoades et al. 2006), some miRNAs are affected less than others by the dcl1–7 mutant. The most extreme case was miR168 which did not decrease at all in dcl1–7 based on our 454 data (Table 2). These results are in agreement with Vaucheret et al. (2006), who reported no decrease in miR168 levels in three different dcl1 partial loss-of-function mutants. This fits well with the model that miR168 levels are not limited by DCL1 activity but are instead controlled by a feedback loop involving AGO1, the target of miR168; AGO1 is hypothesized to both stabilize miR168 and also slice its own mRNA using miR168 as a guide (Vaucheret et al. 2006). The accumulation of miR159 and miR165/166 has also been reported to be somewhat less sensitive to dcl mutations than other miRNAs tested (Park et al. 2002; Reinhart et al. 2002; Kasschau et al. 2003; Kurihara et al. 2006; Vaucheret et al. 2006) and we also observed these subtleties. Finally, members of the miR161 family and miR408 are known to be rather insensitive to the dcl1–7 allele (Allen et al. 2004) and the dcl1–9 allele (Sunkar and Zhu 2004) respectively, results quite consistent with our 454 data. Based on the close recapitulation of published observations with our dcl1 data, it seems likely that other differential accumulation characteristics resulting from our data set represent regulatory characteristics of potential biological significance. These would include miR167, which is down-regulated in rdr2 compared with wild type, and miR172 which is of particularly high abundance in rdr2 and dcl2/3/4 (Table 2). Another miRNA with unusual characteristics is miR169. This miRNA is an outlier in the correlation of rdr2 and dcl2/3/4 (Supplemental Fig. S10), having a very low accumulation in dcl2/ 3/4, with high accumulation in rdr2. Given that miR169 is also increased in rdr6 and encoded by a tandem array of genes, these accumulation results may be due to a secondary level of control by an siRNA-mediated pathway.
Discussion
We have characterized the small RNAs in the Arabidopsis rdr2 mutant and demonstrated that this mutant contains a dramatically altered population of small RNAs. Many fewer heterochromatic siRNAs were observed, and miRNAs and other specialized categories of small RNAs were enriched, facilitating their identification and classification. We applied two complementary sequencing methods, MPSS, which is very deep, and 454, which provides size information; these combined data sets confirmed and greatly extended earlier studies by providing quantitative, genome-wide small RNA data. Endogenous siRNAs corresponding to transposons, retrotransposons, centromere repeats, and pseudogenes virtually disappear in the rdr2 background (Fig. 2; Supplemental Fig. S2), as predicted by prior low-depth analyses (Xie et al. 2004; May et al. 2005). A surprising finding was that some of the small RNAs remaining in rdr2 had significantly higher abundance than in wild type (Supplemental File 1) and derived from repeats that had special characteristics. Other RDR2-independent siRNAs were phased by 21 nt and/or had other features characteristic of ta-siRNAs. Indeed, we found that small RNAs matching to known ta-siRNA loci were dramatically enriched in rdr2 (5.5% by abundance compared with 0.2% in wild type) to an extent even greater than that of miRNAs (51.5% in rdr2 compared with 28.9% in wild type) (Table 3). The enrichment of miRNAs in rdr2 was a tremendous advantage for the identification of new low abundance miRNAs. And finally, the parallel sequencing by 454 of additional libraries from small RNA biosynthetic mutants not only recapitulated published data but also provided new and interesting examples of differential effects on specific miRNAs and siRNAs.
miRNA discovery
While most Arabidopsis miRNAs have been identified by traditional cloning and sequencing of small RNAs, it is unlikely that these screens are saturating for rare or tissue-specific miRNAs. The need for additional methods of miRNA identification led to the development of bioinformatics methods for the prediction of miRNAs (Bonnet et al. 2004; Jones-Rhoades and Bartel 2004; Wang et al. 2004). Most of these computer algorithms rely on evolutionary conservation of miRNA sequences between different species and, therefore, are limited to the detection of only conserved miRNAs, although at least one analysis has relied only on intra-genomic comparisons (Adai et al. 2005). Even these predictions ultimately require either high-throughput or highly sensitive methods for validation. With MPSS and other high-throughput sequencing technologies, the sequencing of small RNAs is no longer a limiting factor in the discovery of novel miRNAs. However, we have demonstrated that by combining these approaches with mutants in which miRNAs are significantly enriched compared with wild type, such as rdr2 and dcl2/ 3/4, we can efficiently delineate the small RNAs as miRNAs, siRNAs, or other categories. Even at relatively low sampling depths, many known miRNAs were observed and their abundance was measured. Compared with wild type, the MPSS data for rdr2 were dramatically simplified and “cleaned up” of siRNAs, making miRNA candidates much easier to identify. Our 454 analysis indicated that the rdr2 and dcl2/3/4 triple mutants are most similar in their small RNA profiles, consistent with the idea that these genes may be in the same pathway involved in heterochromatic siRNA production (for review, see Brodersen and Voinnet 2006) and a mutant of either type (rdr2 and dcl2/3/4) enriches for miRNAs.
