Abstract
MicroRNAs are 20- to 23-nucleotide RNA molecules that can regulate gene expression. Currently >400 microRNAs have been experimentally identified in mammalian genomes, whereas estimates go up to 1000 and beyond. Here we show that many more mammalian microRNAs exist. We discovered novel microRNA candidates using two approaches: testing of computationally predicted microRNAs by a modified microarray-based detection system, and cloning and sequencing of large numbers of small RNAs from different human and mouse tissues. Together these efforts experimentally identified 348 novel mouse and 81 novel human microRNA candidate genes. Most novel microRNAs candidates are not conserved beyond mammals, and ~10% are taxon-specific. Our analyses indicate that the entire microRNA repertoire is not remotely exhausted.
Although the first microRNA (miRNA) was identified >10 yr ago (Lee et al. 1993; Wightman et al. 1993), it was only recently recognized that miRNAs form a major class of ribo-regulators (Lagos-Quintana et al. 2001; Lau et al. 2001; Lee and Ambros 2001). Currently, >400 miRNAs have been described for human (Griffiths-Jones 2004), but estimates based upon computational predictions range between 500 and 1000 (Bentwich et al. 2005; Berezikov et al. 2005, 2006; Xie et al. 2005). miRNAs are transcribed as long precursors (pri-miRNAs) that are processed by Drosha, resulting in an ~70-nucleotide (nt) stem–loop structure (pre-miRNA), are transported to the cytoplasm, and are further processed by the Dicer-containing complex. The resulting 20- to 25-nt mature miRNAs are loaded in the RNA-induced silencing complex (RISC) that can effect gene silencing through sequence- specific base pairing with target mRNAs, resulting in either transcriptional repression or target breakdown (Bartel 2004; Du and Zamore 2005). Although only a limited number of biological targets has been validated experimentally, computational approaches, applying rules based on validated targets, suggest that the currently known set of miRNAs may regulate between 20% and 30% of all transcripts in a vertebrate genome (Lewis et al. 2005; Lim et al. 2005). Indeed, many developmental and disease processes have now been found to be under critical regulation by miRNAs (Alvarez-Garcia and Miska 2005; Plasterk 2006). To obtain better insight into the biological function of this class of small RNA molecules in general and individual miRNAs in particular, it will be essential to identify all miRNAs that are expressed from a genome. Here, we describe the identification of many novel miRNA candidates by following two different experimental approaches. First, we used a modified microarray-based detection system to experimentally confirm computationally predicted miRNAs; secondly, we generated several high-titer cDNA libraries for small RNAs from different human and mouse tissues, followed by sequencing and computational analysis of inserts. Together, these efforts identified 348 novel mouse and 81 novel human microRNA candidate genes and suggested that still more miRNAs exist in genomes.
Results
Microarray-based confirmation of computationally predicted candidates
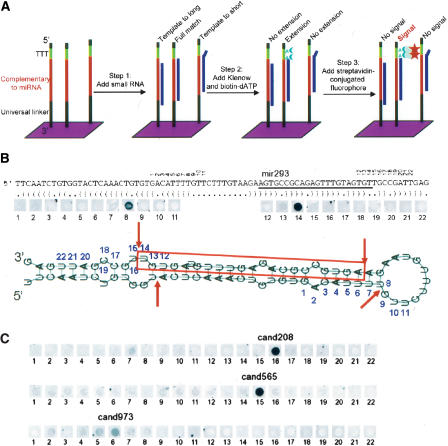
Previously, we reported computational predictions of >800 novel mammalian miRNA candidates that meet stringent phylogenetic conservation profiles and RNA folding criteria (Berezikov et al. 2005). To verify these predicted miRNA candidate genes experimentally, we now employed a modified RNA-primed Array-based Klenow Extension (RAKE) approach (Nelson et al. 2004). This assay is based on the ability of an RNA molecule to function as a primer for Klenow polymerase extension when fully base-paired with a single-stranded DNA molecule (Fig. 1A). As the exact 3′ end of the miRNA should be known for successful extension, and computational predictions are not optimal for predicting the correct start and end of the mature miRNA, we designed a tiling path of probes complementary to both known and predicted miRNA precursors (Fig. 1B). Such a tiling path RAKE assay is less prone to false positives than standard hybridization assays, as it depends on the presence of a fully matching 3′ end of the miRNA and hence distinguishes between miRNA family members that differ in their 3′ sequences. Flanking tiling path probes function as negative controls.
Figure 1.
Schematic representation of the modified RAKE assay (A) and experimental results obtained for known (B) and novel (C) candidate miRNAs. (A) A 44K microarray with a tiling path of 60-mer probes that are attached with their 3′ end to the glass surface was designed for the Agilent platform. Each DNA probe consists of a universal 3′ spacer sequence (black) followed by 22 nt of sequence complementary to the microRNA candidate (red), three thymidine nucleotides (green), and a short universal spacer (black). Unlabeled small RNA is hybridized to these arrays, followed by a Klenow extension reaction in the presence of biotinylated dATP. As miRNAs function as a primer for extension and the thymidines are the only template for extension, a complete 3′-end match is required for biotin incorporation and streptavidin fluorophore-mediated detection in the final step. (B) Schematic representation of the miR-293 pre-miRNA with predicted secondary structure and RAKE results. The mature miRNA (red) and numbers above the sequence indicate the 3′ end that fully matches the respective tiling path probe on the array. The strongest signal in the RAKE assay is obtained for probe 14, confirming the known 3′ end of miR-293. A weaker signal is obtained for probe 8, consistent with sensitive detection of the star sequence, which is produced as a side product from the hairpin structure by Drosha and Dicer nucleases that cut double-stranded RNA with a 2-nt 3′ overhang. The star sequence cannot be detected for all positive miRNAs. (C) RAKE results for three candidate miRNAs confirm the existence of novel miRNAs and identify the 3′ end of the mature miRNAs. For cand208 the star sequence can be detected (probe 7), whereas for cand973 multiple probes are positive with a rapidly decreasing intensity around the predominant probe, most likely representing imprecise 3′ end processing.
Although some rules have been put forward to determine which strand of the stem is preferentially loaded as mature miRNA in the RISC complex (Khvorova et al. 2003; Schwarz et al. 2003), such computational predictions can be done only when the precise ends of the processed miRNA duplex are known. In addition, due to the nature of the hairpin sequence it is often difficult to predict which strand of the genomic DNA encodes a precursor. To take a fully unbiased approach, we designed tiling paths of 11 probes covering each arm of the stem–loop structure, for the sense as well as the antisense genomic sequence, resulting in sets of 44 probes per candidate microRNA gene. Due to G–U pairing allowed in RNA folding and different nucleotide composition of the complementary DNA strand, antisense transcripts do not necessarily fold into stable stem–loop structures, and for such candidates only 22 probes were included. The central position in the tiling path was determined by predicting the most likely Dicer/Drosha processing sites from secondary structure hairpin information.