Prior experimental and computational efforts over the last several years have resulted in the identification of 117 miRNA genes in Arabidopsis which can be grouped into 42 families. The 13 new miRNAs that we report all represent new families that presumably escaped previous discovery because of their low abundance. These new miRNAs increase the total number of Arabidopsis miRNA families by 25%. Eight of the newly described miRNAs are found only in Arabidopsis. For nonconserved miRNAs, it is more difficult to confidently predict targets because the conservation of the target site cannot be used as a filter to remove false positives. Therefore, a highly stringent score (≤2.5) was applied in our target prediction. Potential regulatory targets were found for 10 of the 13 miRNAs. Some of the biological roles of the newly confirmed or predicted targets resemble those of previously identified Arabidopsis miRNAs. At least three of these are bona fide because we could map the cleavage products and we predict that others were simply beneath our threshold of detection. MiR774 targets the mRNA of at least one F-box protein. Combined with six previously identified F-box genes, there are at least seven F-box mRNAs targeted by miRNAs, suggesting that the protein degradation machinery is subject to considerable miRNA regulation (Jones-Rhoades et al. 2006). Our observation that miR773 mediates the cleavage of at least two, and potentially more, members of the CC-NBS-LRR class of putative disease resistance proteins suggests a previously unknown role of miRNAs in plant defense. As new and more sensitive methods for verifying miRNA targets are developed, it will be exciting to see if some of the other interesting putative targets such as the methytransferases in Supplemental Figure S4 can be verified. While, our target predictions focused on protein-coding genes, at least two miRNAs (mir173 and mir390) target precursors of ta-siRNAs; consequently, there may be additional targets for some of these new miRNAs that have not yet been identified.
From our analyses, we have predicted based on our sequence data and RNA gel blot assays that at least 155 miRNAs should exist in the Arabidopsis genome. We believe that this is a conservative estimate. Several groups have shown that RNA gel blots are not sensitive enough to detect many real miRNAs (Grad et al. 2003; Bentwich et al. 2005; Grundhoff et al. 2006). Moreover, if a higher percentage of the candidate miRNAs that were negative by gel blot analysis can be detected by more sensitive detection methods such as PCR, or if we loosen our abundance cutoff to below 10 TPQ, more miRNAs may appear to be bona fide. It should be noted also that inflorescence tissues were used for many of the previous miRNA cloning projects, and, as a result, the set of known miRNAs likely is biased toward those that are expressed in flowers. Therefore, the real number of miRNAs may well exceed our estimate. Interestingly, many of the new miRNAs identified in this study did not have homologs in rice, suggesting that conservation between Arabidopsis and rice may not be one of the best filters for identifying additional new miRNAs. The conservation of miRNA families across plant subgroups such as eudicots, monocots, gymnosperms, and even moss has been reported (Arazi et al. 2005; Axtell and Bartel 2005; Zhang et al. 2006). However, the presence of eight Arabidopsis-specific miRNAs indicates substantial miRNA evolution in the time since the divergence of monocots and dicots, and suggests that miRNA enrichment methods such as our use of the rdr2 mutant may be valuable in many plant species.
RDR2-independent siRNAs
Tandem repeats are prone to epigenetic silencing mediated by RNA interference (Martienssen 2003). Previous studies have shown that several siRNAs corresponding to tandem repeats in the Arabidopsis genome were absent in rdr2 (Xie et al. 2004; May et al. 2005). It has been proposed that tandem repeats can sustain RdRp activity because the first round siRNAs can randomly initiate subsequent rounds of siRNA production and perpetuate the siRNA pool (Martienssen 2003). While this model has not been proven, it is substantiated by our MPSS data indicating that almost all the tandem repeats in the Arabidopsis genome required RDR2 activity to generate siRNAs. However, for some of these tandem repeats, the small RNAs were significantly higher in rdr2 than in wild type (Supplemental File 1). Something about these tandem repeats, perhaps their relatively low quality, may allow these sequences to be silenced independently of RDR2. In this case, other components of the siRNA biogenesis machinery must be involved in the recognition and generation of siRNAs from these specific loci. This suggests that the biogenesis pathway for repeat-associated siRNAs is more complex than initially believed and the production of some repeat-associated siRNAs does not require RDR2 activity.