We designed a custom validation microarray with 44,000 features, covering 259 known mouse miRNAs and 687 novel predicted miRNAs that are conserved between mouse and human, and filled up the array with 200 additional candidates based on stringent randfold criteria (Bonnet et al. 2004) and mouse and rat genome conservation. These arrays were probed with four different sources of small RNAs: mouse embryos at embryonic days 8.5 and 16.5, adult mouse brain, and embryonic stem (ES) cells (Fig. 1C). Mature miRNAs were semi-manually annotated after preprocessing the raw microarray output data using custom scripts. A nonredundant set of 184 of the known miRNAs (71%), 419 of the candidate conserved miRNAs (61%), and 128 of the extra set (64%) were found positive (Supplemental data 1, 2). The confirmed numbers of known and novel miRNAs may still be underestimates due to the limited number of different tissues assayed and the limited length of the tiling path used. Interestingly, for more than half of the known miRNAs the most prominent 3′ end observed in the RAKE assay differed from the annotated form, including several mature miRNAs residing in the other arm of the hairpin, suggesting that originally the star-sequence was annotated (Supplemental data1). In addition, for various candidate and known miRNAs, multiple subsequent probes (2 or 3) resulted in a positive signal, indicating that 3′ end processing of miRNAs is not a completely accurate process at the single nucleotide level. These findings are in line with the observed variation in ends of cloned miRNAs (Aravin and Tuschl 2005).
Manual inspection of microarray signal along the tiling paths in known miRNAs suggests a high specificity of the RAKE assay, since we did not observe signal outside the expected locations (Fig. 1B). To directly estimate the rate of false-positive discovery in RAKE, we tested a number of RAKE-positive candidates by Northern blot analysis (Supplemental Fig. 1). From a selected set of 29 candidates that had a clear signal on RAKE, 22 were found positive on Northerns (76%). For another set of 11 candidates that were classified as “inconclusive” based on RAKE results, 6 (55%) were positive on Northerns (Supplemental Table1), suggesting that among candidates that failed in our RAKE experiments there is still a substantial fraction of real miRNAs. During the course of the experiments, 18 of the 28 miRNA candidates that were confirmed by Northern blot analysis were also identified by other research groups and are currently annotated in miRBase (Poy et al. 2004; Bentwich et al. 2005; Sewer et al. 2005; Xie et al. 2005; Mineno et al. 2006; Wheeler et al. 2006). However, as it will remain difficult to obtain solid numbers on false-negative rates, but more importantly also on false-positive rates of the RAKE assay, we propose to consider all RAKE-positive hairpins as candidate miRNAs until independent additional confirmation (e.g., cloning, Northern, PCR-based amplification) is obtained.
Small RNA cloning and analysis
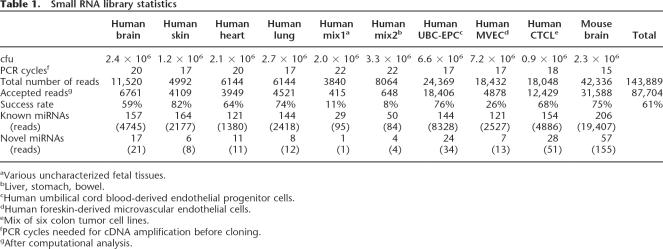
The second approach we pursued is deep sequencing of size- fractionated small RNA libraries of isolated human and mouse tissues. Although it was suggested previously that such efforts had reached near saturation (Lim et al. 2003), only limited numbers of library clones from a selected set of vertebrate tissues have been sequenced. Moreover, our computational predictions and microarray-based confirmations suggested many novel miRNAs yet to be discovered. Therefore, we generated nine high-titer nonconcatamerized libraries of size-fractioned small RNAs from mouse brain and various human fetal tissues (brain, skin, heart, lung, and mixed tissues), primary cells (human umbilical cord blood-derived endothelial progenitor cells, UCB-EPC; human foreskin-derived microvascular endothelial cells, MVEC), and cell lines (mix of six colon tumor cell lines, CTCL) and sequenced in total >140,000 clones (Table 1). After vector and quality trimming, 87,704 inserts >18 bases were recovered.
Table 1.
Small RNA library statistics
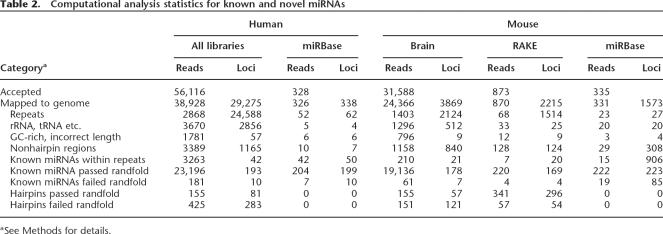
We established a computational pipeline for automated annotation of the cloned sequences, taking into account unique chromosomal position, location in repetitive elements or rRNA, tRNA, snoRNA genes, conservation data from 18 animal genomes (human, chimp, macaque, mouse, rat, dog, cow, chicken, opossum, zebrafish, fugu, tetraodon, Xenopus, Anopheles, bee, fly, worm, Ciona), and secondary structure information using rand- fold (Bonnet et al. 2004). This analysis was applied to the mouse and human cloned fragments, as well as to all known human and mouse miRNAs and the positive candidates identified using the RAKE assay (Table 2; Supplemental Data 3). Two hundred twenty-two out of 335 mouse (66%) and 204 out of 328 human (62%) miRNAs, as deposited in miRBase (Griffiths-Jones 2004), passed the automated filtering and annotation. Most of the miRNAs that were discarded by the computational pipeline overlap with various repeat annotations. For the 873 mouse miRNA sequences that tested positive in RAKE, 312 were rejected ad hoc by the computational analysis for various reasons (Supplemental Data 1). To maintain very stringent criteria for all novel miRNA candidates reported here, we decided to exclude the latter set from further analyses and calculations, although they may represent genuine miRNAs.
Table 2.
Computational analysis statistics for known and novel miRNAs
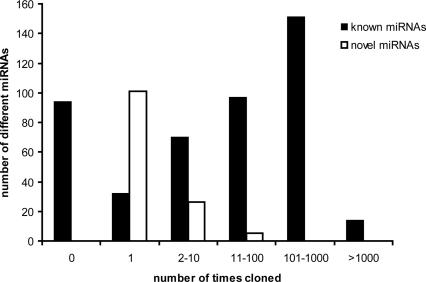
For the sequenced small RNAs, 19,291 mouse (79%) and 23,351 human (60%) clones passed this filtering. Known abundant microRNA sequences dominate this set, but, interestingly, ~0.5% of the reads represent 54 novel mouse and 81 novel human miRNA genes (Supplemental Data 3). Underrepresentation of novel miRNAs in cloning and sequencing experiments can be expected, as there is a strong bias toward highly expressed and abundant miRNAs, which have already been identified in previous low-depth sequencing efforts. As a result, 76% of the novel miRNAs are supported by only a single clone, but it should be noted that 28% of the known miRNAs were also detected only once or not at all in our set of tissues (Fig. 2). It now becomes clear that expression levels of miRNAs may range over more than three orders of magnitude (Neely et al. 2006), which is indeed reflected in our cloning results where the most abundant miRNAs were picked up >3000 times and many others (both known and novel) only once (Supplemental data 4).