Our understanding of the mechanism by which siRNAs are derived from inverted repeats is less clear. siRNA accumulation from inverted-repeat loci is dependent on RDR2 and DCL3 (Xie et al. 2004). While DCL3 clearly functions as the ribonuclease to process dsRNA precursors, it is unclear why RDR2 is essential to this pathway. Another example is siRNA production from constructs used for inverted-repeat post-transcriptional gene silencing (IR-PTGS, typically used for RNAi). Although widely used as a research tool, IR-PTGS remains one of the least understood plant RNA silencing processes. Until recently, no mutant defective in this pathway had been recovered, and IR-transgene-induced siRNA accumulation is not affected by single-gene mutations (for review, see Brodersen and Voinnet 2006). Our analysis of rdr2 by MPSS may provide an explanation for these apparently contradictory observations. In agreement with previous studies, the majority of endogenous inverted repeats, such as the siRNA02 locus (Xie et al. 2004), did not accumulate siRNAs in the absence of RDR2. However, we also identified a group of inverted repeats which produced siRNAs independently of RDR2 (Supplemental File 1). One difference between RDR2-dependent and RDR2-independent inverted repeats is that the latter set tends to have a higher repeat score and larger size of repeat unit. Although it is difficult to rule out alternative hypotheses completely, the simplest interpretation of the data is that RDR2 and DCL3 are required for only a subset of inverted repeats, generally with low scores and relatively short repeat units. In the case of longer and higher scoring inverted repeats, RDR2 activity (and probably DCL3) may not be required, similar to IR-transgenes. One likely scenario is that the high quality dsRNA structures generated from long inverted repeats are subject to the activity of different Dicers (Brodersen and Voinnet 2006). Consistent with this model, recent analyses of combinatorial Dicer knockout mutants indicated that the functions of different Arabidopsis Dicer proteins are highly redundant (Gasciolli et al. 2005; Henderson et al. 2006).
Conclusions
Clearly, analyses of plant miRNAs may achieve a higher level of sensitivity by using an rdr2 background. This approach may prove even more beneficial in other plant species, particularly those with larger and more repeat-laden genomes. The combined deep profiling data from MPSS and full-length sequencing of small RNAs from different genotypes by 454 demonstrate that small RNA sequence libraries are a rich and novel source of data that have yet to be fully exploited in Arabidopsis or any other organism. As sequencing costs drop with the advent of new short-read sequencing technologies, the approaches that we have implemented for the analysis of Arabidopsis mutants are likely to be more broadly applied for experimental investigation of different conditions or mutants.
Methods
Plant growth
All plant material was from Arabidopsis ecotype Col-0. The rdr2, rdr6, dcl1–7, and dcl2/3/4 mutants have been described previously (Peragine et al. 2004; Xie et al. 2004; Henderson et al. 2006). Inflorescence tissue was harvested from plants grown in soil in a growth chamber with 16 h of light for 5 wk. Floral tissue included the inflorescence meristem and early stage floral buds (up to Stage 11/12). Total RNA was isolated using Trizol reagents (Invitrogen). Inflorescence and seedling material were harvested approximately at eight hours into the subjective day.
RNA gel blot analysis
Total RNA was extracted with Trizol (Invitrogen). Low molecular weight (LMW) RNA gel blots, radiolabeled probes for specific small RNAs, and hybridization/wash conditions were described previously (Lu et al. 2005). Blot hybridization analysis was performed as described (Llave et al. 2002a). All blots shown are representative of at least two independent experiments.
Locked nucleic acid (LNA) probes (Valoczi et al. 2004) were used as indicated in the figure legends; these probes were used when the hybridization signal was not detectable using regular oligonucleotides. LNA oligos were obtained from Sigma-Proligo. Hybridization conditions were as described (Valoczi et al. 2004).
MPSS and 454 data generation and analysis
All MPSS sequencing and analysis were performed essentially as described (Brenner et al. 2000a, b; Meyers et al. 2004; Lu et al. 2005). The small RNA libraries were constructed as previously described (Lu et al. 2005; Meyers et al. 2006). The raw and normalized MPSS data are available at http://mpss.udel.edu/at. 454 analysis was performed essentially as described (Margulies et al. 2005). Adapter sequences were identified and removed using local alignments. The summary statistics of the rdr2 and wild-type 454 libraries are described in the text; the dcl1–7 and rdr6 libraries included 12,060 and 16,856 adapter-trimmed small RNA inserts, respectively, and the dcl2/3/4 triple mutant 454 library has recently been described (Henderson et al. 2006).
We compared MPSS signatures with the TIGR annotation version 5.0 (Wortman et al. 2003) and assigned signatures to each location at which a perfect match was found. The number of matches was recorded as the “hits.” As previously described (Lu et al. 2005), we merged the MPSS sequencing runs and calculated a single abundance normalized to “transcripts per quarter million” (TPQ) after the removal of rRNAs, tRNAs, snoRNAs, or snRNAs signatures. Clustering of small RNAs was based on the previously described proximity-based algorithm, with the same setting of a 500-bp window for the clusters that was used in our prior analysis (Lu et al. 2005). Repeat analysis was also performed as described previously (Lu et al. 2005) using a combination of programs including RepeatMasker (http://www.repeatmasker.org/), Einverted and Etandem (Rice et al. 2000).
Accession numbers
Sequence data from this article can be found in the Gene Expression Omnibus database at NCBI (http://www.ncbi.nlm.nih.gov/geo/). The accession numbers for GEO are as follows: Series identifier GSE5343, platform identifier GPL4011, and sample identifiers are GSM120717 (rdr2 MPSS data), and GSM121453 to GSM121457 (454 data). The raw and normalized MPSS data are also available at http://mpss.udel.edu/at, and this Web site allows users to query these data based on physical location, gene identifiers, or by sequence.
Acknowledgments
We thank Mayumi Nakano for her work on the Web interface, Kalyan Vemaraju for his work on the MPSS database, Deborah Charlesworth for discussions of the SRK locus, and several anonymous reviewers for their valuable comments. This work was supported primarily by NSF grants 0439186 and 0548569 (P.J.G. and B.C.M.), with additional support and resources provided by NSF Plant Genome grant 0321437 (B.C.M.), DOE DE-FG02-04ER15541 (P.J.G.), and NIH P20 RR16472-04. I.R.H. was supported by an EMBO Long-term Fellowship. S.E.J. is a Howard Hughes Medical Institute Investigator, and work in S.E.J.’s laboratory was supported by NIH grant GM60398. Research in the Poethig laboratory was supported by NIH grant GM51893.
Footnotes
Supplemental material is available online at www.genome.org.
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5530106.
References
- Adai A., Johnson C., Mlotshwa S., Archer-Evans S., Manocha V., Vance V., Sundaresan V., Johnson C., Mlotshwa S., Archer-Evans S., Manocha V., Vance V., Sundaresan V., Mlotshwa S., Archer-Evans S., Manocha V., Vance V., Sundaresan V., Archer-Evans S., Manocha V., Vance V., Sundaresan V., Manocha V., Vance V., Sundaresan V., Vance V., Sundaresan V., Sundaresan V. Computational prediction of miRNAs in Arabidopsis thaliana . Genome Res. 2005;15:78–91. doi: 10.1101/gr.2908205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E., Xie Z., Gustafson A.M., Sung G.H., Spatafora J.W., Carrington J.C., Xie Z., Gustafson A.M., Sung G.H., Spatafora J.W., Carrington J.C., Gustafson A.M., Sung G.H., Spatafora J.W., Carrington J.C., Sung G.H., Spatafora J.W., Carrington J.C., Spatafora J.W., Carrington J.C., Carrington J.C. Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana . Nat. Genet. 2004;36:1282–1290. doi: 10.1038/ng1478. [DOI] [PubMed] [Google Scholar]
- Allen E., Xie Z., Gustafson A.M., Carrington J.C., Xie Z., Gustafson A.M., Carrington J.C., Gustafson A.M., Carrington J.C., Carrington J.C. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Ambros V., Bartel B., Bartel D.P., Burge C.B., Carrington J.C., Chen X., Dreyfuss G., Eddy S.R., Griffiths-Jones S., Marshall M., Bartel B., Bartel D.P., Burge C.B., Carrington J.C., Chen X., Dreyfuss G., Eddy S.R., Griffiths-Jones S., Marshall M., Bartel D.P., Burge C.B., Carrington J.C., Chen X., Dreyfuss G., Eddy S.R., Griffiths-Jones S., Marshall M., Burge C.B., Carrington J.C., Chen X., Dreyfuss G., Eddy S.R., Griffiths-Jones S., Marshall M., Carrington J.C., Chen X., Dreyfuss G., Eddy S.R., Griffiths-Jones S., Marshall M., Chen X., Dreyfuss G., Eddy S.R., Griffiths-Jones S., Marshall M., Dreyfuss G., Eddy S.R., Griffiths-Jones S., Marshall M., Eddy S.R., Griffiths-Jones S., Marshall M., Griffiths-Jones S., Marshall M., Marshall M., et al. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arazi T., Talmor-Neiman M., Stav R., Riese M., Huijser P., Baulcombe D.C., Talmor-Neiman M., Stav R., Riese M., Huijser P., Baulcombe D.C., Stav R., Riese M., Huijser P., Baulcombe D.C., Riese M., Huijser P., Baulcombe D.C., Huijser P., Baulcombe D.C., Baulcombe D.C. Cloning and characterization of micro-RNAs from moss. Plant J. 2005;43:837–848. doi: 10.1111/j.1365-313X.2005.02499.x. [DOI] [PubMed] [Google Scholar]
- Axtell M.J., Bartel D.P., Bartel D.P. Antiquity of microRNAs and their targets in land plants. Plant Cell. 2005;17:1658–1673. doi: 10.1105/tpc.105.032185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentwich I., Avniel A., Karov Y., Aharonov R., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Avniel A., Karov Y., Aharonov R., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Karov Y., Aharonov R., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Aharonov R., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Barzilai A., Einat P., Einav U., Meiri E., Einat P., Einav U., Meiri E., Einav U., Meiri E., Meiri E., et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- Bonnet E., Wuyts J., Rouze P., Van de Peer Y., Wuyts J., Rouze P., Van de Peer Y., Rouze P., Van de Peer Y., Van de Peer Y. Detection of 91 potential conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important target genes. Proc. Natl. Acad. Sci. 2004;101:11511–11516. doi: 10.1073/pnas.0404025101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani O., Zhu J., Verslues P.E., Sunkar R., Zhu J.K., Zhu J., Verslues P.E., Sunkar R., Zhu J.K., Verslues P.E., Sunkar R., Zhu J.K., Sunkar R., Zhu J.K., Zhu J.K. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis . Cell. 2005;123:1279–1291. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., Johnson M., Bridgham J., Golda G., Lloyd D.H., Johnson D., Luo S., McCurdy S., Foy M., Ewan M., Johnson M., Bridgham J., Golda G., Lloyd D.H., Johnson D., Luo S., McCurdy S., Foy M., Ewan M., Bridgham J., Golda G., Lloyd D.H., Johnson D., Luo S., McCurdy S., Foy M., Ewan M., Golda G., Lloyd D.H., Johnson D., Luo S., McCurdy S., Foy M., Ewan M., Lloyd D.H., Johnson D., Luo S., McCurdy S., Foy M., Ewan M., Johnson D., Luo S., McCurdy S., Foy M., Ewan M., Luo S., McCurdy S., Foy M., Ewan M., McCurdy S., Foy M., Ewan M., Foy M., Ewan M., Ewan M., et al. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat. Biotechnol. 2000a;18:630–634. doi: 10.1038/76469. [DOI] [PubMed] [Google Scholar]
- Brenner S., Williams S.R., Vermaas E.H., Storck T., Moon K., McCollum C., Mao J.I., Luo S., Kirchner J.J., Eletr S., Williams S.R., Vermaas E.H., Storck T., Moon K., McCollum C., Mao J.I., Luo S., Kirchner J.J., Eletr S., Vermaas E.H., Storck T., Moon K., McCollum C., Mao J.I., Luo S., Kirchner J.J., Eletr S., Storck T., Moon K., McCollum C., Mao J.I., Luo S., Kirchner J.J., Eletr S., Moon K., McCollum C., Mao J.I., Luo S., Kirchner J.J., Eletr S., McCollum C., Mao J.I., Luo S., Kirchner J.J., Eletr S., Mao J.I., Luo S., Kirchner J.J., Eletr S., Luo S., Kirchner J.J., Eletr S., Kirchner J.J., Eletr S., Eletr S., et al. In vitro cloning of complex mixtures of DNA on microbeads: Physical separation of differentially expressed cDNAs. Proc. Natl. Acad. Sci. 2000b;97:1665–1670. doi: 10.1073/pnas.97.4.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P., Voinnet O., Voinnet O. The diversity of RNA silencing pathways in plants. Trends Genet. 2006;22:268–280. doi: 10.1016/j.tig.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Chen X. microRNA biogenesis and function in plants. FEBS Lett. 2005;579:5923–5931. doi: 10.1016/j.febslet.2005.07.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasciolli V., Mallory A.C., Bartel D.P., Vaucheret H., Mallory A.C., Bartel D.P., Vaucheret H., Bartel D.P., Vaucheret H., Vaucheret H. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol. 2005;15:1494–1500. doi: 10.1016/j.cub.2005.07.024. [DOI] [PubMed] [Google Scholar]
- Grad Y., Aach J., Hayes G.D., Reinhart B.J., Church G.M., Ruvkun G., Kim J., Aach J., Hayes G.D., Reinhart B.J., Church G.M., Ruvkun G., Kim J., Hayes G.D., Reinhart B.J., Church G.M., Ruvkun G., Kim J., Reinhart B.J., Church G.M., Ruvkun G., Kim J., Church G.M., Ruvkun G., Kim J., Ruvkun G., Kim J., Kim J. Computational and experimental identification of C. elegans microRNAs. Mol. Cell. 2003;11:1253–1263. doi: 10.1016/s1097-2765(03)00153-9. [DOI] [PubMed] [Google Scholar]
- Grundhoff A., Sullivan C.S., Ganem D., Sullivan C.S., Ganem D., Ganem D. A combined computational and microarray-based approach identifies novel microRNAs encoded by human γ-herpesviruses. RNA. 2006;12:733–750. doi: 10.1261/rna.2326106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson A.M., Allen E., Givan S., Smith D., Carrington J.C., Kasschau K.D., Allen E., Givan S., Smith D., Carrington J.C., Kasschau K.D., Givan S., Smith D., Carrington J.C., Kasschau K.D., Smith D., Carrington J.C., Kasschau K.D., Carrington J.C., Kasschau K.D., Kasschau K.D. ASRP: The Arabidopsis Small RNA Project Database. Nucleic Acids Res. 2005;33:D637–D640. doi: 10.1093/nar/gki127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson I.R., Zhang X., Lu C., Johnson L., Meyers B.C., Green P.J., Jacobsen S.E., Zhang X., Lu C., Johnson L., Meyers B.C., Green P.J., Jacobsen S.E., Lu C., Johnson L., Meyers B.C., Green P.J., Jacobsen S.E., Johnson L., Meyers B.C., Green P.J., Jacobsen S.E., Meyers B.C., Green P.J., Jacobsen S.E., Green P.J., Jacobsen S.E., Jacobsen S.E. Dissecting Arabidopsis DICER function in small RNA processing, gene silencing, and DNA methylation patterning. Nat. Genet. 2006;38:721–725. doi: 10.1038/ng1804. [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades M.W., Bartel D.P., Bartel D.P. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades M.W., Bartel D.P., Bartel B., Bartel D.P., Bartel B., Bartel B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- Kasschau K.D., Xie Z., Allen E., Llave C., Chapman E.J., Krizan K.A., Carrington J.C., Xie Z., Allen E., Llave C., Chapman E.J., Krizan K.A., Carrington J.C., Allen E., Llave C., Chapman E.J., Krizan K.A., Carrington J.C., Llave C., Chapman E.J., Krizan K.A., Carrington J.C., Chapman E.J., Krizan K.A., Carrington J.C., Krizan K.A., Carrington J.C., Carrington J.C. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell. 2003;4:205–217. doi: 10.1016/s1534-5807(03)00025-x. [DOI] [PubMed] [Google Scholar]