Figure 2.
The cloning frequency for known and novel mouse and human miRNAs is significantly different. Although many known miRNAs were not picked up in our cloning experiments and about half of the novel miRNAs were identified multiple times, a relatively high fraction of novel miRNA candidates was identified only once, indicating that small RNA sequencing efforts have not been exhausted.
Overlap between approaches
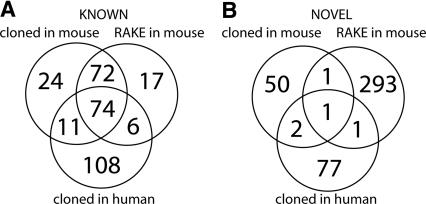
Overlap between the novel miRNAs obtained by the RAKE-based computational verification experiments and the cloning experiment is limited (Fig. 3), potentially reflecting the fundamentally different biases underlying each approach, such as phylogenetic conservation for the RAKE experiments and expression abundance in cloning from tissue samples. Furthermore, different tissue sources that potentially express different miRNAs were used in the various experiments to maximize the chance of finding novel miRNAs. Both the cloning approach and the microarray experiments show that the abundance of miRNAs varies over several orders of magnitude and that different tissues are characterized by a different group of highly expressed miRNAs (Supple- mental data 4). Since the tissues used consist of many different cell types, miRNAs that are highly expressed in only a few cells of a tissue would seem to be expressed at low levels when the whole tissue is assayed and could easily be missed by cloning efforts. However, such miRNAs would be extremely powerful indicators for specific cell types (Wienholds et al. 2005) and, in line with previous reports, could be very valuable, for example, in classifying tumors (Calin et al. 2005; He et al. 2005; Lu et al. 2005).
Figure 3.
Overlap of cloned small miRNAs and RAKE results for known (A) and novel (B) mouse miRNA candidates. Known microRNAs were found to varying degrees in the different experiments, but most are detected in limited numbers of samples in both human and mouse. Most novel microRNA candidates were identified by the combination of computational prediction and RAKE confirmation approach.
To test the presence of RAKE-confirmed miRNAs in our small RNA libraries, we used information about the 3′ ends of miRNA to design primers that overlap with the miRNA sequence and the poly-A tail that was introduced during the cloning process. These oligos can be used in combination with a vector-based primer to amplify specific miRNA clones from total library DNA. From 32 known miRNAs tested, eight were positive in this PCR assay using the mouse brain library as template, indicating that in principle the assay can be used for specific amplification of miRNA clones, although the success rate is only 25% (Supple- mental data 5). From 276 novel RAKE candidates that were tested by PCR, three candidates were positive (MM_25, MM_139, and MM_359). From these, two candidates (MM_25 and MM_139) were also observed in sequenced clones from mouse brain library, and MM_139 appeared to be a homolog of a recently published human miRNA, mir-551b (Cummins et al. 2006). Although confirmation rates are rather low, the results do also indicate that library sequencing is not exhausted and that more extensive sequencing of the libraries may yield more novel miRNAs and increase the overlap between miRNA candidates identified by RAKE and by cloning.
To further assess reliability of the RAKE assay, we compared mouse brain cloning and RAKE data. One hundred eleven known miRNAs were positive on RAKE using RNA from mouse brain and were also cloned from the mouse brain library. In 63 cases (57%), the most frequently cloned sequence had the same 3′ end as that identified in the RAKE experiment, and in 32 cases (29%), the miRNAs RAKE 3′ end corresponded to 3′ ends of some of the cloned inserts, but these were not the most frequently cloned sequences. Only in 16 cases (14%) were there no matching 3′ ends between RAKE and cloned sequences. This analysis demonstrates that, at least for known miRNAs, there is a good agreement between RAKE and cloning data, further substantiating the validity of the RAKE approach.
Discussion
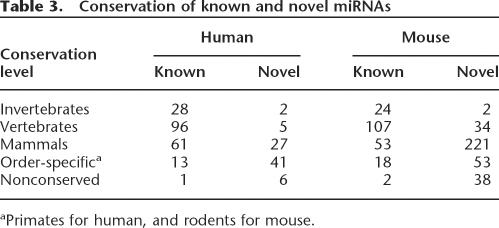
According to the guidelines for miRNA annotation (Ambros et al. 2003), a cloned RNA must have either convincing evolutionary conservation or some other evidence of expression in order to be annotated as a genuine miRNA. Although most of the novel miRNAs presented here are supported by evolutionary conservation and stringent bioinformatic criteria, independent expression evidence is lacking for most of them. To distinguish between miRNAs that are extensively verified by independent experimental assays (e.g., Northern blots) and RNA species that are supported by limited single experiments (e.g., cloned only once, or only found positive in RAKE), but are highly likely to be real miRNAs, we prefer to use the term “miRNA candidate” for the latter class. Taken together, we identified 348 novel mouse (RAKE and cloning) and 81 novel human (cloning only) miRNA candidate genes (Fig. 3). Novel miRNAs in general are expressed at a very low level and constitute <1% of clones in small RNA libraries. At the same time, novel miRNA candidates are less conserved than known miRNAs (Table 3). While the majority of known miRNAs are conserved among invertebrates, vertebrates, and mammals, only a few novel miRNAs are conserved to this extent. Instead, most of the novel miRNAs are conserved only in mammals, primates (for human-derived miRNAs), or rodents (for mouse-derived miRNAs). Seven percent of human and 11% of mouse novel miRNA are species-specific.
Table 3.
Conservation of known and novel miRNAs
Similar to known miRNAs, several of the novel miRNA candidates were found to belong to families that have high sequence similarity, even though this characteristic has already been used to identify miRNAs by systematic genome analysis (Nam et al. 2005). In human, one family was identified that contains only novel miRNA candidates, and eight families were found to contain both known and novel miRNAs (Supplemental Table 2). In mouse, novel miRNA candidates make up 12 novel families and contribute to two known miRNA families.
Genomic clustering is a feature that has been employed for miRNA gene discovery previously (Altuvia et al. 2005), and from our newly identified set of miRNA candidates 19 mouse and 13 human miRNA genes reside in existing or novel (11 and five, respectively) genomic clusters (with 20 kb distance) of multiple miRNAs (Fig. 4; Supplemental Table 3). Interestingly, 10 miRNA candidates are novel members of a large cluster of >30 miRNAs.
Figure 4.
Chromosomal location of human (A) and (B) miRNAs. (Red) Noval miRNA candidates, (green) known miRNAs. Clusterd miRNAs are stacked
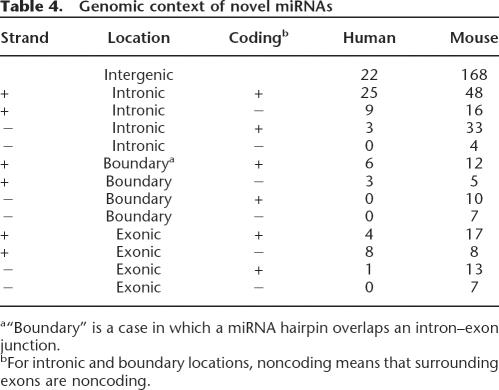
Many known miRNAs reside in introns of protein-coding genes and in introns and exons of noncoding RNAs (Rodriguez et al. 2004). Similarly, 46% of novel human miRNA candidates are intronic (Table 4), and 27% overlap with both coding and non- coding exons, including 11% that overlap exon–intron junctions (“boundary” in Table 4). In mouse, 29%, 10%, and 13% of novel miRNAs are intronic, boundary, or exonic, respectively, and 48% of miRNAs do not overlap with known transcripts. The majority of novel miRNA candidates originate from the same strand as a host gene (66% in human and 73% in mouse).
Table 4.
Genomic context of novel miRNAs
Our results do indicate that miRNA discovery efforts are not exhausted. We have used very stringent parameters for the computational prediction of phylogenetically conserved miRNAs, thereby missing miRNAs that are less conserved or even species- specific. Indeed, using the RAKE assay, we could experimentally detect many novel miRNAs that were predicted without using stringent phylogenetic conservation criteria. Several miRNA candidates that gave inconclusive results on RAKE (and were excluded from further analysis) were still positive by Northern blot analysis, indicating that there are more real miRNAs among RAKE candidates than we were able to confirm. Furthermore, we discarded many RAKE-positive candidates that failed the computational pipeline developed for analysis of sequences from small RNA libraries. We decided to comply with these very stringent filtering criteria for consistency with the cloning approach, and to obtain the most reliable data set, although one should realize that we may have filtered out genuine miRNAs. Indeed, a number of known miRNAs did not pass the pipeline under the selected stringent parameters (Table 2), and several nonpassed RAKE candidates were confirmed by Northern blot analysis (Supplemental Table 1). The 296 miRNA RAKE candidate regions that passed the computational pipeline overlap poorly with our cloning efforts (Fig. 3), with only three miRNAs confirmed by cloning and one additional RAKE candidate recovered from the mouse brain library by directed PCR (Supplemental data 5). Positive results of directed PCR indicate that more RAKE candidates can be confirmed by increasing the sequencing depth of small RNA libraries. At the same time, the limited success rate of the PCR-based confirmation suggests that many RAKE candidates may not be present in the libraries. Since the confirmation rate of RAKE candidates by Northern blot analysis is >50% (Supplemental Fig. 1), it is possible that some real miRNAs are simply not“clonable” by the methods used, for example, due to their physical properties or modifications, and therefore cannot be detected in small RNA libraries. Although we provide strong bioinformatics and expression evidence for 296 RAKE miRNA candidates, for 290 of them we do not have experimental evidence for the exact location of the 5′ end of the mature miRNA sequence, and therefore these miRNAs should still be considered as candidate miRNAs.
Our cloning efforts also suggest that more miRNAs can be discovered in the future. Many known and novel miRNAs were represented by just a single clone in the various libraries (Fig. 2). Near saturation is expected only when the vast majority of miRNAs has been identified multiple times. Both increased depth analysis of existing libraries and cell-type-specific analyses are likely to further increase the number of miRNAs. To minimize the false-discovery rate, we have applied stringent criteria for computational analysis of small RNA sequencing reads that discarded several hundred potential miRNA hairpins along with a number of known miRNAs (Table 2). Future improvements in the bioinformatics analysis combined with experimental methods of independent verification of miRNA candidates may further increase the yield of novel miRNAs from small RNA sequencing efforts. Eventually, a comprehensive inventory of mammalian microRNA genes will facilitate elucidation of biological functions of this class of genes and aid in the identification of the involvement of individual microRNAs in development and disease processes (Alvarez-Garcia and Miska 2005; Hammond 2006; Plasterk 2006).
Methods
Computational prediction of novel microRNAs
All conserved human–mouse microRNAs were deduced from the candidate microRNA set as described in Berezikov et al. (2005). To produce additional candidate microRNA genes, the mouse genome was scanned for potential hairpins with a sliding window of 100 nt, and randfold values were calculated for resulting hairpins (mononucleotide shuffling, 1000 iterations). From a large set of hairpins that have low randfold values but are not necessarily conserved in other species, a subset of 200 was randomly selected to fill up the array.
RAKE microarray design and analysis
The microarray for verification of candidate microRNAs using the RAKE assay was designed as a 44K custom microarray (Agilent Technologies). Probes (60-mer) that are attached to the glass surface with their 3′ end were designed to include a fully matching probe sequence of 22 nt complementary to the predicted microRNA with universal spacers on each side (5′ end, 5′ spacer: CGATCTTT, sequence of 22 nt complementary to the microRNA candidate region [tiling path], 3′ spacer: TAGGGTCCGATAAGG GTCAGTGCTCGCTCTA, 3′ end attached to glass surface). The three Ts in the 5′ spacer function as a template for Klenow- mediated microRNA extension using biotin-dATP. A tiling path of 11 nt was designed to cover the most likely Dicer/Drosha cleavage site determined at 22 nt upstream and downstream from the terminal loop extended to contain at least 11 unpaired nucleotides. For all cases, probes were designed for both arms of the hairpin sequence, and for 648 candidates an additional set of 2 × 11 probes was designed, as the transcript originating from the antisense genomic sequence can also efficiently fold into a stable hairpin structure. All 22/44 probes for a candidate microRNA were located in clusters on the array to exclude regional background effects. Ten different hybridization controls complementary to plant microRNAs (miR-402, UUCGAGGCCUAUU AAACCUCUG; miR-418, UAAUGUGAUGAUGAACUGACCU; miR-167, UGAAGCUGCCAGCAUGATCUGG; miR-416, GGUUCGUACGUACACUGUUCAU; miR-173, UUCGCUUGCA GAGAGAAAUCAC; miR-417, GAAGGUAGUGAAUUUGUUC GAC; miR-163, GAAGAGGACUUGGAACUUCGAU; miR-419, UUAUGAAUGCUGAGGAUGUUGU; miR-405, GAGUUGGGU CUAACCCAUAACU; miR-420, UAAACUAAUCACGGAAAUG CAC) were represented 10 times randomly distributed on the array. Microarrays were scanned on an Agilent scanner model G2565B at 10-μm resolution, and spot identification and intensity determination was done using Agilent Feature Extraction software (Image Analysis version A.7.5.1) with standard settings. To permit manual inspection and annotation of mature microRNA sequences, the raw images and spot intensity data were processed using custom scripts and visualized together with tiling path sequence information (Supplemental data 2). Web- based interfaces were designed for annotation of single experiments and for summarizing all experiments. After manual inspection, all novel mature microRNA sequences that were positive (Supplemental data 1) were fed into the bioinformatics analysis pipeline set up for the evaluation of the cloned small RNAs, to filter out signal originating from repetitive elements and structural RNAs and to find homologous miRNAs in other species (Table 2).