- Kurihara Y., Takashi Y., Watanabe Y., Takashi Y., Watanabe Y., Watanabe Y. The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA. 2006;12:206–212. doi: 10.1261/rna.2146906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusaba M., Dwyer K., Hendershot J., Vrebalov J., Nasrallah J.B., Nasrallah M.E., Dwyer K., Hendershot J., Vrebalov J., Nasrallah J.B., Nasrallah M.E., Hendershot J., Vrebalov J., Nasrallah J.B., Nasrallah M.E., Vrebalov J., Nasrallah J.B., Nasrallah M.E., Nasrallah J.B., Nasrallah M.E., Nasrallah M.E. Self-incompatibility in the genus Arabidopsis Characterization of the S locus in the outcrossing A. lyrataand its autogamous relative A. thaliana . Plant Cell. 2001;13:627–643. [PMC free article] [PubMed] [Google Scholar]
- Lippman Z., Martienssen R., Martienssen R. The role of RNA interference in heterochromatic silencing. Nature. 2004;431:364–370. doi: 10.1038/nature02875. [DOI] [PubMed] [Google Scholar]
- Llave C., Kasschau K.D., Rector M.A., Carrington J.C., Kasschau K.D., Rector M.A., Carrington J.C., Rector M.A., Carrington J.C., Carrington J.C. Endogenous and silencing-associated small RNAs in plants. Plant Cell. 2002a;14:1605–1619. doi: 10.1105/tpc.003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave C., Xie Z., Kasschau K.D., Carrington J.C., Xie Z., Kasschau K.D., Carrington J.C., Kasschau K.D., Carrington J.C., Carrington J.C. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002b;297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- Lu C., Tej S.S., Luo S., Haudenschild C.D., Meyers B.C., Green P.J., Tej S.S., Luo S., Haudenschild C.D., Meyers B.C., Green P.J., Luo S., Haudenschild C.D., Meyers B.C., Green P.J., Haudenschild C.D., Meyers B.C., Green P.J., Meyers B.C., Green P.J., Green P.J. Elucidation of the small RNA component of the transcriptome. Science. 2005;309:1567–1569. doi: 10.1126/science.1114112. [DOI] [PubMed] [Google Scholar]
- Margulies M., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., Berka J., Braverman M.S., Chen Y.J., Chen Z., Braverman M.S., Chen Y.J., Chen Z., Chen Y.J., Chen Z., Chen Z., et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martienssen R.A. Maintenance of heterochromatin by RNA interference of tandem repeats. Nat. Genet. 2003;35:213–214. doi: 10.1038/ng1252. [DOI] [PubMed] [Google Scholar]
- May B.P., Lippman Z.B., Fang Y., Spector D.L., Martienssen R.A., Lippman Z.B., Fang Y., Spector D.L., Martienssen R.A., Fang Y., Spector D.L., Martienssen R.A., Spector D.L., Martienssen R.A., Martienssen R.A. Differential regulation of strand-specific transcripts from Arabidopsis centromeric satellite repeats. PLoS Genet. 2005;1 doi: 10.1371/journal.pgen.0010079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers B.C., Kozik A., Griego A., Kuang H., Michelmore R.W., Kozik A., Griego A., Kuang H., Michelmore R.W., Griego A., Kuang H., Michelmore R.W., Kuang H., Michelmore R.W., Michelmore R.W. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis . Plant Cell. 2003;15:809–834. doi: 10.1105/tpc.009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers B.C., Tej S.S., Vu T.H., Haudenschild C.D., Agrawal V., Edberg S.B., Ghazal H., Decola S., Tej S.S., Vu T.H., Haudenschild C.D., Agrawal V., Edberg S.B., Ghazal H., Decola S., Vu T.H., Haudenschild C.D., Agrawal V., Edberg S.B., Ghazal H., Decola S., Haudenschild C.D., Agrawal V., Edberg S.B., Ghazal H., Decola S., Agrawal V., Edberg S.B., Ghazal H., Decola S., Edberg S.B., Ghazal H., Decola S., Ghazal H., Decola S., Decola S. The use of MPSS for whole-genome transcriptional analysis in Arabidopsis . Genome Res. 2004;14:1641–1653. doi: 10.1101/gr.2275604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers B.C., Souret F.F., Lu C., Green P.J., Souret F.F., Lu C., Green P.J., Lu C., Green P.J., Green P.J. Sweating the small stuff: MicroRNA discovery in plants. Curr. Opin. Biotechnol. 2006;17:139–146. doi: 10.1016/j.copbio.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Nasrallah M.E., Liu P., Nasrallah J.B., Liu P., Nasrallah J.B., Nasrallah J.B. Generation of self-incompatible Arabidopsis thaliana by transfer of two S locus genes from A. lyrata . Science. 2002;297:247–249. doi: 10.1126/science.1072205. [DOI] [PubMed] [Google Scholar]
- Naumann K., Fischer A., Hofmann I., Krauss V., Phalke S., Irmler K., Hause G., Aurich A.C., Dorn R., Jenuwein T., Fischer A., Hofmann I., Krauss V., Phalke S., Irmler K., Hause G., Aurich A.C., Dorn R., Jenuwein T., Hofmann I., Krauss V., Phalke S., Irmler K., Hause G., Aurich A.C., Dorn R., Jenuwein T., Krauss V., Phalke S., Irmler K., Hause G., Aurich A.C., Dorn R., Jenuwein T., Phalke S., Irmler K., Hause G., Aurich A.C., Dorn R., Jenuwein T., Irmler K., Hause G., Aurich A.C., Dorn R., Jenuwein T., Hause G., Aurich A.C., Dorn R., Jenuwein T., Aurich A.C., Dorn R., Jenuwein T., Dorn R., Jenuwein T., Jenuwein T., et al. Pivotal role of AtSUVH2 in heterochromatic histone methylation and gene silencing in Arabidopsis . EMBO J. 2005;24:1418–1429. doi: 10.1038/sj.emboj.7600604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W., Li J., Song R., Messing J., Chen X., Li J., Song R., Messing J., Chen X., Song R., Messing J., Chen X., Messing J., Chen X., Chen X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana . Curr. Biol. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine A., Yoshikawa M., Wu G., Albrecht H.L., Poethig R.S., Yoshikawa M., Wu G., Albrecht H.L., Poethig R.S., Wu G., Albrecht H.L., Poethig R.S., Albrecht H.L., Poethig R.S., Poethig R.S. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis . Genes & Dev. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redei G.P. Arabidopsis as a genetic tool. Annu. Rev. Genet. 1975;9:111–127. doi: 10.1146/annurev.ge.09.120175.000551. [DOI] [PubMed] [Google Scholar]
- Reinhart B.J., Weinstein E.G., Rhoades M.W., Bartel B., Bartel D.P., Weinstein E.G., Rhoades M.W., Bartel B., Bartel D.P., Rhoades M.W., Bartel B., Bartel D.P., Bartel B., Bartel D.P., Bartel D.P. MicroRNAs in plants. Genes & Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades M.W., Reinhart B.J., Lim L.P., Burge C.B., Bartel B., Bartel D.P., Reinhart B.J., Lim L.P., Burge C.B., Bartel B., Bartel D.P., Lim L.P., Burge C.B., Bartel B., Bartel D.P., Burge C.B., Bartel B., Bartel D.P., Bartel B., Bartel D.P., Bartel D.P. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- Rice P., Longden I., Bleasby A., Longden I., Bleasby A., Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Sunkar R., Zhu J.K., Zhu J.K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis . Plant Cell. 2004;16:2001–2019. doi: 10.1105/tpc.104.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valoczi A., Hornyik C., Varga N., Burgyan J., Kauppinen S., Havelda Z., Hornyik C., Varga N., Burgyan J., Kauppinen S., Havelda Z., Varga N., Burgyan J., Kauppinen S., Havelda Z., Burgyan J., Kauppinen S., Havelda Z., Kauppinen S., Havelda Z., Havelda Z. Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res. 2004;32 doi: 10.1093/nar/gnh171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H. Post-transcriptional small RNA pathways in plants: Mechanisms and regulations. Genes & Dev. 2006;20:759–771. doi: 10.1101/gad.1410506. [DOI] [PubMed] [Google Scholar]
- Vaucheret H., Vazquez F., Crete P., Bartel D.P., Vazquez F., Crete P., Bartel D.P., Crete P., Bartel D.P., Bartel D.P. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes & Dev. 2004;18:1187–1197. doi: 10.1101/gad.1201404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H., Mallory A.C., Bartel D.P., Mallory A.C., Bartel D.P., Bartel D.P. AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol. Cell. 2006;22:129–136. doi: 10.1016/j.molcel.2006.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F., Vaucheret H., Rajagopalan R., Lepers C., Gasciolli V., Mallory A.C., Hilbert J.L., Bartel D.P., Crete P., Vaucheret H., Rajagopalan R., Lepers C., Gasciolli V., Mallory A.C., Hilbert J.L., Bartel D.P., Crete P., Rajagopalan R., Lepers C., Gasciolli V., Mallory A.C., Hilbert J.L., Bartel D.P., Crete P., Lepers C., Gasciolli V., Mallory A.C., Hilbert J.L., Bartel D.P., Crete P., Gasciolli V., Mallory A.C., Hilbert J.L., Bartel D.P., Crete P., Mallory A.C., Hilbert J.L., Bartel D.P., Crete P., Hilbert J.L., Bartel D.P., Crete P., Bartel D.P., Crete P., Crete P. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell. 2004;16:69–79. doi: 10.1016/j.molcel.2004.09.028. [DOI] [PubMed] [Google Scholar]