Modified RAKE assay
The original RAKE assay (Nelson et al. 2004) was modified for use with high-density custom-printed microarrays in the Agilent platform. Most importantly, in contrast to most custom-spotted microarrays, custom-printed probes are attached with their 3′ end to the glass surface. This excludes the need for the exonuclease step that was included in the original protocol to reduce background signal from fold-backs of the free 3′ ends of the probes that result in double-stranded DNA structures that can function as a template for the Klenow extension, resulting in nonspecific background signal. Furthermore, hybridization, washing, and incubation conditions were adapted. All hybridization and wash buffers were made fresh from autoclaved stock solutions using DEPC-treated water, filter-sterilized, and pre- heated. Microarray slides and coverslips were pre-washed two times for 2 min at 37°C with preheated wash buffer (2 × SSPE,0.025% N-lauroylsarcosine), followed by 5 min incubation with pre-hybridization buffer (5 × SSPE, 40% formamide, 0.025% N-lauroylsarcosine). Next, the Agilent hybridization chamber was completely filled with hybridization mix, leaving no air bubbles, as the usual air bubble for mixing does not move around at low temperature and with the hybridization mix used. The hybridization mix (750 μL total per slide) consists of 500 μL of1.5 × hybridization buffer (7.5 × SSPE, 60% formamide, 0.0375% N-lauroylsarcosine), 10 μL of spike-in RNA (control plant microRNAs stock: miR-402, 1 × 10−6 M; miR-418, 3.3 × 10−7 M; miR-167, 1 × 10−7 M; miR-416, 3.3 × 10−8 M; miR-173, 1 × 10−8 M; miR-417, 3.3 × 10−9 M; miR-163, 1 × 10−9 M; miR-419, 3.3 × 10−10 M; miR-405, 1 × 10−10 M; miR-420,3.3 × 10−11 M), and 20 μg of small RNA sample (8.5 dpc and16.5 dpc mouse embryo, mouse embryonic stem [ES] cells, and total brain), isolated using the MirVana microRNA isolation kit (Ambion) and supplemented with DEPC-treated water up to 240 μL. The hybridization mix was heated for 5 min to 75°C and cooled on ice before application to the array. The array was incubated overnight at 37°C, followed by four washes of 2 min in wash buffer and one wash for 2 min in 1× Klenow buffer (10 mM Tris pH 7.9, 50 mM NaCl, 10 mM MgCl2, 1 mM DTT, 0.025% N-lauroylsarcosine). For the Klenow extension, an enzyme mix (750 μL total per slide) containing 375 μL of 2× Klenow buffer, 365 μL of DEPC-treated water, 2.5 μL of Klenow Exo− (50,000 U/μL, NEB), and 7.5 μL of biotin-14-dATP (4 μM stock, Perkin Elmer) was applied to the array in a clean incubation chamber and incubated for 1 h at 37°C. Next, the array was washed four times for 2 min with wash buffer and once for 2 min with 1× Klenow buffer. Next, the dye conjugation mix (total volume 750 μL) consisting of 375 μL of 2× Klenow buffer, 368 μL of DEPC treated water, and 20 μL of streptavidin-conjugated Alexa fluor- 647 (2 mg/mL stock, Invitrogen) was applied in a new incubation chamber for 30 min at 37°C, followed by four washes of 2 min at 37°C with wash buffer and five brief dips in DEPC water to remove salts. Slides were dried by centrifugation in a 50-mL tube by spinning for 5 min at 1000 rpm (180 × g).
Northern blot analysis
The small RNA fraction of 16.5 dpc mouse embryos was isolated using the mirVana miRNA isolation kit (Ambion). Two micrograms of small RNA was loaded per lane and separated on 12% denaturing polyacrylamide gels alongside the RNA Decade marker (Ambion), transferred by electroblotting to positively charged nylone membranes (Roche). Blots were hybridized overnight at 37°C with radioactively (32P) labeled DNA oligo probes in modified Church and Gilbert buffer, washed three times with 2× SSC, 0.1% SDS at 37°C, and visualized using phosphoimaging (Typhoon).
Small RNA library construction and sequencing
Nine high-titer small RNA libraries (Table 1) were made by Vertis Biotechnology AG. For human fetal tissue collection, individual permissions using standard informed consent procedures and prior approval of the ethics committee of the University Medical Center Utrecht were obtained. Briefly, the small RNA fraction from adult mouse brain (12 wk), various human fetal tissues (17 wk of development: brain; heart; skin; lung; mix 1: multiple fetal tissues; mix 2: liver, stomach, bowel), UCB-EPC (primary human endothelial progenitor cells, cultured from CD34+ cells purified from human umbilical cord blood), MVEC (primary human fore- skin-derived microvascular endothelial cells), and colon tumor cell lines (mix of six cell lines) were isolated using the mirVana microRNA isolation kit (Ambion), followed by an additional enrichment by excision of the 15- to 30-nt fraction from a polyacrylamide gel. For cDNA synthesis, the RNA molecules in this fraction were first poly A-tailed using yeast poly(A) polymerase followed by ligation of a RNA linker oligo to the 5′ phosphate of the miRNAs. First-strand cDNA synthesis was then performed using an oligo(dT)-linker primer and M-MLV-RNase H- reverse transcriptase. The resulting cDNA was then PCR amplified for 15–22 cycles (depending on the starting material quality and quantity; see Table 1 for details), followed by restriction nuclease treatment, gel purification of the 95- to 110-bp fraction, and cloning in the EcoRI and BamHI sites of the pBSII SK+ plasmid vector. Ligations were electroporated into T1 Phage-resistant TransforMaxTMEC100TM electrocompetent cells (Epicentre), resulting in titers between 1.2 and 3.3 × 106 recombinant clones per library. A total of 83,328 colonies was automatically picked into 384-well plates (Genetix QPix2) containing 75 μL of LB-Amp and grown overnight at 37°C with continuous shaking. All of the following pipetting steps were performed using liquid-handling robots (Tecan Genesis RSP200 with integrated TeMo96 and Velocity11 Vprep with BenchCell 4 ×). Five microliters of culture were transferred to a 384-well PCR plate (Greiner) containing 20 μL of water, and cells were lysed by heating for 15 min at 95°Cin a PCR machine. One microliter of lysed suspension was transferred to a fresh 384-well plate containing 4 μL of PCR mix (final concentrations: 0.2 μM M13 forward, TGTAAAACGACGGC CAGT; 0.2 μM M13 reverse, AGGAAACAGCTATGACCAT; 400 μM of each dNTP; 25 mM tricine; 7.0% glycerol [w/v]; 1.6% DMSO [w/v]; 2 mM MgCl2; 85 mM ammonium acetate pH 8.7; and 0.2 U Taq polymerase in a total volume of 10 μL), and the insert was amplified by 35 cycles of 20 sec at 94°C, 10 sec at 58°C, and 30 sec at 72°C. After adding 30 μL of water, 1 μL of PCR product was directly used for dideoxy sequencing by transferring to a new 384-well PCR plate containing 4 μL sequencing mix(0.027 μL of BigDye terminator mix v3.1 [Applied Biosystems],1.96 μL of 2.5 × dilution buffer [Applied Biosystems], 0.01 μL of sequencing oligo [100 μM stock T7, GTAATACGACTCAC TATAGGGC], and 2 μL of water). Thermocycling was performed for 35 cycles of 10 sec at 94°C, 10 sec at 50°C, 20 sec at 60°C, and final products were purified by ethanol precipitation in 384-well plates as recommended by the manufacturer (Applied Biosystems) and analyzed on ABI3730XL sequencers with a modified protocol for generating ~100-nt sequencing reads.