- Wang X.J., Reyes J.L., Chua N.H., Gaasterland T., Reyes J.L., Chua N.H., Gaasterland T., Chua N.H., Gaasterland T., Gaasterland T. Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol. 2004;5 doi: 10.1186/gb-2004-5-9-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenegger M., Krczal G., Krczal G. Nomenclature and functions of RNA-directed RNA polymerases. Trends Plant Sci. 2006;11:142–151. doi: 10.1016/j.tplants.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Wortman J.R., Haas B.J., Hannick L.I., Smith R.K., Jr., Maiti R., Ronning C.M., Chan A.P., Yu C., Ayele M., Whitelaw C.A., Haas B.J., Hannick L.I., Smith R.K., Jr., Maiti R., Ronning C.M., Chan A.P., Yu C., Ayele M., Whitelaw C.A., Hannick L.I., Smith R.K., Jr., Maiti R., Ronning C.M., Chan A.P., Yu C., Ayele M., Whitelaw C.A., Smith R.K., Jr., Maiti R., Ronning C.M., Chan A.P., Yu C., Ayele M., Whitelaw C.A., Maiti R., Ronning C.M., Chan A.P., Yu C., Ayele M., Whitelaw C.A., Ronning C.M., Chan A.P., Yu C., Ayele M., Whitelaw C.A., Chan A.P., Yu C., Ayele M., Whitelaw C.A., Yu C., Ayele M., Whitelaw C.A., Ayele M., Whitelaw C.A., Whitelaw C.A., et al. Annotation of the Arabidopsis genome. Plant Physiol. 2003;132:461–468. doi: 10.1104/pp.103.022251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Kasschau K.D., Carrington J.C., Kasschau K.D., Carrington J.C., Carrington J.C. Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr. Biol. 2003;13:784–789. doi: 10.1016/s0960-9822(03)00281-1. [DOI] [PubMed] [Google Scholar]
- Xie Z., Johansen L.K., Gustafson A.M., Kasschau K.D., Lellis A.D., Zilberman D., Jacobsen S.E., Carrington J.C., Johansen L.K., Gustafson A.M., Kasschau K.D., Lellis A.D., Zilberman D., Jacobsen S.E., Carrington J.C., Gustafson A.M., Kasschau K.D., Lellis A.D., Zilberman D., Jacobsen S.E., Carrington J.C., Kasschau K.D., Lellis A.D., Zilberman D., Jacobsen S.E., Carrington J.C., Lellis A.D., Zilberman D., Jacobsen S.E., Carrington J.C., Zilberman D., Jacobsen S.E., Carrington J.C., Jacobsen S.E., Carrington J.C., Carrington J.C. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2 doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Allen E., Fahlgren N., Calamar A., Givan S.A., Carrington J.C., Allen E., Fahlgren N., Calamar A., Givan S.A., Carrington J.C., Fahlgren N., Calamar A., Givan S.A., Carrington J.C., Calamar A., Givan S.A., Carrington J.C., Givan S.A., Carrington J.C., Carrington J.C. Expression of Arabidopsis MIRNA genes. Plant Physiol. 2005a;138:2145–2154. doi: 10.1104/pp.105.062943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Allen E., Wilken A., Carrington J.C., Allen E., Wilken A., Carrington J.C., Wilken A., Carrington J.C., Carrington J.C. DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana . Proc. Natl. Acad. Sci. 2005b;102:12984–12989. doi: 10.1073/pnas.0506426102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M., Peragine A., Park M.Y., Poethig R.S., Peragine A., Park M.Y., Poethig R.S., Park M.Y., Poethig R.S., Poethig R.S. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis . Genes & Dev. 2005;19:2164–2175. doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D., Fan B., MacFarlane S.A., Chen Z., Fan B., MacFarlane S.A., Chen Z., MacFarlane S.A., Chen Z., Chen Z. Analysis of the involvement of an inducible Arabidopsis RNA-dependent RNA polymerase in antiviral defense. Mol. Plant Microbe Interact. 2003;16:206–216. doi: 10.1094/MPMI.2003.16.3.206. [DOI] [PubMed] [Google Scholar]
- Zhang B., Pan X., Cannon C.H., Cobb G.P., Anderson T.A., Pan X., Cannon C.H., Cobb G.P., Anderson T.A., Cannon C.H., Cobb G.P., Anderson T.A., Cobb G.P., Anderson T.A., Anderson T.A. Conservation and divergence of plant microRNA genes. Plant J. 2006;46:243–259. doi: 10.1111/j.1365-313X.2006.02697.x. [DOI] [PubMed] [Google Scholar]