PCR-based miRNA verification of library DNA
About 0.6 × 106 cfu from the mouse brain small RNA library was used for inoculation of a 100-mL liquid culture (LB + ampicillin). After overnight growth, plasmid DNA was isolated using mini- columns according to the manufacturer's instructions (Qiagen). Ten nanograms of plasmid DNA was used as a template for PCR using a vector-based forward primer (M13 forward) and miRNA- specific reverse primers that are complementary to the last 14 nt of the candidate miRNA and 8 nt of the poly-A tail that was introduced in the cloning procedure. PCR and sequencing (using T7 primer) conditions were identical to those described above for the small RNA library sequencing.
Computational analysis of cloned small RNAs sequencing reads
Base calling and quality trimming of sequence chromatograms were done by Phred software (Ewing et al. 1998). After masking of vector and adapter sequences, and removing redundancy, inserts of length ≥18 bases were mapped to genomes (ncbi35 assembly for human and ncbim34 assembly for mouse) using MegaBlast software (ftp://ftp.ncbi.nlm.nih.gov/blast/). Not all inserts matched perfectly to a genome, and detailed analysis of non- matching sequences indicated that in most cases the best hit in the genome corresponded to the beginning of the read, with several nucleotides at the 3′ end of the read nonmatching (Supplemental Fig. 2A). The fraction of known miRNAs in perfect and nonperfect reads is similar (Supplemental Fig. 2B), justifying trimming of the nonmatching bases. These nongenomic sequences may be artifacts of the cloning procedure or a result of nontemplated modification of mature microRNAs (Aravin and Tuschl 2005). Such sequences were trimmed at the 3′ end according to the best blast hit to a genome. Next, for every genomic locus matching to an insert, repeat annotations were retrieved from the Ensembl database (http://www.ensembl.org) and tRNA, rRNA, snoRNA regions, and repetitive regions were discarded from further analysis, with the exception of simple and trf repeats, since these repeat annotations overlap with some known miRNAs. Genomic regions containing inserts with 100-nt flanks were retrieved from the Ensembl and RNAshapes programs (Steffen et al. 2006), and were used to find hairpin structures in sliding windows of 80, 100, and 120 nt that folded into hairpins with the abstract shape “[ ],” had a probability of folding >0.8, and contained an insert in one of the hairpin arms.
To find homologous hairpins in other genomes, mature miRNA regions were blasted against human, chimpanzee, macaque, mouse, rat, dog, cow, opossum, chicken, zebrafish, fugu, tetraodon, Xenopus, Anopheles, Drosophila, bee and Ciona genomes. Where available, BLASTZ_NET aligned regions were also retrieved from Ensembl. All hits matching to at least seven continuous nucleotides starting from the 1st, 2nd, or 3rd nucleotide of the mature sequence were extracted and folded using the RNAshapes program with the same parameters as mentioned above. Next, similarity between all potential homologous hairpins and the original hairpin was calculated using RNAforester software (http://bibiserv.techfak.uni-bielefeld.de/rnaforester). If a BLASTZ_NET aligned region folded into a hairpin and had an RNAforester score >0.3, it was assigned as an orthologous hairpin in a particular species; otherwise, the highest scoring hairpin above a score of 0.3 was defined as an ortholog. Next, homologs from different organisms were aligned with the original hairpin by CLUSTAL W (Thompson et al. 1994) to produce a final multiple alignment of the hairpin region. Chromosomal locations of homologous sequences were used to retrieve gene and repeat annotations from the respective species in the Ensembl database. Hairpins that contained repeat/RNA annotations in one of the species, as well as hairpins containing mature regions >25 nt or with GC content >85% were discarded. For the remaining hairpins, randfold values were calculated for every sequence in an alignment using mononucleotide shuffling and 1000 iterations (Bonnet et al. 2004). The cut-off of 0.005 was used for randfold, and only regions that contained a hairpin below this cut-off for at least one species in an alignment were considered as miRNA candidates. Finally, positive hairpins were split into known and novel miRNAs according to annotations. To facilitate these annotations and also to track performance of the pipeline, mature sequences of known microRNAs from miRBase v.8.0 (Griffiths- Jones 2004) were included into the analysis.
ACKNOWLEDGMENTS
We thank Wigard Kloosterman and Rene Ketting for critically reading the manuscript. This work was supported by grants from the Horizon (E.B.) and BioRange (E.C.) programs of the Netherlands Genomics Initiative (NGI). J.V. was supported by a grant from the Dutch Program for Tissue Engineering. R.V. was supported by a grant from the Netherlands Heart Foundations (NHS 2002157).
Footnotes
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5159906.
Supplemental material is available online at www.genome.org. All novel microRNAs described in this study are submitted to the central miRNA repository miRBase, maintained at the Wellcome Trust Sanger Institute, under the ID nos. listed in the Supplemental material.
REFERENCES
- Altuvia Y., Landgraf P., Lithwick G., Elefant N., Pfeffer S., Aravin A., Brownstein M.J., Tuschl T., Margalit H., Landgraf P., Lithwick G., Elefant N., Pfeffer S., Aravin A., Brownstein M.J., Tuschl T., Margalit H., Lithwick G., Elefant N., Pfeffer S., Aravin A., Brownstein M.J., Tuschl T., Margalit H., Elefant N., Pfeffer S., Aravin A., Brownstein M.J., Tuschl T., Margalit H., Pfeffer S., Aravin A., Brownstein M.J., Tuschl T., Margalit H., Aravin A., Brownstein M.J., Tuschl T., Margalit H., Brownstein M.J., Tuschl T., Margalit H., Tuschl T., Margalit H., Margalit H. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res. 2005;33:2697–2706. doi: 10.1093/nar/gki567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Garcia I., Miska E.A., Miska E.A. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- Ambros V., Bartel B., Bartel D.P., Burge C.B., Carrington J.C., Chen X., Dreyfuss G., Eddy S.R., Griffiths-Jones S., Marshall M., Bartel B., Bartel D.P., Burge C.B., Carrington J.C., Chen X., Dreyfuss G., Eddy S.R., Griffiths-Jones S., Marshall M., Bartel D.P., Burge C.B., Carrington J.C., Chen X., Dreyfuss G., Eddy S.R., Griffiths-Jones S., Marshall M., Burge C.B., Carrington J.C., Chen X., Dreyfuss G., Eddy S.R., Griffiths-Jones S., Marshall M., Carrington J.C., Chen X., Dreyfuss G., Eddy S.R., Griffiths-Jones S., Marshall M., Chen X., Dreyfuss G., Eddy S.R., Griffiths-Jones S., Marshall M., Dreyfuss G., Eddy S.R., Griffiths-Jones S., Marshall M., Eddy S.R., Griffiths-Jones S., Marshall M., Griffiths-Jones S., Marshall M., Marshall M., et al. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin A., Tuschl T., Tuschl T. Identification and characterization of small RNAs involved in RNA silencing. Bartel, D.P. 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297. FEBS Lett. 2005;579:5830–5840. doi: 10.1016/j.febslet.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Bentwich I., Avniel A., Karov Y., Aharonov R., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Avniel A., Karov Y., Aharonov R., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Karov Y., Aharonov R., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Aharonov R., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Barzilai A., Einat P., Einav U., Meiri E., Einat P., Einav U., Meiri E., Einav U., Meiri E., Meiri E., et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- Berezikov E., Guryev V., van de Belt J., Wienholds E., Plasterk R.H., Cuppen E., Guryev V., van de Belt J., Wienholds E., Plasterk R.H., Cuppen E., van de Belt J., Wienholds E., Plasterk R.H., Cuppen E., Wienholds E., Plasterk R.H., Cuppen E., Plasterk R.H., Cuppen E., Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Berezikov E., Cuppen E., Plasterk R.H.A., Cuppen E., Plasterk R.H.A., Plasterk R.H.A. Approaches to microRNA discovery. Nat. Genet. 2006;38:S2–S7. doi: 10.1038/ng1794. [DOI] [PubMed] [Google Scholar]
- Bonnet E., Wuyts J., Rouze P., Van de Peer Y., Wuyts J., Rouze P., Van de Peer Y., Rouze P., Van de Peer Y., Van de Peer Y. Evidence that microRNA precursors, unlike other non-coding RNAs, have lower folding free energies than random sequences. Bioinformatics. 2004;20:2911–2917. doi: 10.1093/bioinformatics/bth374. [DOI] [PubMed] [Google Scholar]
- Calin G.A., Ferracin M., Cimmino A., Di Leva G., Shimizu M., Wojcik S.E., Iorio M.V., Visone R., Sever N.I., Fabbri M., Ferracin M., Cimmino A., Di Leva G., Shimizu M., Wojcik S.E., Iorio M.V., Visone R., Sever N.I., Fabbri M., Cimmino A., Di Leva G., Shimizu M., Wojcik S.E., Iorio M.V., Visone R., Sever N.I., Fabbri M., Di Leva G., Shimizu M., Wojcik S.E., Iorio M.V., Visone R., Sever N.I., Fabbri M., Shimizu M., Wojcik S.E., Iorio M.V., Visone R., Sever N.I., Fabbri M., Wojcik S.E., Iorio M.V., Visone R., Sever N.I., Fabbri M., Iorio M.V., Visone R., Sever N.I., Fabbri M., Visone R., Sever N.I., Fabbri M., Sever N.I., Fabbri M., Fabbri M., et al. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- Cummins J.M., He Y., Leary R.J., Pagliarini R., Diaz L.A., Jr., Sjoblom T., Barad O., Bentwich Z., Szafranska A.E., Labourier E., He Y., Leary R.J., Pagliarini R., Diaz L.A., Jr., Sjoblom T., Barad O., Bentwich Z., Szafranska A.E., Labourier E., Leary R.J., Pagliarini R., Diaz L.A., Jr., Sjoblom T., Barad O., Bentwich Z., Szafranska A.E., Labourier E., Pagliarini R., Diaz L.A., Jr., Sjoblom T., Barad O., Bentwich Z., Szafranska A.E., Labourier E., Diaz L.A., Jr., Sjoblom T., Barad O., Bentwich Z., Szafranska A.E., Labourier E., Sjoblom T., Barad O., Bentwich Z., Szafranska A.E., Labourier E., Barad O., Bentwich Z., Szafranska A.E., Labourier E., Bentwich Z., Szafranska A.E., Labourier E., Szafranska A.E., Labourier E., Labourier E., et al. The colorectal microRNAome. Proc. Natl. Acad. Sci. 2006;103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du T., Zamore P.D., Zamore P.D. microPrimerThe biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- Ewing B., Hillier L., Wendl M.C., Green P., Hillier L., Wendl M.C., Green P., Wendl M.C., Green P., Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S. The microRNA registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond S.M. MicroRNAs as oncogenes. Curr. Opin. Genet. Dev. 2006;16:4–9. doi: 10.1016/j.gde.2005.12.005. [DOI] [PubMed] [Google Scholar]
- He L., Thomson J.M., Hemann M.T., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S.W., Hannon G.J., Thomson J.M., Hemann M.T., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S.W., Hannon G.J., Hemann M.T., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S.W., Hannon G.J., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S.W., Hannon G.J., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S.W., Hannon G.J., Goodson S., Powers S., Cordon-Cardo C., Lowe S.W., Hannon G.J., Powers S., Cordon-Cardo C., Lowe S.W., Hannon G.J., Cordon-Cardo C., Lowe S.W., Hannon G.J., Lowe S.W., Hannon G.J., Hannon G.J., et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A., Reynolds A., Jayasena S.D., Reynolds A., Jayasena S.D., Jayasena S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T., Rauhut R., Lendeckel W., Tuschl T., Lendeckel W., Tuschl T., Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lau N.C., Lim L.P., Weinstein E.G., Bartel D.P., Lim L.P., Weinstein E.G., Bartel D.P., Weinstein E.G., Bartel D.P., Bartel D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans . Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee R.C., Ambros V., Ambros V. An extensive class of small RNAs in Caenorhabditis elegans . Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- Lee R.C., Feinbaum R.L., Ambros V., Feinbaum R.L., Ambros V., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14 . Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., Bartel D.P., Burge C.B., Bartel D.P., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lim L.P., Glasner M.E., Yekta S., Burge C.B., Bartel D.P., Glasner M.E., Yekta S., Burge C.B., Bartel D.P., Yekta S., Burge C.B., Bartel D.P., Burge C.B., Bartel D.P., Bartel D.P. Vertebrate microRNA genes. Science. 2003;299 doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Bartel D.P., Linsley P.S., Johnson J.M., Linsley P.S., Johnson J.M., Johnson J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lu J., Getz G., Miska E.A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A., Getz G., Miska E.A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A., Miska E.A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A., Ebert B.L., Mak R.H., Ferrando A.A., Mak R.H., Ferrando A.A., Ferrando A.A., et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Mineno J., Okamoto S., Ando T., Sato M., Chono H., Izu H., Takayama M., Asada K., Mirochnitchenko O., Inouye M., Okamoto S., Ando T., Sato M., Chono H., Izu H., Takayama M., Asada K., Mirochnitchenko O., Inouye M., Ando T., Sato M., Chono H., Izu H., Takayama M., Asada K., Mirochnitchenko O., Inouye M., Sato M., Chono H., Izu H., Takayama M., Asada K., Mirochnitchenko O., Inouye M., Chono H., Izu H., Takayama M., Asada K., Mirochnitchenko O., Inouye M., Izu H., Takayama M., Asada K., Mirochnitchenko O., Inouye M., Takayama M., Asada K., Mirochnitchenko O., Inouye M., Asada K., Mirochnitchenko O., Inouye M., Mirochnitchenko O., Inouye M., Inouye M., et al. The expression profile of microRNAs in mouse embryos. Nucleic Acids Res. 2006;34:1765–1771. doi: 10.1093/nar/gkl096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam J.W., Shin K.R., Han J., Lee Y., Kim V.N., Zhang B.T., Shin K.R., Han J., Lee Y., Kim V.N., Zhang B.T., Han J., Lee Y., Kim V.N., Zhang B.T., Lee Y., Kim V.N., Zhang B.T., Kim V.N., Zhang B.T., Zhang B.T. Human microRNA prediction through a probabilistic co-learning model of sequence and structure. Nucleic Acids Res. 2005;33:3570–3581. doi: 10.1093/nar/gki668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neely L.A., Patel S., Garver J., Gallo M., Hackett M., McLaughlin S., Nadel M., Harris J., Gullans S., Rooke J., Patel S., Garver J., Gallo M., Hackett M., McLaughlin S., Nadel M., Harris J., Gullans S., Rooke J., Garver J., Gallo M., Hackett M., McLaughlin S., Nadel M., Harris J., Gullans S., Rooke J., Gallo M., Hackett M., McLaughlin S., Nadel M., Harris J., Gullans S., Rooke J., Hackett M., McLaughlin S., Nadel M., Harris J., Gullans S., Rooke J., McLaughlin S., Nadel M., Harris J., Gullans S., Rooke J., Nadel M., Harris J., Gullans S., Rooke J., Harris J., Gullans S., Rooke J., Gullans S., Rooke J., Rooke J. A single-molecule method for the quantitation of microRNA gene expression. Nat. Methods. 2006;3:41–46. doi: 10.1038/nmeth825. [DOI] [PubMed] [Google Scholar]
- Nelson P.T., Baldwin D.A., Scearce L.M., Oberholtzer J.C., Tobias J.W., Mourelatos Z., Baldwin D.A., Scearce L.M., Oberholtzer J.C., Tobias J.W., Mourelatos Z., Scearce L.M., Oberholtzer J.C., Tobias J.W., Mourelatos Z., Oberholtzer J.C., Tobias J.W., Mourelatos Z., Tobias J.W., Mourelatos Z., Mourelatos Z. Microarray-based, high-throughput gene expression profiling of microRNAs. Nat. Methods. 2004;1:155–161. doi: 10.1038/nmeth717. [DOI] [PubMed] [Google Scholar]
- Plasterk R.H.A. MicroRNAs in animal development. Cell. 2006;124:877–881. doi: 10.1016/j.cell.2006.02.030. [DOI] [PubMed] [Google Scholar]
- Poy M.N., Eliasson L., Krutzfeldt J., Kuwajima S., Ma X., Macdonald P.E., Pfeffer S., Tuschl T., Rajewsky N., Rorsman P., Eliasson L., Krutzfeldt J., Kuwajima S., Ma X., Macdonald P.E., Pfeffer S., Tuschl T., Rajewsky N., Rorsman P., Krutzfeldt J., Kuwajima S., Ma X., Macdonald P.E., Pfeffer S., Tuschl T., Rajewsky N., Rorsman P., Kuwajima S., Ma X., Macdonald P.E., Pfeffer S., Tuschl T., Rajewsky N., Rorsman P., Ma X., Macdonald P.E., Pfeffer S., Tuschl T., Rajewsky N., Rorsman P., Macdonald P.E., Pfeffer S., Tuschl T., Rajewsky N., Rorsman P., Pfeffer S., Tuschl T., Rajewsky N., Rorsman P., Tuschl T., Rajewsky N., Rorsman P., Rajewsky N., Rorsman P., Rorsman P., et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- Rodriguez A., Griffiths-Jones S., Ashurst J.L., Bradley A., Griffiths-Jones S., Ashurst J.L., Bradley A., Ashurst J.L., Bradley A., Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz D.S., Hutvagner G., Du T., Xu Z., Aronin N., Zamore P.D., Hutvagner G., Du T., Xu Z., Aronin N., Zamore P.D., Du T., Xu Z., Aronin N., Zamore P.D., Xu Z., Aronin N., Zamore P.D., Aronin N., Zamore P.D., Zamore P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Sewer A., Paul N., Landgraf P., Aravin A., Pfeffer S., Brownstein M.J., Tuschl T., van Nimwegen E., Zavolan M., Paul N., Landgraf P., Aravin A., Pfeffer S., Brownstein M.J., Tuschl T., van Nimwegen E., Zavolan M., Landgraf P., Aravin A., Pfeffer S., Brownstein M.J., Tuschl T., van Nimwegen E., Zavolan M., Aravin A., Pfeffer S., Brownstein M.J., Tuschl T., van Nimwegen E., Zavolan M., Pfeffer S., Brownstein M.J., Tuschl T., van Nimwegen E., Zavolan M., Brownstein M.J., Tuschl T., van Nimwegen E., Zavolan M., Tuschl T., van Nimwegen E., Zavolan M., van Nimwegen E., Zavolan M., Zavolan M. Identification of clustered microRNAs using an ab initio prediction method. BMC Bioinformatics. 2005;6 doi: 10.1186/1471-2105-6-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen P., Voss B., Rehmsmeier M., Reeder J., Giegerich R., Voss B., Rehmsmeier M., Reeder J., Giegerich R., Rehmsmeier M., Reeder J., Giegerich R., Reeder J., Giegerich R., Giegerich R. RNAshapes: An integrated RNA analysis package based on abstract shapes. Bioinformatics. 2006;22:500–503. doi: 10.1093/bioinformatics/btk010. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J., Higgins D.G., Gibson T.J., Gibson T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler G., Ntounia-Fousara S., Granda B., Rathjen T., Dalmay T., Ntounia-Fousara S., Granda B., Rathjen T., Dalmay T., Granda B., Rathjen T., Dalmay T., Rathjen T., Dalmay T., Dalmay T. Identification of new central nervous system specific mouse microRNAs. FEBS Lett. 2006;580:2195–2200. doi: 10.1016/j.febslet.2006.03.019. [DOI] [PubMed] [Google Scholar]
- Wienholds E., Kloosterman W.P., Miska E., Alvarez-Saavedra E., Berezikov E., de Bruijn E., Horvitz H.R., Kauppinen S., Plasterk R.H.A., Kloosterman W.P., Miska E., Alvarez-Saavedra E., Berezikov E., de Bruijn E., Horvitz H.R., Kauppinen S., Plasterk R.H.A., Miska E., Alvarez-Saavedra E., Berezikov E., de Bruijn E., Horvitz H.R., Kauppinen S., Plasterk R.H.A., Alvarez-Saavedra E., Berezikov E., de Bruijn E., Horvitz H.R., Kauppinen S., Plasterk R.H.A., Berezikov E., de Bruijn E., Horvitz H.R., Kauppinen S., Plasterk R.H.A., de Bruijn E., Horvitz H.R., Kauppinen S., Plasterk R.H.A., Horvitz H.R., Kauppinen S., Plasterk R.H.A., Kauppinen S., Plasterk R.H.A., Plasterk R.H.A. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Wightman B., Ha I., Ruvkun G., Ha I., Ruvkun G., Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans . Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Xie X., Lu J., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Lu J., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Lindblad-Toh K., Lander E.S., Kellis M., Lander E.S., Kellis M., Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]